Abstract
Transcriptionally silent genes are maintained in inaccessible chromatin. Accessibility of these genes requires their modification by chromatin remodeling complexes (CRCs), which are recruited to promoters by sequence-specific DNA-binding proteins. Early B-cell factor (EBF), which is crucial for B-cell lineage specification, reprograms mb-1 (Ig-α) promoters by increasing chromatin accessibility and initiating the loss of DNA methylation. In turn, this facilitates promoter activation by Pax5. Here, we investigated the roles of ATP-dependent CRCs in these mechanisms. Fusion of EBF and Pax5 with the ligand-binding domain of ERα allowed for 4-hydroxytamoxifen-dependent, synergistic activation of mb-1 transcription in plasmacytoma cells. Knock-down of the SWI/SNF ATPases Brg1 and Brm inhibited transcriptional activation by EBF:ER and Pax5:ER. In contrast, knock-down of the Mi-2/NuRD complex subunit Mi-2β greatly enhanced chromatin accessibility and mb-1 transcription in response to the activators. The reduction of Mi-2β also propagated DNA demethylation in response to EBF:ER and Pax5:ER, resulting in fully unmethylated mb-1 promoters. In EBF- or EBF/Pax5-deficient fetal liver cells, both EBF and Pax5 were required for efficient demethylation of mb-1 promoters. Together, our data suggest that Mi-2/NuRD is important for the maintenance of hypermethylated chromatin in B cells. We conclude that SWI/SNF and Mi-2/NuRD function in opposition to enable or limit the reprogramming of genes by EBF and Pax5 during B-cell development.
Keywords: DNA methylation, mb-1 promoter, Cd19 promoter, chromatin accessibility
The development of B cells from progenitor cells in the bone marrow is controlled by a network of transcriptional regulators (reviewed in 1). Early B-cell Factor (EBF; also known as EBF1/O/E-1/COE1) plays an integral role in this network and has been implicated as a major determinant of the B-cell fate (2, 3). In the absence of EBF, B-cell development is arrested at an early progenitor stage (3, 4). EBF is essential for the rearrangement and expression of Ig (Ig) genes in B cells and is required for expression of the B-cell commitment factor Pax5. EBF and Pax5 synergistically activate transcription of B cell-specific genes including mb-1 (Cd79a), which encodes the Ig-α subunit of the pre-B and B-cell receptors (5). We have proposed that EBF functions as a ‘pioneer’ factor by controlling the epigenetic states of its target genes (6). In response to EBF, the accessibility of mb-1 promoter chromatin is increased, while DNA methylation of the promoter is decreased.
Specific biochemical interactions necessary for the pioneer functions of EBF have not been identified. However, transitions between active and inactive states of chromatin can be mediated by the recruitment of chromatin-remodeling complexes (CRCs) by transcription factors (7). CRCs serve as ‘molecular motors’ that mediate changes in the relative positions of nucleosomes. In this regard, CRCs of the mammalian SWI/SNF (related to yeast switch/sucrose non-Fermenter) subfamily are important for the functions of many tissue-specific transcription factors. SWI/SNF components include the Brahma (Brm/Smarca2) or Brahma-related gene 1 (Brg1/Smarca4) ATPases, which provide the energy to slide or evict nucleosomes. SWI/SNF CRCs are important for controlling differentiation and proliferation in many cell types. In lymphocytes, CRCs have been implicated in activating transcription and antigen receptor rearrangements (8–10).
Mi-2/nucleosome remodeling deacetylase (Mi-2/NuRD) complexes constitute a second class of heterogeneous CRCs that are primarily associated with gene silencing (11–13). Mi-2/NuRD complexes include the SNF2-related ATPase Mi-2β (Chd4) together with histone deacetylases (HDAC1 and HDAC2), histone binding proteins (RbAp46 and RbAp48), metastasis-associated (MTA) proteins 1, 2, or 3 and methyl(CpG)-binding domain 3 (MBD3) proteins (14, 15). Mi-2/NuRD induces chromatin compaction via the coupling of nucleosome sliding and histone deacetylase activities. Moreover, Mi-2/NuRD associates with methylated DNA (16). In these capacities, Mi-2/NuRD has been implicated in the control of both B- and T-cell differentiation (15, 17, 18).
We investigated roles of SWI/SNF and Mi-2/NuRD complexes in the activation of the mb-1 promoter by EBF and Pax5. Here, we show that the activation of mb-1 promoters by EBF and Pax5 is SWI/SNF-dependent. Activation of mb-1 promoters by EBF and Pax5 is restrained by Mi-2/NuRD, which limits the extent of chromatin remodeling and DNA demethylation.
Results
4-OHT-Dependent EBF and Pax5.
We produced retroviral vectors for expression of FLAG-tagged EBF (18–591) or Pax5 (1–381) as fusion proteins with the ligand-binding domain (residues 282–595) of human estrogen receptor α (ER-LBD). The mutated ER-LBD used here is activated by the synthetic estrogen 4-hydroxytamoxifen (4-OHT) (19), but the domain does not include sequences that are important for recruitment of SWI/SNF or Mi-2/NuRD by ERα (20).
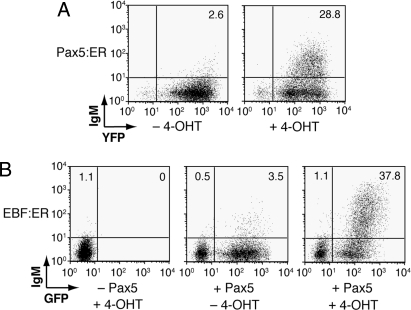
We demonstrated previously that enforced expression of Pax5 is sufficient for activation of endogenous mb-1 gene transcription in the μM.10 subclone of 558LμM plasmacytoma cells (21). On average, each of these cells possesses 1 hypomethylated mb-1 promoter that is accessible to Pax5 in the absence of EBF. Membrane-bound IgM (mIgM) is displayed on μM.10 cells in direct proportion to mb-1 gene transcripts. We expressed Pax5:ER in μM.10 cells using MSCV-Pax5:ER-YFP virus. YFP+ cells were purified using fluorescence activated cell sorting (FACS) and incubated for 48 h without or with 0.5 μM 4-OHT. In the absence of 4-OHT (Fig. 1A), mIgM+ cells were detected at a low frequency (2.6%) and with a low mean fluorescence intensity (MFI) of mIgM staining (MFI = 3.1). Both reflect low levels of mb-1 gene transcripts. In contrast, 4-OHT stimulated mIgM expression on 28.8% of the cells (MFI = 17.9), indicating a significant increase of mb-1 transcripts in response to Pax5:ER.
Fig. 1.
Tamoxifen-dependent activation of mb-1 gene expression. (A) Pax5:ER increases the display of mIgM on μM.10 cells. Cells were infected with Pax5:ER-YFP retroviruses. YFP+ cells were sorted and 0.5 μM 4-OHT was added at 48 h posttransduction. 48 h after the addition of 4-OHT, cell surface expression of mIgM (which directly reflects mb-1 expression) was analyzed by labeling cells with biotinylated anti-IgM and streptavidin-conjugated APC. mIgM was detected using a FACScalibur™ flow cytometer. (B) Synergistic activation of mb-1 transcription by EBF:ER and Pax5 in μM.2 cells. Cells were stably transfected to express EBF:ER (μM.2+EBF:ER cells), then infected with MSCV-Pax5-GFP retroviruses. Cells are shown following incubation with or without 4-OHT and mIgM staining as in A.
To generate cells expressing EBF:ER, we expressed the fusion protein stably in the 558LμM subclone μM.2 (21). Unlike μM.10 cells, μM.2 cells are refractory to activation by Pax5 because their mb-1 promoters are uniformly hypermethylated in relatively inaccessible chromatin (21, 22). Following incubation with 4-OHT (Fig. 1B), μM.2 cells expressing EBF:ER exhibited minimal mIgM expression (1.1% mIgM+ cells; MFI = 3.9). In the absence of 4-OHT, expression of Pax5 in these cells resulted in few mIgM+ cells (3.5% mIgM+ cells; MFI = 4.7). However, in the presence of both EBF:ER and Pax5, incubation with 4-OHT increased mIgM+ cells (37.8%) and the levels of mIgM that they express (MFI = 121). In other experiments, EBF or EBF:ER exhibited nearly identical abilities to increase mIgM synergistically with Pax5 (Fig. S1). Therefore, synergistic activation of mb-1 gene expression by the EBF:ER fusion protein is dependent on 4-OHT and Pax5.
Roles of CRCs in Transcriptional Activation of the mb-1 Gene.
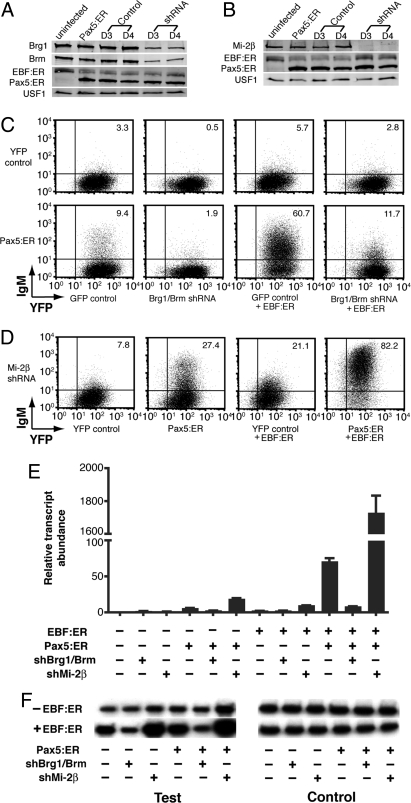
We tested whether transcriptional activation of mb-1 promoters by EBF and Pax5 requires CRCs. For these experiments we used μM.2 cells that stably expressed EBF:ER (μM.2+EBF:ER cells). The cells were infected with control YFP or Pax5:ER-YFP retroviruses. YFP+ cells were collected 48 h after infection. The purified cells were infected to express previously validated shRNAs specific for a sequence that is conserved between Brg1 and Brm transcripts, or specific for mRNA encoding Mi-2β (Chd4; referred to hereafter as Mi-2β) (23). All cells were incubated with 4-OHT for the final 48 h of culture. After expression of shRNAs for 3 or 4 days, Western analysis demonstrated that Brg1 and Brm (Fig. 2A), or Mi-2β (Fig. 2B), proteins were depleted efficiently in the presence of shRNAs. Western blotting also confirmed the expression of EBF:ER and Pax5:ER. Co-expression with Pax5:ER consistently reduced expression of EBF:ER in the presence of 4-OHT.
Fig. 2.
mb-1 gene activation in the context of EBF, Pax5 and knock-down of chromatin remodeling complexes (CRCs). (A) Western detection of Brg1, Brm, EBF:ER, and Pax5:ER in μM.2 or μM.2+EBF:ER cells. Cells were infected sequentially with Pax5:ER-YFP or YFP control retroviruses and Brg1/Brm-specific shRNA-GFP or control GFP retroviruses. GFP+YFP+ cells were purified on day 2 postinfection with GFP/shRNA-GFP viruses. 0.5 μM 4-OHT was added for the final 48 h. Western blotting was performed on the sorted cells at day 3 or 4 of shRNA expression. EBF:ER and Pax5:ER were detected using anti-ERα serum. USF1 served as a loading control. (B) Western detection of Mi-2β, EBF:ER and Pax5:ER in μM.2 or μM.2+EBF:ER cells. Other aspects of this experiment are similar to (A). (C) mIgM expression in response to EBF:ER, Pax:ER without or with Brg1/Brm shRNA. In this and all subsequent experiments, cells were labeled with Cy5-conjugated anti-IgM and detected using a CyAn™ flow cytometer. (D) mIgM expression in response to Mi-2β shRNA without or with EBF:ER, Pax:ER or both factors. (E) Quantitative RT-PCR analysis of mb-1 transcripts for the cells in (C and D). All data were obtained from sorted YFP+GFP+ cells and normalized relative to β-actin transcripts (n = 3) (mean ± SEM.). (F) Accessibility of mb-1 promoter chromatin in response to EBF:ER, Pax5:ER and shRNA. Purified YFP+GFP+ cells were incubated 48 h with 4-OHT. (C and D). Relative cleavage by Sau96I (test; see Fig. 3A for location) was detected using LM-PCR. Total cleavable DNA was measured following digestion of samples to completion (control). Exposure of control gel was one-fifth that of test gel exposure.
We next examined the induction of mIgM on cells expressing EBF:ER, Pax5:ER, and/or shRNAs in the presence of 4-OHT (Fig. 2C). Expression of Pax5:ER alone in μM.2 cells resulted in 9.4% mIgM+ (MFI = 12.6) cells (increased from 3.3% obtained with control retroviruses). In the absence of Pax5:ER, only a small fraction (5.7%; MFI = 4.8) of EBF:ER-expressing cells displayed mIgM in the presence of 4-OHT. In contrast, co-expression of EBF:ER and Pax5:ER in the presence of 4-OHT resulted in 60.7% mIgM+ cells (MFI = 76.2), which, although reduced somewhat in intensity, is similar to results obtained using unmodified EBF and Pax5 (Fig. S1). Next, we examined whether transcriptional activation in response to EBF:ER and/or Pax5:ER is dependent on SWI/SNF. The Brg1/Brm-specific shRNA reduced the percentage of mIgM+ cells induced by either factor, separately or together. In the presence of Pax5:ER, EBF:ER, or both factors together, the shRNA reduced the fraction of mIgM+ cells to 1.9% (MFI = 2.7), 2.8% (MFI = 3.8) or 11.7% (MFI = 7.6), respectively (Fig. 2C). Also, the background expression of mIgM+ was reduced by the shRNA alone (from 3.3% to 0.5%). We conclude that EBF and Pax5 both require SWI/SNF for mb-1 promoter activation.
In contrast to the inhibitory effects of knocking down Brg1 and Brm, the knock-down of Mi-2β enhanced the frequency of mIgM+ cells under all conditions (Fig. 2D). In μM.2 cells alone, the Mi-2β-specific shRNA had only a small effect on the frequency of mIgM+ cells (increased from 3.3% to 7.8%). In the presence of Mi-2β shRNA, Pax5:ER and 4-OHT increased the frequency of mIgM+ cells from 7.8% to 27.4% and the MFI from 5.2 to 38. Therefore, Mi-2β knock-down greatly enhanced induction of mIgM expression mediated by Pax5:ER. The knock-down of Mi-2β also increased the percentage of mIgM+ cells in the presence of EBF:ER (from 5.7% to 21.1%; MFI = 9.3)(Fig. 2 C and D). Strikingly, when μM.2 cells expressed EBF:ER, Pax5:ER and Mi-2β shRNA, mIgM was detected on nearly all infected cells (82.2%; MFI = 484). The MFI of these cells was much greater than that obtained in the presence of EBF:ER, Pax5:ER and normal levels of Mi-2β (MFI = 76.2). These results indicate that Mi-2/NuRD complexes attenuate the activation of mb-1 promoters by Pax5:ER, EBF:ER or by both factors together.
To assess effects of the early B cell-specific factors and CRCs on mb-1 transcription, we isolated RNA from 4-OHT-treated cell cultures and quantitated steady state levels of mb-1 mRNA transcripts using quantitative PCR (qPCR). Relative to uninfected cells (Fig. 2E), mb-1 transcript levels were unchanged in μM.2 cells in the absence of EBF:ER and Pax5:ER, regardless of the status of Brg1/Brm or Mi-2β (1.7- or 1.1-fold with knock-downs, respectively). Expression of Pax5:ER alone resulted in increased mb-1 transcripts (5.7-fold relative to control; P < 0.005) that were dependent on Brg1/Brm (reduced to 2.4-fold in the presence of Brg1/Brm shRNA). As expected from the flow cytometry data, expression of Pax5:ER together with the knock-down of Mi-2β increased transcript levels to 18.4-fold (P < 0.004). In the presence of EBF:ER alone, mb-1 transcription was stimulated weakly (2.3-fold) relative to that in control μM.2 cells. The weak activation by EBF:ER was relatively unaffected by the knock-down of Brg1/Brm (2.2-fold); however, the knock-down of Mi-2β increased activation to 9.8-fold (P < 0.0003). Synergistic effects were observed in the presence of EBF:ER and Pax5:ER, which strongly increased mb-1 transcripts (70.4-fold; P < 0.002). This was greatly reduced by the knock-down of Brg1/Brm (to only 8.0-fold; P < 0.002). Significantly, in the presence of EBF:ER and Pax5:ER, the knock-down of Mi-2β increased mb-1 transcripts 1,727-fold (P < 0.002).
Previously, we concluded that EBF increases the accessibility of mb-1 promoter DNA (22). In turn, this facilitates DNA binding and activation of the promoter by Pax5. To determine whether changes in DNA accessibility are mediated by CRCs, we measured the relative accessibility of a Sau96I restriction site in mb-1 promoters in intact nuclei (the location of the Sau96I site is indicated in Fig. 3A). Nuclei were obtained from 4-OHT-treated cells and incubated with a limiting concentration of Sau96I enzyme (Test). Signals obtained from completely cleaved DNA are shown (Control). As demonstrated in Fig. 2F, mb-1 promoters in un-manipulated μM.2 cells were relatively inaccessible to Sau96I digestion (arbitrarily assessed as 1.0). Knock-downs of Brg1/Brm or Mi-2β affected mb-1 promoter accessibility only slightly in the absence of EBF:ER or Pax5:ER (1.0- or 1.5-fold, respectively). Activation of EBF:ER increased accessibility by 2.9-fold. This activity was decreased by the knock-down of Brg1/Brm (reduced to 1.1-fold). We conclude that EBF-dependent chromatin remodeling is dependent on SWI/SNF. In contrast, the knock-down of Mi-2β increased accessibility induced by EBF:ER (to 5.9-fold). Accessibility was not affected by Pax5:ER alone (1.4-fold). In the presence of Pax5:ER, accessibility was enhanced 2.4-fold by the knock-down of Mi-2β. We conclude that Pax5, by itself, is less able to modulate accessibility than EBF. However, EBF:ER and Pax5:ER increased accessibility in concert (3.8-fold relative to control μM.2 cells). This increase was blocked by the knock-down of Brg1/Brm (reduced to 1.3-fold), but the knock-down of Mi-2β increased accessibility potently when both EBF:ER and Pax5:ER were active (10.4-fold).
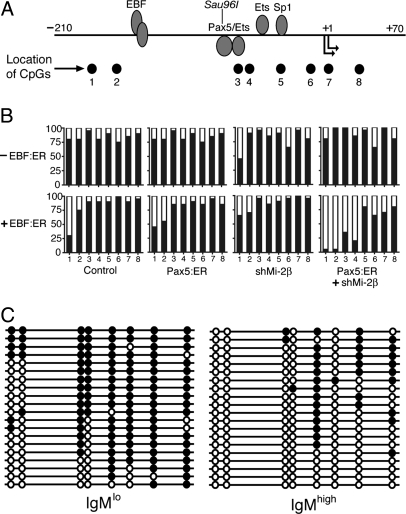
Fig. 3.
Knockdown of Mi-2β increases and propagates EBF- and Pax5-dependent DNA demethylation. (A) Positions of EBF, Pax5/Ets and proximal promoter factor binding sites are indicated relative to CpGs1–8. Binding sites of other factors present in early B cells (Runx1/CBFβ, E2A), but not plasmacytoma cells, are not shown. The location of the Sau96I site used to measure chromatin accessibility is indicated. (B) DNA methylation of mb-1 promoters is decreased synergistically by EBF:ER, Pax5:ER, and Mi-2β shRNA in μM.2 cells. All cells were cultured 48 h in the presence of 4-OHT. Genomic DNA was recovered from μM.2 cells without EBF:ER (−EBF:ER) or μM.2 +EBF:ER cells as shown and converted with sodium bisulfite before amplification and subcloning of mb-1 promoters. The frequency of methylated CpGs at each of the eight CpGs is shown for 20 sequenced clones (black bars). (C) The lack of DNA methylation correlates with high levels of mIgM. μM.2 cells expressing EBF:ER, Pax:ER, and Mi-2β shRNA were sorted to recover the highest and lowest 5% of mIgM+ cells. Bisulfite converted mb-1 promoters were amplified and twenty individual clones were sequenced as in B. Methylated (black circles) and unmethylated (open circles) CpGs are indicated in each individual sequence.
We performed chromatin immunoprecipitation (ChIP) analyses to measure changes in factor occupancy of mb-1 promoters in response to EBF:ER, Pax5:ER and Mi-2β shRNA. As expected from the increase in mb-1 transcripts, ChIP assays detected increased occupancy of mb-1 promoters in μM.2 cells by RNA polymerase II (pol II) in the presence of these inducers (Fig. S2A). We also detected the association of Brg1 with mb-1 promoters, confirming that it is recruited in the course of mb-1 promoter activation. Notably, the knock-down of Mi-2β increased detection of associated Brg1. Increased detection of pol II and Brg1 correlated with reduced levels of histone H3 at mb-1 promoters in cells expressing EBF:ER, Pax5:ER and Mi-2β shRNA (Fig. S2B). This suggests that nucleosomes are repositioned or removed during mb-1 transcriptional activation.
The question arose as to whether other genes are reprogrammed similarly in μM.2 cells by EBF, Pax5 and CRCs. Previous reports demonstrated that activation of murine and human promoters of Cd19 genes, which encode a costimulatory receptor on B cells, is dependent on EBF and Pax5 (24–26). Therefore, we measured Cd19 transcripts in μM.2 cells expressing EBF:ER, Pax5:ER, or both factors in the presence of Brg1/Brm- or Mi-2β-specific shRNAs and 4-OHT (Fig. S3). Similar to the control of mb-1 expression, EBF:ER and Pax5:ER increased endogenous Cd19 transcripts synergistically (23.5-fold; P < 0.03). The increase was ablated completely in the presence of Brg1/Brm shRNA, indicating a requirement for SWI/SNF. Inclusion of Mi-2β shRNA significantly increased Cd19 transcripts (to 52.2-fold; P < 0.07). However, the expression of EBF:ER is increased similarly in cells expressing Mi-2β shRNA. The reduced enhancement of Cd19 versus mb-1 transcripts by Mi-2β shRNA in the presence of EBF and Pax5 may reflect differences in chromatin structure and/or requirements for activation of their respective promoters in μM.2 cells.
Demethylation of mb-1 Promoters in Response to EBF and Pax5 Is Restrained by Mi-2/NuRD.
Our previous studies demonstrated that mb-1 promoter activation is blocked, in part, by DNA methylation because 5-methylcytosine (5-meC) interferes with Pax5's recruitment of Ets transcription factors to bind mb-1 promoters in μM.2 cells (21, 22). Our observation that mb-1 transcripts are increased efficiently by EBF:ER, Pax5:ER and the knock-down of Mi-2β suggested that these conditions result in demethylation of mb-1 promoters. Therefore, we measured the frequency of 5-meC at 8 positions in mb-1 promoters under various conditions in the presence of 4-OHT (Fig. 3 A and B). Bisulfite-treated mb-1 promoter antisense DNA strands were amplified, cloned, and sequenced. Activation of EBF:ER (with or without Pax5:ER) increased demethylation of CpG-1 and CpG-2. Significantly, with expression of EBF:ER, Pax5:ER and Mi-2β shRNA, CpG-1 and CpG-2 were almost completely demethylated, while demethylation of CpG-3 and CpG-4 was substantially increased. Overall, these conditions resulted in the graded loss of methylation from 5′ to 3′ ends of mb-1 promoters.
We next examined patterns of methylation of individual mb-1 promoters in cells exhibiting different levels of mIgM expression. μM.2 cells expressing EBF:ER, Pax5:ER and Mi-2β shRNA were incubated with 4-OHT and stained to detect mIgM. Using FACS, we purified cell populations that expressed maximal (top 5%) or minimal (bottom 5%) mIgM. We sequenced 20 individual bisulfite-treated clones from each of these populations (Fig. 3C). The data confirmed that mb-1 transcription is increased in proportion to the overall demethylation of mb-1 promoters. Only cells expressing high levels of mIgM exhibited extensive demethylation of the Pax5-dependent Ets binding site (overlapping with CpG-3) and other downstream CpGs. CpG-5, which is nested within a site known to bind Sp1 (27), was demethylated with the lowest efficiency. Four of 20 sequences derived from cells expressing high levels of mIgM were demethylated completely. We conclude that EBF:ER and Pax5:ER induce mb-1 promoter demethylation, but they do so inefficiently in the presence of Mi-2/NuRD.
To further investigate the mechanism of mb-1 promoter demethylation in response to EBF and Pax5, we asked whether scheduled DNA replication (i.e., S phase of the cell cycle) is required. We performed experiments similar to Fig. 2D, but in the presence or absence of the DNA cross-linker mitomycin C (MitC). After 48 h incubation with 4-OHT, the generation of mIgM+ cells was unaffected by MitC (Fig. S4A), which reduced DNA synthesis by more than 76% (Fig. S4B). Moreover, mb-1 promoter demethylation in response to EBF:ER, Pax5:ER and Mi-2β shRNA was similar in the absence or presence of MitC (Fig. S4C). Together, these data suggest that mb-1 promoter demethylation is an active process that does not require scheduled DNA replication.
Initiation of mb-1 Promoter Demethylation by EBF in ex Vivo B Cell Progenitors.
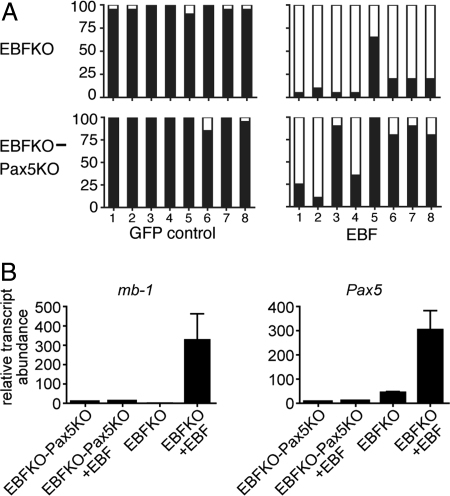
We previously demonstrated that mb-1 promoters are completely unmethylated in pro-B cells and subsequent stages of B-cell development (22), whereas, only hypermethylated mb-1 promoters were detected in B cell progenitors from EBF-deficient mice. Therefore, to study DNA demethylation in cells ex vivo we used cytokine-dependent B cell progenitors from E = 14.5 Ebf1−/− (EBFKO) or Ebf1−/−Pax5−/− (EBFKO-Pax5KO) fetal livers. We generated B220+CD43+CD117+CD28loCD19−CD24−BP-1−IgM− cells, which represent EBFKO or EBFKO-Pax5KO ‘B-biased progenitor’ cell populations. As expected for cells lacking EBF, we detected only hypermethylated mb-1 promoters in EBFKO or EBFKO-Pax5KO cells (Fig. 4A). Next, we infected EBFKO or EBFKO-Pax5KO cells with retroviruses for expression of EBF (18–429) (28). After 4 days, infected cells were purified for analysis of mb-1 promoter DNA methylation. Clones obtained from GFP+ control cells were uniformly hypermethylated. However, expression of EBF (18–429) resulted in nearly complete demethylation of mb-1 promoters in EBFKO cells (except for CpG-5). Although expression of EBF (18–429) in EBFKO-Pax5KO cells resulted in significant demethylation of CpG-1 -2, and -4, demethylation was not propagated to 3′ ends of mb-1 promoters. Notably, EBFKO-Pax5KO cells failed to demethylate CpG-3, which coincides with the Pax5-dependent Ets binding site. Analysis of mb-1 transcripts in EBFKO-Pax5KO cells demonstrated a correlation between inefficient mb-1 promoter demethylation and the absence of mb-1 transcripts (Fig. 4B). However, EBF increased chromatin accessibility in these cells by 3.5-fold (Fig. S5). In contrast, the more complete demethylation of mb-1 promoters in EBFKO cells reconstituted with EBF correlated with greatly increased (327-fold; P < 0.07) mb-1 gene transcription in these cells. EBF (18–429) also increased endogenous Pax5 transcripts (6.7-fold; P < 0.04) in EBFKO cells. Taken together, our data suggest that EBF and Pax5 drive demethylation and transcription of mb-1 promoters synergistically during early B-cell development.
Fig. 4.
Reconstitution of EBF expression initiates mb-1 promoter demethylation in EBF KO and EBFKO-Pax5KO cells. (A) DNA demethylation in response to EBF. Cells were infected with FLAG-tagged EBF (18–429) or empty GFP control virus. At day 4 postinfection, GFP positive cells were sorted for bisulfite treatment and isolation of mb-1 promoter clones for sequencing. Each bar graph represents 20 independent clones. (B) Quantitative PCR of gene expression in uninfected versus EBF (18–429)-infected (+EBF) cells at day 4 postinfection. mb-1 transcripts levels were normalized to those of β-actin. Expression of mb-1 transcripts in EBFKO cells and Pax5 transcripts in EBFKO-Pax5KO cells were each set to 1. Data were generated in triplicate for statistical analysis (mean ± SEM).
Discussion
The specification of developmental programs by transcription factors requires epigenetic changes necessary for the activation of silent genes. Our studies suggest that EBF initiates epigenetic reprogramming in the course of B-cell lineage specification. EBF is expressed at low levels in common lymphoid progenitors (CLPs) and drives their differentiation to become B cells (29). To address how EBF accomplishes this, we have sought to determine how EBF activates individual genes. Our current studies further strengthened a central role for EBF and revealed roles of Pax5. EBF simultaneously promoted chromatin remodeling, DNA demethylation, and transcription of mb-1 promoters in plasmacytoma cells. In concert with EBF, Pax5 was necessary for propagating demethylation of mb-1 promoters and for efficient expression of mb-1 genes. Experiments in B-cell progenitors derived from EBF- or EBF/Pax5-deficient fetal livers confirmed the importance of these factors for epigenetic remodeling in vivo.
We demonstrated that 2 distinct types of CRCs, SWI/SNF, and Mi-2/NuRD, function in opposition to enable or limit activation of the mb-1 promoter by EBF and Pax5. The induction of mb-1 promoter accessibility by EBF:ER is dependent on SWI/SNF, which has been shown to directly mediate accessibility in other contexts (30). Our ChIP studies suggest that SWI/SNF complexes including Brg1 regulate mb-1 promoters directly. The dependence of EBF activity on SWI/SNF has not been documented previously, but Pax5 may recruit SWI/SNF indirectly via its interactions with CBP or with Ada2β within the STAGA co-activator complex (31). It is possible that EBF recruits SWI/SNF via other proteins as well. EBF interacts with the co-activator p300/CBP, which acts as a bridge between transcription factors and SWI/SNF (32, 33). The requirement for SWI/SNF distinguishes pioneer functions of EBF from those of transcription factors that bind and disrupt nucleosomes independently of ATP-dependent chromatin remodeling enzymes, for example, FoxA1 (34).
Similar to other studies suggesting antagonistic functions of SWI/SNF and Mi-2/NuRD (23), our data indicate that Mi-2/NuRD acts as a ‘gatekeeper’ at the mb-1 promoter by maintaining a high threshold for transcriptional activation. Interestingly, EBF:ER and Pax5:ER were not sufficient to support maximal mb-1 transcription in plasmacytoma cells, but the factors potently increased transcription when Mi-2β was greatly reduced. Notably, depletion of Mi-2β allowed for increased transcription in response to Pax5:ER alone. This supports the hypothesis that Mi-2/NuRD restrains the binding of Pax5 to mb-1 promoters. This threshold is partially overcome by EBF, which promotes the repositioning of nucleosomes necessary for enhanced accessibility. We cannot rule out indirect effects of decreasing Mi-2β at other loci, which could activate mb-1 promoters in trans. However, the observed effects on chromatin accessibility of mb-1 promoters are consistent with known functions of Mi-2/NuRD. In addition to its effects on chromatin accessibility, Mi-2/NuRD impedes the propagation of DNA demethylation. This has profound effects on mb-1 expression because demethylation of CpG-3 is necessary for the assembly of Pax5/Ets ternary complexes on mb-1 promoters (21, 22). Following co-expression of EBF, Pax5 and Mi-2β shRNA, complete demethylation of mb-1 promoters was observed in cells expressing the highest levels of mb-1 transcripts.
We observed that, similar to mb-1, Cd19 expression is synergistically activated by EBF and Pax5. However, it is unclear whether Mi-2/NuRD controls Cd19 transcription. However, evidence from the Tagoh laboratory suggested that Cd19 promoters are activated by a stepwise mechanism including 1) chromatin remodeling, 2) binding of EBF, and 3) subsequent binding of Pax5 (26). Similar to mb-1 promoters, transcription of Cd19 promoters does not commence before Pax5 binding.
We used ex vivo B-cell progenitors to confirm the importance of EBF for initiating demethylation of mb-1 promoters in a more physiological context. Our experiments suggested that Pax5 is essential for this process because restoration of EBF expression in EBFKO-Pax5KO cells lacking Pax5 resulted in only partial demethylation of mb-1 promoters. Therefore, new questions arise concerning the role of Pax5 in this mechanism. Pax5 promotes the propagation of DNA demethylation, including demethylation of CpGs flanking its binding site (CpG-3). Pax5 may accomplish this by recruiting STAGA, which has been linked with nucleotide excision repair in other contexts (35). Binding of these complexes may assist with exclusion of Mi-2/NuRD, thus favoring transcriptional activation over the repressed state of the mb-1 promoter.
Similar mechanisms involving the exclusion or removal of Mi-2/NuRD may be important in other contexts of gene activation during the differentiation of B cells and other cell types. Details of this mechanism have yet to be revealed. Is the repressed state imposed by Mi-2/NuRD largely a function of nucleosome compaction, negatively acting histone modifications, or both? Does Mi-2/NuRD repress transcription in B-cell progenitors before the induction of EBF expression, and if so, how is Mi-2/NuRD recruited to promoters? Another question concerns whether the homolog of Mi-2β, Mi-2α, has similar functions in lymphocytes. Evidence has confirmed that components of Mi-2/NuRD complexes change during the course of B-cell differentiation (15). Finally, a central question is how DNA demethylation is achieved in response to EBF and Pax5. Recent studies in zebrafish suggest that DNA deaminase and glycosylase enzymes may be recruited by transcription factors, resulting in the modification and replacement of 5-meC as part of a mechanism related to DNA mismatch repair (36). Future studies will identify enzymes involved in the demethylation process and address their recruitment by EBF and Pax5.
Materials and Methods
Plasmids.
Sequences encoding the Pax5:ER fusion protein were generated by linking residues 1–381 of human Pax5 (provided by Dr. Peter Gruss, Max Planck-Institute, Göttingen, Germany) and residues 282–595 of the 4-OHT-responsive ER-LBD (provided by D. Metzger, IGBMC Recherche, Strasbourg, France) with the linker GALTGALTGAI. The fusion protein sequences were inserted into XhoI sites of MSCV-IRES-GFP or MSCV-IRES-YFP vectors (provided by P. Marrack, National Jewish Health, Denver, CO). FLAG-tagged EBF:ER was made by linking FLAG-EBF (18–591) and the ER-LBD with DAGALTGALTEAI. The fusion protein sequences were inserted into the PmeI site of the Igλ-based expression vector VLPEλ2.13 (37) to make Vλ-FLAG-EBF:ER-Eλ. Retroviral plasmids for expression of Brg1/Brm- or Mi-2β-specific shRNAs were provided by S. T. Smale (UCLA, Los Angeles, CA) (23). The MSCV-FLAG-EBF (18–429)-GFP and MSCV-Pax5-GFP plasmids were reported previously (28). All plasmids were sequenced.
Cell lines, Transfection, Retroviral Infection, and Flow Cytometry.
μM.2 and μM.10 cells were cultured as described previously (21). To create the μM.2 cell line expressing FLAG-EBF:ER, μM.2 cells were electroporated with 3 μg of pPUR (Clontech) and 20 μg of Vλ-FLAG-EBF:ER-Eλ and cultured as previously reported (22). Generation of retroviruses and infection of cells were described previously (21, 22). mIgM was detected using biotinylated anti-IgM (Caltag Laboratories) and streptavidin-conjugated allophycocyanin (APC) and detected using a FACScalibur™ flow cytometer (BD). Alternatively, mIgM was labeled with Cy5-conjugated anti-IgM (Jackson Research Laboratories, West Grove, PA) and detected using a CyAn™ flow cytometer (Dako). ER fusion proteins were induced using 0.5 μM 4-OHT (Sigma-Aldrich). Cell sorting was performed as described (21, 22).
Western Blotting.
Whole cell protein extraction and western blotting were performed as described (32, 33). Rabbit anti-Mi-2β antibody was kindly provided by S. T. Smale (Univ. Calif., Los Angeles, CA). Rabbit ERα- and USF1-specific antibodies were purchased from Santa Cruz Biotechnology. Rabbit anti-Brg1 and mouse anti-Brm were purchased from Upstate USA, Inc. and BD BioSciences, respectively.
Analysis of mRNA Transcripts, DNA Methylation, and Chromatin Accessibility.
Isolation of total RNA and qPCR were reported previously (28). Data were analyzed for significance using Student's 1-tailed t test. Sodium bisulfite treatment, PCR amplification, subcloning, and sequencing of mb-1 promoter antisense strands were performed using 2 × 104 cells as previously described (21). Chromatin accessibility assays were performed as described (22), except that 10 U of Sau96I was used in all test digestion reactions.
Derivation and Maintenance of Fetal Liver B-Cell Progenitors.
Mice were bred and maintained in the Biological Resource Center at National Jewish Health (NJH). All studies were approved by the NJH Institutional Animal Care and Use Committee. Ebf1+/− mice were made by R. Grosschedl (Max Planck-Institute of Immunobiology, Freiburg, Germany) and provided by Y. Zhuang (Duke University, Durham, NC). Pax5+/− mice were made and provided by M. Busslinger (Research Institute of Molecular Pathology, Vienna, Austria). Lineage marker-negative (Lin−) cells were isolated from E = 14.5 Ebf1−/− and Ebf1−/−Pax5−/− fetal livers (4, 38) by negative MACS separation (Miltenyi Biotec) as described (39). Lin− cells were expanded and maintained in the absence of stromal cells in Iscove's modified Dulbecco's medium containing 10% FBS (HyClone), 50 μg/mL gentamicin, 1× GlutaMAX-1 (Invitrogen) and recombinant (r) IL-7 (3.3 ng/mL), rSCF (3.3 ng/mL), and rFLT3 ligand (3.3 ng/mL) (R&D Systems).
Supplementary Material
Acknowledgments.
We thank C. Murre and S. T. Smale for helpful discussions; R. Grosschedl, Y. Zhuang and M. Busslinger for providing mice; and K. Tuttle for excellent technical assistance. This work was supported by National Institutes of Health Grants R01 AI54661, R01 AI56322, and P01 AI22295 (to J.H.) and National Institute of Allergy and Infectious Diseases Training Grant T32-AI007405 (to S.F. and J. R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809485106/DCSupplemental.
References
- 1.Nutt SL, Kee BL. The transcriptional regulation of B-cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Seet CS, Brumbaugh RL, Kee BL. Early B cell factor promotes B lymphopoiesis with reduced interleukin-7 responsiveness in the absence of E2A. J Exp Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medina KL, et al. Defining a regulatory network for specification of the B-cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 5.Sigvardsson M, et al. Early B-cell Factor, E2A, and Pax-5 cooperate to activate the early B cell-specific mb-1 promoter. Mol Cell Biol. 2002;22:8539–8551. doi: 10.1128/MCB.22.24.8539-8551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagman J, Lukin K. Early B-cell Factor ‘pioneers’ the way to B-cell development. Trends Immunol. 2005;26:455–461. doi: 10.1016/j.it.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Fry CJ, Peterson CL. Chromatin remodeling enzymes: Who's on first? Curr Biol. 2001;11:R185–R187. doi: 10.1016/s0960-9822(01)00090-2. [DOI] [PubMed] [Google Scholar]
- 8.Chi TH, et al. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- 9.Gebuhr TC, et al. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T-cell development. J Exp Med. 2003;198:1937–1949. doi: 10.1084/jem.20030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osipovich O, et al. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nat Immunol. 2007;8:809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- 11.Tong JK, et al. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 12.Xue Y, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 14.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- 15.Fujita N, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams CJ, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T-cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Naito T, Gómez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity. 2007;27:723–734. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belandia B, Orford RL, Hurst HC, Parker M. G. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 2002;21:4094–4103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maier H, Colbert J, Fitzsimmons D, Clark DR, Hagman J. Activation of the early B cell-specific mb-1 (Ig-α) gene by Pax-5 is dependent on an unmethylated Ets binding site. Mol Cell Biol. 2003;23:1946–1960. doi: 10.1128/MCB.23.6.1946-1960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maier H, et al. Early B-cell Factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- 23.Ramirez-Carrozzi VR, et al. Selective and antagonistic properties of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozmik Z, Wang S, Dorfler P, Adams B, Busslinger M. The promoter of the CD19 gene is a target for the B cell-specific transcription factor BSAP. Mol Cell Biol. 1992;12:2662–2672. doi: 10.1128/mcb.12.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gisler R, Åkerblad P, Sigvardsson M. A human early B-cell factor-like protein participates in the regulation of the human CD19 promoter. Mol Immunol. 1999;36:1067–1077. doi: 10.1016/s0161-5890(99)00092-9. [DOI] [PubMed] [Google Scholar]
- 26.Walter K, Bonifer C, Tagoh H. Stem cell specific epigenetic priming and B cell-specific transcriptional activation at the mouse Cd19 locus. Blood. 2008;112:1673–1682. doi: 10.1182/blood-2008-02-142786. [DOI] [PubMed] [Google Scholar]
- 27.Travis A, Hagman J, Grosschedl R. Heterogeneously initiated transcription from the pre-B- and B-cell-specific mb-1 promoter: Analysis of the requirement for upstream factor-binding sites and initiation site sequences. Mol Cell Biol. 1991;11:5756–5755. doi: 10.1128/mcb.11.11.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields S, et al. The ‘zinc knuckle’ motif of Early B-cell Factor is required for transcriptional activation of B cell-specific genes. Mol Immunol. 2008;45:3786–3796. doi: 10.1016/j.molimm.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zandi S, et al. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol. 2008;181:3364–3372. doi: 10.4049/jimmunol.181.5.3364. [DOI] [PubMed] [Google Scholar]
- 30.Côté J, Peterson CL, Workman JL. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci USA. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barlev NA, et al. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol Cell Biol. 2003;23:6944–6957. doi: 10.1128/MCB.23.19.6944-6957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao F, McCarrick-Walmsley R, Åkerblad P, Sigvardsson M, Kadesch T. Inhibition of p300/CBP by early B-cell factor. Mol Cell Biol. 2003;23:3837–3846. doi: 10.1128/MCB.23.11.3837-3846.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agalioti T, et al. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 34.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 35.Martinez E, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rai T, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner DH, Jr, et al. Thymocytes are rescued from glucocorticoid-mediated cell death by CD28/CTLA-4 costimulatory interactions with B7–1/B7–2. J Exp Med. 1996;184:1631–1638. doi: 10.1084/jem.184.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urbánek P, Wang Z-Q, Fetka I, Wagner EF, Busslinger M. Complete block of early B-cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 39.Ikawa T, Kawamoto H, Wright LY, Murre C. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity. 2004;20:349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.