Abstract
Epigenetic mutations confer heritable changes in gene expression that are not due to changes in the underlying sequence of the DNA. We identified a spontaneous rice mutant, Epi-d1, that shows a metastable dwarf phenotype. The phenotype is mitotically and meiotically inheritable and corresponds to the metastable epigenetic silencing of the DWARF1 (D1) gene. The silenced state is correlated with repressive histone and DNA methylation marks in the D1 promoter region but is not associated with DNA sequence alterations. Compared with other known epigenetic silenced loci in plants such as paramutable maize alleles and silent Arabidopsis genes, the Epi-d1 silencing phenomenon shows a high level of bidirectional metastable mutability. Epigenetic alleles such as Epi-d1 could thus provide for rapid adaptation under selective conditions.
Epigenetic alleles are defined as those carrying heritable differences in gene expression that are not due to changes in the underlying sequence of the DNA (1). Although epigenetic changes in gene expression are likely to play an important role in evolution, very few epigenetic alleles have thus far been described. In plants, a well-described set of epialleles exist at maize loci that undergo paramutation. Paramutation is defined as an interaction between 2 alleles of a single locus, resulting in a heritable change of one allele that is induced by the other allele (2). Recent studies have suggested that small RNA-mediated gene silencing is important in the maintenance and establishment of the repressed state in paramutation (3, 4). Other epigenetic alleles have been studied in the model plant Arabidopsis thaliana, including at the SUPERMAN, PAI, FWA, and BONSAI loci, and studies of these have contributed to our understanding of gene-silencing mechanisms (5–10).
In this study, we report genetic analysis of a rice epigenetic mutant that affects plant stature. This mutant, named Epi-d1 shows a metastable dwarf phenotype. It was originally isolated as spontaneous dwarf mutant and has been maintained as a breeding material in Kyusyu University >90 years. As a result of genetic analysis on its metastability, we determined the target gene for epigenetic regulation and found that its epigenetic state is bidirectionally mutable, from active to repressed and from repressed to active. Epi-d1 is an epiallele described in rice and also has unique epigenetic characteristics.
Results
Character of Epi-d1 Mutant.
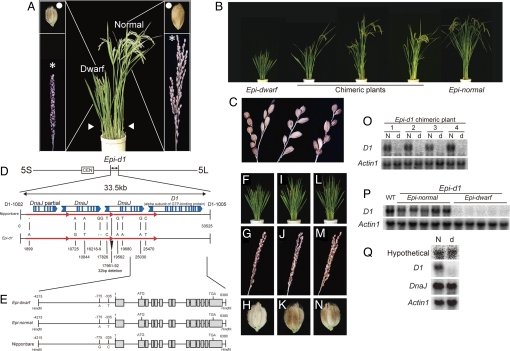
The Epi-d1 mutant is often chimeric, producing both dwarf and normal tillers (vegetative branch shoots) on the same plant (Fig. 1A). Dwarf tillers have short dark-green leaves, compact panicles (inflorescences), and small round grains. On the other hand, the normal tillers have normal features (Fig. 1A). In addition, different Epi-d1 plants show a wide variety of dwarf and normal features, from completely dwarf to completely normal (Fig. 1B). This strongly suggests that the cell fate to be dwarf or normal type is not genetically fixed but can vary epigenetically in the developing plant. The completely dwarf and normal type plants were named Epi-dwarf and Epi-normal, respectively (Fig. 1B Left and Right plants). Although most dwarf tillers have small round grains, and most normal tillers have normal grains, we can find chimeric features also in the panicles (Fig. 1C). These results indicate that cell type (normal or dwarf) frequently changes from the vegetative to reproductive stage.
Fig. 1.
Mapping analysis of Epi-d1. (A) Gross morphology of Epi-d1 mutant (Center) and magnified view of its dwarf panicle and seed in dwarf tillers (Left) and normal panicle and seed in normal tillers (Right). Arrowheads indicate tillers (vegetative branch shoots), asterisks indicate panicles (inflorescences), and white circles indicate grains. (B) Epi-d1 shows varying phenotypes from completely dwarf to completely normal. (C) Chimeric phenotype in panicle. A panicle showing 2 separate rachises with either small round or normal grains (Left). A panicle showing small round and normal grains arranged alternately on the rachis (Center). A panicle showing a random distribution of the 2 grain phenotypes (Right). (D) Epi-d1 was mapped into 59 cM of chromosome 5, then narrowed down to a 33.5-kb region between 2 molecular markers, D1-1002 and D1-1005. In this region, there are 3 8.6-kb tandem repeats (red arrows) and 4 ORFs are predicted by RiceGAAS. There are 11 polymorphisms in this 33.5-kb region between Nipponbare and Epi-d1 mutant. The numbers indicate the positions of polymorphisms in this region. (E) Comparison of sequence of D1 gene between Epi-normal, Epi-dwarf, and Nipponbare. (F–H) The phenotypes of d1 loss-of-function mutant are shown. (I–K) The phenotypes of Epi-dwarf plant are shown. (L–N) The phenotypes of F1 plant derived from cross between Epi-dwarf and d1 are shown. The gross morphologies are shown in F, I, and L; panicles in G, J, and M, and seeds in H, K, and N. (O) Total RNAs were extracted from 4 independent chimeric plants by separating tissues with different phenotypes. RNA blots were hybridized with D1 probe. N and d indicate RNA extracted from normal and dwarf tillers, respectively. (P) Total RNAs were extracted from Epi-normal and Epi-dwarf plants and hybridized with D1 probe. (Q) Total RNAs were extracted from Epi-normal and Epi-dwarf plants and hybridized with probes of D1 and its neighbor genes. Hypothetical and DnaJ gene are located 7 kb downstream and 4 kb upstream of the D1 gene, respectively. N and d indicate RNA extracted from Epi-normal and Epi-dwarf, respectively. Actin1 is hybridized as a control (O–Q).
To clarify the epigenetic regulation of Epi-d1, phenotypic segregation ratios were determined for the progenies of independent chimeric Epi-d1 plants [supporting information (SI) Table S1]. Although we always found a range of phenotypes in the progeny, we observed a strong tendency for seeds collected from normal tillers to give rise to a higher proportion of normal plants and seeds collected from dwarf tillers to give rise to more dwarf plants. As one example, the progeny from normal tillers of chimeric plant 1 segregated 44 normal, 127 chimeric, and 16 dwarf plants, whereas the progeny from the dwarf tillers on this same plant produced 3 normal, 44 chimeric, and 59 dwarf plants (Table S1). We also observed a tendency for small seeds of dwarf tillers to produce a higher proportion of dwarf plants, and normal seed in normal tillers to give rise to a higher proportion of normal plants. We also studied the progenies of 4 independent Epi-normal plants and 4 Epi-dwarf plants (Table S2). The progenies from Epi-normal had 86.2% normal, 9.3% chimeric, and 4.5% dwarf phenotypes (Table S2). In contrast, the progenies from Epi-dwarf had 0.8% normal, 26.6% chimeric, and 72.6% dwarf phenotypes (Table S2). Together, these results show that although the Epi-d1 phenotypes are strongly heritable, the phenotypic transmission is metastable, and the conversion between the 2 phenotypic states occurs bidirectionally.
We next tested weather Epi-d1 showed allelic interactions similar to those of known maize epialleles that undergo paramutation. We first performed reciprocal crosses between Epi-normal and Epi-dwarf plants. All 7 F1 plants had the normal phenotype. In the F2 generation, the progenies of both reciprocal crosses produced normal and dwarf plants in a ratio of ≈3:1 (Table S3). This suggests that the alleles of Epi-normal and Epi-dwarf do not affect each other and are thus not paramutable. These results also show that Epi-normal is dominant, and Epi-dwarf is a recessive allele. To confirm these results, Epi-dwarf plants were further reciprocally crossed with 2 wild-type rice cultivars, IR24 (an indica cultivar) and Taichung65 (a japonica cultivar). The F2 progenies of these crosses also segregated in a ratio of 3:1 (Table S3), suggesting that the Epi-dwarf allele is recessive and that the Epi-dwarf allele does not significantly affect wild-type alleles. These genetic analyses suggest that the metastable phenomenon of Epi-d1 is different from paramutation.
To understand the molecular mechanism of Epi-d1, we performed mapping analysis using 16,000 F2 plants from crosses between Epi-d1 and Kasalath (an indica rice cultivar). This mapping revealed that the Epi-d1 gene was located between molecular markers D1-1002 and D1-1005 within a 33.5-kb region of rice chromosome 5 (Fig. 1D). This candidate region contains 3 8.6-kb tandem repeats, and 4 ORFs encoding 3 DnaJ genes and the DWARF1 (D1) gene, encoding the α-subunit of a GTP-binding protein (Fig. 1D). The recessive loss-of-function of dwarf1 (d1) shows a dwarf phenotype with short dark-green leaves, compact panicles, and small round grains (Fig. 1 F–H) (11, 12). The dwarf characters of Epi-d1 are very similar to that of d1 (Fig. 1 I–K). From the phenotypic resemblance and mapping analysis, we suspected that the dwarf phenotype of Epi-d1 would be caused by defects in the D1 gene. To clarify this, we tested allelism between Epi-dwarf and d1 using complementation crosses. Of 150 F1 plants; 6 showed a chimeric dwarf phenotype, and 144 showed a full dwarf phenotype (Table S4 and Fig. 1 L–N). This strongly indicates that the Epi-dwarf is allelic to d1.
Metastable Phenotype of Epi-d1 Is Due to Metastable Expression of D1 Gene.
Next, we compared sequences of the D1 gene among Epi-normal, Epi-dwarf, and dwarf and normal tillers in a chimeric plant. Sequencing of the D1 locus between −4,215 bp and 6,380 bp relative to the transcription initiation site showed that there were no differences in the Epi-normal and Epi-dwarf DNA sequence (Fig. 1E). In addition, we compared the D1 sequences between Epi-d1 and Nipponbare (a japonica rice cultivar). Although 2 SNPs were detected in the promoter region (Fig. 1E), no mutation was found in the D1 coding region of Epi-d1 (Fig. 1E). To determine whether these SNPs cause the metastable phenotype of Epi-d1, we transformed 3 versions of the 10.5-kb D1 locus corresponding to the Epi-dwarf, Epi-normal, and Nipponbare alleles into a d1 loss-of-function mutant (Fig. 1E). Transformants of all 3 constructs showed a fully normal phenotype (Fig. S1). This indicates that the D1 gene of Epi-d1 encodes a functional allele and that the 2 SNPs detected in the upstream region (−775 and −335) of D1 are not mutations controlling the metastable phenotype of Epi-d1. We also investigated whether a wild-type D1 locus could complement the dwarf phenotype of Epi-dwarf. We transformed the D1 gene cloned from Nipponbare into the Epi-dwarf plants and found that its dwarf phenotypes were fully rescued (Fig. S1). These results strongly confirm that the defect in Epi-d1 is due to a loss of function of the D1 gene. Finally, we sequenced the 33.5-kb candidate region defined by our recombination mapping. Although there were several polymorphisms between Epi-d1 and Nipponbare (Fig. 1D and Fig. S2), when we transformed the fragment containing the 33.5-kb candidate region cloned from Epi-dwarf into a d1 mutant, the transformants also completely rescued the d1 phenotype (Fig. S1). These results suggest that Epi-d1 contains a potentially functional DNA sequence for the D1 gene but that the D1 gene in Epi-dwarf plants is not active.
To determine the cause of the metastable phenotype of Epi-d1, we performed expression analysis of the D1 gene. Northern blot analysis revealed dramatically different expression of D1 in Epi-d1. Although D1 was highly expressed in normal tillers of chimeric plants, the D1 transcript was not detected in dwarf tillers of these same plants (Fig. 1O). We also examined D1 expression in Epi-dwarf and Epi-normal plants and found that the D1 gene was highly expressed in Epi-normal but not in Epi-dwarf (Fig. 1P). These results indicate that the epigenetic phenomenon in Epi-d1 is due to the switching on or off of RNA expression of D1. We conclude that the Epi-d1 mutant is an epigenetic allele of the D1 locus that we have named Epiallele of d1 (Epi-d1).
To determine whether the difference in expression in Epi-d1 occurs in a narrow or broad genomic region around D1, we investigated transcripts of genes located around the D1 gene in Epi-d1. The transcript level of DnaJ located 4 kb upstream of D1 and a hypothetical gene located 7 kb downstream of D1 were equal between dwarf tillers and normal tillers (Fig. 1Q). These results suggest that epigenetic regulation in Epi-d1 only affects the D1 gene. Finally, to determine whether the D1 locus, narrowed down by mapping analysis, is sufficient for the metastable expression of D1, we used introgression to produce a nearly isogenic line (NIL) carrying only the D1 locus of the Epi-d1 chromosome segment in an otherwise Kasalath genome background (Fig. S3a). Resultant NIL-Epi-d1 shows dwarf, chimeric, and normal phenotypes similar to the original Epi-d1 plants (Fig. S3b). This indicates that the D1 locus of Epi-d1 is sufficient to confer the epigenetic D1 expression instability.
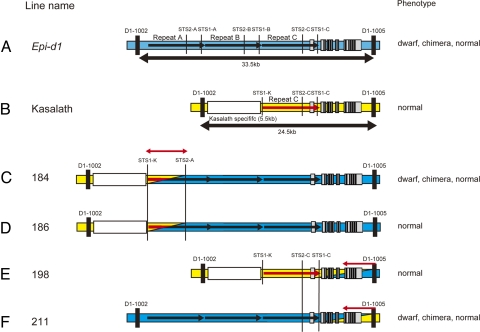
During the mapping analysis, we obtained recombinant plants containing recombination breakpoints within the D1 gene or upstream repeat region (Fig. 2 C–F). In these recombinant plants, lines 184 and 186 have a recombination point between the markers STS1-K and STS2-A on repeat A (Fig. 2 C and D). Although line 184 shows 3 kinds of phenotypes, dwarf, chimera, and normal phenotypes, line 186 shows only normal phenotype. These lines indicate the candidate region of metastability is located within repeat A. On the other hand, lines 198 and 211 have a recombination point between the markers, STS1-C and D1-1005 (Fig. 2 E and F). Because line 198 shows only normal phenotype, and line 211 shows dwarf, chimera, and normal phenotypes, the candidate epigenetic region of Epi-d1 is located upstream from the marker D1-1005, which is also consistent with the interpretation from lines 184 and 186 that the metastability element maps to repeat A. From these results, repeat A seems to be required for phenotypic metastability.
Fig. 2.
Graphical genotypes of recombinant plants. Blue and yellow bars indicate chromosome of Epi-d1 and Kasalath respectively. (A) Graphical genotype of Epi-d1. (B) Graphical genotype of Kasalath. (C–F) Graphical genotypes of recombinant lines 184, 186, 198, and 211 are shown. The phenotypes of these lines are shown on the right of the chromosome.
Metastable D1 Expression Is Caused by Epigenetic Modification.
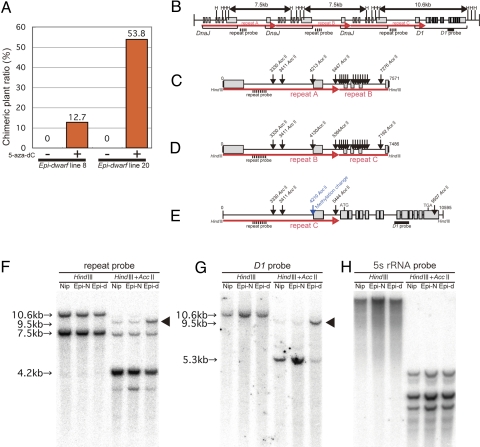
To determine the possible epigenetic marks that regulate the expression state of the D1 gene, we analyzed DNA methylation and histone modification. In the continuous selection of Epi-dwarf plants from Epi-dwarf over several generations, we could obtain Epi-dwarf line 8 and 20, which rarely show normal tissues. We treated seeds derived from these Epi-dwarf lines with 5-aza-2′ deoxycytidine (5-aza-dC), an inhibitor of DNA methylation (13). Although seeds not treated with 5-aza-dC showed no chimeric or normal plants, the 5-aza-dC-treated seeds produced 12–53% chimeric plants (Fig. 3A). This suggests that DNA demethylation promotes the transition from dwarf to normal in Epi-d1 plants. Next, we investigated the DNA methylation level around the D1 locus. When the genomic DNAs were double digested by HindIII and AccII (a methylated DNA-sensitive restriction enzyme that recognizes CGCG but cannot digest methylated CGCG), Nipponbare and normal tillers in Epi-d1 had a 5.3-kb band, whereas a 9.5-kb band was observed in dwarf tillers in Epi-d1 (Fig. 3G). When 5s rRNA and 168bp centromere repeat were hybridized as a probe, there were no difference in Nipponbare and dwarf and normal tillers of Epi-d1 (Fig. 3H and Fig. S4), suggesting that these changes in DNA methylation are specific to the D1 locus. There are 5 AccII sites in the 10.6-kb genomic DNA region between the 2 HindIII sites (Fig. 3E). The detection of a 5.3-kb signal in normal plants indicates that the DNA fragment was digested at the 4210 AccII site and the 9507 AccII site (Fig. 3E). The detection of a 9.5-kb signal in dwarf tillers indicates that the DNA fragment was digested with the 0 HindIII site and the 9507 AccII site (Fig. 3 E and G). The methylation status at the 4210 AccII site therefore differs between dwarf and normal tillers (Fig. 3E). The 4210 AccII site is at position −2 bp relative to the D1 transcription initiation site, and thus may be in a region critical for D1 promoter activity. DNA methylation of the upstream repeat region was also examined by using a repeat probe for analysis (Fig. 3 B–D). Southern hybridization using this probe showed 2 bands with HindIII, 10.6 kb and 7.5 kb (Fig. 3F). The 10.6-kb fragment contains the D1 gene, and the 7.5-kb fragment corresponds to both repeat A and B (Fig. 3 B–D). When the genomic DNA was double digested by HindIII and AccII, a 9.5-kb band was enriched in dwarf tillers by using the repeat probe (Fig. 3F), again consistent with hypermethylation of the 4210 AccII site. Consistently, a 4.2-kb band was strongly detected in all plants, although it was weaker in dwarf plants than in normal plants (Fig. 3F), consistent with hypermethylation of the 4210 AccII site. The observation that the 4.2-kb band was present in all genotypes (Fig. 3F), indicates that the 3330 and 3411 AccII sites in both 7.5-kb fragments from junctions between repeat A to B and between repeat B to C (Fig. 3 C and D) are consistently hypermethylated. These results suggest that variation in methylation at the AccII site in the promoter of the D1 gene strongly correlate with D1 silencing.
Fig. 3.
DNA methylation analysis of Epi-d1. (A) The appearance of chimeric plants from Epi-dwarf seeds treated with inhibitor of DNA methylation, 5-aza-2′ deoxycytidine. Values in the graph indicate the ratio of chimeric plants from Epi-dwarf (percentage). (B) Restriction map of the candidate region of Epi-d1 gene. HindIII sites are described as H. (C and D) The restriction map of first and second 7.5-kb region digested by 2 HindIII sites. (E) The restriction map of 10.6-kb region contains the D1 gene. (F and G) The probes used are indicated as broken line and simple line, respectively. (C–E) Arrows indicate the sites of DNA methylation sensitive restriction enzyme, AccII (CGCG). (F–H) The genomic DNA of Nipponbare (Nip), Epi-normal (Epi-N), and Epi-dwarf (Epi-d) were digested by HindIII or double digested by HindIII and AccII. Digested DNAs were hybridized with repeat probe (F) and D1 probe (G). 5s rRNA probe was also hybridized as a control (H).
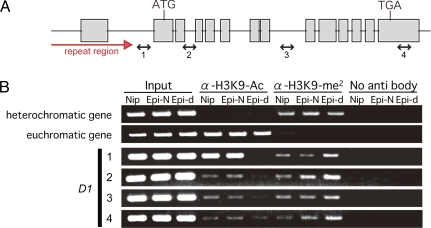
To investigate the relationship between metastable D1 expression and chromatin modification, we performed ChIP analysis using acetylated K9 of histone H3 (Ac-H3K9), a mark usually associated with active genes, and dimethylated K9 of histone H3 (dime-H3K9), a mark usually associated with silent genes (14). We could successfully use primers designed over the D1 gene (Fig. 4A); however, because of the highly conserved repeat sequence, we could not design primers specific for the upstream region of D1. We detected accumulation of Ac-H3K9 and reduced dime-H3K9 at the D1 locus in Epi-normal (Fig. 4B). Conversely, accumulation of dime-H3K9 and reduction of Ac-H3K9 were detected at the D1 locus in Epi-dwarf (Fig. 4B). These results are consistent with the expression pattern and DNA methylation of D1, and suggest that repressive chromatin marks are correlated with the silent Epi-d1 state.
Fig. 4.
ChIP assay of D1 gene. (A, 1–4) The regions amplified to detect the chromatin state are indicated as arrows. (B) Nucleosomes were precipitated by anti-acetylated histone H3K9 (α-H3K9-Ac) and anti-dimethylated histone H3K9 (α-H3K9-me2) Input and no antibody were also amplified as positive and negative controls, respectively. The precipitated nucleosomes of Nipponbare (Nip), Epi-normal (Epi-N), and Epi-dwarf (Epi-d) were amplified by using primers of heterochromatic gene, euchromatic gene, and D1 gene.
Discussion
In summary, Epi-d1 represents a metastable epigenetic mutant of the D1 locus that is associated with DNA methylation and repressive histone marks. Epi-d1 is not only an epigenetic allele described in rice but also exhibits unique qualities compared with previously described epigenetic loci. Given that both the silent and active state of D1 show metastability, and given that allelic interactions are not observed between alleles in these different expression states, the Epi-d1 phenomenon is unlike paramutation. Furthermore, we did not observe allelic interactions between the silent Epi-d1 alleles and incoming transgenic alleles (Fig. S1). This is different from Arabidopsis epigenetic alleles of the FWA locus, where silencing can be transmitted from the silent endogenous locus to cause efficient silencing of incoming FWA transgenes (15). Most strikingly, the transcriptional initiation site of D1 was differentially hypermethylated in Epi-d1, even though identical sequences in the upstream repeats did not show methylation variation between Nipponbare, Epi-normal, and Epi-dwarf epialleles (Fig. 3). Thus, the factor(s) repressing D1 expression in Epi-dwarf do not affect DNA sequences sharing homology with the target of repression. These observations suggest that Epi-d1 silencing is a uniquely cis-limited epigenetic phenomenon, which may be controlled by unique epigenetic mechanisms. Finally, the observations that very stable Epi-dwarf plants can be selected over multiple generations suggests that epigenetic phenotypes such as Epi-d1 could provide plants with a repertoire of quickly selectable traits that change much more quickly than those caused by difference in DNA sequence.
Materials and Methods
Mapping and NIL Production.
To map Epi-d1, F2 plants from cross between Epi-dwarf and the indica strain Kasalath were used. ≈16,000 F2 plants from this cross were used for positional cloning of Epi-d1. To create NILs of Epi-d1, F1 plants (Epi-dwarf/Kasalath) were back-crossed with Kasalath 4 times. NIL were selected from the BC4F2 generation (i.e., Epi-dwarf/Kasalath/Kasalath/Kasalath/Kasalath/Kasalath) by using MAS (16, 17). The phenotypes of all plants were determined after grains ripened. Genomic DNA was extracted from each F2 population and NIL by using the TPS method for mapping and NIL production. For the TPS method, ≈2-cm lengths of rice leaf tips were harvested and ground by using a MultiBead Shocker (Yasui Kikai) in TPS buffer [100 mM Tris·HCl (pH 8.0), 1 M KCl, 10 mM EDTA]. After centrifugation, the supernatant was recovered, and an equal volume of isopropyl alcohol was added. Isopropyl alcohol-insoluble material was recovered by centrifugation, and the pellet was washed with 75% ethanol. The pellet was then dried and dissolved in TE [10 mM Tris·HCl (pH 8.0), 1 mM EDTA]. The purified DNA samples were then genotyped by using molecular markers. PCR-based markers, including simple-sequence repeat (SSR) markers (www.gramene.org/) (18, 19), cleaved amplified polymorphic sequence (CAPS) markers (20), and SNPs, which were identified by comparing the genomic DNA sequences of each parent, were used for mapping and NIL production.
Transgenic Analysis.
A construct containing the D1 gene of Nipponbare or Epi-d1 was introduced to a d1 loss-of-function mutant line, HO541, and Epi-dwarf. The D1 gene in the BAC clone constructed from Nipponbare genomic DNA, B022G02, and from Epi-d1 genomic DNA, DC15F12 (constructed by the National Institute of Agrobiological Sciences) were digested with HindIII and inserted into the HindIII site of a pBluescript vector. The clone was redigested with HindIII. The fragment of ≈10.6 kb was fused into the HindIII site of the binary vector pBI-Hm12, which contains a hygromycin-resistance gene, kindly provided by Hiroyuki Hirano (Tokyo University, Tokyo). The 33.5-kb candidate D1 locus was constructed from DC15F12 DNA partially digested by BamHI and cloned into BamHI site of binary vector pYLTAC7 (provided by RIKEN BioResouce Center, Ibaraki, Japan). The binary vector was introduced into Agrobacterium tumefaciens strain EHA105 by electroporation, and rice plants were transformed with this strain.
Construction of a BAC Clone and Sequencing.
To determine the sequence of the candidate region of Epi-d1, a rice BAC library was constructed from young leaves of Epi-d1 based on the protocol reported by the Arizona Genomics Institute (www.genome.arizona.edu/information/protocols/index.html). The library (host: Escherichia coli strain DH10B; vector: pIndigoBAC5; cloning site: BamHI) contains 19,439 clones with an average insert size of 157 kb, which is estimated to cover the haploid genome ≈7 times. In total, 5 markers (d1–8, d1–17, D1-exon9–10, d1–11, and d1–12) were applied for screening of BAC clones with the PCR method using pooled DNA from the whole BAC library. Each positive BAC clone was used for end-sequencing using an ABI 3730xl DNA Analyzer (Applied Biosystems) according to a previously described method (23).
RNA Extraction and Expression Analysis.
Total RNAs (10 μg) isolated from leaves by using the TRIzol reagent (Invitrogen) were separated on a 1% agarose gel and transferred to Hybond n + membranes (Amersham Pharmacia). The membrane was hybridized with a 32P-labeled partial D1 cDNA fragment of 929 bp, which was amplified by using the primers D1-cDNA-probe-U: 5′-TTTGATGAGGCAGAACTTAG-3′ and D1-cDNA-probe-L: 5′-AGCGTCTCATGCTCTCATC-3′, which correspond to regions in exon 5 and the 3′ untranslated region, respectively. The probes were 32P-labeled by using the Bca BEST Labeling Kit (Takara). The membrane was hybridized at 65 °C in a hybridization solution of 5× SSC (1× SSC = 0.15 M NaCl and 0.015 M sodium citrate), 5× Denhardt's solution (1× Denhardt's solution = 0.02% ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 0.5% SDS, 10% dextran sulfate, and 0.1 mg/mL denatured salmon sperm DNA. DNA probes were labeled with 32P-dCTP. The membrane was washed twice with 2× SSC and 0.1% SDS at 65 °C for 20 min and once with 0.2× SSC and 0.1% SDS at 65 °C for 10 min. The membranes were exposed to a Fuji imaging plate, and the image was visualized by using a BAS2000 imaging analyzer (Fuji Photo Film).
5-Aza-2′Deoxycytidine Treatment.
Seeds were sterilized with 2.5% NaClO for 30 min and washed 5 times in sterile distilled water. The seeds were then placed on MS medium [4.6 g/L MS plant salt mixture (cat. no. 392–00591; Nihon Pharmaceutical), 5 mg/L nicotinic acid, 10 mg/L pyridoxine hydrochloride, 10 mg/L thiamine hydrochloride, 2 mg/L glycine, 100 mg/L myo-inositol and 1% agar] with or without 70 mg/L 5-aza-2′deoxycytidine (5-aza-dC) and grown under dark conditions at 30 °C for 1 week. After treatment with 5-aza-dC, seeds were transferred to water and grown under light conditions at 30 °C for 3 days and then transferred to the nursery bed and grown in greenhouse for 1 month. The seedlings were then transplanted to the paddy field.
DNA Gel Blot Analyses.
Rice genomic DNA was isolated from leaf tissue by using the ISOPLANT DNA isolation kit (Nippon Gene), and 1 μg was digested with suitable restriction enzymes, transferred onto Hybond n + membranes (Amersham) under alkaline conditions, and analyzed. Hybridization was performed at 65 °C in 0.25 M Na2HPO4, 1 mM EDTA, and 7% SDS. Filters were washed twice in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 65 °C for 15 min and once in 0.2× SSC and 0.1% SDS at 65 °C for 15 min. Isotope probes were amplified and 32P-labeled by using the Bca Best Labeling kit (Takara). The centromere-specific repeat probes were adopted from the centromere-specific sequences, RCS2, designed by Wu et al. (24).
ChIP Assay.
Approximately 50 g of 2-week-old etiolated rice seedlings were ground to fine powder with liquid nitrogen, resuspended in 40 mL of TBS [0.01 M Tris·HCl (pH 7.5), 3 mM CaCl2, 2 mM MgCl2 with 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and proteinase inhibitors (Complete; Roche Molecular Biochemicals)] with 0.5% Tween 40. The suspension was filtered through Miracloth (cat. no. 475855; Calbiochem) on ice. Nuclei were pelleted by centrifugation at 600 × g for 10 min at 4 °C and resuspended in 1× TBE with 25% sucrose. Nuclei were further purified by centrifugation at 1,500 × g for 20 min at 4 °C through a 1× TBE with a 25%/50% discontinuous sucrose gradient. Purified nuclei were then digested with 0.9 units of micrococcal nuclease (USB Corp.) in 4.5 mL of digestion buffer [0.32 M sucrose, 50 mM Tris·HCl (pH 7.5), 4 mM MgCl2, 1 mM CaCl2, 0.1 mM PMSF] at 37 °C for 10 min. After digestion was stopped by adding 45 μL of 0.5 M EDTA, the nucleosome was purified by centrifugation at 15,000 × g for 10 min at 4 °C, and the first supernatant was kept on ice. The pellet was further incubated with lysis buffer [1 mM Tris·HCl (pH 7.5), 0.2 mM EDTA, 0.2 mM PMSF, and proteinase inhibitors] on ice for 1 h and centrifuged as before, and the 2 supernatants were pooled. The resulting oligonucleosome suspension was precleared by incubation with a 1:1,000 dilution of preimmune serum and 10% protein A-Sepharose (cat. no. 17-1279-02GE Healthcare) at 4 °C for 4 h, and centrifuged at 250 × g for 5 min at 4 °C. The supernatant was used immediately for immunoprecipitation. Equal volumes of the input fraction and incubation buffer [50 mM NaCl, 20 mM Tris·HCl (pH 7.5), 5 mM EDTA, 0.1 mM PMSF, and protease inhibitors] were incubated with 5 μg of antibodies at 4 °C overnight.
Normal rabbit serum was then added as no antibody. Immune complexes were captured by incubation with 25% vol/vol protein A-Sepharose at 4 °C for 2 h. At the end of the incubation, the protein A-Sepharose was washed stepwise in buffer A [50 mM Tris·HCl (pH 7.5), 10 mM EDTA] containing 50, 100, and 150 mM NaCl 3 times. Bound immune complexes (bound fraction) were eluted with 2 vol of incubation buffer containing 1% SDS. DNA was extracted from the supernatant by phenol/chloroform extraction and resuspended in TE buffer (pH 8.0). The antibodies and primers used in this assay were anti-dimethyl histone H3 K9 antibody (cat. no. ab7312-100; Abcam), anti -acetyl histone H3 K9 antibody (cat. no. ab4441-50; Abcam).
The primers of C-kinase substrate and 30 designed by Nagaki et al. (25) were adopted as control for euchromatic and heterochromatic genes, respectively. For additional materials and methods, see SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. P. J. Green, B. C. Meyers, and D. H. Jeong for helpful suggestions, Dr. K. Nagaki and H. Tsuji for providing a detailed protocol for the ChIP assay. This work was supported in part by Grant-in-Aid for Scientific Research on Priority Areas 17027012 and a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Integrated research project for plant, insect and animal by using genome technology IP1003). S.E.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901942106/DCSupplemental.
References
- 1.Kakutani T. Epi-alleles in plants: Inheritance of epigenetic information over generations. Plant Cell Physiol. 2002;43:1106–1111. doi: 10.1093/pcp/pcf131. [DOI] [PubMed] [Google Scholar]
- 2.Chandler V, Stam M. Chromatin conversations: Mechanisms and implications of paramutation. Nat Rev Genet. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- 3.Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 4.Hale C, Stonaker J, Gross S, Hollick J. A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 2007;5:e275. doi: 10.1371/journal.pbio.0050275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindroth A, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 6.Jackson J, Lindroth A, Cao X, Jacobsen S. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 7.Zilberman D, Cao X, Jacobsen S. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 8.Chan S, Henderson I, Jacobsen S. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 9.Kinoshita Y, et al. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 2007;49:38–45. doi: 10.1111/j.1365-313X.2006.02936.x. [DOI] [PubMed] [Google Scholar]
- 10.Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 11.Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc Natl Acad Sci USA. 1999;96:10284–10289. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujisawa Y, et al. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA. 1999;96:7575–7580. doi: 10.1073/pnas.96.13.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang S, Pikaard C. Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J Biol Chem. 2005;280:796–804. doi: 10.1074/jbc.M409053200. [DOI] [PubMed] [Google Scholar]
- 14.Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol. 2007;10:528–533. doi: 10.1016/j.pbi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Chan S, Zhang X, Bernatavichute Y, Jacobsen S. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yano M, Sasaki T. Genetic and molecular dissection of quantitative traits in rice. Plant Mol Biol. 1997;35:145–153. [PubMed] [Google Scholar]
- 17.Yano M. Genetic and molecular dissection of naturally occurring variation. Curr Opin Plant Biol. 2001;4:130–135. doi: 10.1016/s1369-5266(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 18.McCouch S, et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) DNA Res. 2002;9:257–279. doi: 10.1093/dnares/9.6.257. [DOI] [PubMed] [Google Scholar]
- 19.Ware D, et al. Gramene, a tool for grass genomics. Plant Physiol. 2002;130:1606–1613. doi: 10.1104/pp.015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konieczny A, Ausubel F. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 21.Hood E, Helmer G, Fraley R, Chilton M. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J Bacteriol. 1986;168:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 23.Katagiri S, et al. End sequencing and chromosomal in silico mapping of BAC clone derived from an indica rice cultivar, Kasalath. Breed Sci. 2004;54:273–279. [Google Scholar]
- 24.Wu J, et al. A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell. 2002;14:525–535. doi: 10.1105/tpc.010274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaki K, et al. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.