Abstract
In general, the strict preservation of broad-scale structure is thought to be critical for maintaining the precisely tuned functionality of vertebrate genomes, although nearly all vertebrate species undergo a small number of programmed local rearrangements during development (e.g., remodeling of adaptive immune receptor loci). However, a limited number of metazoan species undergo much more extensive reorganizations as a normal feature of their development. Here, we show that the sea lamprey (Petromyzon marinus), a jawless vertebrate, undergoes a dramatic remodeling of its genome, resulting in the elimination of hundreds of millions of base pairs (and at least one transcribed locus) from many somatic cell lineages during embryonic development. These studies reveal the highly dynamic nature of the lamprey genome and provide the first example of broad-scale programmed rearrangement of a definitively vertebrate genome. Understanding the mechanisms by which this vertebrate species regulates such extensive remodeling of its genome will provide invaluable insight into factors that can promote stability and change in vertebrate genomes.
Keywords: development, lamprey, rearrangement, chordate, Petromyzon
A few species are known to undergo extensive genomic rearrangements during the specification of different cell lineages [e.g., sciarid flies (1, 2), some copepods (3, 4), and some roundworms (5)] or nuclear lineages [e.g., ciliates (6, 7)] (8). These rearrangements result in the selective removal of repetitive sequences (9–11), entire chromosomes (5), or single-copy genes (12, 13). Notably, only one chordate group (hagfish) is thought to undergo similar large-scale rearrangements and possesses certain repetitive elements in its germline that undergo diminution in somatic tissues, sometimes exhibiting a reduction in chromosome number (9–11). The unique genome biologies of these organisms are fascinating because changes that are tightly regulated in these exceptional genomes are reminiscent of the dysregulated structural changes that give rise to cancers or other genomic disorders (14, 15). In this context, developmentally regulated rearrangements hold great potential for studying the factors that promote genome stability and change in normally developing somatic cells and the means by which alterations in genome structure contribute to the differentiation of cell lineages (8).
Here, we report new data that reveal extensive programmed genome rearrangement in the sea lamprey, an important model system for studying basal vertebrate biology. Several complementary lines of evidence indicate that hundreds of megabases of DNA are specifically removed from somatic cell lineages. Here, we demonstrate the existence of a specific DNA sequence (called Germ1) that is highly abundant in germline but substantially rarer in many, if not all, somatic tissues. Fluorescence in situ hybridization reveals that Germ1 is present at a high copy number on several chromosomes in meiotic germline but dramatically reduced in soma. Estimates of relative copy number indicate that the loss of Germ1 is tightly regulated and occurs during early embryonic development at the blastula/gastrula transition. At a broader scale, we observe major differences in DNA content (> 20% of the genome, or 0.5 billion base pairs) between somatic and germline cell lineages using flow cytometry. Experimentally validated computational screens for other germline-limited sequences uncovered several additional sequences that are lost during developmental restructuring of the lamprey genome, including one sequence that is homologous to SPOPL (speckle-type POZ protein-like) genes, which is transcribed in lamprey testes and lost during embryonic development. The presence of predictable and extensive reorganization events within the genome of the lamprey, coupled with its high fecundity (16) and amenable embryology (17–20), present a uniquely tractable system for understanding the dynamics of genome stability and the consequences of reorganization in the context of “normal” vertebrate development and cell biology.
Results
Global Changes in DNA Content.
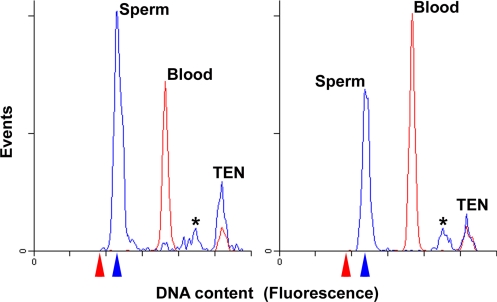
To assay for the possibility of genome rearrangement at the genome-wide scale, we estimated the total nuclear DNA content of postmeiotic testes (predominantly germline) and blood (somatic) nuclei via flow cytometry. Genome size estimates for sperm and blood nuclei were highly reproducible across individuals and preparations, but the estimated DNA content of sperm nuclei (1C = 2.31 pg, 95% C.I. ± 0.07 pg, n = 4) was substantially larger than those of erythrocyte (1C = 1.82 pg, 95% C.I. ± 0.02 pg, n = 4), kidney (1C = 1.87 pg, 95% C.I. ± 0.03 pg, n = 4), and liver (1C = 1.96 pg, 95% C.I. ± 0.06 pg, n = 4) nuclei from the same individuals. Moreover, comparison of flow histograms from germline and soma reveal that these nuclei form highly discrete distributions [Fig. 1; note that somatic cells appear to contribute a very small number of nuclei to testes preparations (Left, see also Fig. S1)]. Our estimates of nuclear DNA content indicate a ≈0.5 gigabase (>20%) difference in genome size between germline (sperm) and soma (1C value for blood).
Fig. 1.
Germline nuclei contain more DNA than somatic nuclei. Flow-cytometric analyses of nuclear DNA content reveal major differences in nuclear DNA content between germline (spermatids) and somatic tissues. Sperm and blood nuclei were isolated from the same four individuals. This figure shows sperm and blood traces from two different animals. Blue and red arrows mark the relative sizes of 1C sperm (2.31 pg) and 2C blood (1C = 1.82 pg) genomes, relative to arbitrary fluorescence units. Nuclei from each tissue and individual were measured in separate runs along with trout erythrocyte nuclei (TEN) as internal standards. An asterisk marks a peak corresponding to 2C spermatogonia or sperm doublets. (Left) The small blue peak under the (red) blood peak likely indicates a minor amount of somatic cells in the testes sperm preparation (see also Fig. S1). (Right) Somatic cells are less apparent in the sperm trace.
Screening for Germline-Specific Signatures.
To assay for structural differences between germline and somatic genomes, we hybridized restricted DNAs from germline and somatic tissues (sperm and blood, respectively) using an abundant sequence element (rpt200) that was detected in lamprey whole genome shotgun reads (21). This element was selected as a marker of “random” genome segments, because it was distributed across numerous genomic contigs and was highly abundant in the whole genome shotgun (WGS) dataset. These Southern hybridization assays revealed several bands that differed in size or intensity between germline and somatic DNAs (Fig. S2), viz., they represent a clear signature of genome rearrangement. One of these fragments (Germ1) appeared to be greatly enriched in germline DNA and was specifically targeted for further study. Replication of this same Southern hybridization assay using tissue-specific genomic DNAs isolated from additional individuals revealed the presence of Germ1 in testes genomic DNAs (largely germline) and the substantial reduction of the fragment from five somatic tissues (blood, liver, kidney, tail fin, and muscle) of diverse embryonic origin (Fig. 2). Thus, the Germ1 fragment appears to be uniquely and reproducibly enriched in the germline, relative to several somatic tissues.
Fig. 2.
Germ1. (A) Sequence analysis: Germ1 was isolated by screening a size-selected (9–10 kb) HindIII library (from sperm DNA) for presence of the rpt200 probe. Sequencing of this fragment revealed that this particular germline-enriched element is internally composed of several different repetitive elements. Analysis of somatic depth of coverage reveals that much of the sequence is abundantly represented by somatic whole genome shotgun reads, similar to the functional rDNAs. Notably though, the 5′ region (not homologous to 28S RNA) is relatively rarer. Labels show the position of sequence elements and molecular probes: FISH, the FISH probe corresponding to the somatically rare region (see Fig. S3); RT, the real-time probe for Germ1 (note that this probe overlaps the boundary between somatically-rare and 28S rDNA-like regions); RT (control), the real-time probe for 28S rDNA (note that the forward primer matches only rDNA, and the reverse primer matched both sequences) (see Fig. 4). (B) Structural differences between germline and soma are widespread and reproducible. To test whether fragments that were identified as sperm specific in our initial screen are truly unique to germline, rpt200 was used to probe Southern blots of respective genomic DNAs that were isolated from testes and several somatic tissues of two animals. Fragment sizes were estimated on the basis of electrophoretic migration of molecular mass markers (Low Range PFG Marker; New England Biolabs) that were run in adjacent lanes. T, testes; B, blood; L, liver; K, kidney; F, fin; M, muscle.
Nucleotide Sequence of Germ1.
To characterize the nucleotide sequence of Germ1, we first isolated clones containing Germ1 from a size-selected (9–10 kb) library of HindIII-digested sperm DNA, then sequenced the fragment using a combination of random and targeted sequencing. Alignments against the National Center for Biotechnology Information nonredundant databases reveal several identifiable features within Germ1 (Fig. 2). Germ1 contains perfect matches to 18S rDNA of Petromyzon marinus (GI: 174596) and the terminal 1,136 bases of 28S rDNA (GI: 3603300); however, it contains no matches to the first 2,989 bases of the 28S rDNA subunit or any of the known transcribed spacers. A third region shows strong alignment to ReO-6 from medaka (blastx; E-value = 9e-166; GI: 18157526) and several related non-LTR (long terminal repeat) retrotransposable elements from teleost fishes. Notably spermatocytes are typically translationally silent and do not actively synthesize ribosomes; therefore that these additional sequences could reflect specific amplification of the ribosomal gene copies in sperm seems unlikely (22). Fluorescence in situ hybridization experiments (below) also argue strongly against this possibility.
Alignments to the ≈6× WGS sequence for lamprey liver DNA (21) revealed that sequences with similarity to a majority of the Germ1 fragment are very common in the somatic genome (Fig. 2). However, one region is dramatically underrepresented, relative to the majority of the fragment, and includes one of the two HindIII restriction sites that are necessary to generate the Germ1 fragment. Notably, the change in coverage depth occurs precisely at the alignment breakpoint between Germ1 and rDNA. It is interesting to speculate that this somatically underrepresented region might participate in the differential retention of Germ1-like elements relative to canonical rDNA genes.
Germ1 in Germline and Soma.
To further test our conjecture that Germ1-like sequences are localized to germline chromosomes and lost from somatic tissues, the entire Germ1 clone and a subregion corresponding to the somatically rare region (Fig. 2) were hybridized individually to germline (testes) and somatic (gill epithelium) metaphases (Fig. 3, Fig. S3, and Fig. S4) that were isolated from the same individual. These hybridizations revealed that Germ1-like fragments are abundant in the lamprey germline at meiosis I and are distributed across several chromosomes, with numerous copies apparently arrayed in dense tandem repeats. In stark contrast to the situation in the germline, FISH on somatic (gill) metaphases identified Germ1-like fragments on only a single chromosome pair. Fluorescence in situ hybridization on mature spermatozoa and interphase nuclei from immature (mitotic) testes revealed patterns consistent with meiotic metaphase hybridizations (Fig. S5). Likewise, FISH on interphase gill nuclei gave patterns that were consistent with metaphase spreads from the same tissue (Fig. S5). Together these studies represent nearly incontrovertible evidence that this family of sequences experiences substantial rearrangement/reduction in somatic lineages.
Fig. 3.
The distribution of Germ1-like sequences differs strikingly between germline and somatic genomes. Germ 1 strongly hybridizes to eight germline (testes) chromosomes (meiotic metaphase I), whereas hybridization to somatic (gill) chromosomes (mitotic metaphase) from the same animal is limited to a single chromosome pair. These hybridizations show clear changes in genome structure and Germ1 copy number.
Tracking Changes in Germ1 during Development.
The fact that several tissues of diverse embryonic origin possess fewer copies of Germ1-like sequences is taken as evidence that reductions might occur early in development, perhaps during the initial establishment of somatic lineages from their germline progenitors. To gain insight into the developmental context of these changes, real-time PCR was used to measure the relative abundance of Germ1 and canonical 28S rDNA over the first 2 weeks of embryonic development (Fig. 4). These measurements revealed a dramatic change in the relative copy number of Germ1 between 2 and 3 days postfertilization (Fig. 4). This pattern of copy number reduction during early embryonic development is consistent with the observed depletion of Germ1 from a majority of somatic tissues. Presumably, the two strongly hybridizing somatic chromosomes (Fig. 3 Right and Fig. S3) carry the functional rDNAs, which show strong sequence homology with portions of Germ1, although possibly Germ1 and canonical rDNAs are interspersed across all clusters and that they are randomly retained on one chromosome pair. Taken with the above studies, real-time assays indicate that Germ1 undergoes loss during development (i.e., it is not amplified in the germline) and that a majority of this loss takes place during a brief window at approximately the transition from blastula to gastrula.
Fig. 4.
Variation in abundance of Germ1 over embryonic development. The abundance of Germ1 was measured relative to 28S rDNA. Data are plotted relative to the log ratio of these two fragments in sperm DNA. These measurements indicate that loss of the Germ1 fragment occurs between 2 and 3 days postfertilization, at approximately the transition from blastula to gastrula. Real-time probes were designed to specifically detect Germ1 or canonical rDNAs; their locations are shown in Fig. 2. A timeline of major embryological events (16) is superimposed on this plot. Corresponding Tahara (34) stages are (formatted as day:stage): D2:11, D3:14, D4:18, D6:21, D8:22, D10:23, D12:24, D14:25.
Other Losses.
We reasoned that identification of other low-copy elements that are lost from the lamprey genome might shed additional insight into the biological function of developmentally regulated rearrangement in lamprey and perhaps ultimately the mechanism by which these rearrangements occur. Densitometry measurements of FISH-labeled chromosomes indicate that ≈7% of the genome is associated with Germ1/rDNA-like sequence, less than the 20% reduction detected in our cytometry experiments. In an attempt to identify other sequences that are specific to lamprey germline, we performed alignments between a small set (n = 3,072) of paired BAC end sequences from sperm DNA and a large dataset (n = 18,506,949) of WGS sequences that were generated from the liver DNA of a different animal (21). Polymerase chain reaction primers were developed for 7 BACs that yielded long reads (>400 bp) for both read ends and further yielded no strong alignment to the WGS database. Five of these primer pairs yielded reproducible and correctly sized amplicons in the germline (sperm or testes) but produced no amplicon in somatic tissues (blood, liver, kidney, muscle, or tail fin) (Fig. S6). One of the two nonvalidating fragments is related (≈90% identity) to a repeat class that is abundant in both germline and soma, and thus a large number of PCR fragments was generated in both lineages. The other primer pair yielded a single strong fragment of expected size in both germline and soma, presumably reflecting a coverage gap in somatic WGS sequence. Interestingly, one of the eliminated sequences (corresponding to clone PMAY-409L01) shows similarity (blastx, bitscore = 88.6, E = 2e−16, 83% identity) to SPOPL genes from human and several other metazoans. This germline-specific gene is expressed in the testes of juvenile and adult lampreys and is lost during embryogenesis, although more gradually than Germ1 (Fig. 5). Results from our screen of germline BAC ends therefore indicate that many other low-copy sequences and genes are altered in a similar fashion to Germ1.
Fig. 5.
A lamprey SPOPL homolog is present and expressed in the germline but lost from soma during embryonic development. (A) Polymerase chain reactions were performed using primers that were targeted to a lamprey SPOPL homolog and an internal control (IC). Templates were genomic DNA (gDNA) from sperm (S) and blood (B), cDNA from adult (A) and juvenile (J) lamprey, RNAs from these same samples, and diH2O (X). Amplifications of genomic DNAs show that the gene is present in sperm but substantially reduced in blood. Amplifications of cDNAs reveal expression of the gene in juvenile and adult testes. Failure to amplify fragments from source RNAs rules out the possibility that contaminating genomic DNAs contributed to the amplification of SPOPL fragments from cDNAs. Amplified fragments are flanked by size standards (M, 1 kb Plus DNA Ladder; Invitrogen). (B) Variation in abundance of SPOPL over embryonic development. The abundance of SPOPL was measured relative to a single-copy gene HMG. Data are plotted relative to the log ratio of these two fragments in sperm DNA. The value for blood is plotted but is not distinguishable from zero. These measurements indicate that loss of SPOPL occurs during early development and is already apparent by day 2 postfertilization. A timeline of major embryological events (16) is superimposed on this plot. See Fig. 4 for corresponding Tahara (34) stages.
Discussion
The above studies reveal the dynamic nature of the lamprey genome. Somatic cells necessarily arise from the germline but contain much less DNA and lack defined sequence elements that are present in the germline. These results reveal a major difference in the genome biology of jawed and jawless vertebrates, although jawed vertebrates have not been extensively surveyed. Notably, if the ostensibly similar developmentally programmed rearrangements in hagfish and lamprey are found to share a common evolutionary origin, then this dynamic genome biology can be traced to a point very near (perhaps including) the ancestor of all extant vertebrates.
On the basis of the available data, lampreys appear to undergo reproducible genomic alterations that, in magnitude, are not typically tolerated by vertebrate genomes. Despite this striking difference in genome biology, decades of research have demonstrated that many of the basal features of embryonic development, cell biology, and gene content are largely conserved between these two major vertebrate lineages. On the basis of the evidence at hand, we surmise that programmed genome reorganization results in biologically important changes in the lamprey's genome architecture. Moreover, we expect that some of the molecular players that participate in reorganization of the lamprey genome will have homologs in “higher” vertebrates that mediate (albeit in a different manner) genome rearrangements.
The observation of widespread and regulated changes in the lamprey genome is taken as evidence that these changes play an important role in the lamprey's biology. In this regard, it is interesting to note some changes take place quite early in development and result in the loss of transcripts (SPOPL homolog) or other potentially functional (rDNA-like) sequences. One possible explanation for the patterns of loss during early development is that it permits the maintenance of genes in the germline that are deleterious, or dispensable, in the context of most somatic tissues. That SPOP family members are associated with nuclear phenotypes, apoptosis, and regulatory modifications of DNA is also worth noting (23). Although speculation that the eliminated SPOPL homolog plays a role in programmed rearrangement is premature, the gene is certainly an obvious candidate for future studies.
As yet, essentially nothing is known regarding the cellular mechanisms that result in the programmed loss of DNA during embryonic development. However, it is interesting to speculate that the developmental restructuring of the lamprey genome might be related to highly reproducible artifacts that arise when the TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP–biotin nick end-labeling) assay is used to detect apoptosis-associated DNA breakage. Nearly every nucleus shows strong TUNEL labeling during the first few weeks of embryonic development (Fig. S7 and Fig. S8). Logically, programmed death of every cell is not consistent with embryonic development or survival. Moreover, alternate assays for apoptosis do not indicate widespread cell death (19) (Fig. S7 and Fig. S8). This raises the possibility that abundant DNA breaks observed during embryogenesis are functionally related to developmentally regulated deletions. That DNA breaks are visible well after the majority of Germ1 sequences are lost suggests that other changes may be regulated to occur later in development or that the breakdown products of earlier excision events persist into later embryonic stages. One could envisage that excision events also may result in the generation of unique sequences in somatic cell lineages. If excisions are interstitial, then resolution of broken DNA ends will result in the joining of sequences that were once separated by some distance. Thus, loss of DNA could result in the generation of coding, promoter, enhancer, etc. sequences that impart unique functionality to somatic cell lineages. Ongoing work to characterize the somatic derivatives of germline alterations and dissect the precise changes that lead to DNA loss are necessary to generate more realistic hypotheses as to the biological mechanism and function of developmentally regulated DNA loss in the lamprey.
All somatic tissues surveyed show consistent patterns of loss for Germ1 and five other germline-specific sequences. However, some tissue-specific variation is apparent among flow-cytometry estimates of genome size. Although these tissue-specific differences are smaller than the germline-soma difference, they do suggest the possibility of variation in genome content among somatic tissues, possibly resulting from later rearrangement events (or alternatively, tissue-specific differences in the relationship between DNA content and propidium iodide fluorescence). Notably none of these tissues represents a single pure lineage, and the presence of different cell lineages in liver (e.g., erythrocytes, hepatocytes, and connective tissues), with structurally different genomes, might contribute to the current difficulties that have been experienced in assembling long contigs from the existing lamprey WGS dataset. We are currently pursuing methods that can more precisely detect and characterize potential differences among somatic lineages.
Because of its deep evolutionary relationship with other vertebrate groups, the lamprey provides critical insight into the biology of the vertebrate ancestor. The results presented here suggest that considering the germline genome structure and how somatic changes alter this structure when using lamprey as a model for comparative developmental and genomic studies will be especially important. With ongoing progress toward complete sequencing of the lamprey somatic genome (21) and the recent development of several techniques for performing genetic manipulations of lamprey embryos (17–20), we anticipate that the lamprey genome will prove fertile ground for identifying mechanisms that mediate the differential development of cell lineages and participate in large-scale rearrangement and stabilization of vertebrate genomes.
Materials and Methods
Animals.
All animals were obtained from the Lake Michigan population, via the Great Lakes Fisheries Commission under Washington State permit number 4865–5-08. Within 12 h of receipt, animals were euthanized by immersion in MS-222 (150 μg/mL) and dissected, and tissues immediately were fixed or snap-frozen for subsequent experiments.
Flow Cytometry.
Nuclei for flow cytometry were isolated from snap-frozen tissues by homogenizing ≈0.5 g of tissue in 10 mL of homogenization buffer (20 mM Hepes, 10 mM EDTA, 1 mM spermidine, 0.5% Nonidet P-40 detergent, pH 7.6). This homogenate was adjusted to 70% ethanol, and nuclei were fixed for 30 min on ice. These suspensions were then centrifuged at 500 × g for 15 min, after which fixed nuclei were resuspended in PBS with 50 mM EDTA. Before flow-cytometric analysis, fixed nuclei were pelleted, resuspended in propidium iodide staining buffer [Solution A (24)], and incubated on ice for 15 min. Replicate flow-cytometric measurements were obtained for four individuals and four tissues (sperm, blood, kidney, and liver) using a FACSort flow cytometer (Becton Dickinson). Estimates of nuclear DNA content were based on comparison to an internal standard (trout erythrocyte nuclei) and were averaged across four captures of 1,000 events, which were temporally interleaved across individuals and tissues.
Southern Blots.
Genomic Southern blots were probed with an abundant endogenous repetitive element that was identified within the lamprey WGS dataset. Genomic Southern blots were prepared by incubating high-molecular-mass DNAs with restriction enzyme (HindIII or XhoI; Invitrogen) overnight at 37 °C. Blood and sperm samples that were used in the initial identification of Germ1 were prepared from agarose-embedded cells (25). Genomic DNAs from testes, blood, and other tissues were isolated using standard phenol/chloroform extraction (26), and 5 μg was digested at a total volume of 50 μL using manufacturer-specified buffers and reaction conditions. Restriction-digested DNAs were separated by electrophoresis on a CHEF DRII (Bio-Rad) pulsed-field apparatus (0.5–3s switch time, 6 V/cm, 120° angle, for 16 h), then transferred to a positively charged nylon membrane (Hybond-N+; Amersham Biosciences).
The Southern blots were hybridized with the element rpt200, using standard methods (26). Oligonucleotide primers were designed for rpt200 (rpt200.f, GAAATGCATGTGCACTCAAAA; rpt200.r, ATGGGGTTGAATGCTTTTTG). This element was amplified from blood genomic DNA using standard PCR conditions [0.5 ng of DNA, 50 ng of each primer, 1.2 mM MgCl2, 0.3 U Taq polymerase, 1× PCR buffer (Promega), and 200 mM each of dATP, dCTP, dGTP, and dTTP; thermal cycling at 94 °C for 4 min; 25 cycles of 94 °C for 45 s, 60 °C for 45 s, 72 °C for 30 s; and 72 °C for 7 min]. The amplified fragment was purified via filtration (YM50; Microcon), and the sequence was verified by direct sequencing and radiolabeled using [α-32P]dCTP incorporation (NEBlot Kit, N1500; New England Biolabs).
Isolation and Sequencing of Germ1.
Sperm genomic DNA was restriction-digested with HindIII and size-separated as described above for Southern hybridizations. The portion of the gel corresponding to ≈8–16 kb was excised, and DNA was isolated via electroelution in 1/2× TBE (4 °C, 100 V, 4 h). Electroeluted DNAs were dialyzed against 1/2× TE for 12 h and then ligated into pCC1BAC vector (Epicentre). Ligation products were electroporated into E. coli (Top10; Invitrogen) and grown overnight on selective media (LB media with 12.5 μg/mL chloramphenicol) supplemented with 1.5% bacto agar. Individual colonies were robotically picked into eight 384-well plates and grown overnight in selective media supplemented with 10% glycerol. These cultures were spotted onto nylon, and the remaining culture was maintained as frozen stock for later use.
Nylon membranes were probed with radiolabeled rpt200 as described above to identify clones of the correct size that also contained the probe sequence that was originally used in their identification. In total, some 150 positive clones were identified by hybridization. Four of these clones were end-sequenced to test for variation among clones (all were identical with the exception that one possessed an additional 185 bp, resulting from restriction digestion at an adjacent site 5′ of Fig. 2), and one clone was sequenced via several rounds of primer walking and dye-terminator sequencing (primer sequences are given in Table S1).
Sequence Alignment to the WGS Dataset and Analysis of Germ1 Coverage Depth.
We aligned Germ1 and a small collection of sperm BAC-end sequences to the WGS sequence for lamprey. Traces for sperm BAC-end and WGS reads were downloaded from National Center for Biotechnology Information Trace Archives (www.ncbi.nlm.nih.gov/Traces/trace.cgi?), quality trimmed to Q20 using phred (27, 28), vector trimmed using phrap (29), and aligned using megablast (30). In both cases, high-identity alignments (≥95% identity and ≥400-bp alignment length) were recorded and used for downstream analysis. High-identity alignments were selected to reduce the contribution of deep homologs and unlinked transposable elements to the depth of coverage plots for this already highly repetitive element. For analysis of Germ1 coverage depth, we extracted the start and stop sites of these alignments and tabulated the number of sequences that represented each 100-bp interval, over the length of Germ1. Because we used relatively long alignments for this analysis, transitions between differentially represented sequence elements (i.e., rare vs. abundant regions of Germ1) exhibit artifactual reduction in alignment depth (edge effects: Fig. 2).
Fluorescence in situ Hybridization.
Chromosomes were prepared from lamprey testes and gill by first disaggregating the tissues in hypotonic KCl (75 mM) via gentle grinding in a Dounce homogenizer. Single cells were allowed to swell in suspension for 1 h, prefixed by adding an equal volume of 3:1 methanol/glacial acetic acid (Farmer's solution), then fixed through three changes of Farmer's solution. Suspensions of fixed cells were dropped onto microscope slides and permitted to air dry at room temperature. Fluorescence in situ hybridization experiments were performed using a plasmid subclone containing the Germ1 fragment or a PCR-amplified subregion of this clone. Probes were directly labeled by nick translation with Cy3-dUTP (Perkin-Elmer), as described in ref. 31, with minor modifications. Briefly, 500 ng of labeled probe was used for the FISH experiments; hybridization was performed at 37 °C in 2× SSC, 50% (vol/vol) formamide, 10% (wt/vol) dextran sulfate, and 3 μg of sonicated salmon sperm DNA, in a volume of 15 μL. Posthybridization washing was at 60 °C in 0.1× SSC (three times, high stringency). Nuclei were simultaneously DAPI-stained. Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). DAPI and Cy3 fluorescence signals, detected with specific filters, were recorded separately as gray scale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software. The somatically rare region was amplified from the Germ1 clone via PCR [0.1 ng of plasmid DNA, 50 ng of each primer (GERM1.F2, TGTTACTTCAGAGCATAGAGAGGA; GERM1.R2, TTGATAACACGGGGTGGAGT), 1.2 mM MgCl2, 0.3 U Taq polymerase, 1× PCR buffer, and 200 mM each of dATP, dCTP, dGTP, and dTTP; thermal cycling at 94 °C for 4 min; 33 cycles of 94 °C for 15 s, 65 °C for 15 s, 72 °C for 15 s; and 72 °C for 7 min].
To estimate the maximal fraction of the germline genome that can be accounted for by Germ1-like sequence (including rDNAs), we used ImageJ (http://rsb.info.nih.gov/ij/) to generate a mask for labeled regions (i.e., red in Fig. 3). The DAPI intensity of these regions was compared with that of the entire spread to estimate the fraction of the genome that corresponds to the Germ1 label.
Real-Time PCR.
Real-time PCR was performed to estimate changes in the abundance of genomic copies of Germ1 and SPOPL during embryogenesis. The rate of amplification of these two fragments was compared with a copy-number-matched control probe that was included in a replicate reaction. For Germ1, oligonucleotide primers were designed to unique regions of Germ1 (GERM1-RT.F.1, GTCGAAAGAAGCCGAGTACC) and 28S rDNA (28S-RT.F.1, CGGCGGGAGTAACTATGAC) and a region common to both (G1–28S-RT.R.1, CAGTGGGAATCTCGTTCATC). Primers GERM1-RT.F.1 and G1–28S-RT.R.1 were used to detect Germ1, and primers 28S-RT.F.1 and G1–28S-RT.R.1 were used to detect 28S rDNA. For SPOPL, amplifications were performed with gene-specific primers (SPOPL.F.1, GTATGCATTAGAGAGGTTGAAGGTG; SPOPL.R.1,TGCCTGTTTATTAGAAAGTCAATGG), and relative copy number was estimated on the basis of amplification from primers that were designed to a nonaltered control gene (HMG: HMG.F.1, TTTGCTAGCGTTCGTTTCCT; HMG.R.1, TAATGCGGACAATCCGTACA). Genomic DNAs from sperm, blood, and embryos were isolated using standard phenol/chloroform extraction (26). Real-time PCR was performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems), using the SYBR GreenER qPCR SuperMix, ≈1 ng of DNA, and 50 ng of each primer. Thermal cycling conditions were: 10 min of initial denaturation at 95 °C, followed by 99 cycles of 95 °C for 15 seconds and 65 °C for 20 seconds. All reactions were run in 6 replicate pairs of test (Germ1) and control (28S rDNA) for each DNA sample.
Screening for Additional Germline-Limited Fragments.
Oligonucleotide primer pairs were designed to amplify genomic regions that were present in a sample of sperm BACs but had no corresponding sequence in the WGS dataset (≥95% identity and ≥400-bp alignment length, as above). Traces for sperm BAC-end and WGS reads were downloaded from National Center for Biotechnology Information Trace Archives (www.ncbi.nlm.nih.gov/Traces/trace.cgi?), quality trimmed to Q20 using phred (27, 28), vector trimmed using phrap (29), and aligned using megablast (32). Oligonucleotides were designed using Primer3 (33) and used to prime PCR reactions from multiple DNA samples under standard amplification conditions [1 ng of DNA (or 0.5 μL of cDNA or 1 μL of source RNA), 50 ng of each primer, 1.2 mM MgCl2, 0.3 U Taq polymerase, 1× PCR buffer, and 200 mM each of dATP, dCTP, dGTP, and dTTP; thermal cycling at 94 °C for 4 min; 33 cycles of 94 °C for 15 s, 65 °C for 15 s, 72 °C for 15 s; and 72 °C for 7 min]. These reactions also included a second pair of oligonucleotides at ½ the concentration of target primers (25 nM) that were designed to amplify a single (gene-encoding) region of the lamprey somatic genome (P. marinus assembly V3 Contig10620.3: F, ACCTGTACGAAGCCATGTCC; R, CCGAGTTCTCCAAGAAGTGC). The DNAs used in these reactions were extracted from multiple tissues (testes, blood, liver, kidney, muscle, and tail fin) that were collected from two individuals (Animals 4 and 5), using standard phenol/chloroform extraction (26). PolyA-primed cDNAs were generated for testes via reverse transcription (SuperScript III; Invitrogen) of RNAs that were isolated from juvenile (early parasitic phase) and adult (late parasitic/prespawning phase) lampreys (RNeasy; Qiagen). Primers target to BAC ends and the lamprey SPOPL homolog are provided in Table S1.
Supplementary Material
Acknowledgments.
Catherine Peichel and Barbara Trask provided valuable discussions that greatly improved the quality of the manuscript. Tatjana Sauka-Spengler and Nil Ratan Saha provided reagents and supplemental TUNEL data. Marianne Bronner-Fraser and James Winton provided laboratory space and infrastructure for lamprey husbandry at the California Institute of Technology and Western Fisheries Research Center, respectively. Members of the C.T.A. laboratory assisted with various phases of this project. Whole genome shotgun sequence data that were used in this study were produced by the Genome Center at Washington University School of Medicine in St. Louis. This project was supported by National Institutes of Health Grant GM079492 (to C.T.A.) and National Science Foundation Grant MCB-0719558 (to C.T.A.). J.J.S. received support under Institutional Ruth L. Krischstein National Research Service Award T32-HG00035 and National Research Service Award 1-F32-GM087919-01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GQ215662 and GQ257741).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902358106/DCSupplemental.
References
- 1.Sanchez L, Perondini ALP. Sex determination in sciarid flies: A model for the control of differential X-chromosome elimination. J Theor Biol. 1999;197:247–259. doi: 10.1006/jtbi.1998.0868. [DOI] [PubMed] [Google Scholar]
- 2.Goday C, Esteban MR. Chromosome elimination in sciarid flies. Bioessays. 2001;23:242–250. doi: 10.1002/1521-1878(200103)23:3<242::AID-BIES1034>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 3.Drouin G. Chromatin diminution in the copepod Mesocyclops edax: Diminution of tandemly repeated DNA families from somatic cells. Genome. 2006;49:657–665. doi: 10.1139/g06-022. [DOI] [PubMed] [Google Scholar]
- 4.Degtyarev SV, et al. DNA sequences eliminated during chromatin diminution from somatic cell chromosomes of Cyclops kolensis. Dokl Biochem Biophys. 2002;384:148–151. doi: 10.1023/a:1016068029880. [DOI] [PubMed] [Google Scholar]
- 5.Bachmann-Waldmann C, Jentsch S, Tobler H, Muller F. Chromatin diminution leads to rapid evolutionary changes in the organization of the germline genomes of the parasitic nematodes A. suum and P. univalens. Mol Biochem Parasitol. 2004;134:53–64. doi: 10.1016/j.molbiopara.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Duret L, et al. Analysis of sequence variability in the macronuclear DNA of Paramecium tetraurelia: A somatic view of the germline. Genome Res. 2008;18:585–596. doi: 10.1101/gr.074534.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao MC, Chao JL. RNA-guided DNA deletion in Tetrahymena: An RNAi-based mechanism for programmed genome rearrangements. Annu Rev Genet. 2005;39:537–559. doi: 10.1146/annurev.genet.39.073003.095906. [DOI] [PubMed] [Google Scholar]
- 8.Zufall RA, Robinson T, Katz LA. Evolution of developmentally regulated genome rearrangements in eukaryotes. J Exp Zoolog B Mol Dev Evol. 2005;304:448–455. doi: 10.1002/jez.b.21056. [DOI] [PubMed] [Google Scholar]
- 9.Kubota S, et al. Highly repetitive DNA families restricted to germ cells in a Japanese hagfish (Eptatretus burgeri): A hierarchical and mosaic structure in eliminated chromosomes. Genetica. 2001;111:319–328. doi: 10.1023/a:1013751600787. [DOI] [PubMed] [Google Scholar]
- 10.Goto Y, Kubota S, Kohno S. Highly repetitive DNA sequences that are restricted to the germline in the hagfish Eptatretus cirrhatus: A mosaic of eliminated elements. Chromosoma. 1998;107:17–32. doi: 10.1007/s004120050278. [DOI] [PubMed] [Google Scholar]
- 11.Kubota S, Ishibashi T, Kohno S. A germline restricted, highly repetitive DNA sequence in Paramyxine atami: An interspecifically conserved, but somatically eliminated, element. Mol Gen Genet. 1997;256:252–256. doi: 10.1007/s004380050567. [DOI] [PubMed] [Google Scholar]
- 12.Spicher A, Etter A, Bernard V, Tobler H, Muller F. Extremely stable transcripts may compensate for the elimination of the gene fert-1 from all Ascaris lumbricoides somatic cells. Dev Biol. 1994;164:72–86. doi: 10.1006/dbio.1994.1181. [DOI] [PubMed] [Google Scholar]
- 13.Etter A, Bernard V, Kenzelmann M, Tobler H, Muller F. Ribosomal heterogeneity from chromatin diminution in Ascaris lumbricoides. Science. 1994;265:954–956. doi: 10.1126/science.8052853. [DOI] [PubMed] [Google Scholar]
- 14.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 15.Ye CJ, Liu G, Bremer SW, Heng HH. The dynamics of cancer chromosomes and genomes. Cytogenet Genome Res. 2007;118:237–246. doi: 10.1159/000108306. [DOI] [PubMed] [Google Scholar]
- 16.Hardisty MW. The Biology of Lampreys. New York: Academic; 1971. [Google Scholar]
- 17.Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev Cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Kusakabe R, Kuratani S. Evolutionary perspectives from development of mesodermal components in the lamprey. Dev Dyn. 2007;236:2410–2420. doi: 10.1002/dvdy.21177. [DOI] [PubMed] [Google Scholar]
- 19.Nikitina N, Sauka-Spengler T, Bronner-Fraser M. Dissecting early regulatory relationships in the lamprey neural crest gene network. Proc Natl Acad Sci USA. 2008;105:20083–20088. doi: 10.1073/pnas.0806009105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusakabe R, Tochinai S, Kuratani S. Expression of foreign genes in lamprey embryos: An approach to study evolutionary changes in gene regulation. J Exp Zoolog B Mol Dev Evol. 2003;296:87–97. doi: 10.1002/jez.b.11. [DOI] [PubMed] [Google Scholar]
- 21.Washington University Genome Sequencing Center. Washington University School of Medicine. [Accessed January 9, 2009]; Available at http://genome.wustl.edu/pub/organism/Other_Vertebrates/Petromyzon_marinus/assembly/Petromyzon_marinus-3.0/
- 22.Kalt MR, Gall JG. Observations on early germ cell development and premeiotic ribosomal DNA amplification in Xenopus laevis. J Cell Biol. 1974;62:460–472. doi: 10.1083/jcb.62.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchler-Bauer A, et al. CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arumuganathan K, Earle ED. Estimation of nuclear DNA contents in plants by flow cytometry. Plant Mol Biol Rep. 1991;9:229–241. [Google Scholar]
- 25.Amemiya CT, Ota T, Litman GW. Construction of P1 artificial chromosome (PAC) libraries from lower vertebrates. In: Lai E, Birren B, editors. Analysis of Nonmammalian genomes. San Diego: Academic; 1996. pp. 223–256. [Google Scholar]
- 26.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 27.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 28.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 29.Green P. Seattle: University of Washington; 1994. Phrap: Phragment Assembly Program. Available at www.phrap.org/phredphrapconsed.html. [Google Scholar]
- 30.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 31.Lichter P, et al. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz SA, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Clifton, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 34.Tahara Y. Normal stages of development in the lamprey Lampetra reissneri (Dybowski) Zoolog Sci. 1988;5:109–118. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.