Abstract
The central enzyme responsible for human cytomegalovirus (HCMV) DNA synthesis is a virally encoded DNA polymerase that includes a catalytic subunit, UL54, and a homodimeric accessory subunit, UL44, the presumptive HCMV DNA polymerase processivity factor. The structure of UL44 is similar to that of the eukaryotic processivity factor proliferating cell nuclear antigen (PCNA), which interacts with numerous other proteins required for faithful DNA replication. We sought to determine whether, like PCNA, UL44 is capable of interacting with multiple DNA replication proteins and, if so, whether these proteins bind UL44 at the site corresponding to where multiple proteins bind to PCNA. Initially, several proteins, including the viral DNA replication factors UL84 and UL57, were identified by mass spectrometry in immunoprecipitates of UL44 from infected cell lysate. The association of UL44/UL84, but not UL44/UL57, was confirmed by reciprocal coimmunoprecipitation of these proteins from infected cell lysates and was resistant to nuclease treatment. Yeast two-hybrid analyses demonstrated that the substitution of residues in UL44 that prevent UL44 homodimerization or abrogate the binding of UL54 to UL44 do not abrogate the UL44/UL84 interaction. Reciprocal glutathione-S-transferase (GST) pulldown experiments using bacterially expressed UL44 and UL84 confirmed these results and, further, demonstrated that a UL54-derived peptide that competes with UL54 for UL44 binding does not prevent the association of UL84 with UL44. Taken together, our results strongly suggest that UL44 and UL84 interact directly using a region of UL44 different from the UL54 binding site. Thus, UL44 can bind interacting replication proteins using a mechanism different from that of PCNA.
Protein-protein interactions orchestrate the process of DNA synthesis. Most replicative DNA polymerases include at least two interacting components, a catalytic subunit responsible for the polymerization of DNA and a processivity subunit. The processivity subunit of the polymerase holds the catalytic subunit on DNA while DNA polymerization takes place, thereby permitting long-chain DNA synthesis. One of the best-described processivity factors is proliferating cell nuclear antigen (PCNA) of eukaryotic DNA polymerases δ and ɛ (23, 26). PCNA is a head-to-tail homotrimer which, with the aid of clamp loader proteins, forms a toroidal ring around DNA, creating an internal channel of sufficient diameter to accommodate the DNA duplex (15). Studies of PCNA by numerous laboratories have described a large number of interactions between PCNA and proteins that participate in and abet DNA synthesis (reviewed in references 23 and 26). Of particular interest is that many of these proteins bind PCNA at a specific site, inserting a conserved hydrophobic domain (a PCNA-interacting protein [PIP] box) into a hydrophobic pocket lying beneath an interdomain connector loop of PCNA. PIP box proteins bind and dissociate from PCNA at different times during DNA replication when the need for their function arises (23, 26). Moreover, the trimeric structure of PCNA makes it possible for three PIP box proteins to bind PCNA simultaneously.
The human cytomegalovirus (HCMV) DNA polymerase consists of a catalytic subunit, UL54, and an accessory subunit, UL44. UL44 interacts with UL54 and DNA and stimulates long-chain DNA synthesis (21, 39) and thus very likely serves as a processivity factor. UL44 forms a head-to-head homodimer (1) that binds DNA without the aid of clamp loaders yet wraps around DNA akin to PCNA (14). While UL44 shows little or no sequence homology with PCNA, there is striking structural similarity between UL44 and PCNA monomers (1, 2). Furthermore, similar to the binding of PIP box proteins to PCNA, the binding of UL54 to UL44 relies on hydrophobic interactions between the UL54 carboxyl terminus and a hydrophobic pocket beneath the connector loop of UL44 that corresponds to the site of PIP box protein binding to PCNA (1, 19, 20). However, there are subtle, but important, differences in the mechanism of UL54 binding to UL44 compared to that of PIP box protein binding to PCNA. For example, a leucine residue in UL54 (Leu1227) that is essential to the UL54/UL44 interaction (19) occupies a position in the hydrophobic crevice of UL44 that has no known counterpart in interactions of PIP box protein binding to PCNA (1). As UL44 is a homodimer, UL54 can, in principle, bind one monomer of UL44 while the UL54 binding site on the other UL44 monomer is free for other proteins to bind.
Several viral proteins other than UL44 and UL54 are required for viral DNA replication. These proteins include a trimeric helicase-primase complex (UL70, UL102, and UL105) and a single-stranded DNA (ssDNA)-binding protein (UL57) (25). The UL36 to UL38 loci also appear to be necessary for DNA synthesis (27, 28, 32, 34), as does the transcriptional activator IE2-p86 (32), although the function(s) of the proteins encoded by these loci in DNA synthesis are as yet unclear. The virus also encodes a DNA glycosylase, UL114, which appears to be important for efficient DNA synthesis (30). An additional viral protein involved in viral DNA synthesis is UL84. Although the deletion of the UL84 gene from the viral genome has been reported to impair viral DNA replication (40), the function of UL84 remains unclear. It has been suggested that UL84 acts as a viral origin-binding protein, as it has been found at the viral origin of replication oriLyt, using chromatin immunoprecipitation (IP) assays (6). Also, UL84 exhibits some homology with DEXD/H box proteins (8), some of which are origin-binding proteins. It has yet to be formally demonstrated, however, that UL84 is required for the initiation of DNA synthesis from oriLyt.
Upon the initiation of this project, two viral proteins, other than UL54 (1, 10, 19, 20, 39), had been reported to interact with UL44: UL114, the viral DNA glycosylase (30), and UL97, the viral protein kinase (16, 24), neither of which participates directly in viral DNA synthesis. We sought to determine if, like PCNA, UL44 is capable of interacting with multiple proteins involved in viral DNA replication and whether these proteins bind UL44 at the site corresponding to that in PCNA that binds PIP box proteins.
MATERIALS AND METHODS
Cells and viruses.
The human foreskin fibroblast (HFF) cell line Hs29 was obtained from the American Type Culture Collection (ATCC; Manassas, VA). HCMV strain AD169 was used in all experiments and was propagated in HFF cells as previously described (13).
IP of UL44 from HFF cells for mass spectrometry analysis.
HFF (1 × 106) were infected with HCMV AD169 at a multiplicity of infection (MOI) of 3 or mock infected. At 72 h postinfection, the cells were washed once with phosphate-buffered saline and harvested into 500 μl of buffer EBC2 (50 mM Tris [pH 8.0], 300 mM NaCl, 0.5% NP-40). After incubation on ice at 4°C for 15 min, each lysate was clarified by centrifugation at 10,000 × g for 15 min. Seventy-five microliters of each lysate was mixed with 50 μl of a 50% slurry of protein A-Sepharose beads (Zymed) and incubated with rotation for 1 h at 4°C. After centrifugation to remove beads was performed, 5 μl of anti-UL44 monoclonal antibody (MAb) 28-21 (5) (generously provided by W. Britt, University of Alabama—Birmingham) conjugated to protein A-Sepharose beads was added to each lysate and incubated at 4°C with rotation for 6 h. The beads were then washed four times with 1 ml EBC2 buffer and then resuspended in Laemmli buffer (17). Each IP was resolved on a 10% sodium dodecyl sulfate (SDS) polyacrylamide gel that was subsequently silver stained using a SilverSnap for MS kit (Pierce) according to the manufacturer's instructions. The bands indicated in Fig. 1 were submitted for liquid chromatography-tandem mass spectrometry to the Taplin Biological Mass Spectrometry Facility, Harvard Medical School.
FIG. 1.
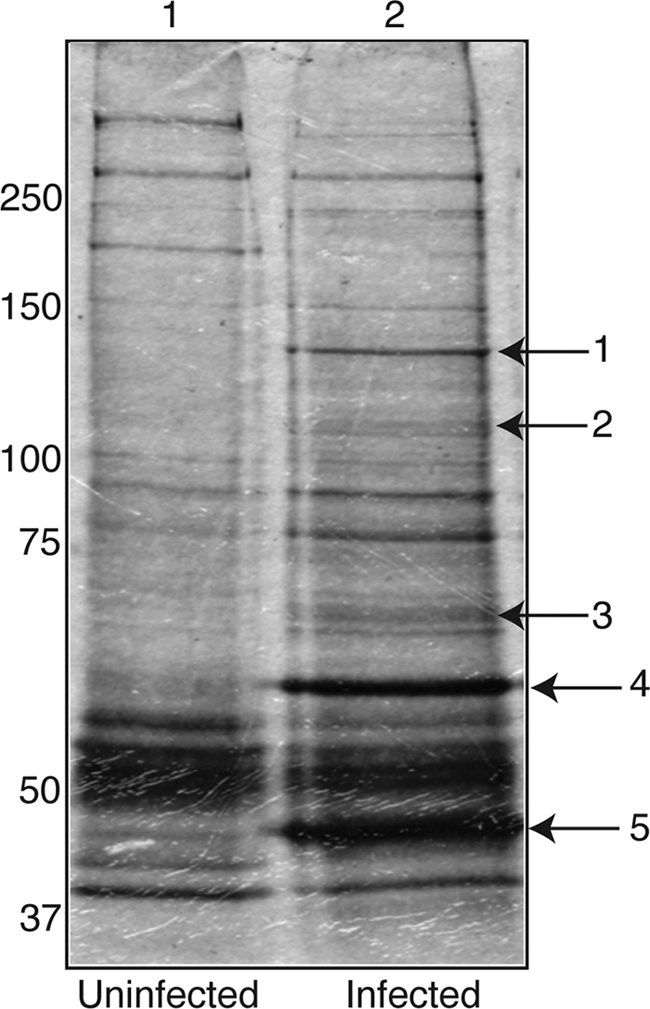
IP of proteins associated with UL44 from infected cells. Lysates from uninfected HFF cells or HFF cells infected with HCMV AD169 (MOI, 3) were prepared, and IP was performed using a MAb recognizing UL44. Immunoprecipitated proteins were separated on a 10% polyacrylamide gel and silver stained. Lane 1, IP from uninfected cells; lane 2, IP from infected cells. The protein bands extracted from the gel for analysis by mass spectrometry are indicated by arrows. The positions of molecular mass markers in kDa are indicated to the left.
Reciprocal co-IPs.
HFF (3 × 105) were infected with HCMV AD169 at an MOI of 3 or mock infected and resuspended in 250 μl of EBC2 buffer. After the lysate was clarified, 20 μl protein A-Sepharose beads and 5 μg of the appropriate isotype control antibody (Bethyl Laboratories) were added and the mixture was incubated at 4°C with rotation for 3 h. For the IP of UL44, after centrifugation to remove beads, 20 μl of protein A-Sepharose beads was added with either 5 μg isotype control antibody or 5 μg UL44 MAb (Virusys). After incubation was completed overnight at 4°C with rotation, the beads were spun down and the supernatant removed. The beads were washed four times with 1 ml of EBC2 buffer and resuspended in 20 μl of Laemmli buffer. For the IP of UL84 and UL57 from infected cells, the same method was employed, utilizing 5 μg of either anti-UL84 MAb (7) (a generous gift from G. Pari, University of Nevada—Reno) or anti-UL57 MAb (Virusys), except that cells were resuspended in IP lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 1 mM EDTA) and the beads were washed with Tris-buffered saline before resuspension in Laemmli buffer. Where indicated, 400 U benzonase (Novagen) or 25 μg/ml ethidium bromide was added after clarification of the lysate by centrifugation. Where indicated, the supernatant used in the IP was mixed 1:1 (vol/vol) with 6× gel loading buffer, and 50 μl was analyzed on a 0.8% agarose gel containing 100 μg/ml ethidium bromide.
Western blotting.
The Western blotting of proteins separated on 10% SDS polyacrylamide gels was carried out as described elsewhere (37), using MAbs recognizing UL44 or UL57 (both from Virusys and used at a 1:1,000 dilution) or UL84 (7) as the primary antibodies. Ten microliters of each IP was analyzed with 10 μl of the infected cell lysate. Anti-mouse TruBlot antibody conjugated to horseradish peroxidase (HRP; eBioscience), which recognizes the native (not denatured) form of mouse antibody, was used to detect primary antibodies except in the experiment for which the results are depicted in Fig. 4, where goat anti-mouse HRP-conjugated antibody (Southern Biotech) was used to detect the anti-UL57 MAb. An ECL Western blotting chemiluminescence substrate kit (Pierce) was used to detect HRP-conjugated antibodies in all cases.
FIG. 4.
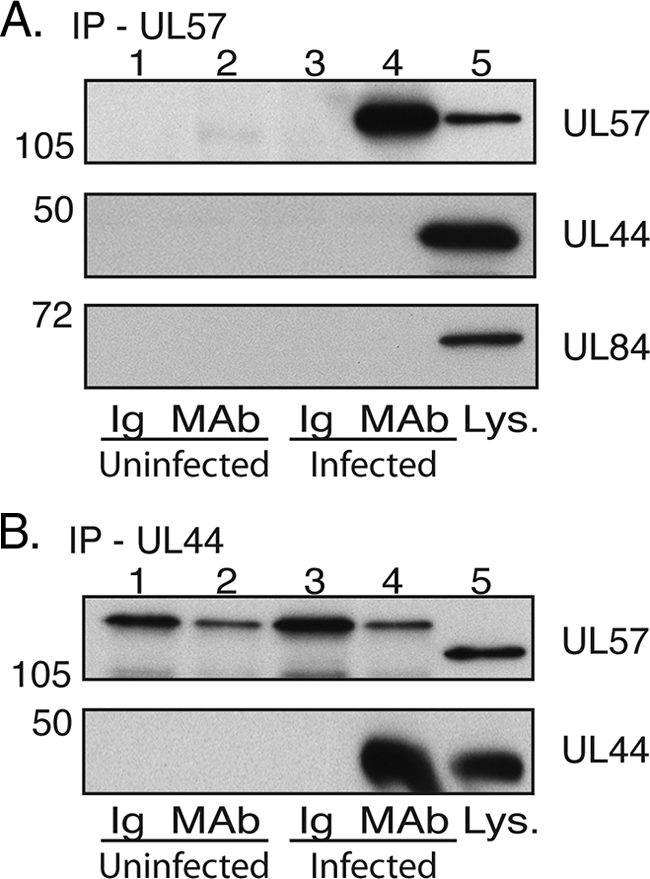
IP of UL44 and UL57 from infected cell lysate. Lysates from uninfected HFF cells or HFF cells infected with HCMV AD169 (MOI, 3) were prepared and precleared with the relevant control immunoglobulin (Ig). IP was then carried out with either MAbs recognizing UL57 (A) or UL44 (B) or a control antibody of the same isotype as the MAb used (Ig). Immunoprecipitated proteins were analyzed by Western blotting using MAbs recognizing UL57, UL44, or UL84, as indicated. Lanes 1 and 2, uninfected cells immunoprecipitated with Ig and MAb, respectively; lanes 3 and 4, infected cells immunoprecipitated with Ig and MAb, respectively; lane 5, infected cell lysate (Lys.). The positions of molecular mass markers in kDa are indicated to the left of the panels.
Plasmids.
To generate the pGBT9-UL84 plasmid, which encodes the UL84 protein fused in frame to the DNA-binding domain (Gal4BD; amino acids 1 to 147) of Saccharomyces cerevisiae Gal4 (Gal4BD-UL84 hybrid), the UL84 gene coding sequence was amplified from HCMV AD169 genomic DNA by PCR, using primers 5′-CACGATGAATTCATGCCACGCGTCGACCCC-3′ (forward) and 5′-TTCCACCTGCAGTTAGAGATCGCCGCAGACC-3′ (reverse), and cloned into the EcoRI/PstI sites of pGBT9 (Clontech) downstream from the Gal4BD sequence. The pGBT9-UL54 plasmid, in which UL54 is fused to Gal4BD (Gal4BD-UL54 hybrid), was created by amplifying the UL54 gene coding sequence from pRSET-Pol (a gift from P. F. Ertl, GlaxoSmithKline, Stevenage, United Kingdom) by PCR with primers 5′-CACGATGAATTCATGTTTTTCAACCCG-3′ (forward) and 5′-TTCCACCCCGGGTCAACAGCATTCGTGCGC-3′ (reverse) and cloning the PCR fragment into the EcoRI/SmaI sites of pGTB9. To construct the plasmid encoding the Gal4AD-UL44 fusion protein, where UL44 is fused to the C terminus of the GAL4-activating domain (Gal4AD; amino acids 768 to 881), the UL44 gene coding sequence was amplified from pRSET44 (also a gift from P. F. Ertl, GlaxoSmithKline, Stevenage, United Kingdom) by PCR with primers 5′-CACGATGGATCCTTATGGATCGCAAGACGC-3′ (forward) and 5′-TTCCACCTCGAGCTAGCCGCACTTTTGC-3′ (reverse) and cloned into the BamHI/XhoI sites of pACT2 (Clontech), yielding the pACT-UL44 plasmid. The UL44 mutants were obtained using the QuikChange mutagenesis kit (Stratagene), amplifying the pACT-UL44 plasmid with primer pairs containing the appropriate nucleotide change(s). A list of the mutagenic primers and their sequences can be found in reference 36. Finally, to generate the pRSET-UL84 plasmid, the UL84 gene coding sequence was amplified from HCMV AD169 genomic DNA by PCR using primers 5′-CACGATAGATCTATGCCACGCGTCGACCCC-3′ (forward) and 5′-TTCCACAGATCTTTAGAGATCGCCGCAGACC-3′ (reverse) and cloned into the BglII site of pRSETA (Invitrogen).
Mutations in pRSET-UL44 to generate pRSET-UL44I135A and pRSET-UL44ΔC290 were generated using the QuikChange mutagenesis kit (Stratagene) and the primer sets listed in reference 36. pGEXUL84 was generated by the PCR amplification of the UL84 gene sequence in plasmid pZIP13 (27) (a kind gift from David Anders, Wadsworth Institute, New York State Department of Health) using primers 5′-CGCGCGCGAATTCATGCCACGCGTCGACCCCAACCTT-3′ (forward) and 5′-GCGGCGGCGTCCTCGAGCTTAGAGATCGCCGCAGACCATG-3′ (reverse). The PCR product was cloned into the EcoRI and XhoI sites of pGEX-6P-1 (GE Lifesciences).
Yeast two-hybrid assays.
The growth media and standard methods used for manipulating the yeast cells were as described by Rose et al. (33). The yeast strain S. cerevisiae Y190 (MATa ura3-52 his3-200 ade2-101 lys2-801 trp1- 901 leu2-3,112 gal4Δ gal80Δ cyhr2; LYS::GAL1UAS-HIS3TATA-HIS3 URA3::GAL1UAS-GAL1TATA-lacZ; Clontech) was transformed with plasmid DNA expressing Gal4BD or Gal4AD fusion proteins by the lithium acetate method of Schiestl and Gietz (35). The transformed cells were assayed for expression of the lacZ reporter gene by β-galactosidase (β-Gal) filter assays as described previously (4). In these assays, β-Gal expression was scored as follows: + for blue color detected after 2 to 3 h of incubation and − for no signal detected after 24 h of incubation.
Proteins and peptides.
The recombinant glutathione S-transferase (GST)-UL44, GST-UL44ΔC290 (GST fused to a truncated form of UL44 lacking the protein's carboxyl-terminal 143 residues), and GST-UL84 fusion proteins and GST were purified from Escherichia coli BL21(DE3)pLysS (Novagen) harboring pD15-UL44, pD15-UL44ΔC290, pGEXUL84, or pD15-GST, respectively, as described previously (19). Peptides corresponding to the C-terminal 22 residues of UL54 and the C-terminal 18 residues of UL30, respectively, were synthesized at the Department of Biological Chemistry & Molecular Pharmacology Biopolymers Facility, Harvard Medical School. The molecular masses of the peptides were determined by matrix-assisted laser desorption-time of flight mass spectrometry and corresponded to the expected values. The peptide concentrations were confirmed by quantitative amino acid analysis.
GST pulldown assays.
The in vitro transcription-translation of the pRSET plasmids described above was performed using the TNT T7 coupled transcription-translation system (Promega) in the presence of [35S]methionine (Amersham Pharmacia Biotech) according to the manufacturer's suggestion. For the GST pulldown assays with GST-UL44ΔC290 and mutants derived from it, fusion proteins (2.7 nmol) were incubated in a final volume of 550 μl with either 50 μl of in vitro-translated UL54 (2 h on ice) or with 50 μl of in vitro-translated UL84 (1 h at 37°C) in binding buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 0.1 mM EDTA, 2 mM dithiothreitol [DTT]), containing 5 μl of RNAce-It RNase Cocktail (Stratagene) and 50 U of benzonase (Sigma), and then loaded onto 0.2-ml glutathione columns. Where indicated, 25 μM UL54 peptide was added to the reaction mixture. The columns were washed with 5 ml of wash buffer A (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 10% glycerol, 0.1 mM EDTA, 2 mM DTT, 0.5% NP-40, and 0.5% Triton X-100) in the UL54-UL44 binding assays or with 5 ml of wash buffer B (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 0.1 mM EDTA, 2 mM DTT, 0.5% NP-40, and 0.5% Triton X-100) in the UL84-UL44 binding assays. The bound proteins were then eluted with wash buffer A or B containing 15 mM glutathione in the UL54-UL44 or UL84-UL44 binding assays, respectively. Experiments using GST-UL84 were carried out in the same fashion except that 40 μl of in vitro-translated protein was incubated with 100 μg of GST-UL84 in each reaction in the presence of 200 U benzonase (2 h on ice). Where indicated, 50 μM of either UL54 or UL30 peptide was added to the reaction. The proteins were visualized by SDS-polyacrylamide gel electrophoresis (PAGE) and autoradiography.
RESULTS
Identification of proteins immunoprecipitating with UL44 from HCMV-infected cells.
In an initial effort to identify proteins associated with UL44 during HCMV infection, mock-infected and infected cell lysates were prepared 72 h postinfection and IP was performed using an MAb recognizing UL44. Viral DNA replication is known to be under way at this point in the virus replication cycle (25); therefore, it was thought that this procedure might detect proteins associated with UL44 that are involved in viral DNA synthesis. The immunoprecipitated proteins were separated onto an SDS polyacrylamide gel, which was then silver stained (Fig. 1). A number of bands could be seen in the lane containing the IP from infected cell lysate (lane 2) that could not be seen in the lane containing the IP from mock-infected cell lysate (lane 1). Of these bands, those most easily visualized were extracted from the gel and subjected to liquid chromatography-tandem mass spectrometry analysis to determine the proteins present. The proteins identified in each band indicated in Fig. 1 are shown in Table 1. As expected, UL44 was detected in the most prominent band (band 5). (It was also detected in band 4, which could be due to the existence of higher-molecular-weight forms of UL44, e.g., from posttranslational modifications, or due to the incomplete resolution of UL44 during electrophoresis.) In each of the other major bands, several proteins, viral and cellular, were identified, and in each case, peptides corresponding to HCMV proteins were observed more often than any other protein. Several viral structural proteins, the major capsid protein UL86 (band 1) and the tegument proteins US22 and UL83 (bands 3 and 4, respectively) were detected, as were proteins involved in viral DNA synthesis: UL57 (band 2), the major ssDNA-binding protein, and UL84 (band 3), which has been suggested to be a viral origin-binding protein (6). We also observed peptides corresponding to the viral alkaline nuclease UL98 (band 4). While it is possible that UL44 interacts with any of the viral or cellular proteins we found, we chose to focus our studies on the possible interactions between UL44 and those proteins known to be involved in viral DNA synthesis, UL57 and UL84.
TABLE 1.
Selected proteins in an IP of UL44 from HCMV-infected cells
| Banda | Protein identified | GenBank no. | Species | No. of peptidesb | % Coveragec |
|---|---|---|---|---|---|
| 1 | HCMV major capsid protein | P16729 | 16 | 12.9 | |
| Dead box protein 9 | Q08211 | Human | 3 | 2.1 | |
| 2 | HCMV ssDNA-binding protein UL57 | P17147 | 7 | 6.3 | |
| AP-3 complex subunit 1 | O00203 | Human | 6 | 5.7 | |
| Regulator nonsense transcript 1 | Q92900 | Human | 1 | 0.9 | |
| Pre-mRNA processing factor | O75400 | Human | 1 | 0.9 | |
| 3 | HCMV tegument protein US22 | P09722 | 8 | 11.8 | |
| HCMV replication factor UL84 | P16727 | 5 | 8.0 | ||
| Stress-70 protein | P38646 | Human | 3 | 5.4 | |
| Lamin-A/C | P02545 | Human | 1 | 2.6 | |
| 4 | HCMV tegument protein pp65 | P06725 | 35 | 58.6 | |
| HCMV Pol accessory subunit UL44 | P16790 | 8 | 19.9 | ||
| Ig gamma chain | P01867 | Murine | 5 | 18.6 | |
| Ig alpha chain | P01878 | Murine | 4 | 16.0 | |
| Heterogeneous nuclear ribonucleoprotein K | P61978 | Human | 3 | 9.1 | |
| Ub cross-reactive protein | P05161 | Human | 2 | 13.4 | |
| HCMV alkaline nuclease UL98 | P16789 | 2 | 2.9 | ||
| 5 | HCMV Pol accessory subunit UL44 | P16790 | 60 | 93.8 |
The bands indicated by the arrows in Fig. 1 were analyzed by liquid chromatography-tandem mass spectrometry.
Number of peptides from each protein identified.
Percentage of the total protein sequence that is represented by the peptides identified.
Reciprocal co-IP of UL44 and UL84 from infected cells.
To extend the observations made from our initial IP (Fig. 1 and Table 1), reciprocal co-IPs were performed. IP was carried out on infected or uninfected cell lysates using either an MAb recognizing UL84 (Fig. 2A), an MAb recognizing UL44 (Fig. 2B), or a control antibody of the same isotype as the accompanying MAb (Fig. 2A and B). Immunoprecipitated proteins were examined by Western blotting using antibodies recognizing UL84 and UL44 (Fig. 2). A species of the size of UL84 and reacting with anti-UL84 MAb could be observed in infected cell lysates and in the proteins immunoprecipitated from the infected cell lysate with either anti-UL44 or anti-UL84 MAb but not in uninfected cell lysate, the proteins immunoprecipitated from uninfected cell lysate, or lysates immunoprecipitated with isotype control antibody (Fig. 2A and B, top panels). Similarly, a species of the size of UL44 and reacting with UL44 MAb could be found in infected cell lysates and the proteins immunoprecipitated from infected cell lysate with anti-UL84 or anti-UL44 MAb but not in any other sample (Fig. 2A, middle panel, and B, bottom panel). These results provide further evidence for an association of UL44 and UL84 in the lysates of infected cells.
FIG. 2.
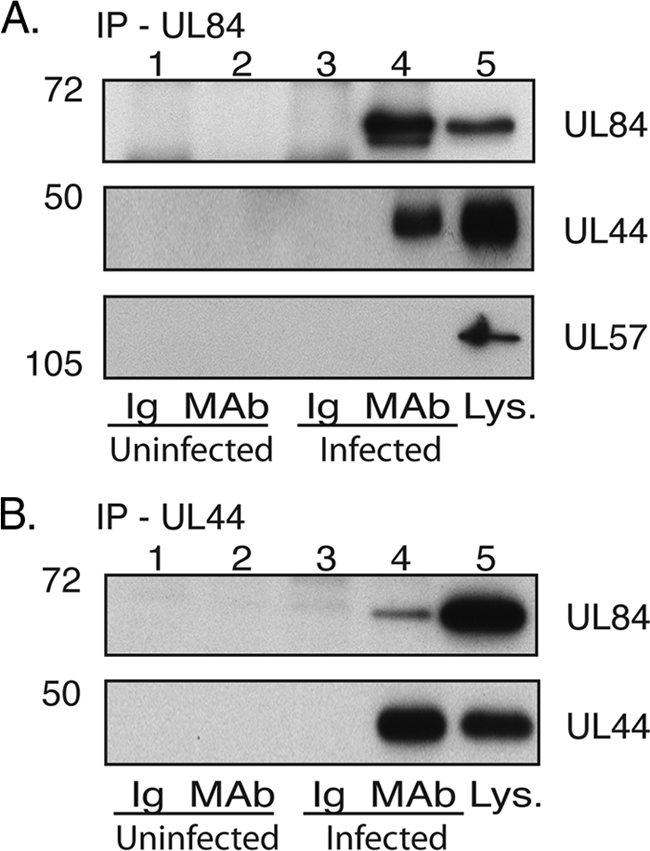
IP of UL44 and UL84 from infected cell lysate. Lysates from uninfected HFF cells or HFF cells infected with HCMV AD169 (MOI, 3) were prepared and precleared with the relevant control immunoglobulin (Ig). IP was then carried out with a MAb recognizing either UL84 (A) or UL44 (B) or a control antibody of the same isotype as the MAb used (Ig). Immunoprecipitated proteins were analyzed by Western blotting using MAbs recognizing UL84, UL44, or UL57, as indicated. Lanes 1 and 2, uninfected cells immunoprecipitated with Ig or MAb, respectively; lanes 3 and 4, infected cells immunoprecipitated with Ig or MAb, respectively; lane 5, infected cell lysate (Lys.). The positions of molecular mass markers in kDa are indicated to the left of the panels.
We noted that while a single form of UL84 was observed in infected cell lysate, in IPs using UL84 MAb from infected cell lysates, a second, faster-migrating species could also be observed (Fig. 2A). UL84 can be phosphorylated (8, 12) and ubiquitinated (11), but it remains unknown if the different UL84 species observed represent differences in the posttranslational modification of the protein. However, during IP with UL44 MAb, only the slower-migrating form of UL84 was detected by Western blotting, suggesting that UL44 interacts with this form of UL84.
The association of UL44 and UL84 does not require nucleic acid.
It has been demonstrated elsewhere that UL84 is capable of binding DNA and RNA (6) and that UL44 binds to DNA (20, 22, 39). DNA-binding proteins can associate during IP due to their adjacent binding on DNA rather than due to protein-protein interactions (18, 38). To determine if nucleic acid is required for the UL44/UL84 association, IP using an anti-UL44 MAb or an isotype control antibody was performed as before in the presence or absence of benzonase, a nonspecific nuclease, and the IPs were probed on Western blots with either anti-UL44 or anti-UL84 MAb (Fig. 3A). No UL44 or UL84 was detected in IPs using the control antibody (Ig). However, these proteins were detected in IPs from infected cell lysate using an anti-UL44 MAb in either the absence or presence of benzonase (Fig. 3A). To confirm the action of benzonase on nucleic acid, the cell lysates analyzed in Fig. 3A were examined on an ethidium bromide-stained agarose gel (Fig. 3B). In the absence of benzonase, the robust staining of nucleic acid could be seen (lane 2), whereas in the presence of benzonase (lane 1), other than some staining at the dye front of the gel, there was no more staining than in a no-sample control (lane 3), indicating that benzonase had efficiently degraded the nucleic acid in the cell lysate. IP was also performed in the presence of the DNA intercalating agent ethidium bromide, which has been shown to abrogate protein binding to DNA (18, 38). The co-IP of UL44 and UL84 could again be observed (data not shown), further confirming that protein binding to DNA is not required for the association of UL44 and UL84 during IP.
FIG. 3.
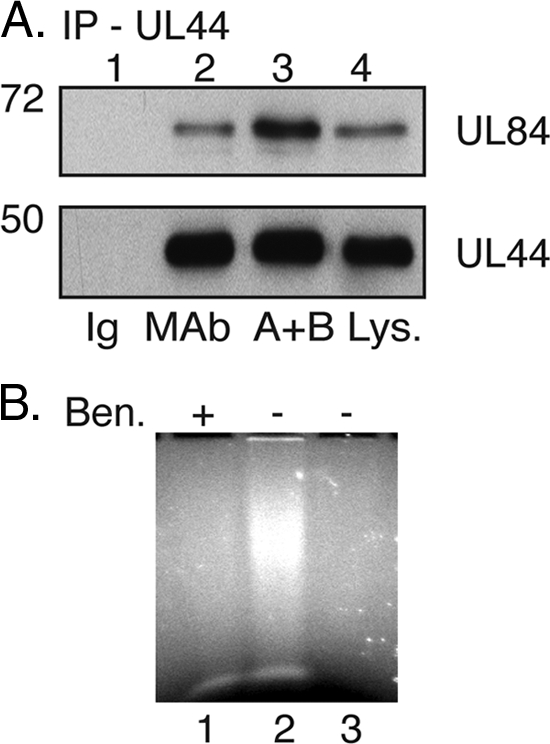
IP of UL44 from infected cell lysate in the presence of nuclease. (A) Infected cell lysate was immunoprecipitated as in Fig. 2 with control immunoglobulin (Ig; lane 1) or anti-UL44 MAb in the absence (lane 2) or the presence of benzonase (A+B; lane 3). Proteins from the IP were analyzed by Western blotting using MAb recognizing UL84 (top panel) or UL44 (bottom panel). Infected cell lysate (Lys.) is shown in lane 4. The positions of molecular mass markers in kDa are indicated to the left of the panels. (B) Samples used in the IP depicted in panel A were run out onto an ethidium bromide-stained 0.8% agarose gel. Lane 1, IP in the presence (+) of benzonase (panel A, lane 3); lane 2, IP in the absence (−) of benzonase (panel A, lane 2); lane 3, no sample.
Next, we examined the possible association of UL44 and UL57 in infected cells by reciprocal co-IP. IP was performed on uninfected or infected cell lysate using a MAb recognizing UL57 or a control antibody of the same isotype (Fig. 4A). Immunoprecipitated proteins were examined by Western blotting using MAbs recognizing UL57 or UL44. UL57 could be observed in infected cell lysates and in the proteins immunoprecipitated from infected cell lysate with an anti-UL57 MAb but not in any other sample. UL44 could be detected in infected cell lysates but was not detected in any other sample. IP was also performed on uninfected or infected cell lysate using an antibody recognizing UL44 or a control antibody of the same isotype (Fig. 4B). Immunoprecipitated protein was examined by Western blotting using antibodies recognizing UL44 and UL57. Again, UL44 could be observed in infected cell lysate and in the proteins immunoprecipitated from infected cell lysate with anti-UL44 MAb but not in the other samples. UL57 could be detected in infected cell lysate but not in any other sample. Taken together, these data indicate that under the conditions used, there was no detectable association of UL44 and UL57 in infected cell lysates, so the characterization of this interaction was not pursued any further.
Finally, we examined the IPs, for which the results are depicted in Fig. 2 and 4, for a possible association of UL57 and UL84. When the products of the UL84 IP were examined by Western blotting using an anti-UL57 MAb, while UL57 could be readily detected in infected cell lysate, it could not be detected in the UL84 IP, even upon overexposure of the film to the blot (Fig. 2A and data not shown). Similarly, while UL84 could be readily detected in infected cell lysates, it could not be detected in any UL57 IP, again, even upon overexposure of the film to the blot (Fig. 4A and data not shown). Taken together, these data provide no evidence that UL84 and UL57 associate in the infected cell. Regardless, the failure to detect the DNA-binding protein UL57 in either the UL84 IP or the UL44 IP further argues that the association of UL44 and UL84 that we have detected is not simply due to the simultaneous binding of the proteins to DNA during IP.
In summary, while we could not confirm an association between UL44 and UL57 or UL84 and UL57, our results do indicate an association between UL44 and UL84 in the infected cell lysates that, by multiple criteria, does not require the presence of nucleic acid.
Interaction of UL84 and UL44 mutants in yeast two-hybrid assays.
To further investigate the association between UL44 and UL84, their interaction was studied in yeast two-hybrid assays. UL84 or UL54 was fused to the Gal4BD, while the wild-type UL44 sequence was fused to the Gal4AD. Combinations of these constructs were introduced into yeast and tested for their ability to activate a β-Gal reporter gene, which contains Gal4 binding sites in its promoter. Gal4BD-UL84, Gal4BD-UL54, and Gal4AD-UL44 could not stimulate β-Gal expression alone, but β-Gal expression could be observed when Gal4AD-UL44 was in the presence of either Gal4BD-UL54 or Gal4BD-UL84, indicating UL44/UL54 interactions, as expected, and UL44/UL84 interactions (Table 2). Similar results were obtained in reverse combinations (i.e., in yeast two-hybrid assays with UL44 fused to Gal4BD and UL84 or UL54 fused to Gal4AD [data not shown]). These data extend our observations of an association between UL44 and UL84 in infected cell lysates to intact yeast cells.
TABLE 2.
Yeast two-hybrid assaysa
| DNA-binding domain fusion protein | Activation domain fusion protein | β-Gal expressionb |
|---|---|---|
| Gal4BD-UL84 | − | |
| Gal4AD-UL44 | − | |
| Gal4BD-UL84 | Gal4AD-UL44 | + |
| Gal4AD-UL44P85G | − | |
| Gal4BD-UL84 | Gal4AD-UL44P85G | + |
| Gal4AD-UL44L86A/L87A | − | |
| Gal4BD-UL84 | Gal4AD-UL44L86A/L87A | + |
| Gal4AD-UL44F121A | − | |
| Gal4BD-UL84 | Gal4AD-UL44F121A | + |
| Gal4AD-UL44I135A | − | |
| Gal4BD-UL84 | Gal4AD-UL44I135A | + |
| Gal4BD-UL54 | − | |
| Gal4BD-UL54 | Gal4AD-UL44 | + |
| Gal4BD-UL54 | Gal4AD-UL44P85G | + |
| Gal4BD-UL54 | Gal4AD-UL44L86A/L87A | + |
| Gal4BD-UL54 | Gal4AD-UL44F121A | + |
| Gal4BD-UL54 | Gal4AD-UL44I135A | − |
UL84 or UL54 was fused to Gal4BD, and wild-type or mutant UL44 proteins were fused to Gal4AD and assayed for interaction by β-Gal filter assays.
+, blue color detected after 2 to 3 h of incubation; −, no signal detected after 24 h of incubation.
We also used the yeast two-hybrid assay to assess the interaction of UL84 with mutants of UL44. We previously have described mutations that alter residues at the interface of the UL44 monomers, which either interfere (L86A/L87A, F121A) or do not interfere (P85A) with homodimerization in vitro and in cells (1, 36). These mutations were introduced into the Gal4 activating domain construct Gal4AD-UL44, and their ability to activate the expression of the reporter gene β-Gal in the presence of Gal4 binding domain constructs Gal4BD-UL54 and Gal4BD-UL84 was tested. β-Gal expression was observed when each of the mutants was tested with either Gal4BD-UL54 or Gal4BD-UL84 (Table 2), demonstrating that none of these mutations abrogates the interaction between UL44 and UL54 or UL84. Thus, neither the altered residues nor the dimerization of UL44 appear to be required for the binding of UL84. We also tested the ability of UL84 to bind a UL44 mutant containing a substitution that abrogates binding to UL54. As expected (19), we could not observe an interaction between Gal4BD-UL54 and Gal4AD fused to UL44 containing the I135A substitution (Gal4AD-UL44I135A). An interaction between Gal4BD-UL84 and Gal4AD-UL44I135A could, however, be observed (Table 2), suggesting that UL84 binding to UL44 differs from the binding of UL54 to UL44.
Interaction of UL84 and UL44 mutants in GST pulldown experiments.
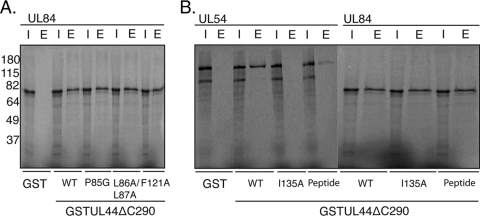
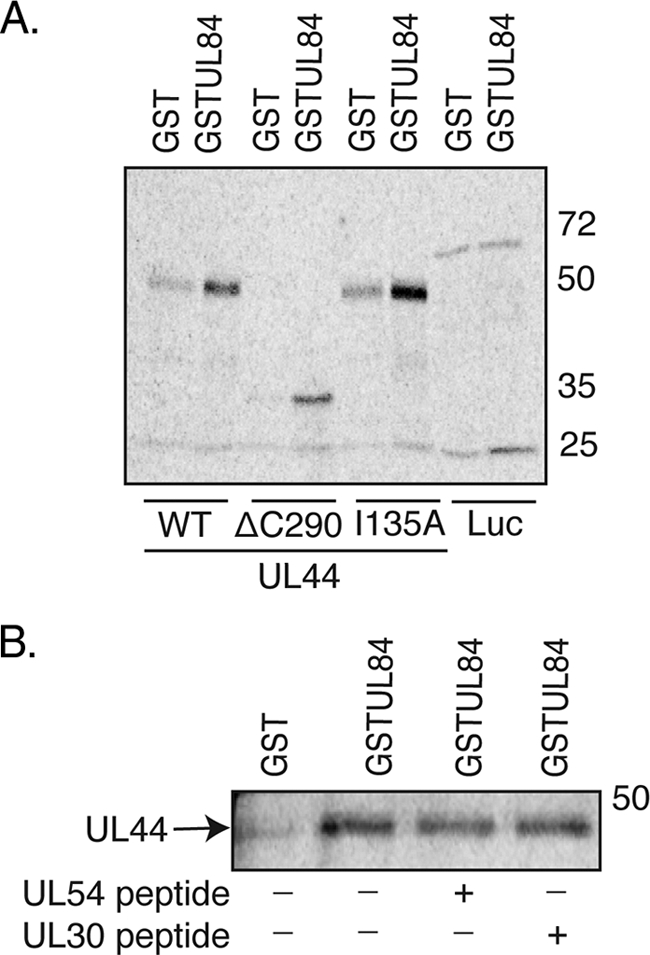
We next sought to confirm and extend the observations made in the yeast two-hybrid experiments (Table 2) by testing the ability of UL84 to bind mutants of UL44 in GST pulldown assays. The GST fusion proteins of UL44 consisted of GST fused to a truncated form of UL44 lacking the protein's carboxyl-terminal 143 residues, UL44ΔC290 (GST-UL44ΔC290). (The carboxyl terminus of full-length UL44 is readily cleaved in E. coli [2]; therefore, generating substantial quantities of full-length protein is problematic.) Unsubstituted GST-UL44ΔC290 or GST-UL44ΔC290 containing substitution P85A, L86A/L87A, or F121A (GST-UL44ΔC290P85A, GST-UL44ΔC290L86A/L87A, and GST-UL44ΔC290F121A, respectively) was incubated with radiolabeled UL84 generated by in vitro transcription-translation in the presence of [35S]methionine. As a negative control, GST alone was incubated with radiolabeled UL84. All binding reactions were performed in the presence of benzonase. After incubation, the reactions were passed over glutathione columns, and after the washing of the columns, bound proteins were eluted from the columns in the presence of excess glutathione. Aliquots of the proteins loaded on the column (input) and the eluted proteins from each column were analyzed by SDS-PAGE and autoradiography (Fig. 5A). Radiolabeled UL84 was bound by GST-UL44ΔC290 but not by GST alone. Thus, the interaction between UL84 and GST-UL44ΔC290 requires the UL44 component of the fusion protein. UL84 was also bound by GST-UL44ΔC290P85A, GST-UL44ΔC290L86A/L87A, and GST-UL44ΔC290F121A. As the final two mutants are defective for dimerization, the results indicate that the dimerization of UL44 most likely is not a requirement for the binding of UL84 in vitro, confirming the yeast two-hybrid results. Furthermore, as the GST fusion proteins used in this experiment lack the carboxyl terminus of UL44, this region of UL44 is not necessary for the association of UL84 with UL44.
FIG. 5.
Interaction of UL84 with mutant GST-UL44 proteins. (A) GST pulldown assays were performed in which GST or GST-UL44 fusion proteins were incubated with radiolabeled UL84 and passed over a glutathione column. The GST fusion proteins used in each reaction are noted below the figure. The input (I) and protein eluted by glutathione (E) from each reaction are shown. (B) GST pulldown assays were repeated as described for panel A. Radiolabeled UL54 and UL84 were passed over a glutathione column in the presence and absence of a peptide corresponding to the extreme carboxyl terminus of UL54. The GST proteins used in each reaction are noted below the figure. The input (I) and protein eluted by glutathione (E) from each reaction are shown. The positions of molecular mass markers in kDa are indicated to the left of the panels.
GST-UL44ΔC290 containing the I135A substitution was also tested and found to bind radiolabeled UL84 in GST pulldown assays (Fig. 5B). As a control, the ability of radiolabeled UL54 to bind the various GST fusion proteins or GST alone was tested (Fig. 5B). As previously observed (19), GST-UL44ΔC290 bound radiolabeled UL54, but no binding was detected using GST alone or GST-UL44ΔC290I135A. Thus, residue I135 of UL44, which is required for interaction with UL54, is not required for interaction with UL84. We then assayed the ability of radiolabeled UL54 or UL84 to bind GST-UL44ΔC290 in the presence or absence of a peptide corresponding to the extreme carboxyl terminus of UL54 (UL54 peptide), which binds UL44 (1). Notably, less radiolabeled UL54 bound to GST-UL44ΔC290 in the presence of UL54 peptide compared to that binding to GST-UL44ΔC290 in the absence of the UL54 peptide, as expected (Fig. 5B). In contrast, no difference in the amount of radiolabeled UL84 binding to GST-UL44ΔC290 in the presence or absence of UL54 peptide was observed, indicating that the UL54 peptide does not compete with UL84 for binding to GST-UL44ΔC290. This provides further evidence that UL84 does not bind UL44 at the UL54 binding site.
Next, we performed reciprocal GST pulldown experiments in which we tested the ability of GST or a fusion protein of full-length UL84 protein fused to GST (GST-UL84) to bind radiolabeled UL44 and various mutant forms of UL44, again in the presence of benzonase. The proteins that bound to and could be eluted from each column (Fig. 6A) were analyzed by SDS-PAGE and autoradiography. Substantially more full-length radiolabeled UL44 bound to GST-UL84 than to GST, indicating a requirement for the UL84 portion of the fusion protein for efficient binding (Fig. 6A). UL44ΔC290 bound to a similar extent to GST-UL84 as full-length protein (and with less binding to GST), suggesting that the carboxyl terminus of UL44 does not meaningfully influence binding to UL84 (Fig. 6A). Additionally, full-length UL44 containing the I135A substitution bound efficiently to GST-UL84 and at levels greater than its binding to GST, indicating that this substitution had little, if any, effect on the UL44/UL84 interaction even in the context of the full-length protein (Fig. 6A). As a negative control, radiolabeled luciferase (Luc) was incubated with either GST or GST-UL84 and passed over glutathione columns. Compared to the binding of radiolabeled wild-type or mutant UL44 to GST-UL84, little binding of Luc to either GST or GST-UL84 could be observed. Moreover, no difference in Luc binding to GST or GST-UL84 was detected (Fig. 6A), indicating that the UL84 component of GST-UL84 does not nonspecifically bind radiolabeled protein. The binding of radiolabeled UL44 to GST-UL84 was also assayed in GST pulldown assays in the presence or absence of UL54 peptide (1) or a peptide corresponding to the extreme carboxyl terminus of the herpes simplex virus type 1 (HSV-1) polymerase catalytic subunit UL30 (29). Neither the UL54 nor the UL30 peptide effectively inhibited the binding of radiolabeled UL44 to GST-UL84, although there may have been a slight decrease in UL44 binding to UL84 in the presence of the UL54 peptide (Fig. 6B). The UL54 and UL30 peptides used in the experiment for which the results are shown in Fig. 6B were active in inhibiting their respective protein-protein interactions (Fig. 5 and data not shown). Taken together, the experiments for which the results are depicted in Fig. 5 and 6 indicate that the binding of UL84 to UL44 does not require the carboxyl terminus of UL44 and occurs at a site different from that used in the binding of UL54 to UL44.
FIG. 6.
Interaction of GST-UL84 with mutants of UL44. (A) GST or GST-UL84 was incubated with radiolabeled UL44 or radiolabeled luciferase (Luc) and passed over a glutathione column. The protein eluted by glutathione from each column is shown. The GST fusion protein used in each reaction is noted above the figure, and the radiolabeled protein used in each reaction is noted below the figure. (B) GST or GST-UL84 was incubated with radiolabeled UL44 in the presence (+) or the absence (−) of peptides corresponding to the extreme carboxyl termini of UL54 or HSV-1 UL30 and passed over a glutathione column. The protein eluted by glutathione from each column is shown. The GST protein used in each reaction is noted above the figure. The positions of molecular mass markers in kDa are indicated to the right of the panels.
DISCUSSION
We have previously found striking similarities not only between the structure of UL44 and its counterpart in the eukaryotic cell, PCNA (2), but also between the binding of the HCMV DNA polymerase catalytic subunit UL54 to UL44 and the binding of cellular proteins to PCNA (1, 19, 20), revealing a possible mechanism which could allow the binding of proteins other than UL54 to UL44 during viral DNA replication. In our current study, we have demonstrated an association between UL44 and the HCMV DNA replication factor UL84 in infected cell lysates, in intact yeast cells, and in vitro. The association of the two proteins does not require nucleic acid. Our data taken together strongly suggest that the interaction between UL44 and UL84 is direct. Moreover, based on both mutational studies and peptide inhibition experiments, we find that UL84 does not interact with UL44 at the site of UL54 binding to UL44.
When our study was nearly complete, evidence for an association between UL44 and UL84 was outlined by Gao and coworkers (11). Proteins immunoprecipitated from infected cell lysate using a UL84 MAb were separated by 2-D gel electrophoresis, and a number of viral and cellular proteins, including UL44, were identified by mass spectrometry. Additionally, UL44 and UL84 could be detected in IPs from the lysates of cells overexpressing tagged versions of these proteins. Our study confirms and extends the observations made by Gao et al. (11) by demonstrating an association of UL44 and UL84 in infected cell lysate by reciprocal co-IP, by thoroughly documenting that this association does not require nucleic acid, by analyzing this interaction in yeast and in vitro, and by studying the binding of UL44 mutants to UL84.
As UL84 can interact with UL44 mutants that are impaired for dimerization, the binding of UL84 to UL44 does not, therefore, appear to require the surface formed by UL44 dimerization. This raises the possibility that two UL84 molecules can bind both of the monomers of the UL44 dimer simultaneously during viral DNA synthesis. Interestingly, it has been suggested from studies of tagged versions of UL84 that UL84 can dimerize (7); however, it has not been established whether untagged UL84 dimerizes in the infected cell.
The possible associations of UL57 and UL44 and of UL57 and UL84 were also examined. These studies were prompted, in part, by reports of interactions between PCNA and the eukaryotic ssDNA-binding protein replication protein A (9) and an interaction between the HSV-1 ssDNA-binding protein ICP8 and the processivity subunit UL42 (38) on the one hand, and of HSV-1 ICP8 and origin-binding protein UL9 on the other (3). Although we cannot exclude the possibility that the HCMV counterparts to these interactions occur in the infected cell but were not revealed by the experimental conditions used here, it is also possible that there are aspects of HCMV DNA replication that differ from these other systems. Additionally, while ICP8 and UL57 are direct homologues in sequence and known function, UL84 has yet to be demonstrated to have a role in the initiation of DNA synthesis at oriLyt and may therefore not be functionally homologous to HSV-1 UL9.
The carboxyl terminus of UL44 has been recently reported to be necessary for the binding of the viral uracil DNA glycosylase UL114 to UL44 (31). However, it is not clear if UL114 and UL44 interact directly. UL114 can be found to co-IP with UL44 from infected cell lysate, but a requirement for nucleic acid for this interaction was not ruled out (30, 31). In vitro, an association of these two proteins was observed in the presence of DNA (31); no association between the two proteins could be detected in yeast two-hybrid assays (31; E. Sinigalia and A. Loregian, unpublished data). The evidence for a role of the carboxyl terminus of UL44 in association with UL114 rests mainly on assays measuring association in the presence of DNA and assays of the stimulation of uracil DNA glycosylase activity. In the latter case, increasing amounts of full-length UL44 resulted in the decreased stimulation of uracil DNA glycosylase activity relative to the stimulation of enzyme activity in the presence of UL44 lacking its carboxyl terminus. Full-length UL44 protein prepared in E. coli aggregates nonspecifically under conditions of low ionic strength (20), such as those used in the study mentioned above (31). Similarly, we observed the nonspecific binding of full-length UL44 to GST but much less nonspecific binding of UL44 lacking its carboxyl terminus to GST. It is possible that full-length UL44 interacts nonspecifically with UL114 and that the removal of the carboxyl terminus decreases these nonspecific interactions, rather than this segment being required for specific interactions.
If the report on the UL44/UL114 interaction discussed above (31) is taken at face value, then taken together with previous work on the UL54-UL44 interaction (19, 20), the results presented here indicate that there could be at least three regions on each monomer of the UL44 dimer available for the binding of viral DNA replication proteins: (i) the hydrophobic region corresponding to the PIP box protein binding site in PCNA which is required for UL54 binding, (ii) the UL44 carboxyl terminus, and (iii) the UL84 binding site. Our results indicate that, similar to PCNA (23, 26), UL44 possesses the ability to bind multiple proteins simultaneously during viral DNA synthesis, although by a different mechanism. Although UL54 interacts with a hydrophobic region of UL44 corresponding to the PIP box protein binding site in PCNA and uses an aromatic residue akin to those found in PIP boxes, it does not contain the PIP box consensus sequence (QXXHXXAA) or the 310 helix found in PCNA-PIP box protein structures (1). Similarly, UL84 does not contain a PIP box consensus sequence. As we have demonstrated here, UL44 may interact with its binding partners using mechanisms that differ from PCNA; therefore, a PIP-box-like sequence may not be present or required in proteins interacting with UL44. It should be interesting to elucidate these mechanisms. Finally, although UL44 and PCNA are structurally homologous (2), the mechanism of UL44 binding to DNA (14), the mechanism of UL54 binding to UL44 (1), and the results we present here indicate notable evolutionary divergence between UL44 and PCNA. This observation leads, in turn, to the suggestion that there may be more as-yet-unknown functions of UL44 and PCNA that the two proteins do not share.
Acknowledgments
We are very grateful for the generous gifts of the pRSET plasmids from Peter Ertl (GlaxoSmithKline) and plasmid pZIP13 from David Anders (Wadsworth Institute, New York State Department of Health) plus the UL84 MAb from Greg Pari (University of Nevada—Reno) and UL44 antibody 28-21 from William Britt (University of Alabama). We are also indebted to Ross Tomaino (Taplin Biological Mass Spectrometry Facility, Harvard Medical School) for his efforts and invaluable advice.
This work was supported in part by NIH grants AI019838 and AI026077 to D.M.C., an award from the William Randolph Hearst Fund to B.L.S., and MURST ex 60% and Progetto di Ricerca di Ateneo 2007 (grant no. CPDA074945) to A.L.
Footnotes
Published ahead of print on 20 May 2009.
REFERENCES
- 1.Appleton, B. A., J. Brooks, A. Loregian, D. J. Filman, D. M. Coen, and J. M. Hogle. 2006. Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44. J. Biol. Chem. 2815224-5232. [DOI] [PubMed] [Google Scholar]
- 2.Appleton, B. A., A. Loregian, D. J. Filman, D. M. Coen, and J. M. Hogle. 2004. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell 15233-244. [DOI] [PubMed] [Google Scholar]
- 3.Boehmer, P. E., M. C. Craigie, N. D. Stow, and I. R. Lehman. 1994. Association of origin binding protein and single strand DNA-binding protein, ICP8, during herpes simplex virus type 1 DNA replication in vivo. J. Biol. Chem. 26929329-29334. [PubMed] [Google Scholar]
- 4.Breeden, L., and K. Nasmyth. 1985. Regulation of the yeast HO gene. Cold Spring Harb. Symp. Quant. Biol. 50643-650. [DOI] [PubMed] [Google Scholar]
- 5.Britt, W. J., and L. Vugler. 1987. Structural and immunological characterization of the intracellular forms of an abundant 68,000 Mr human cytomegalovirus protein. J. Gen. Virol. 681897-1907. [DOI] [PubMed] [Google Scholar]
- 6.Colletti, K. S., K. E. Smallenburg, Y. Xu, and G. S. Pari. 2007. Human cytomegalovirus UL84 interacts with an RNA stem-loop sequence found within the RNA/DNA hybrid region of oriLyt. J. Virol. 817077-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colletti, K. S., Y. Xu, S. A. Cei, M. Tarrant, and G. S. Pari. 2004. Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication. J. Virol. 789203-9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colletti, K. S., Y. Xu, I. Yamboliev, and G. S. Pari. 2005. Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J. Biol. Chem. 28011955-11960. [DOI] [PubMed] [Google Scholar]
- 9.Davies, A. A., D. Huttner, Y. Daigaku, S. Chen, and H. D. Ulrich. 2008. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell 29625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ertl, P. F., and K. L. Powell. 1992. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J. Virol. 664126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, Y., K. Colletti, and G. S. Pari. 2008. Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J. Virol. 8296-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, Y., and G. S. Pari. 2009. Interaction of human cytomegalovirus pUL84 with casein kinase 2 is required for oriLyt-dependent DNA replication. J. Virol. 832393-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamil, J. P., and D. M. Coen. 2007. Human cytomegalovirus protein kinase UL97 forms a complex with the tegument phosphoprotein pp65. J. Virol. 8110659-10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komazin-Meredith, G., R. J. Petrella, W. L. Santos, D. J. Filman, J. M. Hogle, G. L. Verdine, M. Karplus, and D. M. Coen. 2008. The human cytomegalovirus UL44 C clamp wraps around DNA. Structure 161214-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishna, T. S., X. P. Kong, S. Gary, P. M. Burgers, and J. Kuriyan. 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 791233-1243. [DOI] [PubMed] [Google Scholar]
- 16.Krosky, P. M., M. C. Baek, W. J. Jahng, I. Barrera, R. J. Harvey, K. K. Biron, D. M. Coen, and P. B. Sethna. 2003. The human cytomegalovirus UL44 protein is a substrate for the UL97 protein kinase. J. Virol. 777720-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 896958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loregian, A., B. A. Appleton, J. M. Hogle, and D. M. Coen. 2004. Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44. J. Virol. 78158-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loregian, A., B. A. Appleton, J. M. Hogle, and D. M. Coen. 2004. Specific residues in the connector loop of the human cytomegalovirus DNA polymerase accessory protein UL44 are crucial for interaction with the UL54 catalytic subunit. J. Virol. 789084-9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loregian, A., R. Rigatti, M. Murphy, E. Schievano, G. Palu, and H. S. Marsden. 2003. Inhibition of human cytomegalovirus DNA polymerase by C-terminal peptides from the UL54 subunit. J. Virol. 778336-8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loregian, A., E. Sinigalia, B. Mercorelli, G. Palu, and D. M. Coen. 2007. Binding parameters and thermodynamics of the interaction of the human cytomegalovirus DNA polymerase accessory protein, UL44, with DNA: implications for the processivity mechanism. Nucleic Acids Res. 354779-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maga, G., and U. Hubscher. 2003. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 1163051-3060. [DOI] [PubMed] [Google Scholar]
- 24.Marschall, M., M. Freitag, P. Suchy, D. Romaker, R. Kupfer, M. Hanke, and T. Stamminger. 2003. The protein kinase pUL97 of human cytomegalovirus interacts with and phosphorylates the DNA polymerase processivity factor pUL44. Virology 31160-71. [DOI] [PubMed] [Google Scholar]
- 25.Mocarski, E. S., Jr., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott, Williams & Wilkins, New York, NY. [Google Scholar]
- 26.Moldovan, G. L., B. Pfander, and S. Jentsch. 2007. PCNA, the maestro of the replication fork. Cell 129665-679. [DOI] [PubMed] [Google Scholar]
- 27.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 676979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 672575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilger, B. D., C. Cui, and D. M. Coen. 2004. Identification of a small molecule that inhibits herpes simplex virus DNA polymerase subunit interactions and viral replication. Chem. Biol. 11647-654. [DOI] [PubMed] [Google Scholar]
- 30.Prichard, M. N., H. Lawlor, G. M. Duke, C. Mo, Z. Wang, M. Dixon, G. Kemble, and E. R. Kern. 2005. Human cytomegalovirus uracil DNA glycosylase associates with ppUL44 and accelerates the accumulation of viral DNA. Virol. J. 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranneberg-Nilsen, T., H. A. Dale, L. Luna, R. Slettebakk, O. Sundheim, H. Rollag, and M. Bjoras. 2008. Characterization of human cytomegalovirus uracil DNA glycosylase (UL114) and its interaction with polymerase processivity factor (UL44). J. Mol. Biol. 381276-288. [DOI] [PubMed] [Google Scholar]
- 32.Reid, G. G., V. Ellsmore, and N. D. Stow. 2003. An analysis of the requirements for human cytomegalovirus oriLyt-dependent DNA synthesis in the presence of the herpes simplex virus type 1 replication fork proteins. Virology 308303-316. [DOI] [PubMed] [Google Scholar]
- 33.Rose, M. D., R. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 707398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16339-346. [DOI] [PubMed] [Google Scholar]
- 36.Sinigalia, E., G. Alvisi, B. Mercorelli, D. M. Coen, G. S. Pari, D. A. Jans, A. Ripalti, G. Palu, and A. Loregian. 2008. Role of homodimerization of human cytomegalovirus DNA polymerase accessory protein UL44 in origin-dependent DNA replication in cells. J. Virol. 8212574-12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strang, B. L., and N. D. Stow. 2005. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 7912487-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 785856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiland, K. L., N. L. Oien, F. Homa, and M. W. Wathen. 1994. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 34191-206. [DOI] [PubMed] [Google Scholar]
- 40.Xu, Y., S. A. Cei, A. R. Huete, and G. S. Pari. 2004. Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth. J. Virol. 7810360-10369. [DOI] [PMC free article] [PubMed] [Google Scholar]