Abstract
The recent, unexpected discovery of a functional DNA methylation system in the genome of the social bee Apis mellifera underscores the potential importance of DNA methylation in invertebrates. The extent of genomic DNA methylation and its role in A. mellifera remain unknown, however. Here we show that genes in A. mellifera can be divided into 2 distinct classes, one with low-CpG dinucleotide content and the other with high-CpG dinucleotide content. This dichotomy is explained by the gradual depletion of CpG dinucleotides, a well-known consequence of DNA methylation. The loss of CpG dinucleotides associated with DNA methylation also may explain the unusual mutational patterns seen in A. mellifera that lead to AT-rich regions of the genome. A detailed investigation of this dichotomy implicates DNA methylation in A. mellifera development. High-CpG genes, which are predicted to be hypomethylated in germlines, are enriched with functions associated with developmental processes, whereas low-CpG genes, predicted to be hypermethylated in germlines, are enriched with functions associated with basic biological processes. Furthermore, genes more highly expressed in one caste than another are overrepresented among high-CpG genes. Our results highlight the potential significance of epigenetic modifications, such as DNA methylation, in developmental processes in social insects. In particular, the pervasiveness of DNA methylation in the genome of A. mellifera provides fertile ground for future studies of phenotypic plasticity and genomic imprinting.
Keywords: comparative genomics, phenotypic plasticity
DNA methylation occurs in the genomes of a wide array of bacteria, plants, fungi, and animals (1, 2). In particular, the methylation of cytosine bases represents an important epigenetic mark that affects gene expression in diverse taxa (1, 3). Despite the phylogenetically widespread and ancient origin of DNA methylation, genomic patterns of methylation show considerable variation (2); for example, whereas vertebrate genomes tend to show extensive levels of DNA methylation, many invertebrate genomes display reduced or minimal levels of methylation (1, 2, 4). Variation in genome methylation patterns is of great interest because it suggests that the role of DNA methylation is not strictly conserved among species. Thus, information on the nature and extent of DNA methylation in diverse taxa continues to be a valuable resource for exploring the role of this DNA modification in eukaryotes (2, 3, 5).
Recent research has identified a functional DNA methylation system in a social insect, the honeybee, Apis mellifera (6). Social insects are among the most successful of animal taxa (7, 8). Their success stems from the cooperative behaviors displayed by society members. In particular, members of social insect colonies belong to different castes, which undertake distinct tasks (9); for example, the defining feature of hymenopteran social insects (ants, some bees, and some wasps) is a reproductive division of labor, whereby individuals of the queen caste reproduce while members of the worker caste defend the nest, forage, and rear the young. This division of individuals into alternate castes represents a key evolutionary transition that allowed social insects to come to dominate many terrestrial ecosystems (10, 11).
Remarkably, DNA methylation appears to be directly associated with the differentiation of castes in A. mellifera (12, 13). Kucharski et al. (12) demonstrated that down-regulation of a key DNA methyltransferase (Dnmt3) in developing A. mellifera larvae resulted in profound changes in caste developmental trajectories. Accordingly, DNA methylation may represent an important mechanism facilitating the evolution of social systems (14).
Despite the potential importance of DNA methylation, the genome-wide patterns of methylation within the A. mellifera genome remain poorly understood. This is unfortunate, because knowledge of the patterns of DNA methylation in the A. mellifera genome is critical to assessing its role and significance in this species (6, 12). Moreover, linking molecular changes such as DNA methylation with the evolution and development of social phenotypes remains one of the major challenges in understanding sociality (15, 16). In this study, we investigated the nature of DNA methylation in A. mellifera by analyzing global patterns of methylation using computational methods and comparing them with experimental results in A. mellifera and other species. We found that DNA methylation is widespread and has played a critical role in A. mellifera genome evolution, and that it is associated with important developmental processes, including caste formation.
Results
Depletion of CpG Dinucleotides Suggests Widespread Gene Methylation in A. mellifera.
We used “normalized” CpG content (CpGO/E) to infer the pattern of DNA methylation in A. mellifera. CpGO/E is a robust measure of the level of DNA methylation on an evolutionary time scale due to specific mutational mechanisms of methylated cytosines (17–20). In brief, methylated cytosines are hypermutable due to their vulnerability to spontaneous deamination, which causes a gradual depletion of CpG dinucleotides from methylated regions over time (21). Consequently, genomic regions that are subject to heavy germline DNA methylation (hypermethylated) lose CpG dinucleotides over time and have lower-than-expected CpGO/E. In contrast, regions that undergo little germline DNA methylation (hypomethylated) maintain high CpGO/E. This measure has been successfully used to indirectly measure historical DNA methylation levels (19–22). In particular, the pattern of DNA methylation inferred from CpGO/E corresponds well to the actual pattern of DNA methylation in such diverse taxa as human and sea squirt (19, 20).
We first examined the distribution of CpGO/E in several insect genomes. We focused on analyses of genes, because the annotation of other genomic regions (e.g., intergenic regions and noncoding functional elements) in insect genomes other than Drosophila melanogaster is far from complete.
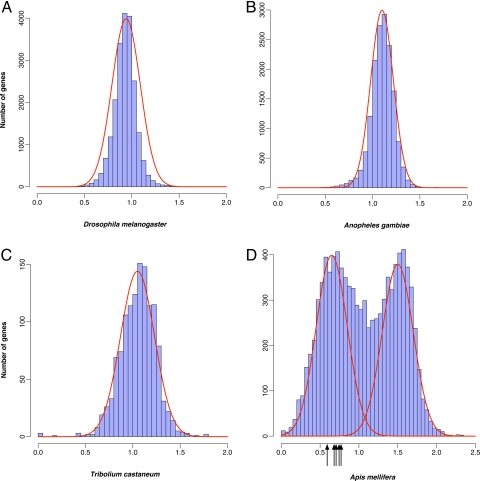
The level of methylation is low in D. melanogaster, and its genome lacks critical DNA methyltransferases (2, 23). Accordingly, the CpGO/E in D. melanogaster genes has an approximately normal distribution with a mean around 1 (Fig. 1A). Analyses of other published insect genomes, including Tribolium castaneum and Anopheles gambiae, yield similar patterns (Fig. 1 B and C). Thus, genes in these insects exhibit little evidence of DNA methylation according to mutational decay of CpG dinucleotides.
Fig. 1.
Contrasting patterns of DNA methylation in A. mellifera genes and those of other insects, as measured by CpGO/E. The y-axis depicts the number of genes with the specific CpGO/E values given on the x-axis. The distribution of CpGO/E in D. melanogaster (A), A. gambiae (B), and T. castaneum genes (C) all show unimodal distribution, reflecting a relative lack of DNA methylation in these species. In contrast, the distribution of CpGO/E in A. mellifera genes (D) is bimodal, likely demonstrating the effects of DNA methylation of CpG dinucleotides (see text). The arrows show the position of the 5 genes [GB16767 (CpGO/E = 0.56), GB19399 (0.66), GB18099 (0.67), GB12504 (0.75), and XP_001121083 (0.71)] found to be methylated in a previous study (6). Note that we could not map the gene GB15223 using our experimental procedure.
In contrast, we find that the CpGO/E of A. mellifera genes exhibits a striking bimodal pattern that is best explained by a mixture of 2 distinct distributions (Fig. 1D; see Materials and Methods). The CpGO/E of approximately half of the A. mellifera genes has a distribution with a remarkably high mean of 1.50 (SD = 0.20), similar to the genomic background (see also ref. 24). Surprisingly, the other half of the A. mellifera genes have a distinct distribution with a mean much lower than the genome average (mean = 0.55, SD = 0.20; Fig. 1D). Low CpGO/E is a signature of DNA methylation, which is the only mechanism known to selectively target CpG dinucleotides in animal genomes. Hereinafter, we refer to the genes belonging to the first category as “high-CpG” genes and those within the latter category as “low-CpG” genes.
Given that the nucleotide composition of the A. mellifera genome is highly heterogeneous (24, 25), we examined whether the observed bimodality in CpG content arises from a bias in nucleotide composition. Previous studies have shown a positive correlation between GC content and CpGO/E (17, 26, 27). We also find that the GC content and CpG content are strongly correlated in the A. mellifera genome (Kendall's correlation coefficient, τ = 0.32; P < 10−15). Thus, it is possible that the distribution of CpG content reflects the influence of GC content. To explore this possibility, we investigated the distribution of normalized GpC content (GpCO/E). GpC dinucleotides have the same C and G composition as CpG dinucleotides but are not targeted by DNA methylation (6, 28). For this reason, GpCO/E often is used as an indicator of nucleotide composition bias while controlling for the influence of DNA methylation (22, 29).
We find that the distribution of GpCO/E in A. mellifera is unimodal (Fig. S1). The GpCO/E distribution in D. melanogaster is unimodal as well, as expected (results not shown). Moreover, analyses of all other dinucleotides in A. mellifera clearly show that bimodality is exclusive to CpG dinucleotides (Fig. S1). These findings indicate that the observed bimodality of CpGO/E in A. mellifera genes stems from the difference in levels of germline DNA methylation on an evolutionary time scale; hypermethylated genes exhibit CpG depletion, whereas hypomethylated genes have high CpG content.
Further support for the link between CpG content and the level of DNA methylation comes from an analysis of CpGO/E profiles of genes in a distantly related invertebrate, Ciona intestinalis. C. intestinalis is the only invertebrate whose genomic pattern of DNA methylation has been experimentally investigated to date (19, 30), and its CpGO/E level has been shown to correspond to the actual level of DNA methylation (19). Furthermore, A. mellifera genes shown to be methylated in a previous study (6) are all found in the low-CpG class, as predicted by the proposed model (Fig. 1D).
To explore whether DNA methylation is widespread in genomic regions other than genes, we analyzed the CpGO/E distribution of the entire A. mellifera genome, as well as putative promoter regions (500 base pairs or 1,000 base pairs upstream of transcription start sites), untranslated regions, and transposable elements. Our analyses demonstrate that the strong bimodality of CpGO/E is unique to amino acid–encoding sequences [supporting information (SI) text and Figs. S2 and S3]. Only coding sequences harbor substantial portions of the low-CpG class, bearing evolutionary signatures of DNA methylation. These results are consistent with the observation that CpG methylation in A. mellifera is found predominantly in exons (6). The pattern of CpG depletion in A. mellifera introns is bimodal as well (Fig. S2), suggesting that some introns are methylated; however, the signal of bimodality is clearer when whole gene sequences (exons and introns) are analyzed, as expected if exons are primary targets of DNA methylation (6).
Low-CpG and High-CpG Genes Are Functionally Distinct.
The observed “bimodality” of A. mellifera genes, which represents an intragenic evolutionary signature of methylation, correlates with gene function; genes found in low-CpG and high-CpG classes are involved in specific biological processes (31). Specifically, the low-CpG and high-CpG classes are enriched with distinct Gene Ontology (GO) categories (Table 1). Low-CpG genes, predicted to be hypermethylated in the germlines, are significantly enriched for terms related to metabolism and ubiquitous housekeeping functions of gene expression and translation (Table 1 and Table S1). In contrast, high-CpG genes, which are predicted to be hypomethylated in the germlines, exhibit a striking and significant enrichment of terms associated with various developmental processes, cellular communication, and adhesion (Table 1 and Table S1).
Table 1.
Distinctive functional enrichment of low-CpG and high-CpG genes
| CpG class | GO biological process term | Accession | Fold enrichment in class | Significance* |
|---|---|---|---|---|
| Low-CpG | Macromolecule metabolic process | GO:0043170 | 1.13 | 3.91e-14 |
| Low-CpG | Cellular metabolic process | GO:0044237 | 1.09 | 1.04e-11 |
| Low-CpG | Metabolic process | GO:0008152 | 1.08 | 1.20e-10 |
| Low-CpG | Primary metabolic process | GO:0044238 | 1.08 | 8.05e-09 |
| Low-CpG | Cellular process | GO:0009987 | 1.04 | 2.83e-08 |
| Low-CpG | Nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process | GO:0006139 | 1.17 | 2.85e-08 |
| Low-CpG | Gene expression | GO:0010467 | 1.18 | 3.15e-08 |
| Low-CpG | RNA processing | GO:0006396 | 1.37 | 1.05e-07 |
| Low-CpG | Biopolymer metabolic process | GO:0043283 | 1.12 | 1.42e-06 |
| Low-CpG | RNA metabolic process | GO:0016070 | 1.19 | 2.28e-06 |
| High-CpG | Multicellular organismal process | GO:0032501 | 1.32 | 1.20e-19 |
| High-CpG | Cell communication | GO:0007154 | 1.37 | 4.10e-16 |
| High-CpG | Organ development | GO:0048513 | 1.41 | 1.52e-11 |
| High-CpG | System development | GO:0048731 | 1.35 | 1.54e-11 |
| High-CpG | Signal transduction | GO:0007165 | 1.35 | 1.71e-11 |
| High-CpG | Multicellular organismal development | GO:0007275 | 1.28 | 2.92e-11 |
| High-CpG | Biological adhesion | GO:0022610 | 1.77 | 7.40e-11 |
| High-CpG | Cell adhesion | GO:0007155 | 1.77 | 7.40e-11 |
| High-CpG | Anatomic structure development | GO:0048856 | 1.30 | 9.39e-11 |
| High-CpG | Developmental process | GO:0032502 | 1.23 | 1.99e-09 |
The top 10 significantly enriched terms for low-CpG and high-CpG classes are shown; for a complete list, see Table S1. GO biological process term enrichment is based on 1,781 D. melanogaster orthologs of A. mellifera high-CpG genes (1,230 with GO annotation) and 2,531 D. melanogaster orthologs of A. mellifera low-CpG genes (1,713 with GO annotation).
*Significance is denoted by a Benjamini correction for multiple testing.
Genes Whose Expression Is Strongly Biased Toward Specific Castes Are Enriched in the High-CpG Class.
Social insect development is marked by a remarkable level of phenotypic plasticity. In particular, many hymenopteran social insect females can develop into distinctive queen and worker castes from identical genomes. Recent studies suggest that DNA methylation may regulate caste differentiation in A. mellifera, by silencing crucial genes involved in caste formation (12, 14). If DNA methylation is truly involved in caste development, then genes that are overexpressed in a specific caste, or “caste-specific” genes, may show preferential enrichment in low-CpG or high-CpG genes. We tested this prediction using a data set from a recent study that identified differential gene expression in brains of queens and sterile workers (32).
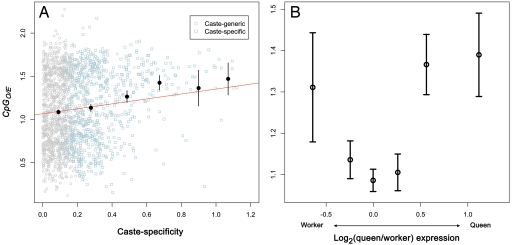
We first examined whether genes that were identified as caste-specific (at a 5% significance level) tend to be biased toward a specific CpG-content class (low-CpG or high-CpG). We find that caste-specific genes tend to harbor more high-CpG genes than expected based on the distribution of caste-generic genes (i.e., genes that are not differently expressed between the castes; Table 2). The enrichment of high-CpG genes increases with the bias toward caste-specific expression (Table 2; Fig. 2). Moreover, the degree of caste-specificity [measured as the absolute value of log2(queen/worker) gene expression] is significantly positively correlated with CpGO/E (Spearman's rank correlation, rs = 0.1405; P = 2.80e-09; Fig. 2A).
Table 2.
Caste-specific genes, which are differentially expressed between the queen and worker castes, are significantly overrepresented in the high-CpG class compared with caste-generic genes, whose expression patterns are not significantly different between the 2 castes
| Gene expression class | Significance threshold* | High-CpG class | Low-CpG class | χ2P value† | CpGO/E, mean ± SEM (median) | Wilcoxon P value‡ |
|---|---|---|---|---|---|---|
| Caste-generic | 474 | 488 | 1.0895 ± 0.0135 (1.0577) | |||
| Caste-specific | P < .05 | 457 | 354 | .0034 | 1.1633 ± 0.0149 (1.2094) | .0003 |
| Caste-specific | P < .01 | 294 | 207 | .0008 | 1.1837 ± 0.0187 (1.2663) | 4.39e-05 |
| Caste-specific | P < .001 | 158 | 75 | 5.35e-07 | 1.2439 ± 0.0260 (1.3637) | 1.07e-07 |
| Caste-specific | P < .0001 | 75 | 19 | 2.96e-08 | 1.3274 ± 0.0352 (1.4042) | 1.07e-07 |
The significance of the tests increases (i.e., P values decrease) as the significance threshold for genes considered caste-specific becomes more stringent.
*Significance threshold for caste-specific genes differentially expressed by queens and sterile workers in a pairwise comparison.
†P values from Pearson's χ2 test of pairwise comparisons of the distribution among high-CpG and low-CpG classes of caste-specific genes versus the caste-generic class, after Yates's correction.
‡P values of Wilcoxon's rank-sum test with continuity correction from pairwise comparisons of CpGO/E values for caste-specific genes versus caste-generic genes.
Fig. 2.
Caste-specific genes tend to have high CpGO/E. (A) Caste-specificity [measured as the absolute value of log2(queen/worker) gene expression] is correlated with CpGO/E (Spearman's rank correlation, rs = 0.1405; P = 2.80e-09). Mean values of CpGO/E for equal windows of caste specificity are shown as black dots with 95% confidence interval error bars. Ten outliers beyond caste-specificity values of 1.2 are excluded from the figure, but are included in calculations of correlation and model fitting. Points in the scatterplot are divided into caste-generic and caste-specific classes according to significant differences in expression between queens and workers (32). (B) The relationship between the values of log2-gene expression ratios between castes and CpGO/E values shows that the enrichment of high-CpG genes holds for genes that are either queen-specific or worker-specific. Genes expressed more highly in workers have log2-ratios < 0, whereas those expressed more highly in queens have log2-ratios > 0. The y-axis shows the mean and 95% confidence intervals of each group. As the log2-expression ratios between castes become more extreme (either side of the x-axis), CpGO/E tends to become more elevated.
We further expanded our analyses to genes implicated in A. mellifera caste differentiation identified by previous studies of gene expression (33–38). Again we found that caste-specific genes overwhelmingly belong to the high-CpG class (Table 3). Note that caste-specific genes are not necessarily those implicated solely in developmental processes; many of these genes perform basic biological functions (Table 3).
Table 3.
Genes identified as caste-specific from previous studies of gene expression and caste development in A. mellifera tend to belong (23 of 28; P < .005) to the hypomethylated class (high-CpG)
| Gene/gene family | Function | Caste-biased expression | CpGO/E class | Reference |
|---|---|---|---|---|
| AmIF-2mt translation initiation factor | Translation of mitochondrial-encoded mRNAs | Higher in queen larvae | 0/1 high-CpG | (33) |
| AmILP-2 insulin-like peptide | Regulation of growth/metabolism | Higher in workers than queens from second instar onward | 1/1 high-CpG | (38) |
| AmInR putative insulin-like peptide receptor family | Regulation of growth/metabolism | Higher in worker adults | 2/2 high-CpG | (34) |
| amTOR (target of rapamycin) | Regulation of growth/metabolism | Higher in queen 3rd instar larvae, but not 5th instar larvae (RNAi linked to worker fate) | 0/1 high-CpG | (37) |
| Hexamerin family | Storage of amino acids for use in metamorphosis or by adults | Either more highly expressed in queen or worker larvae (based on 2 empirically analyzed genes) | 3/4 high-CpG | (36) |
| vitellogenin | Yolk protein | Higher in queen adults | 1/1 high-CpG | (34) |
| Yellow/major royal jelly protein family | Sex-specific reproductive maturity among other functions | Primarily more highly expressed in workers, but some more highly expressed in queens (diverse tissue-dependent expression patterns) | 16/18 high-CpG | (35) |
Discussion
The genomic distribution of CpGO/E in A. mellifera stands in a sharp contrast to that in D. melanogaster, T. castaneum, and A. gambiae (Fig. 1). In particular, approximately half of A. mellifera genes belong to a distinctive low-CpG class (Fig. 1D). Given that (i) methylation in A. mellifera is exclusive to CpG dinucleotides (6), (ii) only CpG content exhibits bimodal distribution (Fig. S1), and (iii) deamination of methylated CpGs to TpG (or CpA in the complementary strand) causes a GC-to-AT mutational bias in diverse taxa (21, 22), these observations implicate DNA methylation in the origin of CpG bimodality. As far as we are aware, no other molecular mechanism is known to influence CpG dinucleotides exclusively and is unique to the A. mellifera genome compared with other sequenced insect genomes.
Our results suggest a unique influence of DNA methylation in A. mellifera evolution that may help explain important genome characteristics. For instance, the A. mellifera genome is known for its overall low and heterogeneous distribution of GC content (24, 25). An earlier study also detected the presence of a mutational bias toward A and T nucleotides (AT) in GC-poor regions of A. mellifera genes (25); however, the nature of such a mutational process remains unknown. Here we show that CpGO/E exhibits a striking bimodality and is strongly correlated with GC content in A. mellifera genes. These observations point to a link between the mutational bias toward AT and the depletion of CpG dinucleotides resulting from DNA methylation.
We also propose that, in addition to the mutational bias decreasing CpG content in low-CpG genes, other molecular mechanisms are operating to increase or maintain CpG content in high-CpG genes. The CpGO/E of high-CpG genes is higher than that of other dinucleotides and exceeds the value of 1.0 expected under random association of C and G nucleotides (Fig. S1). Thus, a process that conserves or even increases CpG dinucleotides against mutational depletion may exist in the honeybee genome, especially in high-CpG genes. The presence and nature of such processes in the A. mellifera genome should be addressed in future studies.
We have demonstrated that a substantial number of A. mellifera genes harbor evolutionary signatures of DNA methylation. This leads to the question of the functional significance of DNA methylation in A. mellifera. One potential role of DNA methylation is genomic imprinting, an epigenetic mechanism through which the expression of a gene is influenced by the parent from which it is inherited. In mammalian systems, DNA methylation is implicated in genomic imprinting (1, 39, 40). Social insects, especially those belonging to the haplodiploid Hymenoptera (social bees, social wasps, and ants), provide another intriguing context in which imprinting may play an important role in mediating a wide array of behaviors (41–43). We predict that imprinted genes, which should bear epigenetic marks (i.e., methylation) in the germlines, preferentially belong to the hypermethylated low-CpG class. Because our results demonstrate that nearly half of A. mellifera genes belong to the low-CpG class, many genes are candidates for studies of imprinting in A. mellifera. In this respect, it is of great interest to note that DNA methylation is widespread in haplodiploid hymenopteran social insects (44). Thus, information on CpG depletion for specific sets of genes in social insects provides fertile ground for future imprinting studies in a comparative context.
Our analyses indicate that methylation targets primarily gene bodies (exons and introns) in the A. mellifera genome. Moreover, methylated and nonmethylated regions coexist. Such a pattern is qualitatively similar to that found in echinoderms (e.g., sea urchin) and urochordates (e.g., sea squirt) (2, 19, 45). In the sea squirt (C. intestinalis), where genomic methylation has been examined in detail, it has been proposed that the primary role of DNA methylation is to suppress spurious transcription of genes that are broadly expressed across tissues with intermediate expression levels (2, 19). Our observation that genes that tend to be methylated are involved in basic biological processes (Table 1) supports this idea.
We found that low-CpG and high-CpG classes are populated with genes belonging to distinctive functional categories (Table 1). Low-CpG genes often are involved in metabolic processes and nucleotide processing, which can be considered basic biological processes. In contrast, a high proportion of high-CpG genes, which are predicted to be hypomethylated, are involved in development. This finding is particularly intriguing when considered along with the results of recent studies implicating DNA methylation in the regulation of phenotypic plasticity in social insects (12, 14).
Interestingly, we found that genes that are overexpressed in a specific caste are found more frequently in the hypomethylated class (Tables 2 and 3; Fig. 2). But it is noteworthy that not all caste-specific genes are found in the high-CpG class (Table 3); for example, genes associated with metabolism are frequently differentially expressed between castes (46–48) but overrepresented in the low-CpG class. Thus, the enrichment of caste-specific genes in the high-CpG class is particularly striking.
Previous studies in A. mellifera also have uncovered associations among cis-regulatory motifs, social behavior, and caste development (46, 49), indicating that cis-regulatory elements represent a putative global control mechanism for caste-specific gene expression. The significance of cis-regulatory elements, coupled with the finding that methylation can regulate caste fate (12), gives rise to the possibility that methylation interacts with regulatory elements to differentiate developmental pathways. But methylation of cis-regulatory elements themselves may not be a major mechanism underlying caste differences in A. mellifera, because our results suggest that methylation is limited primarily to gene bodies (Fig. S2).
Why are caste-specific genes preferentially found in the high-CpG class? We hypothesize that high-CpG genes in A. mellifera generally are more prone to epigenetic modulation than low-CpG genes. Large-scale analyses of methylation patterns in mammals repeatedly show that a subset of high-CpG promoters, particularly those associated with developmental processes, exhibit significant epigenetic flexibility, meaning that they are methylated in some tissues or developmental stages but not in others (50, 51). Furthermore, a class of mammalian genes with high-CpG promoters achieves complex, tissue-specific gene expression via pliable transcriptional regulation (N.E. and S.V.Y., unpublished data). Our observation that caste-specific genes tend to be enriched in the high-CpG class agrees with the aforementioned findings in mammals and may share similar underlying molecular mechanisms. Caste-specific genes must be activated or inactivated based on environmental input to proceed along different developmental paths; the high-CpG content of caste-specific genes may facilitate such modulation, similar to the role played by some high-CpG promoters in mammalian genomes.
The pattern of DNA methylation in insect genomes varies greatly (4). We have found that the genome of A. mellifera can be divided into 2 distinct classes based on the level of CpG depletion. Several pieces of evidence suggest that DNA methylation is the causative mechanism behind the observed bimodality. In particular, our prediction correctly assigns all genes identified as methylated in a previous study (6) to the low-CpG class. Our results suggest that DNA methylation regulates development, as seems to be the case in numerous other taxa (1, 39, 52, 53). In fact, DNA methylation is believed to play a critical role in caste differentiation (12). Our analyses of caste-specific genes provide support for this idea, but future studies and experimental verification of caste-specific gene expression and DNA methylation are needed.
The social Hymenoptera are ideal for studying the evolution and development of phenotypic plasticity, because the order comprises diverse taxa with multiple independent evolutionary origins of specialized queen and worker castes (54). The study of A. mellifera provides an important first look into the genome of a social hymenopteran insect (24), but the genomes of many social insects and related species are likely to be sequenced within the next 10 years (55). Comparative genomic analyses of evolutionary methylation signatures and experimental verification will more fully elucidate the evolutionary history and functional roles of DNA methylation in this important group.
Materials and Methods
Genome Sequences and Annotations.
Genome sequences and gene annotations of A. mellifera, A. gambiae, and D. melanogaster were downloaded from the University of California Santa Cruz genome browser (genome builds apimel2, anoGam1, and dm3). The genome sequence and gene annotation of T. castaneum was downloaded from BeetleBase (www.beetlebase.org). Repetitive elements were annotated using the RepeatMasker program.
Measurement of CpGO/E and Tests for Bimodality.
CpGO/E is a metric of depletion of CpG dinucleotides, normalized by G and C nucleotide content (GC content) of the specific region of interest. The CpGO/E for each gene is defined as
where PCpG, PC, and PG are the frequencies of CpG dinucleotides, C nucleotides, and G nucleotides, respectively, estimated from each gene (Dataset S1). Here a gene was defined as all exons (both coding sequences and untranslated exons) and introns.
The unimodality or bimodality of CpGO/E distributions was tested using the NOCOM software package. In brief, this software uses an expectation maximization algorithm to fit the data to both unimodal and bimodal distribution models and finds the maximum likelihood values (L0 for unimodal models and and L1 for bimodal models). The statistic G2 = 2 [ln(L1) − ln(L0)], which approximately follows a χ2 distribution with 2 degrees of freedom, can be used to test whether a bimodal distribution provides a better fit to the data than a unimodal distribution. The cutoff value between high-CpG genes and low-CpG genes was determined by plotting curves based on the NOCOM means of 0.55 (SD = 0.20) and 1.50 (SD = 0.20) and determining their point of intersection (1.08; Fig. 1D).
GO Biological Process Term Enrichment.
Because GO annotation (31) is limited in A. mellifera, annotations of orthologs in D. melanogaster were used for GO term analysis. To identify orthologous proteins between A. mellifera and D. melanogaster, Refseq RNA nucleotide accessions for A. mellifera sequences were converted to protein GI identifiers using the gene2refseq database from the National Center for Biotechnology Information (NCBI) ftp site (http://www.ncbi.nlm.nih.gov/ftp/), and D. melanogaster orthologs of A. mellifera genes were downloaded from the Roundup database of orthology (56), which uses the reciprocal smallest distance algorithm. A divergence threshold of 0.8 and a BLAST E-value cutoff of 1e-10 were used for ortholog identification. A total of 4,312 orthologous gene pairs between A. mellifera and D. melanogaster were obtained for further analysis.
GO biological process term enrichment was determined by comparing orthologs of low-CpG and high-CpG genes separately with a background composed of both low-CpG and high-CpG orthologs using the DAVID bioinformatics database functional annotation tool (57). A Benjamini multiple-testing correction of the EASE score (a modified Fisher exact P value) was used to determine statistical significance of gene enrichment (58).
Differential Gene Expression Between Honeybee Queen and Worker Castes.
Differential gene expression in brains of A. mellifera adult queens and sterile workers was determined using cDNA microarray analyses by Grozinger et al. (32). A list of BAGEL normalized expression levels (59) and P values for expression differences between queens and sterile workers was obtained from C.M. Grozinger. Gene identifiers for microarray data were converted to RNA nucleotide accessions using the gene_info and gene2refseq databases from the NCBI ftp site (http://www.ncbi.nlm.nih.gov/ftp/; Dataset S2).
Supplementary Material
Acknowledgments.
We thank Tim Nowack for analyzing the whole genome data and C.M. Grozinger for readily corresponding and providing data from previous research. This study was supported by an Alfred P. Sloan Research Fellowship (to S.V.Y.) and National Science Foundation Grant DEB 0640690 (to M.A.D.G. and S.V.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900301106/DCSupplemental.
References
- 1.Klose RJ, Bird AP. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- 4.Field LM, Lyko F, Mandrioll M, Pranter G. DNA methylation in insects. Insect Mol Biol. 2004;13:109–115. doi: 10.1111/j.0962-1075.2004.00470.x. [DOI] [PubMed] [Google Scholar]
- 5.Schaefer M, Lyko F. DNA methylation with a sting: An active DNA methylation system in the honeybee. BioEssays. 2007;29:208–211. doi: 10.1002/bies.20548. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Functional CpG methylation system in a social insect. Science. 2006;314:645–647. doi: 10.1126/science.1135213. [DOI] [PubMed] [Google Scholar]
- 7.Strassmann JE, Queller DC. Insect societies as divided organisms: The complexities of purpose and cross-purpose. Proc Natl Acad Sci USA. 2007;314:645–647. doi: 10.1073/pnas.0701285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson EO. The Insect Societies. Cambridge, MA: Harvard Univ Press; 1971. [Google Scholar]
- 9.Oster GF, Wilson EO. Caste and Ecology in the Social Insects. Princeton, NJ: Princeton Univ Press; 1978. [PubMed] [Google Scholar]
- 10.Keller L. Levels of Selection in Evolution. Princeton, NJ: Princeton Univ Press; 1999. [Google Scholar]
- 11.Maynard Smith J, Szathmary E. The Major Transitions in Evolution. Oxford: Oxford Univ Press; 1998. [Google Scholar]
- 12.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 13.Maleszka R. Epigenetic integration of environmental and genomic signals in honey bees. Epigenetics. 2008;3:188–192. doi: 10.4161/epi.3.4.6697. [DOI] [PubMed] [Google Scholar]
- 14.Moczek AP, Snell-Rood EC. The basis of bee-ing different: The role of gene silencing in plasticity. Evol Dev. 2008;10:511–513. doi: 10.1111/j.1525-142X.2008.00264.x. [DOI] [PubMed] [Google Scholar]
- 15.Goodisman MAD, Kovacs JL, Hunt BH. Functional genetics and genomics in ants (Hymenoptera: Formicidae): The interplay of genes and social life. Myrmecol News. 2008;11:107–117. [Google Scholar]
- 16.Robinson GE, Grozinger CM, Whitfield CW. Sociogenomics: Social life in molecular terms. Nat Rev Genet. 2005;6:257–271. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- 17.Elango N, Kim S-H, Program NCS, Vigoda E, Yi S. Mutations of different molecular origins exhibit contrasting patterns of regional substitution rate variation. PLoS Comput Biol. 2008;4:e1000015. doi: 10.1371/journal.pcbi.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki MM, Kerr ARW, De Sousa D, Bird A. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 2007;17:625–631. doi: 10.1101/gr.6163007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 21.Bird A. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elango N, Yi S. DNA methylation and structural and functional bimodality of vertebrate promoters. Mol Biol Evol. 2008;25:1602–1608. doi: 10.1093/molbev/msn110. [DOI] [PubMed] [Google Scholar]
- 23.Urieli-Shoval S, Gruenbaum Y, Sedat J, Razin A. The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett. 1982;146:148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- 24.The Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee, Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jørgensen FG, Schierup MH, Clark AG. Heterogeneity in regional GC content and differential usage of codons and amino acids in GC-poor and GC-rich regions of the genome of Apis mellifera. Mol Biol Evol. 2006;24:611–619. doi: 10.1093/molbev/msl190. [DOI] [PubMed] [Google Scholar]
- 26.Duret L, Galtier N. The covariation between TpA deficiency, CpG deficiency, and G+C content of human isochores is due to a mathematical artifact. Mol Biol Evol. 2000;17:1620–1625. doi: 10.1093/oxfordjournals.molbev.a026261. [DOI] [PubMed] [Google Scholar]
- 27.Fryxell KJ, Zuckerkandl E. Cytosine deamination plays a primary role in the evolution of mammalian isochores. Mol Biol Evol. 2000;17:1371–1383. doi: 10.1093/oxfordjournals.molbev.a026420. [DOI] [PubMed] [Google Scholar]
- 28.Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 29.Fryxell KJ, Moon W-J. CpG mutation rates in the human genome are highly dependent on local GC content. Mol Biol Evol. 2005;22:650–658. doi: 10.1093/molbev/msi043. [DOI] [PubMed] [Google Scholar]
- 30.Simmen MW, et al. Nonmethylated transposable elements and methylated genes in a chordate genome. Science. 1999;283:1164–1167. doi: 10.1126/science.283.5405.1164. [DOI] [PubMed] [Google Scholar]
- 31.Ashburner M, et al. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grozinger CM, Fan YL, Hoover SER, Winston ML. Genome-wide analysis reveals differences in brain gene expression patterns associated with caste and reproductive status in honey bees (Apis mellifera) Mol Ecol. 2007;16:4837–4848. doi: 10.1111/j.1365-294X.2007.03545.x. [DOI] [PubMed] [Google Scholar]
- 33.Corona M, Estrada E, Zurita M. Differential expression of mitochondrial genes between queens and workers during caste determination in the honey bee, Apis mellifera. J Exp Biol. 1999;202:929–938. doi: 10.1242/jeb.202.8.929. [DOI] [PubMed] [Google Scholar]
- 34.Corona M, et al. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc Natl Acad Sci USA. 2007;104:7128–7133. doi: 10.1073/pnas.0701909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drapeau MD, Albert S, Kucharski R, Prusko C, Maleszka R. Evolution of the yellow/major royal jelly protein family and the emergence of social behavior in honey bees. Genome Res. 2006;16:1385–1394. doi: 10.1101/gr.5012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans JD, Wheeler DE. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc Natl Acad Sci USA. 1999;96:5575–5580. doi: 10.1073/pnas.96.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel A, et al. The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS One. 2007;2:e509. doi: 10.1371/journal.pone.0000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler DE, Buck N, Evans JD. Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol Biol. 2006;15:597–602. doi: 10.1111/j.1365-2583.2006.00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 40.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 41.Haig D. Intragenomic conflict and the evolution of eusociality. J Theor Biol. 1992;156:401–403. doi: 10.1016/s0022-5193(05)80683-6. [DOI] [PubMed] [Google Scholar]
- 42.Haig D. The kinship theory of genomic imprinting. Annu Rev Ecol Syst. 2000;31:9–32. [Google Scholar]
- 43.Queller D. Theory of genomic imprinting conflict in social insects. BMC Evol Biol. 2003;3:15. doi: 10.1186/1471-2148-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kronforst MR, Gilley DC, Strassmann JE, Queller DC. DNA methylation is widespread across social hymenoptera. Curr Biol. 2008;18:R287–R288. doi: 10.1016/j.cub.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 45.Tweedie S, Charlton J, Clark V, Bird A. Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol Cell Biol. 1997;17:1469–1475. doi: 10.1128/mcb.17.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cristino ADS, et al. Caste development and reproduction: A genome-wide analysis of hallmarks of insect eusociality. Insect Mol Biol. 2006;15:703–714. doi: 10.1111/j.1365-2583.2006.00696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans JE, Wheeler DE. Expression profiles during honeybee caste determination. Genome Biol. 2000;2:1. doi: 10.1186/gb-2000-2-1-research0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolschin F, Amdam G. Comparative proteomics reveal characteristics of life-history transitions in a social insect. Proteome Sci. 2007;5:10. doi: 10.1186/1477-5956-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinha S, Ling X, Whitfield CW, Zhai C, Robinson GE. Genome scan for cis-regulatory DNA motifs associated with social behavior in honey bees. Proc Natl Acad Sci USA. 2006;103:16352–16357. doi: 10.1073/pnas.0607448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–771. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 53.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 54.Hughes WOH, Oldroyd BP, Beekman M, Ratneiks FLW. Ancestral monogamy shows kin selection is key to the evolution of eusociality. Science. 2008;320:1213–1216. doi: 10.1126/science.1156108. [DOI] [PubMed] [Google Scholar]
- 55.Smith CR, Toth AL, Suarez AV, Robinson GE. Genetic and genomic analyses of the division of labour in insect societies. Nat Rev Genet. 2008;9:735–748. doi: 10.1038/nrg2429. [DOI] [PubMed] [Google Scholar]
- 56.DeLuca TF, et al. Roundup: A multi-genome repository of orthologs and evolutionary distances. Bioinformatics. 2006;22:2044–2046. doi: 10.1093/bioinformatics/btl286. [DOI] [PubMed] [Google Scholar]
- 57.Dennis G, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 58.Hosack D, Dennis G, Sherman B, Lane H, Lempicki R. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Townsend JP, Hartl DL. Bayesian analysis of gene expression levels: Statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0071. research0071.0071–0071.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.