Abstract
M33, encoded by murine cytomegalovirus (MCMV), is a member of the UL33 homolog G-protein-coupled receptor (GPCR) family and is conserved across all the betaherpesviruses. Infection of mice with recombinant viruses lacking M33 or containing specific signaling domain mutations in M33 results in significantly diminished MCMV infection of the salivary glands. To determine the role of M33 in viral dissemination and/or infection in other tissues, viral infection with wild-type K181 virus and an M33 mutant virus, ΔM33BT2, was characterized using two different routes of inoculation. Following both intraperitoneal (i.p.) and intranasal (i.n.) inoculation, M33 was attenuated for infection of the spleen and pancreas as early as 7 days after infection. Following i.p. inoculation, ΔM33BT2 exhibited a severe defect in latency as measured by a diminished capacity to reactivate from spleens and lungs in reactivation assays (P < 0.001). Subsequent PCR analysis revealed markedly reduced ΔM33BT2 viral DNA levels in the latently infected spleens, lungs, and bone marrow. Following i.n. inoculation, latent ΔM33BT2 viral DNA was significantly reduced in the spleen and, in agreement with results from i.p. inoculation, did not reactivate from the spleen (P < 0.001). Furthermore, in vivo complementation of ΔM33BT2 virus replication and/or dissemination to the salivary glands and pancreas was achieved by coinfection with wild-type virus. Overall, our data suggest a critical tissue-specific role for M33 during infection in the salivary glands, spleen, and pancreas but not the lungs. Our data suggest that M33 contributes to the efficient establishment or maintenance of long-term latent MCMV infection.
Since the discovery of the G protein-coupled receptors (GPCRs) encoded by the betaherpesviruses, there has been intense speculation on the biological role these viral proteins play during infection (15, 16, 22). Human cytomegalovirus (HCMV), a betaherpesvirus, is a ubiquitous pathogen that asymptomatically infects humans and establishes a long-term persistent infection. HCMV is life-threatening, however, to immunocompromised individuals, such as neonates, AIDS patients, and transplant recipients. HCMV, similar to a number of herpesviruses, encodes viral genes that are predicted to impact virus-host interactions that may promote efficient long-term infection of the host. The CMVs encode genes for proteins that potentially enhance viral dissemination and replication and promote immune evasion by mimicry of host functions that influence the conditions of primary infection, the virus-specific immune response, and even long-term host control of persistent or latent infection (reviewed in references 1, 44, and 68).
HCMV encodes four GPCRs (UL33, UL78, US28, and US27) which share homology to host chemokine receptors (16). This suggests that these virally encoded chemokine receptors may function similarly to their cellular receptor counterparts. Chemokines are chemoattractant cytokines that bind and activate chemokine receptors that are on the surfaces of cells. Host chemokine receptors then mediate the activation of cellular signaling pathways and cell migration to sites of inflammation by transmitting signals through G proteins (56, 70). In humans, approximately 50 chemokines and 20 chemokine receptors have been identified, many of which have close homologs in mice and other species (39). Chemokines are divided into two classes, lymphoid chemokines, which are constitutively expressed and involved in lymphoid tissue organization, and inflammatory chemokines, which are induced following infection and part of the inflammatory response (21, 39, 51). Growing evidence indicates that chemokines play a critical role in the host response to infection and inflammation during both the innate and adaptive immune responses (26), thus suggesting that the betaherpesviruses have “hijacked” the chemokine receptors from the host genome to subvert or alter these responses during infection. Besides chemokine receptors, HCMV also encodes a CXC chemokine (UL146) that induces the migration of neutrophils (48); a second CXC chemokine homolog (UL147) whose function is not yet known; a viral CC chemokine (UL131) that is critical for infection of macrophages, endothelial cells, and epithelial cells (25, 57, 73); and a RANTES decoy protein (72). A CC chemokine (vMCK or m131/129) is also encoded by murine CMV (MCMV), and a homolog in rat CMV ([RCMV] r131) that promotes monocyte-associated viremia (20, 37, 59, 60). The MCMV m131/129 chemokine was shown to recruit myelomonocytic progenitors from the bone marrow, perhaps to facilitate cell-type-specific viremia (46). Clearly, the CMVs have invested a great deal of effort into manipulating or subverting the host chemokine system, thus making it reasonable to speculate that these viral members of the chemokine system play an important role during CMV pathogenesis.
Of the HCMV-encoded GPCRs, US28 has been well characterized in vitro and functions as a bona fide chemokine receptor, whereas much less is known about the receptor activity of US27, UL33, and UL78. US28 binds and sequesters CC chemokines, induces smooth-muscle cell migration, and constitutively activates signaling pathways (5, 7-9, 42, 52, 64, 67, 71). US28 and US27 are found only in primate CMVs, whereas both UL33 and UL78 are highly conserved across all betaherpesvirus genomes, suggesting an important evolutionary function for UL33 and UL78 during CMV infection. Two other betaherpesviruses, human herpesviruses 6 and 7 (HHV6 and HHV7), encode homologs to the UL33 and UL78 receptors, U12 and U51, respectively. The U12 receptors of HHV6 and HHV7 (34, 45, 66) and the HHV6-encoded U51 receptor (22) exhibit chemokine binding activity. UL33, along with its rodent CMV homologs, M33 (MCMV) and R33 (RCMV), constitutively activates signaling pathways (13, 23, 71). M33 induces smooth-muscle cell migration (39), similar to US28-mediated smooth-muscle cell migration (64). Thus, members of the UL33 family potentially function during viral infection by modulating or influencing the composition of leukocytes at sites of infection, the migration of infected cells or infiltrating leukocytes, or modulation of intracellular signaling pathways.
Due to the species specificity of CMV, the in vivo role of the HCMV-encoded GPCRs cannot be addressed. However, the importance of UL33 and UL78 for viral dissemination and virulence in vivo has been indicated by disruption of the viral homologs in MCMV and RCMV (6, 19, 36, 47). Disruption of the UL33, M33, and R33 genes demonstrated that they are dispensable for replication in vitro, indicating that the UL33 family members are not required for replication or cell entry in at least some cell types (6, 19, 40). Infection of mice with M33-deficient MCMV or infection of rats with R33-deficient RCMV results in highly attenuated viruses and diminished infection of the salivary glands. The RCMV R33 protein also appears to play a role in virulence since rats infected with an R33 deletion virus had a lower mortality rate (6). More recently, constitutive M33-mediated activation of signaling pathways was shown to be essential for MCMV infection of salivary glands (14). Significantly, the UL33 protein partially rescued the defect in salivary gland infection attributed to disruption of M33, indicating the evolutionary conservation of function between the HCMV (UL33) and MCMV (M33) chemokine receptor homologs.
In this paper, the role of M33 is further investigated using two routes of infection to assess viral dissemination and viral replication kinetics at different tissue sites, the numbers of infected cells following infection, and the possibility that M33 plays a role during latent infection. In addition to the critical role that M33 plays in salivary gland infection, this study reveals that M33 is important for MCMV infection of the spleen and the pancreas but not the lungs. Significantly, our studies provide preliminary evidence that disruption of M33 leads to reduced latent viral load in the spleen, lungs, and bone marrow, perhaps due to defects in the establishment and/or maintenance of latent infection. Lastly, we demonstrate that the tissue defects observed during acute infection with an M33 mutant virus (ΔM33BT2) can be complemented in vivo when mice are coinfected with ΔM33BT2 virus and wild-type MCMV. Taken together, our findings indicate that M33 plays a critical tissue-specific role during acute MCMV infection and, importantly, contributes to the efficient establishment or maintenance of latent MCMV infection.
MATERIALS AND METHODS
Cells and virus.
NIH 3T3 cells (ATCC CRL1658) were grown in Dulbecco's modified Eagle's medium (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 7.5% sodium bicarbonate, 4 mM HEPES, 2 mM l-glutamine, and gentamicin in a humidified 5% CO2 incubator at 37°C. Parent stocks of the wild-type MCMV K181 (Perth strain) and the rescue viruses M33R F2.1 and M33R D6.1 were prepared in NIH 3T3 cells from a salivary gland-derived virus stock. The M33 mutant virus, ΔM33BT2, does not replicate in the salivary gland, so the mutant virus stocks were grown in NIH 3T3 cells and are tissue culture-derived stocks (19). Isolation and characterization of the rescue viruses were previously described (14). No differences were observed in tissue culture-derived virus titer yield or plaque morphology for parent K181, ΔM33BT2, and rescue viruses. Virus titers were determined by plaque assay on NIH 3T3 cells. All virus stocks were stored at −70°C, and titers were determined again before viruses were used experiments.
Mice and infection.
Female BALB/c mice were purchased from Jackson Laboratories (Bar Harbor, ME). Five-week-old mice were infected with MCMV and maintained under specific-pathogen-free conditions at Cincinnati Children's Hospital Medical Center. Mice were infected using two routes of inoculation, intraperitoneal (i.p.) or intranasal (i.n.). For i.p. infection, mice were infected with 1 × 106 to 2 × 106 PFU. For i.n. infection, the mice were anesthetized with Avertin (2,2,2-tribromoethanol) and then infected with 5 × 105 PFU or 1 × 106 PFU (40-μl volume) as previously described (11). At various times after infection, mice were sacrificed (n = 4 mice/group), tissues were collected, and 10% tissue sonicates were prepared for virus titration by plaque assay. Single-cell suspensions from lymph nodes and spleens were prepared for infectious center assays. In some studies, peritoneal exudate cells (PEC) were isolated by peritoneal lavage. Bone marrow cells (BMC) were isolated from femurs of infected mice and washed with medium. Spleen cells enriched for monocyte/macrophages were obtained following a 3-h adherence to plastic at 37°C in a CO2 incubator, washed three times with medium, and collected using a cell scraper.
Virological methods.
For plaque assays, dilutions of virus stocks, 10% mouse tissue sonicates, and sonicated leukocyte cell suspensions were adsorbed onto 70% confluent NIH 3T3 monolayers for 1 h at 37°C and then overlaid with a 1:1 mixture of carboxymethyl cellulose (CMC) and 2× modified Eagle's medium, as previously described (11). At 6 days, the CMC overlay was removed, and the monolayers were fixed with methanol and stained with Giemsa to determine the number of plaques. Infectious center assays were prepared as previously described (11). Briefly, spleens and lymph nodes were homogenized and passed through a cell strainer, and serial dilutions of leukocytes were plated onto NIH 3T3 monolayers containing 0.5 ml of medium. After overnight incubation at 37°C, the wells were carefully overlaid with 2× CMC as described above for plaque assays and incubated at 37°C in 5% CO2. The overlay was removed after 6 days and fixed and stained with Giemsa. The number of plaques observed in the infectious center assay represents the number of infected leukocytes that harbored virus and infected the underlying fibroblast monolayer upon coculture, thus allowing the number of infected cells to be enumerated.
Reactivation assays.
Explant reactivation assays of spleens and lungs isolated from latently infected mice were established as previously described (12, 62). Briefly, mice were infected with 1 × 106 PFU of K181, ΔM33BT2, and M33R F2.1 viruses by both i.p. and i.n. inoculation. At various times after infection when virus was no longer replicating (50 to 110 days postinfection [dpi]), mice were sacrificed (n = 4 mice/group unless otherwise stated), and the salivary gland, spleen, and lungs were collected. The salivary glands were sonicated, and titers were determined to detect whether this tissue site harbored persistent replicating virus. The spleens and lungs were minced, and primary cultures were established as previously described (12, 62). Briefly, the tissues were divided into three parts, and each tissue part was placed into a well of a six-well tissue culture plate containing 5 ml of medium. After each tissue part was minced and prior to incubation at 37°C and 5% CO2, two pieces of tissue per well (six pieces total per tissue) were collected from the primary culture using dissection tools pretreated with DNA Away (Molecular BioProducts). The tissue pieces were pooled for DNA extraction and PCR analysis. The cultures were followed for 6 weeks, and culture supernatants were collected weekly and sonicated, and titers were determined in plaque assays to detect the presence of reactivating virus. To exclude mice with persistent, productive infection in the latency studies, mice that had replicating virus in the salivary glands were not included. Spleen and lung supernatants in the explant reactivation assays which were positive at day 7 postexplant were considered to harbor low levels of infectious virus and thus were excluded from the explant analysis.
PCR analysis.
Tissues were isolated from infected and uninfected mice using dissection tools pretreated with DNA Away (Molecular BioProducts). DNA was isolated from cell suspensions (∼5 × 106 cells) following tissue homogenization using a QIAamp DNA Mini Kit (Qiagen 51306) according to manufacturer's protocol. The ie1 gene was amplified by PCR using previously described primers (27). The primers were CH-16 (5′-TACAGGACAACAGAACGCTC-3′) and CH-17 (5′-CCTCGAGTCTGGAACCGAAA-3′), which amplify a 310-bp ie1 fragment. β-Actin primers were used as controls for the presence of amplifiable DNA (32). Each PCR contained 200 ng of purified DNA, 100 pmol of each primer, and Promega Master Mix (Promega) in a total volume of 25 μl. PCR was performed using an Eppendorf Mastercycler, and the temperature cycling profile began with an initial step of 94°C for 2 min, followed by annealing at 55°C for 45 s and elongation at 72°C for 1 min. An additional 34 cycles was done at 98°C for 45 s and the same annealing and elongation times. An MCMV viral DNA control, pARK25, which harbors the entire MCMV genome as a bacmid, was used as a positive control for ie1 amplification and specificity (kindly provided by Alec Redwood, University of Western Australia) (54). To determine the limit of detection of genome copy number, the pARK25 construct was serially diluted into uninfected tissue DNA. The limit of detection was determined to be ∼200 copies of viral genome. Nested PCR was performed using primers SY1 and SY2 using the reaction conditions above and as previously described (38), with 5 μl of the first-round PCR with the SY1 and SY2 primers serving as the template for the second-round PCR with the CH16 and CH17 primers. A nonspecific band which was amplified only at low copy numbers of the viral DNA control plasmid and in some tissue DNA samples with the CH16 and CH17 primers was eliminated by increasing the annealing temperature to 65°C (as is pointed out in the figure legends).
In vivo complementation studies.
Five-week-old BALB/c mice were infected i.p. with 2 × 106 PFU of ΔM33BT2 or 1 × 106 PFU of M33R F2.1 (M33+) or were coinfected i.p. with 2 × 106 PFU of ΔM33BT2 in combination with 1 × 106 PFU of M33R F2.1. At 14 dpi, the salivary glands, pancreas, and spleens were harvested, and 10% tissue homogenates were prepared by sonication. Titers of the tissue samples were determined on NIH 3T3 cells by plaque assay. After 6 days, one half of the CMV overlay was removed, and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution was added to each well to stain for lacZ-expressing plaques, as previously described (60, 62). The plates were then stained with Giemsa to enumerate the number of white plaques in the samples.
Statistical analysis.
All data shown are expressed as means plus standard errors or as means plus standard deviations, as stated in the figure legends. Statistical analysis was performed using analysis of variance with Bonferonni posttest analysis to compare wild-type virus with test virus groups (GraphPad Instat Program). P values of <0.05 were considered to indicate a significant difference.
RESULTS
M33 is required for efficient infection of the salivary glands, spleen, and pancreas following i.p. inoculation.
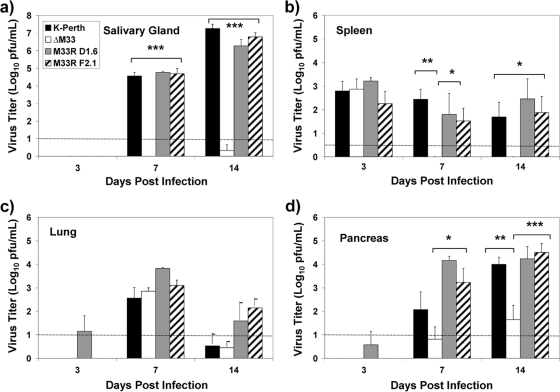
Following infection, MCMV disseminates widely, and infectious virus is found in a number of tissues, including the spleen, lung, liver, kidney, pancreas, adrenal gland, and salivary glands (2, 12, 18, 31, 62). Following i.p. infection with MCMV, the salivary gland is seeded following the second phase of viremia (18) and reaches peak titer levels at 14 dpi, with viral replication lasting for at least 1 month. Previously, it was shown that disruption of M33 resulted in an attenuated virus infection following i.p. inoculation, with dramatically reduced titers in the salivary glands and a trend toward reduced titers in the spleen (19). More recent data demonstrated the critical role of M33-mediated signaling during MCMV infection of the salivary gland (14). To determine whether M33 is required for infection of other tissues, 5-week-old BALB/c mice were inoculated by the i.p. route with 2 × 106 PFU of K181, ΔM33BT2, and two rescue viruses, M33R D1.6 and M33R F2.1 (repaired for the M33 disruption and described in reference 14). The inclusion of the two independent rescue viruses derived from ΔM33BT2 was important to confirm that any differences with respect to K181 were due to the disruption of M33 and not due to adventitious mutations elsewhere in the viral genome. K181 and the rescue viruses are herein referred to collectively as wild-type viruses, and they behaved similarly (P > 0.05). We hypothesized that following i.p. inoculation, the peritoneal cavity was a primary site of infection. To determine whether ΔM33BT2 and the wild-type viruses replicate to similar levels at the primary sites of infection, PEC were collected at 3 dpi, and virus titers were quantified by plaque assay. Both parent K181 and ΔM33BT2 yielded approximately the same virus titers in the PEC (log10 4.46 ± 0.11 PFU/ml for K181 and log10 4.25 ± 0.13 PFU/ml for ΔM33BT2; P > 0.05), indicating that ΔM33BT2 was not impaired for replication at the initial site of infection (62). Although it cannot be ruled out that residual input virus was also detectable, virus titers from PEC have been shown to increase between 2 and 3 dpi (R. Cardin, unpublished data). PEC were also analyzed by infectious center assays, and both wild-type virus infection and ΔM33BT2 infection yielded similar numbers of infected cells in the peritoneal cavity at 3 dpi (data not shown). It was next determined whether a disruption in M33 resulted in a defect in viral dissemination and/or infection of the salivary glands and other tissues such as the spleen, liver, pancreas, and lungs. As shown in Fig. 1, ΔM33BT2 titers in the salivary glands were more than 1,000-fold lower than wild-type viruses at 7 and 14 dpi (P < 0.001), which is similar to previous reports (14, 19). In the spleen at 3 dpi, virus titers were similar for ΔM33BT2 and the wild-type viruses (Fig. 1). In contrast, by 7 dpi, ΔM33BT2 titers in the spleen were at least 10-fold lower than those of K181 and both rescue viruses (P < 0.05). Similar results were observed at 14 dpi. Importantly, the rescue virus titers were similar to the parent virus titer (P > 0.05), indicating that decreased virus titers in the spleen were due to the disruption of the M33 gene. In the liver, titers were too low in these studies to enable reliable comparisons between the groups (data not shown) although following infection with a higher inoculum of 3 × 106 PFU, both K181 and ΔM33BT2 virus titers were similar in the liver at 3 dpi (3.5 to 4.0 log10 PFU/ml).
FIG. 1.
Virus titers of wild-type viruses and ΔM33BT2 in the salivary glands (a), spleen (b), lungs (c), and pancreas (d) following i.p. inoculation. Five-week-old BALB/c mice were inoculated i.p. with 2 × 106 PFU of K181 (K-Perth), ΔM33BT2 (ΔM33), M33R D1.6, and M33R F2.1. Titers of tissue sonicates were determined at 3, 7, and 14 dpi by plaque assay on NIH 3T3 cells. The mean virus titer (log10 PFU/ml) and standard error are shown for K181, ΔM33, and M33R F2.1 (n = 12) for a total of three separate experiments. The M33R D1.6 virus was included in one study and is presented as mean virus titer and standard deviation (n = 4). The limit of detection for the plaque assay is shown by the dotted line. Virus titers depicted below the limit of detection indicate where samples were negative at the limit of detection (e.g., 10−1 dilution) but positive for virus in undiluted samples. The P value is shown in the diagram: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Dissemination of virus to the pancreas and lungs was observed by 7 dpi, indicating that these tissues were secondary sites of infection following i.p. inoculation. As shown in Fig. 1, peak virus replication in the lungs was observed for all of the virus groups at day 7, followed by decline of virus levels close to the limits of detection by 14 dpi. No significant differences between K181 and ΔM33BT2 virus titers in the lungs were observed following i.p. inoculation, indicating that M33 was not required for either virus dissemination to the lungs or virus replication in the lungs. In contrast, decreased ΔM33 infection was detected in the pancreas. As shown in Fig. 1, the titers of ΔM33BT2 in the pancreas were 10- to 100-fold lower than those of the wild-type viruses. At 7 dpi, ΔM33BT2 titers were detectable at lower levels overall than titers of the wild-type viruses although only a trend in statistical significance was observed. By 14 dpi, however, ΔM33BT2 titers in the pancreas were significantly reduced compared to the wild-type viruses (P < 0.01). These data indicate a role for M33 in either dissemination to the pancreas or as a determinant of virus replication in the pancreas.
M33 is required for efficient infection of the salivary glands and spleen following i.n. inoculation.
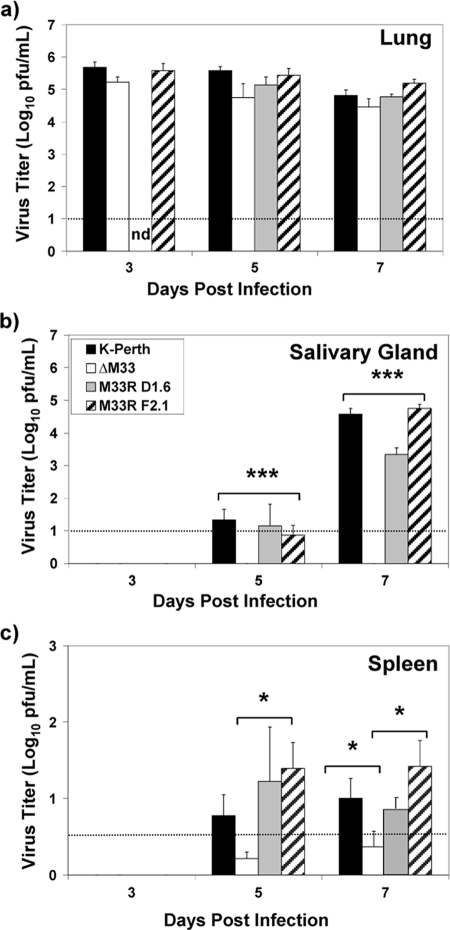
We next investigated whether an alternative route of infection other than i.p. inoculation could be used to track MCMV dissemination and infection. Although the i.p. route of infection is commonly used for MCMV studies, a disadvantage of i.p. inoculation is that virus could potentially be directly deposited onto tissues such as the spleen and liver in the peritoneal cavity (62). Following the i.n. route of infection, however, the lungs would be inoculated as a primary site of infection, and tissues such as the spleen and liver would be more distal secondary sites of infection. To initiate our studies, mice were anesthetized, and 1 × 106 PFU of virus was administered by the i.n. route. As shown in Fig. 2, replicating virus was detected in the lungs at 3 dpi, demonstrating that the lungs were efficiently infected with MCMV by this route. No significant differences were detected between the ΔM33BT2 titers and the wild-type virus titers (P > 0.05) for each of the time points. A similar result was observed with a lower inoculum of 5 × 105 PFU (data not shown). Although conclusions about differences in viral clearance cannot be made since later time points were not included in our analysis, these results demonstrated that disruption of M33 did not significantly reduce the level of virus replication in the lungs up to 7 days following i.n. inoculation, in agreement with the previous observation following i.p. inoculation.
FIG. 2.
Virus titers of wild-type viruses and ΔM33BT2 in the salivary glands, lungs, and spleen following i.n. inoculation. Five-week-old BALB/c mice were inoculated i.n. with 1 × 106 PFU of K181 (K-Perth), ΔM33BT2 (ΔM33), M33R D1.6, and M33R F2.1. Titers of tissue sonicates were determined at 3, 5, and 7 dpi. The mean virus titer (log10 PFU/ml) and standard error are shown for K181, ΔM33, and M33R F2.1 (n = 12) for a total of three separate experiments. The M33R D1.6 virus was included in one study and is presented as mean virus titer and standard deviation (n = 4). The limit of detection for the plaque assay is shown by the dotted line. Virus titers depicted below the limit of detection indicate where samples were negative at the limit of detection (e.g., 10−1 dilution) but positive for virus in undiluted samples. The P value is shown in the diagram: *, P < 0.05; **, P < 0.01; ***, P < 0.001. ND, not done.
We next measured dissemination of MCMV from the lungs to other tissue sites. Following i.n. inoculation, virus was undetectable in the pancreas and close to the limit of detection in the liver (data not shown). As shown in Fig. 2, virus was detectable in the salivary glands and spleen after 5 dpi, indicating that these tissues are secondary sites of infection following i.n. inoculation. In the salivary glands, ΔM33BT2 was undetectable at both 5 and 7 dpi, whereas wild-type viruses reached high titers (P < 0.001), similar to the results following i.p. inoculation. In the spleen, ΔM33BT2 titers were significantly lower than those of wild-type viruses by 7 dpi (P < 0.05). Although ΔM33BT2 titers in the spleen were low following i.n. inoculation, these results agree with the reduced ΔM33BT2 spleen titers observed following i.p. inoculation. Similarly, the i.n. route of inoculation also confirmed the requirement for M33 during salivary gland infection, whereas infection of the lungs did not require M33 function. Notably, the i.n. route of infection provides a picture of dissemination kinetics from a single primary site of infection, the lung, to distal secondary sites of infection.
Disruption of M33 alters the numbers of infected cells in the spleen.
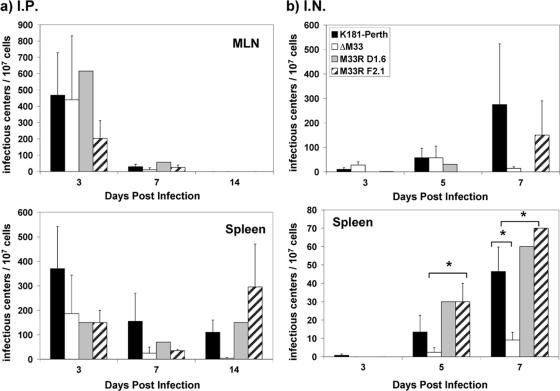
Decreased ΔM33BT2 virus titers in the tissues could be due to decreased viral replication or decreased numbers of infected cells. To detect viral infection at the cellular level, we used a coculture or infectious center assay for MCMV, where each plaque that develops in the underlying fibroblast monolayer represents virus that originated from one infected cell (11, 62). To begin our analysis of the numbers of infected cells, we determined whether MCMV could be detected in the regional draining lymph nodes of the lung, the mediastinal lymph nodes (MLN), similar to our observations with murine gammaherpesvirus-68 (11). This could then serve as a lymphoid site that would provide insights into dissemination following both i.p. and i.n. infection. As shown from this analysis in Fig. 3, MCMV was detected in the MLN as early as 3 dpi by both the i.p. and i.n. routes although to different levels. The numbers of ΔM33BT2-infected cells and wild-type virus-infected cells in the spleen and MLN were similar at 3 dpi following i.p. inoculation. By 14 dpi, the number of ΔM33BT2-infected cells in the spleen was eightfold lower than the number of wild-type virus-infected cells although not significantly different. The route of inoculation influenced the overall total numbers of infected cells detected in the MLN and spleen although the total numbers of leukocytes in the spleen and MLN were similar between the groups over the 14-day infection time and between studies (data not shown). Virus-infected cells were also detected in the MLN and spleen following i.n. inoculation. In the MLN, no significant differences were detected between any of the viruses. In contrast, reduced numbers of ΔM33BT2-infected cells were detected in the spleen at both 5 dpi (P < 0.05) and 7 dpi (P < 0.05) following i.n. inoculation. By 7 dpi, increased numbers of infected cells in the spleen were detected for all of the groups, suggesting that virus was disseminating to or replicating in the spleen. Similar to the results with i.p. inoculation, the total numbers of leukocytes in the MLN and spleens following i.n. inoculation did not vary significantly (data not shown). Overall, we found higher numbers of wild-type virus-infected cells in the spleen than of ΔM33BT2 virus-infected cells following both routes of inoculation.
FIG. 3.
Virus-infected cell numbers following i.p. and i.n. inoculation. Five-week-old BALB/c mice were inoculated i.p. with 2 × 106 PFU or i.n. with 1 × 106 PFU of K181 (K-Perth), ΔM33BT2 (ΔM33), M33R D1.6, and M33R F2.1. Dilutions of cell suspensions from the MLN and spleens were cocultured with NIH 3T3 cells as described in Materials and Methods. Results are depicted as the number of infectious centers/1 × 107 cells. The mean and standard error are shown for K181, ΔM33, and M33R F2.1 (n = 3) from a total of three separate experiments (four mice/group; pooled samples). M33R D1.6 was included for analysis in only one study. The P values are shown in the diagram: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
M33 is required for efficient establishment of latency in the spleen and lung.
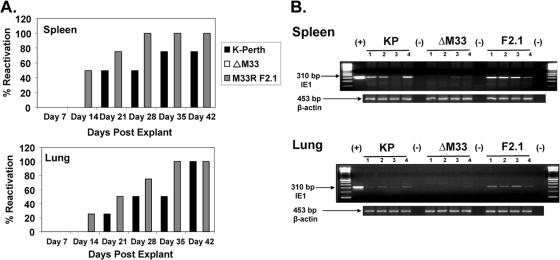
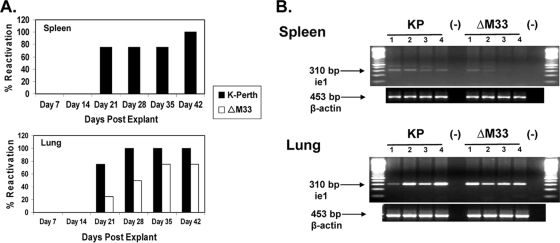
As our acute i.p. infection results demonstrated, M33 deficiency significantly reduced the levels of infectious virus in the spleen at 7 and 14 dpi. To determine whether this acute infection phenotype in the spleen affected latency in the spleen, we performed explant reactivation assays. Mice were inoculated i.p. with 1 × 106 PFU of parent virus, the ΔM33BT2 virus, and the rescue virus M33R F2.1, and at 90 dpi, the salivary gland, spleen, and lungs were collected and analyzed individually for each mouse (four mice/group). The salivary glands and lungs were sonicated and were negative for infectious virus by plaque assay, indicating that these tissues did not harbor persistent replicating virus (data not shown). The spleens were minced, and primary cultures were established as previously described and cultured for 6 weeks (12). Reactivating virus was detected at 14 days postexplant for both K181 (50%) and M33R F2.1 (75%). Significantly, ΔM33BT2 did not reactivate virus (0%) from the spleen cultures and after 6 weeks was still negative for reactivation, whereas 100% reactivation from the spleens was observed for the wild-type viruses (data not shown).
Since ΔM33BT2 virus did not show a defect in lung titers during acute infection, we next compared reactivation from the spleen and lungs to determine whether the defect in latency was also detected in the lungs. A second reactivation assay was performed on both latently infected spleens and lungs isolated from mice infected 50 days earlier with K181, ΔM33BT2, and M33R F2.1 (four mice/group). The salivary glands were negative for infectious virus at 50 dpi, indicating that the mice did not harbor persistently replicating virus at this site (data not shown). As shown in Fig. 4A, the wild-type viruses reactivated 75 to 100% from the spleens and the lungs, whereas ΔM33BT2 did not reactivate from either explanted tissue, indicating that ΔM33BT2 was impaired for latent infection in both the spleen and lungs. Remarkably, unlike results in the spleen, ΔM33BT2 replicated to similar levels as wild type in the lungs following i.p. inoculation, suggesting that the inability to reactivate ΔM33BT2 virus from the lungs was not a consequence of reduced ΔM33BT2 replication in the lung during acute infection.
FIG. 4.
Reduced latent ΔM33BT2 DNA levels lead to reduced reactivation from spleens and lungs following i.p. inoculation. Five-week-old BALB/c mice were inoculated i.p. with 1 × 106 PFU of K181 (K-Perth), ΔM33BT2 (ΔM33), or M33R F2.1. At 50 dpi, spleens and lungs were collected and analyzed for reactivation from latency or detection of latent viral DNA. (A) Spleens and lungs were minced and cultured for 6 weeks in explant reactivation assays. Detection of virus in the cultures was determined weekly by plaque assay. (B) Tissue DNA was isolated as described in Materials and Methods and screened for the presence of MCMV ie1 by PCR using primers CH16 and CH17. Ethidium bromide-stained agarose gels of amplified PCR products are shown: 100-bp DNA ladder, positive MCMV control (pARK25; +), DNA samples from K181 (KP), ΔM33, and M33R F2.1 (F2.1), and negative PCR control (−). Amplification of β-actin is shown below to control for DNA isolation. Results shown are from one study (four mice/group).
Reduced ΔM33BT2 DNA levels in latently infected spleen and lungs.
A defect in virus reactivation from latency in the explant reactivation assay could indicate that ΔM33BT2 is defective in reactivation or does not efficiently establish or maintain a latent infection in the spleen or lungs. To further explore these possibilities, PCR analysis was undertaken on both the spleen and lung cells collected from the tissues isolated at 50 dpi. Following the mincing of the tissues and just prior to incubation of the explant cultures, six small pieces of spleen tissue and lung tissue from each animal were collected (two tissue pieces per well) to isolate whole-cell DNA as described in Materials and Methods. The ie1 sequence was amplified using primers previously used to evaluate latency in the spleen. As shown in Fig. 4B, viral DNA was detected in the spleens and lungs of mice infected with the wild-type viruses, as shown by amplification of the 310-bp ie1 DNA product. In contrast, viral DNA was detected in only two of four spleens (lanes 3 and 4) and one of four lungs (lane 4) of the ΔM33BT2-infected mice. A larger nonspecific DNA product was observed above the specific 310-bp band in some DNA samples, which is consistent with previously reported observations (38), and when control pARK25 DNA was added to uninfected spleen and lung samples to determine the limits of detection.
To further determine whether we could detect viral DNA in the negative ΔM33BT2-infected DNA samples, nested PCR was performed (38). All DNA samples isolated from wild-type virus-infected mice and DNA samples from ΔM33BT2-infected mice which were initially positive by first-round PCR yielded the expected nested PCR product. Nested PCR analysis of the negative ΔM33BT2 DNA samples amplified one faint product from one mouse spleen and one product from a mouse lung that were both initially negative in the 50-dpi DNA samples from ΔM33BT2-infected mice (data not shown). Overall, a total of four separate explant reactivation studies were performed on mice following i.p inoculation, and these are summarized in Table 1. In previous studies, recombinant viruses encoding lacZ were shown to reactivate at levels similar to those of wild-type viruses, indicating that expression of β-galactosidase from the viral genome does not interfere with virus reactivation in the explant assay (12, 62). Significantly, all of the explant assays demonstrated that ΔM33BT2 shows a severely diminished capacity to reactivate from latency in an explant reactivation assay. This may be a consequence of reduced establishment or maintenance of latency since there was a reduced ability to detect viral DNA in the spleen and lungs of mice infected i.p. with ΔM33BT2 compared to wild-type virus.
TABLE 1.
Summary of virus reactivation from latently infected spleen and lungs
| Inoculation route and virus strain | Reactivation in the indicated tissue at 42 days postexplant (no. of mice positive/total no. of mice [% reactivation])c
|
Salivary gland titer (log10 PFU/ml)d | |
|---|---|---|---|
| Spleen | Lung | ||
| i.p. groupa | |||
| K181 | 15/16 (93.8) | 7/8 (87.5) | <1.0 |
| ΔM33BT2 | 1/16 (6.25)*** | 0/8 (0)*** | <1.0 |
| M33R F2.1 | 11/12 (91.7) | 4/4 (100) | <1.0 |
| i.n. groupb | |||
| K181 | 5/6 (83.3) | 6/6 (100) | <1.0 |
| ΔM33BT2 | 0/6 (0)* | 5/6 (83.3) | <1.0 |
For each virus, mice were inoculated with 1 × 106 to 2 × 106 PFU.
For each virus, mice were inoculated with 1 × 106 PFU.
Day 42 explant reactivation results are from four separate experiments for i.p. studies (three to four mice/group) and from two separate experiments for i.n. studies (two mice/group and four mice/group). The tissues from i.p. studies were collected at days 50, 90 (two experiments), and 110 postinfection, and the tissues from i.n. tissues were from days 65 and 92 postinfection. Tissues were divided into three parts and minced, and explant reactivation assays were performed. For each tissue, one well positive for plaques was considered positive for reactivation; for wild-type viruses, at least two of three wells were positive. ***, P < 0.001; *, P < 0.05.
Titers of virus in salivary glands were used to determine whether mice were latently infected. All salivary gland tissues in all studies were negative for virus at the −1 dilution (the limit of detection, 10 PFU/ml tissue homogenate).
Route of inoculation influences ΔM33BT2 latency in the lungs.
To determine whether the defect in latency of the ΔM33BT2 virus was dependent on the route of inoculation, mice were inoculated by the i.n. route with 1 × 106 PFU of K181 and ΔM33BT2, and at 92 dpi, explant cultures of spleens and lungs were evaluated for reactivation of virus. A portion of spleen and lung tissue was collected for isolation of genomic DNA. As shown in Fig. 5A, K181 reactivated 100% from the spleens and lungs of latently infected mice following i.n. inoculation. In contrast, ΔM33BT2 did not reactivate from any spleen cultures, which is consistent with the observed defect in the reactivation assays following i.p. inoculation. Unexpectedly, ΔM33BT2 showed a different reactivation phenotype for the latently infected lungs following i.n. inoculation, with 75% reactivation after 6 weeks. In agreement, PCR analysis demonstrated that both the K181 latently infected lungs and the ΔM33BT2 latently infected lungs harbored detectable viral DNA, as shown in Fig. 5B. In the spleens from ΔM33BT2-infected mice, at least two of the four samples (lanes 1 and 2) had detectable ie1 products following PCR amplification. Even so, ΔM33BT2 did not reactivate from these spleens in the explant reactivation assay, perhaps due to a lower latent DNA load. A second explant reactivation assay was performed at 65 dpi following i.n. inoculation, and, in agreement with the previous assay results, K181 reactivated from 50% of the spleens and 100% of the lungs, whereas ΔM33BT2 reactivated from 0% of the spleens and 100% of the lungs. The combined results from the various explant reactivation studies are summarized in Table 1.
FIG. 5.
Different latent ΔM33BT2 DNA loads in the spleen and lungs following i.n. inoculation leads to reduced reactivation from the spleen but not the lungs. Five-week-old BALB/c mice were inoculated i.n. with 1 × 106 PFU of K181 (K-Perth) or ΔM33BT2 (ΔM33). At 92 dpi, spleens and lungs were collected and analyzed for reactivation from latency or detection of latent viral DNA. (A) Spleens and lungs were minced and cultured for 6 weeks in explant reactivation assays. Detection of virus in the cultures was determined weekly by plaque assay. (B) Tissue DNA was isolated as described in Materials and Methods and screened for the presence of MCMV ie1 by PCR using primers CH16 and CH17. Ethidium bromide-stained agarose gel of amplified PCR products are shown: 100-bp DNA ladder, DNA samples from K181 (KP) and ΔM33, and negative PCR control (−). Amplification of β-actin is shown below to control for DNA isolation. Results shown are from one study (four mice/group).
M33 is required for efficient establishment of latency in the bone marrow.
MCMV establishes latent infection in peritoneal macrophages and in BMC (3, 38, 49), so we next evaluated the viral DNA loads at these two sites at a time when latency is detectable at these sites. For this evaluation, 5-week-old BALB/c mice were inoculated i.p. with 1 × 106 PFU of M33R F2.1 and ΔM33BT2; at 50 dpi, PEC were collected by peritoneal lavage, and BMC were collected from the femurs of the infected mice. The cells were pooled, and DNA was extracted from 5 × 106 cells and analyzed by PCR. As shown in Fig. 6, the expected ie1 product was detected in the bone marrow from M33R F2.1-infected mice, whereas viral DNA was not detectable in the bone marrow of ΔM33BT2-infected mice. This result was confirmed in a second study in which BMC were isolated at 70 dpi from mice previously infected by the i.p. route with 1 × 106 PFU of M33R F2.1 and ΔM33BT2. Similarly, no ΔM33BT2 viral DNA was detected in the BMC by PCR analysis (data not shown). In contrast, latent viral DNA was detected in the peritoneal cavity of mice infected with both viruses. Taken together, these results provide the first preliminary evidence that disruption of M33 impacts the latent viral load in known sites of MCMV latency, such as the spleen, lungs, and bone marrow. The mechanism remains to be determined, but possible explanations include decreased dissemination to or decreased replication at these sites (thereby reducing the opportunity to establish latency) or a reduced ability to establish or maintain latent infection within these sites. Interestingly, the ΔM33BT2 viral DNA load in the lungs and PEC, sites of primary infection following i.n. and i.p. infection, respectively, was similar to wild-type latent viral load, perhaps indicating that the effect of M33 on latent viral load at these sites can be bypassed by direct inoculation of virus onto resident cells where MCMV can establish latency. However, an important caveat of this study is that the methods used for detection of viral DNA did not distinguish between viral genomes that had established a state of latency and viral DNA that may have persisted in the absence of latency (e.g., from residual virus particles or abortive infection).
FIG. 6.
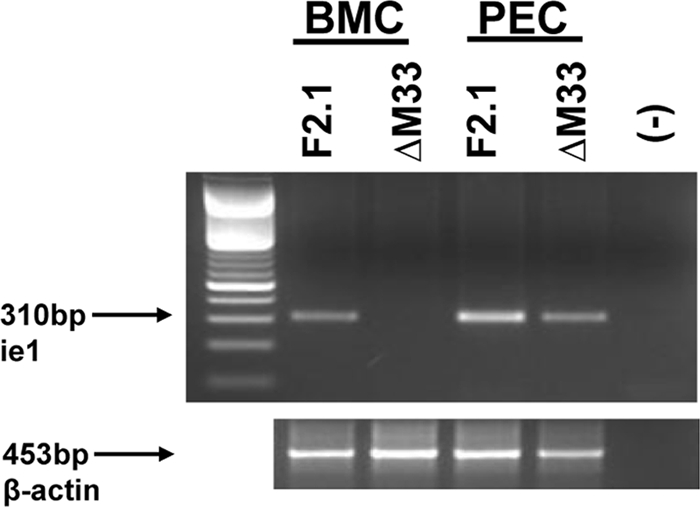
Reduced ΔM33BT2 latency in BMC and PEC following i.p. inoculation. Five-week-old BALB/c mice were inoculated i.n. with 1 × 106 PFU of M33R F2.1 or ΔM33BT2 (ΔM33). At 50 dpi, BMC and PEC were collected and analyzed for the presence of latent viral DNA. DNA was isolated from 5 × 106 pooled cells and screened for the presence of MCMV ie1 by PCR using primers CH16 and CH17. Ethidium bromide-stained agarose gel of amplified PCR products are shown: 100-bp DNA ladder, DNA samples from M33R F2.1 (F2.1) and ΔM33, and negative PCR control (−). Amplification of β-actin is shown below to control for DNA isolation. Results shown are from one study (four mice/group).
In vivo complementation of ΔM33BT2 virus.
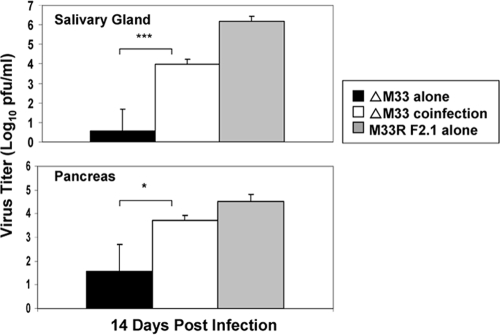
HCMV UL33 partially rescues the ΔM33BT2 defect in salivary gland infection since a UL33 substitution mutant virus partially restored the salivary gland titers (14). Several examples of trans-complementation of attenuated MCMV by coinfection with wild-type MCMV have been reported (17, 58). We next investigated whether coinfection of mice with ΔM33BT2 along with wild-type virus (expressing M33) would functionally complement the defect in the salivary glands, spleen, and pancreas. For this analysis, we took advantage of the “blue-versus-white” detection afforded by the insertion of the lacZ gene into the M33 locus to distinguish the virus titers of the ΔM33BT2 virus from the wild-type virus titers. As shown in Fig. 7, at 14 dpi following i.p. inoculation, salivary glands and pancreas from mice infected with ΔM33BT2 showed the expected reduced virus titers. Coinfection with the rescue virus M33R F2.1 significantly increased the numbers of lacZ-expressing ΔM33BT2 plaques by 595-fold in the salivary glands (P = 0.001), as shown by staining with X-Gal (60, 62). As these data show, wild-type M33 expression was sufficient to significantly restore the salivary gland infection, albeit not to wild-type levels. Coinfection with M33R F2.1 also increased the numbers of blue ΔM33BT2 plaques in the pancreas by 4.9-fold (P < 0.05). In the spleen at 14 dpi, where wild-type levels of infectious virus were much lower than in the salivary glands or pancreas, ΔM33BT2 plaques were not detected (data not shown). These data indicate that the ΔM33BT2 defect in the salivary gland and pancreas can be functionally rescued by wild-type virus coinfection.
FIG. 7.
Complementation of ΔM33BT2 in the salivary glands and pancreas following coinfection. Five-week-old BALB/c mice were inoculated i.p. with either 2 × 106 PFU of ΔM33BT2 (ΔM33) alone, 2 × 106 PFU of M33R F2.1 alone, or 2 × 106 PFU of ΔM33BT2 in combination with 1 × 106 of M33R F2.1 (ΔM33 coinfection). Titers of tissue sonicates were determined at 14 dpi by plaque assay on NIH 3T3 cells. The monolayers were stained with X-Gal to detect blue ΔM33BT2 plaques and then with Giemsa to detect white M33R F2.1 plaques. The mean X-Gal-positive virus titer (log10 PFU/ml) and standard deviation (n = 4) are shown from one study. The P value is shown in the diagram: *, P < 0.05; ***, P = 0.001.
DISCUSSION
The biological significance of the CMV-encoded GPCR homologs can be fully addressed only during in vivo infection, and MCMV infection of mice offers a relevant biological system in which to dissect the role of the CMV-encoded receptor homologs during virus-host interactions. Some possible roles for the CMV-encoded GPCR homologs include the following: (i) as a coreceptor for tissue-specific or cell-type-specific infection; (ii) as a viral chemokine receptor involved in viral dissemination, e.g., the migration or trafficking of infected cells in vivo; (iii) as an activator or modulator of signaling pathways important for viral replication or cell survival; and (iv) as a modulator of the immune response to mediate immune evasion, e.g., by sequestering chemokines and dampening the recruitment of immune cells to sites of infection (reviewed in references 5, 39, and 68). Although M33 has not yet been shown to bind or sequester chemokines, it activates signaling pathways and induces migration of smooth-muscle cells in vitro (14, 41, 63, 71).
The in vivo function of the UL33 receptor family members was first investigated by deletion of the MCMV and RCMV homologs, M33 and R33, which demonstrated that they were required for salivary gland infection (6, 19). More recently, M33-induced G protein-coupled signaling activity was shown to be important for salivary gland infection (14). However, it was not clear whether the defective replication phenotype was restricted to salivary glands or whether this phenotype was associated with a general defect in virus replication, dissemination from primary to secondary sites of infection, or avoidance of immune clearance. In this report, we provide further elucidation of the roles played by M33 during MCMV pathogenesis and latency. Importantly, we demonstrate the following points. First, disruption of M33 results in tissue-specific defects in virus replication. Thus, whereas ΔM33BT2 replication in the lung was similar to that of the wild-type virus, the M33 null virus was attenuated to various degrees for replication in the spleen, pancreas, and salivary glands. These tissue-specific phenotypes were detected following two different routes of inoculation. Second, there did not appear to be a generalized dissemination defect for ΔM33BT2. In particular, dissemination of ΔM33BT2 to the lung was similar to that of wild-type virus. Third, disruption of M33 was associated with inefficient establishment or maintenance of MCMV latent infection, as shown by reduced viral DNA levels and reduced reactivation from latency from the spleen and lungs.
The finding that replication in the lungs in the absence of M33 was similar to wild-type viruses, following both routes of inoculation, suggests that the phenotypes attributed to M33 are due to tissue-specific, rather than general, effects. MCMV infects a broad variety of cell types, and the predominant cell types supporting virus replication vary between different tissues. Therefore, M33 may promote virus replication and/or cell-cell spread in certain cell types and hence is required for efficient virus replication in some tissues but not others. M33-mediated signaling may contribute to the tissue-specific replication defects since MCMV mutants which fail to express M33, or express M33 deficient in signaling activity, are highly attenuated for replication in the salivary glands (14, 19). Interestingly, we found that M33 is required for efficient infection of the pancreas. The observed defect in replication in the pancreas, although not as severe as that in salivary glands, may potentially result from similarities between the cell types infected in these two tissues. Both the salivary glands and the pancreas share similar cell types and lineages, including acinar epithelial cells and ductal epithelial cells, which are susceptible to MCMV infection, and this could indicate that M33 activates a common signaling pathway important for MCMV replication in both of these glandular tissues (29).
ΔM33BT2 infection of the spleen following i.p. inoculation presented a distinct infection profile. At 3 dpi, ΔM33BT2 titers were equivalent to the wild-type virus titers. Strikingly, 4 days later, ΔM33BT2 titers were below detectable levels, suggesting that M33 is not required for initial infection of the spleen but is required for continued efficient replication or perhaps avoidance of immune clearance at this site. ΔM33BT2 was also attenuated in the spleen following i.n. inoculation, further emphasizing the requirement for M33 during spleen infection. This defect in spleen replication is most likely not due to lacZ expression since previous studies showed that lacZ-tagged viruses replicated to high titers in the spleen, and X-Gal-staining cells were detected in the perifollicular areas of the spleen (12, 62). Thus, the dramatic decline in ΔM33BT2 titers could indicate that M33 is required for replication in splenocytes or for cell-to-cell spread within the spleen. Alternatively, GPCR-mediated signaling may promote cell survival, and in the absence of M33-mediated signaling, the ΔM33BT2-infected cells could undergo enhanced apoptosis (69). Furthermore, it is possible that M33 contributes to immune evasion during infection; thus, in the absence of M33, increased clearance of infected cells by the immune response could occur. However, a role for M33-mediated immune evasion in the lung is not apparent since the peak titers and clearance kinetics of ΔM33BT2 were similar to those of the wild type, suggesting that immune control of ΔM33BT2 in the lungs was similar to that of wild-type virus. Further studies are required to address these possible mechanisms.
To further explore the correlation of dissemination of infected cells, we used an assay that was previously used by us and others to evaluate MCMV viremia and murine gammaherpesvirus-infected cells in the spleens and lymph nodes (11, 62, 65). In our current studies, we did not find evidence of a role for M33 during viral dissemination. We employed two different routes of inoculation to compare virus spread from primary sites of infection, the peritoneal cavity and lungs, to secondary sites. There did not appear to be a general deficiency in dissemination following i.p. inoculation since ΔM33BT2 disseminated efficiently to the lungs, spleen, and MLN at levels similar to those of wild-type viruses at early times after infection. It is possible that not all infected cells yielded plaques in the infectious center assays, thus leading to underestimations of the numbers of infected cells. We cannot rule out a tissue-specific defect, however, which could impact dissemination. Theoretically, a cell type important for dissemination to the salivary gland or other tissues may require M33 for efficient transmission. The importance of specific cell types to dissemination has been demonstrated by the recent finding that cell types which are not the predominant source of infectious virus may be the major source of disseminated virus (i.e., virus that replicated in hepatocytes, despite high titers, was relatively unimportant for dissemination, whereas virus derived from endothelial cells was important) (58). It is also possible that MCMV dissemination to some sites may operate through different cell types, such as shown for m131/129-mediated recruitment of myelomonocytic progenitors from the bone marrow (46), or, as recently reported, that MCMV disseminates to the MLN as free virus particles following i.p. inoculation (30). This mechanism of dissemination would not necessarily reveal a role for M33 cell surface-mediated receptor function, at least during primary dissemination. It remains to be determined whether M33 plays a role in secondary viremia and dissemination of infected cells (18, 58, 62).
Like HCMV, MCMV establishes latency in a number of tissues, including the spleen, lungs, and bone marrow (4, 28, 31, 38, 50). Furthermore, monocytes and macrophages, along with hematopoietic progenitor cells, are considered cellular sites of latency for both MCMV and HCMV (10, 24, 33, 38, 43, 49, 61). Latency is defined as the ability to reactivate virus or detect viral genome in the absence of infectious virus (55). The more rapid decline of ΔM33BT2 spleen titers raised the question whether ΔM33BT2 latent infection in the spleen was altered. The explant reactivation assay provided the first intriguing result, demonstrating that ΔM33BT2 was severely deficient in reactivation from the spleens and lungs of latently infected mice following i.p. inoculation even though virus titers in the spleen at 3 dpi and in the lung at 7 dpi were similar for ΔM33BT2 and the wild-type viruses (Fig. 1). As PCR analysis demonstrated, the spleens, lungs, and bone marrow from ΔM33BT2-infected mice harbored reduced levels of detectable latent viral DNA compared to the wild-type-infected tissues. Defective establishment of ΔM33BT2 latency in the lungs is particularly interesting in light of the normal levels of virus titers in the lungs. It is possible that M33 contributes to or facilitates the infection of a particular cell type that is important for latency in the lung but not for infectious virus production in the lungs. We are currently characterizing the wild-type and ΔM33BT2 infection of specific cell types in the lungs and spleen to determine whether this is the case for both of these tissues. Further analysis is under way to determine whether M33 is required for infection of the bone marrow compartment early during infection. It is possible that a latency defect in the bone marrow compartment over time leads to a decline in latently infected cells at other tissue sites. In the spleen, it will be necessary to distinguish between a specific defect in the establishment or maintenance of latency versus a consequence of a generalized reduction in replication and dissemination. Alternatively, if M33 contributes to immune evasion, it will be necessary to demonstrate increased immune surveillance and fewer numbers of latently infected cells in the absence of M33 expression during infection.
The lung is considered a major site of MCMV latency (4). In contrast to the results following i.p. inoculation, ΔM33BT2 viral DNA was readily detected in the lungs following i.n. inoculation, and the virus reactivated from the lungs following explant, although to approximately fivefold lower levels and with delayed kinetics, suggesting lower levels of reactivating virus. Considering the 100-fold higher levels of virus replication in the lungs following i.n. compared with i.p. inoculation, the proposed defect in ΔM33BT2 latency may have been rescued by a higher level of infection of resident lung cells permissive for latent infection. However, as with i.p. inoculation, ΔM33BT2 did not efficiently establish latency in the spleens following i.n. inoculation. Wild-type virus was still efficiently reactivated from the spleen, despite lower levels of replication compared to the i.p. route and relatively low levels of viral DNA detected at the time of tissue harvest. Thus, disruption of M33 impacts latency in the spleen for both routes of inoculation. A similar effect for inoculation route was shown for the murine gammaherpesvirus latency-associated protein, M2, and its role during latent infection in the spleen (35). Overall, the premise that the conditions of primary infection define the load of latent genome in tissues and the risk of CMV recurrences holds true as measured by PCR analysis and reactivation assays (53). Further studies are under way to quantify viral DNA load during the early (establishment) phase of latency to determine whether ΔM33BT2 does not efficiently establish latency in particular cell types or whether ΔM33BT2 latency levels are similar to the level of wild-type virus at early times during latency but is not maintained.
Lastly, we found that we could rescue the defect in ΔM33BT2 infection of the salivary glands and pancreas by coinfection studies. A phenomenon referred to as trans-complementation occurs in vivo when mice are coinfected with an attenuated virus and a wild-type virus (17, 58). In very elegant studies, trans-complementation was shown to occur by the coinfection of the same cell by the two different viruses, resulting in the rescue of the mutant virus for the specific phenotype by the wild-type virus. We reasoned that if the attenuation of ΔM33BT2 was due to lacZ expression (e.g., enhanced immune clearance), trans-complementation by wild-type virus would not occur since lacZ would still be expressed in cells coinfected with ΔM33BT2 and wild-type virus. Using a mixed infection approach, we found complementation of the ΔM33BT2 virus by coinfection with a wild-type MCMV. Impressively, we found that wild-type virus dramatically increased the ΔM33BT2 titers in the salivary glands and, to a lesser extent, in the pancreas. Interestingly, trans-complementation was not detected in the spleen. These results support the hypothesis that ΔM33BT2 lacks a function required for infection of specific tissues and that the observed attenuation did not result from lacZ expression. It remains to be determined whether complementation of ΔM33BT2 by the wild-type virus (presumably via coinfection within cells) promoted either dissemination to or replication in the salivary gland and pancreas.
In summary, the studies presented here identify two new tissues, the spleen and pancreas, that require M33 for efficient MCMV infection in vivo. We also show preliminary data suggesting that M33 contributes to or plays a role in the efficient establishment of latency. This is an intriguing result since host GCPRs are multifunctional proteins which exhibit activities in signal transduction and activation of gene expression, migration and trafficking of cells, survival and apoptosis, homeostasis of immune cells, and potentially modulation of immune responses. Thus, it will be important to delineate the role of the MCMV-encoded GPCR in both tissue-specific viral replication and the establishment or maintenance of latent infection. Studies are currently under way using defined mutant viruses (14) to determine whether M33-mediated signaling pathways play a role during MCMV latency and to identify the cell types that are involved in the ΔM33BT2 phenotypes described in the current study. In light of the data presented, it is intriguing to speculate that the cytomegaloviruses have hijacked the UL33 family of CC chemokine receptors to modulate viral dissemination and tissue-specific infection as well as long-term latent infection.
Acknowledgments
We thank Rosemary Rochford for critical reading of the manuscript and Andrea Sewell and Nicholas Farley for technical assistance.
R.D.C. was supported by grant AHA 0665211B. H.E.F. and N.D.-P. were supported by grants from the NHMRC (456137) and Queensland Health.
Footnotes
Published ahead of print on 13 May 2009.
REFERENCES
- 1.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bale, J. F., Jr., and M. E. O'Neil. 1989. Detection of murine cytomegalovirus DNA in circulating leukocytes harvested during acute infection of mice. J. Virol. 63:2667-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balthesen, M., L. Dreher, P. Lucin, and M. J. Reddehase. 1994. The establishment of cytomegalovirus latency in organs is not linked to local virus production during primary infection. J. Gen. Virol. 752329-2336. [DOI] [PubMed] [Google Scholar]
- 4.Balthesen, M., M. Messerle, and M. J. Reddehase. 1993. Lungs are a major organ site of cytomegalovirus latency and recurrence. J. Virol. 675360-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beisser, P. S., C. S. Goh, F. E. Cohen, and S. Michelson. 2002. Viral chemokine receptors and chemokines in human cytomegalovirus trafficking and interaction with the immune system. CMV chemokine receptors. Curr. Top. Microbiol. Immunol. 269203-234. [DOI] [PubMed] [Google Scholar]
- 6.Beisser, P. S., C. Vink, J. G. Van Dam, G. Grauls, S. J. Vanherle, and C. A. Bruggeman. 1998. The R33 G protein-coupled receptor gene of rat cytomegalovirus plays an essential role in the pathogenesis of viral infection. J. Virol. 722352-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billstrom, M. A., G. L. Johnson, N. J. Avdi, and G. S. Worthen. 1998. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J. Virol. 725535-5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billstrom, M. A., L. A. Lehman, and G. Scott Worthen. 1999. Depletion of extracellular RANTES during human cytomegalovirus infection of endothelial cells. Am. J. Respir. Cell Mol. Biol. 21163-167. [DOI] [PubMed] [Google Scholar]
- 9.Bodaghi, B., T. R. Jones, D. Zipeto, C. Vita, L. Sun, L. Laurent, F. Arenzana-Seisdedos, J. L. Virelizier, and S. Michelson. 1998. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J. Exp. Med. 188855-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brautigam, A. R., F. J. Dutko, L. B. Olding, and M. B. Oldstone. 1979. Pathogenesis of murine cytomegalovirus infection: the macrophage as a permissive cell for cytomegalovirus infection, replication and latency. J. Gen. Virol. 44349-359. [DOI] [PubMed] [Google Scholar]
- 11.Cardin, R. D., J. W. Brooks, S. R. Sarawar, and P. C. Doherty. 1996. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J. Exp. Med. 184863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardin, R. D., J. M. Boname, G. B. Abenes, S. A. Jennings, and E. S. Mocarski. 1993. Reactivation of murine cytomegalovirus from latency, p. 65-74. In S. Michelson and S. A. Plotking (ed.), Multidisciplinary approaches to understanding cytomegalovirus disease. Elsevier, Amsterdam, The Netherlands.
- 13.Casarosa, P., Y. K. Gruijthuijsen, D. Michel, P. S. Beisser, J. Holl, C. P. Fitzsimons, D. Verzijl, C. A. Bruggeman, T. Mertens, R. Leurs, C. Vink, and M. J. Smit. 2003. Constitutive signaling of the human cytomegalovirus-encoded receptor UL33 differs from that of its rat cytomegalovirus homolog R33 by promiscuous activation of G proteins of the Gq, Gi, and Gs classes. J. Biol. Chem. 27850010-50023. [DOI] [PubMed] [Google Scholar]
- 14.Case, R., E. Sharp, T. Benned-Jensen, M. M. Rosenkilde, N. Davis-Poynter, and H. E. Farrell. 2008. Functional analysis of the murine cytomegalovirus chemokine receptor homologue M33: ablation of constitutive signaling is associated with an attenuated phenotype in vivo. J. Virol. 821884-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154125-169. [DOI] [PubMed] [Google Scholar]
- 16.Chee, M. S., S. C. Satchwell, E. Preddie, K. M. Weston, and B. G. Barrell. 1990. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature 344774-777. [DOI] [PubMed] [Google Scholar]
- 17.Cicin-Sain, L., J. Podlech, M. Messerle, M. J. Reddehase, and U. H. Koszinowski. 2005. Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected host. J. Virol. 799492-9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins, T. M., M. R. Quirk, and M. C. Jordan. 1994. Biphasic viremia and viral gene expression in leukocytes during acute cytomegalovirus infection of mice. J. Virol. 686305-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis-Poynter, N. J., D. M. Lynch, H. Vally, G. R. Shellam, W. D. Rawlinson, B. G. Barrell, and H. E. Farrell. 1997. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J. Virol. 711521-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleming, P., N. Davis-Poynter, M. Degli-Esposti, E. Densley, J. Papadimitriou, G. Shellam, and H. Farrell. 1999. The murine cytomegalovirus chemokine homolog, m131/129, is a determinant of viral pathogenicity. J. Virol. 73:6800-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goh, C. S., A. A. Bogan, M. Joachimiak, D. Walther, and F. E. Cohen. 2000. Co-evolution of proteins with their interaction partners. J. Mol. Biol. 299283-293. [DOI] [PubMed] [Google Scholar]
- 22.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 20929-51. [DOI] [PubMed] [Google Scholar]
- 23.Gruijthuijsen, Y. K., P. Casarosa, S. J. Kaptein, J. L. Broers, R. Leurs, C. A. Bruggeman, M. J. Smit, and C. Vink. 2002. The rat cytomegalovirus R33-encoded G protein-coupled receptor signals in a constitutive fashion. J. Virol. 761328-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn, G., R. Jores, and E. S. Mocarski. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 953937-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn, G., M. G. Revello, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 7810023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedrick, J. A., and A. Zlotnik. 1996. Chemokines and lymphocyte biology. Curr. Opin. Immunol. 8343-347. [DOI] [PubMed] [Google Scholar]
- 27.Henry, S. C., and J. D. Hamilton. 1993. Detection of murine cytomegalovirus immediate early 1 transcripts in the spleens of latently infected mice. J. Infect. Dis. 167950-954. [DOI] [PubMed] [Google Scholar]
- 28.Henry, S. C., K. Schmader, T. T. Brown, S. E. Miller, D. N. Howell, G. G. Daley, and J. D. Hamilton. 2000. Enhanced green fluorescent protein as a marker for localizing murine cytomegalovirus in acute and latent infection. J. Virol. Methods 8961-73. [DOI] [PubMed] [Google Scholar]
- 29.Hisatomi, Y., K. Okumura, K. Nakamura, S. Matsumoto, A. Satoh, K. Nagano, T. Yamamoto, and F. Endo. 2004. Flow cytometric isolation of endodermal progenitors from mouse salivary gland differentiate into hepatic and pancreatic lineages. Hepatology 39667-675. [DOI] [PubMed] [Google Scholar]
- 30.Hsu, K. M., J. R. Pratt, W. J. Akers, S. I. Achilefu, and W. M. Yokoyama. 2009. Murine cytomegalovirus displays selective infection of cells within hours after systemic administration. J. Gen. Virol. 9033-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudson, J. B. 1979. The murine cytomegalovirus as a model for the study of viral pathogenesis and persistent infections. Arch. Virol. 621-29. [DOI] [PubMed] [Google Scholar]
- 32.Hummel, M., Z. Zhang, S. Yan, I. DePlaen, P. Golia, T. Varghese, G. Thomas, and M. I. Abecassis. 2001. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J. Virol. 754814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ibanez, C. E., R. Schrier, P. Ghazal, C. Wiley, and J. A. Nelson. 1991. Human cytomegalovirus productively infects primary differentiated macrophages. J. Virol. 656581-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isegawa, Y., Z. Ping, K. Nakano, N. Sugimoto, and K. Yamanishi. 1998. Human herpesvirus 6 open reading frame U12 encodes a functional beta-chemokine receptor. J. Virol. 726104-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacoby, M. A., H. W. Virgin, and S. H. Speck. 2002. Disruption of the M2 gene of murine gammaherpesvirus 68 alters splenic latency following intranasal, but not intraperitoneal, inoculation. J. Virol. 761790-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaptein, S. J., P. S. Beisser, Y. K. Gruijthuijsen, K. G. Savelkouls, K. W. van Cleef, E. Beuken, G. E. Grauls, C. A. Bruggeman, and C. Vink. 2003. The rat cytomegalovirus R78 G protein-coupled receptor gene is required for production of infectious virus in the spleen. J. Gen. Virol. 842517-2530. [DOI] [PubMed] [Google Scholar]
- 37.Kaptein, S. J., K. W. van Cleef, Y. K. Gruijthuijsen, E. V. Beuken, L. van Buggenhout, P. S. Beisser, F. R. Stassen, C. A. Bruggeman, and C. Vink. 2004. The r131 gene of rat cytomegalovirus encodes a proinflammatory CC chemokine homolog which is essential for the production of infectious virus in the salivary glands. Virus Genes 2943-61. [DOI] [PubMed] [Google Scholar]
- 38.Koffron, A. J., M. Hummel, B. K. Patterson, S. Yan, D. B. Kaufman, J. P. Fryer, F. P. Stuart, and M. I. Abecassis. 1998. Cellular localization of latent murine cytomegalovirus. J. Virol. 7295-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liston, A., and S. McColl. 2003. Subversion of the chemokine world by microbial pathogens. Bioessays 25478-488. [DOI] [PubMed] [Google Scholar]
- 40.Margulies, B. J., H. Browne, and W. Gibson. 1996. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology 225111-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melnychuk, R. M., P. Smith, C. N. Kreklywich, F. Ruchti, J. Vomaske, L. Hall, L. Loh, J. A. Nelson, S. L. Orloff, and D. N. Streblow. 2005. Mouse cytomegalovirus M33 is necessary and sufficient in virus-induced vascular smooth muscle cell migration. J. Virol. 7910788-10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michelson, S., P. Dal Monte, D. Zipeto, B. Bodaghi, L. Laurent, E. Oberlin, F. Arenzana-Seisdedos, J. L. Virelizier, and M. P. Landini. 1997. Modulation of RANTES production by human cytomegalovirus infection of fibroblasts. J. Virol. 716495-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell, B. M., A. Leung, and J. G. Stevens. 1996. Murine cytomegalovirus DNA in peripheral blood of latently infected mice is detectable only in monocytes and polymorphonuclear leukocytes. Virology 223198-207. [DOI] [PubMed] [Google Scholar]
- 44.Mocarski, E. S., Jr. 2002. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol. 10332-339. [DOI] [PubMed] [Google Scholar]
- 45.Nakano, K., K. Tadagaki, Y. Isegawa, M. M. Aye, P. Zou, and K. Yamanishi. 2003. Human herpesvirus 7 open reading frame U12 encodes a functional beta-chemokine receptor. J. Virol. 778108-8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noda, S., S. A. Aguirre, A. Bitmansour, J. M. Brown, T. E. Sparer, J. Huang, and E. S. Mocarski. 2006. Cytomegalovirus MCK-2 controls mobilization and recruitment of myeloid progenitor cells to facilitate dissemination. Blood 10730-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliveira, S. A., and T. E. Shenk. 2001. Murine cytomegalovirus M78 protein, a G protein-coupled receptor homologue, is a constituent of the virion and facilitates accumulation of immediate-early viral mRNA. Proc. Natl. Acad. Sci. USA 983237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Penfold, M. E., D. J. Dairaghi, G. M. Duke, N. Saederup, E. S. Mocarski, G. W. Kemble, and T. J. Schall. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. USA 969839-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollock, J. L., R. M. Presti, S. Paetzold, and H. W. t. Virgin. 1997. Latent murine cytomegalovirus infection in macrophages. Virology 227:168-179. [DOI] [PubMed] [Google Scholar]
- 50.Pollock, J. L., and H. W. Virgin. 1995. Latency, without persistence, of murine cytomegalovirus in the spleen and kidney. J. Virol. 691762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Power, C. A. 2003. Knock out models to dissect chemokine receptor function in vivo. J. Immunol. Methods 27373-82. [DOI] [PubMed] [Google Scholar]
- 52.Randolph-Habecker, J. R., B. Rahill, B. Torok-Storb, J. Vieira, P. E. Kolattukudy, B. H. Rovin, and D. D. Sedmak. 2002. The expression of the cytomegalovirus chemokine receptor homolog US28 sequesters biologically active CC chemokines and alters IL-8 production. Cytokine 1937-46. [DOI] [PubMed] [Google Scholar]
- 53.Reddehase, M. J., M. Balthesen, M. Rapp, S. Jonjic, I. Pavic, and U. H. Koszinowski. 1994. The conditions of primary infection define the load of latent viral genome in organs and the risk of recurrent cytomegalovirus disease. J. Exp. Med. 179185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Redwood, A. J., M. Messerle, N. L. Harvey, C. M. Hardy, U. H. Koszinowski, M. A. Lawson, and G. R. Shellam. 2005. Use of a murine cytomegalovirus K181-derived bacterial artificial chromosome as a vaccine vector for immunocontraception. J. Virol. 792998-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roizman, B., and A. E. Sears. 1987. An inquiry into the mechanisms of herpes simplex virus latency. Annu. Rev. Microbiol. 41543-571. [DOI] [PubMed] [Google Scholar]
- 56.Rollins, B. J. 1997. Chemokines. Blood. 90:909-928. [PubMed] [Google Scholar]
- 57.Ryckman, B. J., M. A. Jarvis, D. D. Drummond, J. A. Nelson, and D. C. Johnson. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 80710-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sacher, T., J. Podlech, C. A. Mohr, S. Jordan, Z. Ruzsics, M. J. Reddehase, and U. K. Koszinowski. 2008. The major virus-producing cell type during murine cytomegalovirus infection, the hepatocyte, is not the source of virus dissemination in the host. Cell Host Microbe 3:263-272. [DOI] [PubMed] [Google Scholar]
- 59.Saederup, N., S. A. Aguirre, T. E. Sparer, D. M. Bouley, and E. S. Mocarski. 2001. Murine cytomegalovirus CC chemokine homolog MCK-2 (m131-129) is a determinant of dissemination that increases inflammation at initial sites of infection. J. Virol. 759966-9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saederup, N., Y. C. Lin, D. J. Dairaghi, T. J. Schall, and E. S. Mocarski. 1999. Cytomegalovirus-encoded beta chemokine promotes monocyte-associated viremia in the host. Proc. Natl. Acad. Sci. USA 9610881-10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinzger, C., and G. Jahn. 1996. Human cytomegalovirus cell tropism and pathogenesis. Intervirology 39302-319. [DOI] [PubMed] [Google Scholar]
- 62.Stoddart, C. A., R. D. Cardin, J. M. Boname, W. C. Manning, G. B. Abenes, and E. S. Mocarski. 1994. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J. Virol. 686243-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Streblow, D. N., C. N. Kreklywich, P. Smith, J. L. Soule, C. Meyer, M. Yin, P. Beisser, C. Vink, J. A. Nelson, and S. L. Orloff. 2005. Rat cytomegalovirus-accelerated transplant vascular sclerosis is reduced with mutation of the chemokine-receptor R33. Am. J. Transplant. 5436-442. [DOI] [PubMed] [Google Scholar]
- 64.Streblow, D. N., C. Soderberg-Naucler, J. Vieira, P. Smith, E. Wakabayashi, F. Ruchti, K. Mattison, Y. Altschuler, and J. A. Nelson. 1999. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell 99511-520. [DOI] [PubMed] [Google Scholar]
- 65.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 66.Tadagaki, K., K. Nakano, and K. Yamanishi. 2005. Human herpesvirus 7 open reading frames U12 and U51 encode functional beta-chemokine receptors. J. Virol. 797068-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vieira, J., T. J. Schall, L. Corey, and A. P. Geballe. 1998. Functional analysis of the human cytomegalovirus US28 gene by insertion mutagenesis with the green fluorescent protein gene. J. Virol. 728158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vischer, H. F., R. Leurs, and M. J. Smit. 2006. HCMV-encoded G-protein-coupled receptors as constitutively active modulators of cellular signaling networks. Trends Pharm. Sci. 2756-63. [DOI] [PubMed] [Google Scholar]
- 69.Vlahakis, S. R., A. Villasis-Keever, T. Gomez, M. Vanegas, N. Vlahakis, and C. V. Paya. 2002. G protein-coupled chemokine receptors induce both survival and apoptotic signaling pathways. J. Immunol. 1695546-5554. [DOI] [PubMed] [Google Scholar]
- 70.von Andrian, U. H., and C. R. Mackay. 2000. T-cell function and migration. Two sides of the same coin. N. Engl. J. Med. 3431020-1034. [DOI] [PubMed] [Google Scholar]
- 71.Waldhoer, M., T. N. Kledal, H. Farrell, and T. W. Schwartz. 2002. Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities. J. Virol. 768161-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, D., W. Bresnahan, and T. Shenk. 2004. Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc. Natl. Acad. Sci. USA 10116642-16647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, D., and T. Shenk. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 7910330-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]