Abstract
Transcriptional silencing is a crucial process that is mediated through chromatin structure. The histone deacetylase Sir2 silences genomic regions that include telomeres, ribosomal DNA (rDNA) and the cryptic mating-type loci. Here, we report an unsuspected role for the enzyme Gas1 in locus-specific transcriptional silencing. GAS1 encodes a β-1,3-glucanosyltransferase previously characterized for its role in cell wall biogenesis. In gas1 mutants, telomeric silencing is defective and rDNA silencing is enhanced. We show that the catalytic activity of Gas1 is required for normal silencing, and that Gas1's role in silencing is distinct from its role in cell wall biogenesis. Established hallmarks of silent chromatin, such as Sir2 and Sir3 binding, H4K16 deacetylation, and H3K56 deacetylation, appear unaffected in gas1 mutants. Thus, another event required for telomeric silencing must be influenced by GAS1. Because the catalytic activity of Gas1 is required for telomeric silencing, Gas1 localizes to the nuclear periphery, and Gas1 and Sir2 physically interact, we propose a model in which carbohydrate modification of chromatin components provides a new regulatory element that may be critical for chromatin function but which is virtually unexplored in the current landscape of chromatin analysis.
Keywords: acetylation, beta-glucan, chromatin, S. cerevisiae, telomeres
The formation of silent chromatin leads to the transcriptional repression of regions of the genome. In the yeast Saccharomyces cerevisiae, these regions include the telomeres, ribosomal DNA (rDNA), and the cryptic mating-type loci (the HM loci, HML, and HMR), all of which require the silent information regulator 2 protein (Sir2), an NAD+-dependent protein deacetylase (reviewed in ref. 1). Sir2 is the founding member of the conserved sirtuin deacetylase family (2), with Sir2 directing its activity toward lysine 16 of histone H4 (H4K16) to promote silent chromatin formation (reviewed in ref. 3). There is also evidence that deacetylation of lysine 56 of histone H3 (H3K56) by Sir2 (4), or the sirtuins Hst3 and Hst4 (5), enables silencing at the telomeres and HM loci. Sir2 functions in the SIR complex with Sir3 and Sir4 to silence at telomeres, and also at the HM loci, where an additional protein, Sir1, also participates (1). However, within the rDNA, the Sir2 containing RENT complex acts independently of the other Sir proteins (1). Distinctions among the 3 silenced regions led to the hypothesis that different mechanisms of silent chromatin formation and regulation exist for each region.
The basic model for silent chromatin formation at telomeres and HM loci involves recruitment of the SIR complex by DNA binding proteins, followed by Sir2-mediated histone deacetylation and additional SIR complex spreading. Hypoacetylation of histones enables silent chromatin spreading in the absence of Sir2 deacetylase activity, but is not sufficient for full silencing (6). Instead, deacetylase activity must be targeted to the silenced loci through Sir3 or some other means (7). Although early studies focused on Sir2-mediated histone deacetylation, silent chromatin formation is also regulated through histone methylation and ubiquitination (reviewed in ref. 8), indicating that histone deacetylation alone is not sufficient for silencing. Multiple biochemical activities are recognized to contribute to chromatin function, and more are likely to emerge. The identification of GAS1 by synthetic genetic array (SGA) analysis as an interactor with genes encoding nuclear functions provides one such new candidate activity.
In the cell wall, Gas1 is an abundant protein anchored via glycophosphatidylinositol (GPI) (9). Gas1 β-1,3-glucanosyltransferase activity catalyzes formation and maintenance of chains of β-1,3-glucan (reviewed in ref. 10). The modification occurs on proteins to which mannose residues have first been attached through serine or threonine residues (10). GAS1 deletion mutants have cell wall defects, including reduced viability, thermal sensitivity, and sensitivity to cell wall disrupting compounds (11). GAS1 is broadly conserved in fungi and has 4 homologs in yeast (10). GAS1, GAS3, and GAS5 are expressed in vegetatively growing cells, whereas GAS2 and GAS4 are expressed meiotically during sporulation (12).
GAS1 surfaced in SGA screens using mutants with established nuclear functions. For example, a conditional allele of ORC2 uncovered synthetic sickness with gas1Δ (13). ORC2 encodes a component of the DNA replication origin recognition complex that has a separable function in promoting silencing of the HM loci (reviewed in ref. 14). GAS1 also exhibited a growth defect with deletion of EAF1 or a conditional allele of ESA1 (15), which both encode subunits of the nucleosomal acetylation of H4 (NuA4) complex (reviewed in ref. 16) that functions in silencing at the telomeres and rDNA (17). The basis for these interactions has not been pursued, but they suggest that GAS1 and ORC2, ESA1, and EAF1 may contribute to parallel processes that are critical for cellular function and viability. These might involve Gas1's established role at the cell wall, or may point to a previously unsuspected role for Gas1 in nuclear or chromatin function.
We show here that Gas1 participates in transcriptional silencing in a manner separable from its established function at the cell wall. In gas1Δ mutants, no change in silencing was observed at the HM loci, telomeric silencing was disrupted, and rDNA silencing was enhanced. Key features of silent chromatin, including Sir2 and Sir3 binding at telomeres and deacetylation of H4K16 and H3K56, are comparable in wild-type and gas1Δ strains. Analysis of enzymatically inactive gas1 mutants showed that β-1,3-glucanosyltransferase activity itself is required for transcriptional silencing at the telomeres. Further analysis showed that Gas1 may act through modification of Sir2 or other interacting factors. These findings thus reveal a new nuclear role for a carbohydrate modification enzyme in transcriptional silencing.
Results
Deletion of GAS1 Causes Decreased Telomeric Silencing and Increased rDNA Silencing.
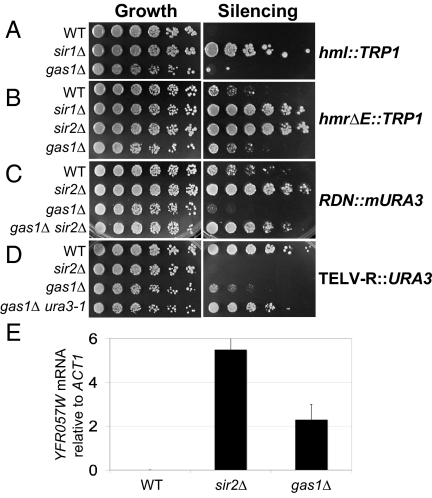
Based on the interactions between GAS1 and transcriptional silencing genes, we tested if GAS1 participated in silencing. Assays of HM loci silencing in gas1Δ strains with TRP1 reporters integrated at HML (Fig. 1A) and HMR (Fig. 1B) showed no defect in silencing compared with sir1Δ or sir2Δ strains, which are defective in HM silencing. Likewise, silencing of an independent reporter gene, ADE2, integrated at HMR was comparable in wild-type and gas1Δ strains (supporting information (SI) Fig. S1A).
Fig. 1.
GAS1 functions in transcriptional silencing. (A) Deletion of GAS1 does not affect HML silencing. All growth plates in A–D are synthetic complete (SC) medium. Wild-type (WT) (LPY13659), sir1Δ (LPY13660), and gas1Δ (LPY13661) strains with a HML::TRP1 reporter were plated on SC lacking tryptophan (SC-trp). Increased growth on SC-trp indicates defective silencing. (B) Deletion of GAS1 does not affect HMR silencing. WT (LPY4912), sir1Δ (LPY4958), sir2Δ (LPY4980), and gas1Δ (LPY13665) strains with an hmrΔE::TRP1 reporter were assayed as in A. Levels of WT silencing differ at these TRP1 HM loci reporters due to differences in the structure of the reporter. The HML::TRP1 reporter (47) contains a TRP1 reporter at HML whereas hmrΔE::TRP1 contains a mutated silencer (48). (C) Deletion of GAS1 causes increased silencing at the rDNA. WT (LPY2446), sir2Δ (LPY2447), gas1Δ (LPY10074), and gas1Δ sir2Δ (LPY10078) with an mURA3 NTS1 rDNA reporter were assayed for silencing on SC plates lacking uracil (SC-ura). Increased growth on SC-ura indicates defective silencing (see Fig. 3A for location of this reporter in the rDNA repeat). (D) Deletion of GAS1 causes a telomeric silencing defect. WT (LPY4916), sir2Δ (LPY10397), and gas1Δ (LPY10362) with a URA3 telomeric reporter on chromosome V-R, and a gas1Δ control strain (LPY10129) with no telomeric reporter (gas1Δ ura3–1), to monitor gas1Δ 5-FOA sensitivity, were plated on SC containing 5-FOA. Decreased growth on 5-FOA indicates defective silencing. (E) Expression of an endogenous telomeric gene, YFR057W, is increased in gas1Δ. cDNAs from WT (LPY1029), sir2Δ (LPY12660), and gas1Δ (LPY10358) strains were analyzed by quantitative PCR, with the bars representing YFR057W cDNA signal minus control reactions without reverse transcriptase, normalized to ACT1.
To determine whether GAS1 influenced silencing in the rDNA, strains with reporters integrated within the rDNA repeat were constructed. A gas1Δ strain with a URA3 reporter integrated at nontranscribed spacer 1 (NTS1) near the 5S rRNA gene of a single rDNA repeat (18) had enhanced silencing compared with wild-type (Fig. 1C). This effect is Sir2 dependent because gas1Δ sir2Δ was as defective as sir2Δ in silencing of the rDNA (Fig. 1C). The increase in silencing seen in gas1Δ is specific for the NTS1 region, as an ADE2-CAN1 reporter in the 25S rRNA gene, which is only moderately subject to Sir2-dependent silencing (19), did not show any change in silencing (Fig. S1B).
To assess telomeric silencing in gas1Δ mutants, a URA3 telomeric reporter on chromosome V-R (20) was evaluated. In this assay, URA3 expression is monitored on medium containing 5-FOA, a suicide substrate for cells expressing URA3. Both sir2Δ and gas1Δ mutants expressed this telomeric reporter gene, whereas wild-type cells had intact telomeric silencing (Fig. 1D). In control gas1Δ strains lacking the reporter, no 5-FOA sensitivity was seen (Fig. 1D). The gas1Δ silencing defect was also observed with a chromosome VII-L URA3 telomeric reporter (Fig. S2A) and with a chromosome V-R ADE2 telomeric reporter (Fig. S2B), indicating that the silencing defect occurs at multiple telomeres and is promoter and gene independent.
In addition to the reporter assays, transcription of the normally silenced YFR057W gene at telomere VI-R was assayed by reverse transcription-coupled quantitative PCR. YFR057W RNA was undetectable in wild-type cells, whereas transcription was readily detected in both the gas1Δ mutant and sir2Δ control (Fig. 1E). Thus telomeric silencing in gas1Δ is defective compared with wild type by 2 independent assays. Yet, some telomeric silencing must be intact because the defects of gas1Δ mutants are somewhat less severe than for sir2Δ mutants.
The locus-specific silencing phenotypes of gas1Δ cells are unusual in that loss of GAS1 function causes loss of silencing at telomeres, but increased silencing at the rDNA, with no effect on HM silencing. This constellation of phenotypes is atypical of the nearly 300 genes previously reported to influence silencing. Thus, GAS1's functions may represent a molecular contribution not yet studied in silent chromatin.
Telomeric Silencing Function Is Not a General Property of Proteins with Roles in Cell Wall Biogenesis.
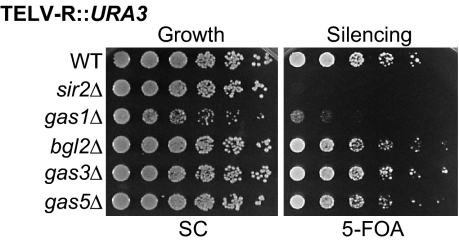
In the same way that multiple proteins have roles in silencing, many different enzymes also function in cell wall formation. To determine whether the gas1Δ defect in telomeric silencing is a general property of proteins with these functions, deletions of 3 genes that contribute enzymatically to the cell wall were constructed with the telomeric reporter. BGL2 encodes an endo-β-1,3-glucanase involved in cell wall construction and remodeling (21), and GAS3 and GAS5 are homologs of GAS1 that also encode β-1,3-glucanosyltransferases (10). Not one of these mutants disrupted telomeric silencing (Fig. 2). Thus, telomeric silencing defects are not a general characteristic of genes involved in maintenance of the cell wall, including those encoding comparable enzymatic activities, but instead are specific to gas1Δ mutants.
Fig. 2.
The telomeric silencing function of GAS1 is not shared with other cell wall genes. WT (LPY4916), sir2Δ (LPY10397), gas1Δ (LPY10362), bgl2Δ (LPY13094), gas3Δ (LPY12337), and gas5Δ (LPY12348) strains were assayed for telomeric silencing as in Fig. 1D.
Well-Defined Hallmarks of Silent Chromatin Are Intact in gas1Δ Mutants.
The telomeric silencing defect in gas1Δ may be caused by a defect in the amount or formation of silent chromatin. At the molecular level, the gas1Δ telomeric silencing defect is not a consequence of Sir expression, as gas1Δ microarray expression analysis reveals normal transcription of SIR2, SIR3, and SIR4 (22), and direct immunoblotting showed that Sir2 and Sir3 levels were not decreased in gas1Δ (Fig. S3).
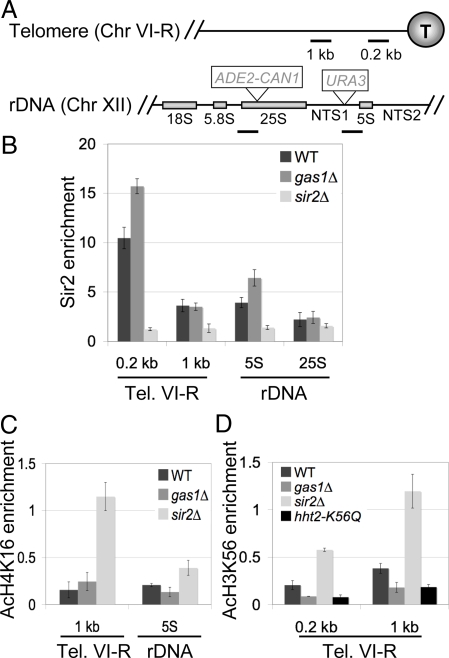
A distinct possibility was that the silencing phenotypes result from a change in Sir protein occupancy at the silenced region. To test this, Sir2 binding in gas1Δ strains was evaluated by chromatin immunoprecipitation (ChIP) experiments using telomere VI-R and rDNA-specific primers (Fig. 3A). We observed that Sir2 occupancy at telomere VI-R in gas1Δ was marginally increased relative to wild type (Fig. 3B). Therefore, decreased Sir2 occupancy does not explain the defective telomeric silencing seen in gas1Δ mutants. Likewise, Sir3 occupancy at the telomeres is unaffected in gas1Δ mutants (Fig. S4A). Furthermore, Sir3 localizes to telomeric foci in gas1Δ mutants, indicating that telomere clustering is normal (Fig. S4B).
Fig. 3.
Key features of silent chromatin are unaltered in gas1Δ mutants. (A) Map of primer sites used for ChIP. Chromosome VI-R primers amplify regions 0.2 kb and 1 kb from the end of the telomere. Chromosome XII primers amplify regions near the 25S rRNA and 5S rRNA genes. Also shown are ADE2-CAN1 and URA3 reporter locations for rDNA silencing assays. (B) Sir2 occupancy in gas1Δ is increased slightly at the telomere and 5S rDNA. ChIP of Sir2 was done in WT (LPY5), sir2Δ (LPY11), and gas1Δ (LPY10129) strains. Input and IP DNA were analyzed with primers shown in A and the nonspecific locus ACT1. Sir2 enrichment at the telomere and rDNA was normalized to ACT1. (C) H4K16 is deacetylated at the telomere and rDNA in gas1Δ. ChIP of acetylated H4K16 (AcH4K16) was done in the same strains as (B). AcH4K16 enrichment at the telomere and rDNA was normalized to the Chr. V intergenic region. (D) H3K56 is deacetylated at the telomere in gas1Δ. ChIP of acetylated H3K56 (AcH3K56) was done in the same strains as B, and as a negative control, hht2-K56Q (LPY13166). AcH3K56 enrichment at the telomere was normalized to the intergenic region.
An increase in Sir2 binding may explain gas1Δ's increase in rDNA silencing. Modestly elevated Sir2 binding was seen near 5S in the rDNA (Fig. 3B), a region of the rDNA repeat including NTS1, where the increase in silencing was observed (Fig. 1B). At the 25S rDNA, there was no change in Sir2 occupancy (Fig. 3B), a location in the rDNA that showed no difference in rDNA silencing for gas1Δ mutants (Fig. S1B). Therefore, occupancy of Sir2 in the rDNA parallels the strength of rDNA silencing observed in gas1Δ mutants.
Because a modest increase of Sir2 occupancy in gas1Δ was also observed at telomeres but yielded defective silencing, we considered the possibility that histone acetylation profiles were altered in gas1Δ. To address this, ChIP experiments were performed for acetylation of the Sir2 histone targets H4K16 and H3K56 at silenced loci. No increase in H4K16 acetylation was observed in gas1Δ at the telomere or 5S rRNA gene (Fig. 3C). Further, no increase in H3K56 acetylation was detected in gas1Δ (Fig. 3D). In contrast, the sir2Δ mutant displayed increased levels of acetylation at both sites (Fig. 3 C and D). Consistent with these findings, in vitro NAD+-dependent deacetylase assays with purified GST-Sir2 showed that deacetylase activity was unaffected by addition of purified Gas1 (Fig. S4C). Thus 5 well-established molecular criteria for telomeric silencing are intact in gas1Δ: Sir2 and Sir3 remain telomere bound, Sir3 localizes to telomeric foci, and both H4K16 and H3K56 remain deacetylated. Therefore, a different GAS1-dependent event in silent chromatin must be disrupted.
Gas1 Silencing Function Is Potentially Mediated Through Its Interaction with Sir2.
In a genome-wide survey, it was reported that GFP-Gas1 surprisingly localizes to the nuclear periphery in addition to its more expected localization at the cell wall, mitochondria, and endoplasmic reticulum (23). Direct examination of GFP-Gas1 showed staining within each cell that coincided with the outer edge of the DAPI nuclear staining, confirming the genome-wide result (Fig. S5A). The nuclear periphery is a clearly relevant location for a fraction of the protein to reside for its function in transcriptional silencing, potentially as a nuclear membrane-associated protein.
Because in the localization study GFP was fused to the Gas1 C terminus, proximal to the GPI anchoring position, we tested whether the tag interfered with proper function of the protein. We found that GFP-Gas1 strains were fully competent for telomeric silencing and had no growth defects at high temperature (Fig. S5B). Therefore it appeared that the tagged protein was functional and there was no reason to consider the reported localization to be spurious.
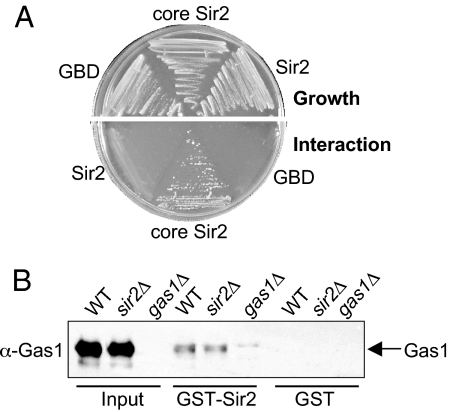
Two converging observations of Gas1-SIR complex physical interactions prompted us to further examine Gas1's connection with the nuclear Sir2 protein. First, in a high-throughput identification of protein complexes by mass spectrometry, Gas1 associated with the SIR complex component Sir3 (24). Second, in a Sir2 two-hybrid screen, Gas1 was found as an interacting protein (25).
Two-hybrid analysis showed an interaction between a GBD-core Sir2 construct containing the catalytic deacetylase domain (residues 244–457) and a GAD-Gas1 construct containing a portion of its catalytic domain (residues 163–407) (Fig. 4A). These constructs contained only a portion of Sir2 and Gas1 to accommodate the possibility that full-length Sir2 bait protein can be repressive (25) and because smaller domain-based protein fragments are observed to increase detection and sensitivity of two-hybrid interactions (26). No interaction was seen when GAD-Gas1 was cotransformed with a GBD vector or GBD-Sir2 construct containing nearly full-length Sir2 (Fig. 4A). Thus, two-hybrid analysis revealed a specific interaction between constructs containing the catalytic domains of Sir2 and Gas1. Previous studies showed that neither SIR3 nor SIR4 were required for the Sir2-Gas1 interaction (25). Of note, however, is that the Sir2-Gas1 interaction was enhanced in the sir4Δ two-hybrid strain (25). These observations suggest competition between Gas1 and Sir4 for Sir2 binding and indicate that the interaction does not require integrity of the SIR complex.
Fig. 4.
Sir2 interacts with Gas1 by two-hybrid and GST pull-down. (A) GBD-core Sir2 interacts with GAD-Gas1. The constructs GBD vector (pLP956), GBD-core Sir2 (pLP1073), and GBD-Sir2 (pLP1074) were expressed from 2μ TRP1 plasmids. GAD-Gas1 (pLP1205) was expressed from a 2μ LEU2 plasmid. The two-hybrid strain (LPY3374) was transformed with pairs of these plasmids to form LPY7251 (with pLP956, pLP1205), LPY7251 (with pLP1073, pLP1205), and LPY7253 (with pLP1074, 1205). The growth control plate is SC-leu-trp medium. The interaction plate is this medium also lacking histidine and adenine, to simultaneously monitor for GAL1-HIS3 and GAL2-ADE2 reporter activation. Growth on this plate indicates a physical interaction between GBD-core Sir2 and GAD-Gas1. (B) GST-Sir2 physically interacts with Gas1. GST (pLP1302) and GST-Sir2 (pLP1275) were purified and incubated with whole-cell extracts from strains used in Fig. 3B. Bound protein was analyzed by immunobloting for Gas1 (125 kDa).
GST affinity experiments were performed to validate the Sir2-Gas1 two-hybrid interaction with full-length proteins. Recombinant GST-Sir2 was incubated with whole-cell extracts from wild-type, sir2Δ, or gas1Δ yeast strains. GST-Sir2 specifically bound Gas1 both in the presence and absence of endogenous Sir2 (Fig. 4B). A catalytically inactive Sir2, Sir2-H364Y, also interacted with Gas1 by GST pull-down, showing that the proteins interact despite the loss of Sir2 deacetylase activity (Fig. S6A). This confirmation of the Sir2-Gas1 physical interaction with full-length proteins implies that the effect of Gas1 on silencing may be mediated through its interaction with members of the SIR complex, especially Sir2. Of note is that GST-Sir2 pulled down only a fraction of Gas1, indicating that both proteins participate in other complexes independently of one another, consistent with Gas1's additional localization beyond the nuclear periphery and the previous characterization of Sir2 as a member of the SIR and RENT complexes.
Gas1's Enzymatic Activity Is Required for Transcriptional Silencing.
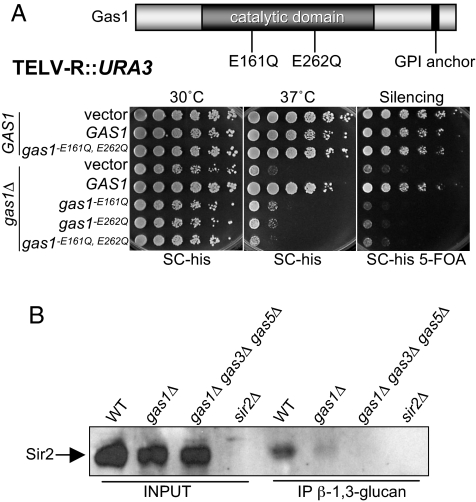
Gas1's physical association with Sir2 suggested a direct role for Gas1 in silencing. Therefore, we tested whether the enzymatic activity of Gas1 contributes to silencing. Previous studies identified 2 amino acid residues critical for Gas1 β-1,3-glucanosyltransferase activity, E161 and E262, that map to its catalytic domain (27, 28). Importantly, Gas1 protein with glutamine substitutions of these 2 catalytic residues remains structurally intact, yet its enzymatic activity is destroyed (27).
The gas1-E161Q and gas1-E262Q mutants were assayed for growth and telomeric silencing. Sensitivity to high temperature was observed in the mutant strains, confirming that the enzymatically inactive gas1 mutants exhibited the classic cell wall defect of gas1Δ (Fig. 5A). The catalytically inactive Gas1 mutant proteins were also completely defective in telomeric silencing, indicating that the enzymatic activity is necessary for silencing (Fig. 5A). When the double-point mutant (gas1-E161Q, E262Q) was expressed in a wild-type background, growth at high temperature and telomeric silencing was not compromised, demonstrating that the mutations were not dominant (Fig. 5A). Further, the Gas1-Sir2 interaction was not disrupted in the gas1 catalytically inactive mutant, as demonstrated by GST affinity (Fig. S6A). Thus, although Gas1's catalytic activity is required for its silencing function, it does not promote interaction with Sir2.
Fig. 5.
Gas1 enzymatic activity is required for transcriptional silencing and is linked to Sir2. (A) gas1 enzymatically inactive point mutants are defective in telomeric silencing. The diagram of Gas1 shows the location of the E161Q and E262Q mutations in the catalytic domain. Dark bar at the C terminus of Gas1 indicates GPI anchor position. WT (LPY4916) strains were transformed with the 2μ HIS3 plasmids: vector (pLP359), GAS1 (pLP2091), and gas1-E161Q, E262Q (pLP2117). Transformed WT strains are LPY13554, LPY13559, and LPY13562. The gas1Δ strain (LPY10362) was transformed with 2μ HIS3 plasmids: pLP359, pLP2091, pLP2093 (gas1-E161Q), pLP2094 (gas1-E262Q), and pLP2117. Transformed gas1Δ strains are LPY13563 and LPY13568-LPY13571. Growth was examined on SC plates lacking histidine (SC-his). Silencing was examined on SC-his containing 5-FOA (SC-his 5-FOA). Growth at elevated temperature was examined at 37 °C. (B) Anti-β-1,3-glucan immunoprecipitates Sir2. β-1–3-glucan immunoprecipitations were performed in extracts from wild-type (LPY5), gas1Δ (LPY10129), and gas1Δ gas3Δ gas5Δ (LPY13543) strains overexpressing SIR2 (pLP349) and from sir2Δ (LPY11) expressing a vector construct (pLP135). Transformed strains are LPY13545, LPY13549, LPY13553, and LPY13546, respectively. Immunoprecipitated material was analyzed by immunoblot for Sir2 (65 kDa).
To determine if Gas1's established function in cell wall biogenesis is separable from its role in silencing, telomeric silencing of gas1Δ was examined in the presence of sorbitol. Sorbitol is an osmotic stabilizing agent capable of rescuing many types of mutants with thermosensitive cell lytic phenotypes (29, 30). Sorbitol rescues the temperature sensitivity of gas1Δ mutants (Fig. S6B). However, sorbitol did not rescue gas1Δ telomeric silencing, demonstrating a separation of function between Gas1 actions at the cell wall and silencing (Fig. S6B). The inability of sorbitol to rescue gas1Δ telomeric silencing defects supports the idea that GAS1's effect on transcriptional silencing is not a simple consequence of its role in the cell wall and provides evidence that silencing is a separate nuclear role for Gas1.
The observations that Gas1 enzymatic activity is required for silencing (Fig. 5A) and that GFP-Gas1 localizes to the nuclear periphery (23) (Fig. S5A) raise the possibility that Gas1 enzymatically modifies chromatin factors, such as Sir proteins or histones. This would result in attachment and elongation of β-1,3-glucan to nuclear protein substrates, which has not previously been observed. To probe for cellular substrates involved in silencing, an antibody directed against β-1,3-glucan (31) was used in immunoprecipitations that were then analyzed for the presence of Sir2 protein. No signal was observed in the sir2Δ control strain, yet Sir2 was immunoprecipitated by anti-β-1,3-glucan, demonstrating that endogenous Sir2 or proteins with which it is complexed were recognized (Fig. 5B). Importantly, in gas1Δ cells, significantly less Sir2 was pulled down, and Sir2 was undetectable in the gas1Δ gas3Δ gas5Δ strain that is likely to have no residual β-1,3-glucanosyltranferase activity (Fig. 5B). Sir2 was also detected in β-1,3-glucan immunoprecipitations in a sir3Δ sir4Δ strain, suggesting that the SIR complex was not required for this potential Sir2 modification or association with other substrate (Fig. S6C). These findings support the possibility that posttranslational modification by β-1,3-glucan of Sir2 or other chromatin components may provide a new mechanism for regulation of locus-specific transcriptional silencing.
Discussion
We report here that the glucanosyltransferase Gas1 participates in locus-specific transcriptional silencing. The role of Gas1 in silencing is mediated by its β-1,3-glucanosyltransferase activity and may act through its physical interaction with Sir2 or other bridging partners (Fig. S7). Importantly, Gas1's function in silencing is separable from its previously described role in cell wall biogenesis, demonstrating an unsuspected nuclear function for this enzyme.
The surprising yet compelling observations that Gas1 is enzymatically active in the nucleus and functions in transcriptional silencing are supported by several independent lines of evidence. First, the β-1,3-glucanosyltranferase activity of Gas1 is required for silencing (Fig. 5A). In addition, sorbitol rescues gas1Δ osmotic sensitivity, but not silencing defects (Fig. S6B), consistent with a role for Gas1 beyond cell wall biogenesis, such as in the nucleus, where silencing occurs. Gas1 localization to the nuclear periphery (23) (Fig. S5A) strengthens the notion that the Sir2-Gas1 physical interaction is relevant in vivo, as a subset of silencing proteins reside in the same nuclear compartment. Indeed, Sir2 was found in immunoprecipitates with an antibody directed against β-1,3-glucan, the subunit transferred by Gas1 catalysis (Fig. 5B), demonstrating that the modification is physically associated with Sir2, either directly or bridged through other substrate proteins.
The regulation of transcriptional silencing is complex, with numerous proteins implicated in the processes of forming silent chromatin and restricting its spread into euchromatic regions. Modifications of histones and other chromatin proteins are fundamental for the appropriate regulation of silent chromatin (reviewed in refs. 1, 8, 32). Posttranslational modifications of nonhistone silencing proteins have also been observed. For example, the SIR complex structural component Sir3 is acetylated and phosphorylated, and these modifications contribute to its function in transcriptional silencing (33, 34). Other less well-studied posttranslational modifications are likely to contribute to transcriptional silencing. The discovery of such modifications will be facilitated by the characterization of new catalytically active proteins with roles in silencing, as reported here.
A Distinct Role for GAS1 in Transcriptional Silencing.
Silencing assays in gas1Δ mutants revealed telomeric silencing defects, increased rDNA silencing, and intact HM silencing. This combination of phenotypes is unique. Deletion of the MAP kinase pathway genes BCK1 and SLT2 results in defective telomeric silencing and enhanced rDNA silencing, but also causes enhanced HMR silencing (35). Slt2 phosphorylates Sir3 (35, 36). During stress response, hyperphosphorylation of Sir3 can decrease telomeric silencing (36), and under normal conditions, Sir3 phosphorylation strengthens telomeric silencing (34). Our data point to modification of the SIR complex by a previously unsuspected activity.
A recent study provides another link between Gas1 and chromatin modification, as gas1Δ and a number of other mutants are implicated in regulation of total acetylation levels of histone H3 and H4 (37). Decreased levels of tetraacetylated H3 and H4 were reported for gas1Δ mutants when assayed by mass spectrometry (37). However, in directed studies with isoform-specific histone antibodies for multiple acetylated lysines, including H4K16, which is required for telomeric silencing, we detected no differences in acetylation in gas1Δ mutants (Fig. S8). Therefore, it remains unresolved if specific changes in acetylation or other histone modifications underlie the silencing phenotypes of gas1Δ mutants.
A Role for Carbohydrate Modification in Chromatin Function.
The modification of Sir2 or other chromatin components by Gas1 raises a new possibility for regulating Sir2 enzymatic activity downstream of Sir2 binding and histone deacetylation, perhaps by influencing the recruitment of other factors crucial for silent chromatin formation. It is possible that the interaction between Sir2, Gas1, and β-1,3-glucan is bridged by another factor that Sir2 contacts, yet it is unlikely that such bridging factors are solely members of the SIR complex, because Sir2 was also recovered in β-1,3-glucan immunoprecipitations in sir3Δ sir4Δ strains (Fig. S6C). Future studies should establish whether other Sir2-interacting proteins, such as histones and other chromatin components, mediate the interaction between Sir2 and β-1,3-glucan, or are themselves substrates for Gas1.
Precedents are known for posttranslational carbohydrate modifications of proteins that affect transcription. Early studies demonstrated glycosylation of histones in Tetrahymena (38) and complex carbohydrate modification of vertebrate high-mobility group proteins (39). More recently, O-linked beta-N-acetylglucosamine (O-GlcNAc) modification at serine and threonine residues has been found on both cytoplasmic and nuclear proteins in all plants and animals. Indeed, in mammalian cells, O-GlcNAcylation of nuclear proteins is critical to cellular processes, including signaling, cell cycle progression, and transcription, and these O-GlcNAcylated proteins are tied to diabetes and neurodegeneration (reviewed in ref. 40). Furthermore, the RNA polymerase II transcription factor Sp1 is O-GlcNAcylated (41). In vitro studies established that Sp1's modification appeared required for its role in transcriptional activation, but not in template loading, analogous to the in vivo observations reported here for chromatin in GAS1-dependent silencing. A number of Sp1-related DNA-binding factors are also glycosylated, illustrating a potentially common mode of regulation (reviewed in ref. 42). Further exploration of the in vivo roles of carbohydrate-modifying enzymes in transcriptional silencing should provide new insights into the diverse regulatory mechanisms for silencing and other chromatin-dependent processes.
Materials and Methods
Yeast Strains and Plasmids.
Strains are listed in Table S1. Plasmids are listed in Table S2 and described in SI Materials and Methods. Yeast strains were constructed with standard methods (43, 44) with deletions using primers in Table S3.
mRNA Quantification.
RNA was prepared using an RNeasy kit (Qiagen). Reverse transcription was performed with a TaqMan kit (ABI), with random hexamer priming. cDNA was diluted 125- to 500-fold before real-time PCR on a DNA Engine Opticon 2 (MJ Research) with primers in Table S3.
Chromatin Immunoprecipitation.
Experiments were performed as described (44). Immunoprecipitation (IP) mixtures were incubated overnight at 4 °C with anti-Sir2 (45), anti-Sir3 (34), anti-AcH4K16 (Upstate), or anti-AcH3K56 (Active Motif). DNA in input and IP samples was quantified by real-time PCR. Primers used are in Table S3. For Sir2, values are IP/input normalized to ACT1 IP/input. For AcH4K16, values are IP/input normalized to the intergenic region IP/input. For AcH3K56, values are IP/input normalized to the intergenic region IP/input.
Two-Hybrid and GST-Affinity Studies.
The Sir2 two-hybrid screen (25, 44) used the reporter strain PJ69–4A (LPY3374) (46). It was cotransformed with pLP1205 (GAD-Gas1) and pLP956 (pGBD-C1), pLP1073 (GBD-core Sir2), or pLP1074 (GBD-Sir2). The GST-affinity binding assays were described previously (44). Samples were probed with a 1:10,000 dilution of anti-Gas1 (9) (gift from C. Sütterlin). Horseradish peroxidase-coupled anti-rabbit secondary (Promega) was used at 1:10,000 and detected using enhanced chemiluminescence reagents (PerkinElmer).
Anti-β-1,3-Glucan Immunoprecipitation.
Cultures were grown to an A600 of 0.8 were lysed with glass beads in Sir2 IP lysis buffer (44). Four micrograms anti-β-1,3-glucan (31) (Biosupplies Australia) was added to cell extract and incubated at 4 °C for 3 h. One hundred microliters of a 50% slurry of Protein G Sepharose (GE Healthcare) was then added and incubated for 1 h at 4 °C. Beads were washed once with Sir2 IP buffer, and twice with wash buffer (50 mM Hepes [pH 7.5], 150 mM NaCl, 1 mM EDTA). Immunoblotting was as above, with a 1:5,000 dilution of anti-Sir2.
Supplementary Material
Acknowledgments.
We especially thank S.N. Garcia for early contributions to this work and observations underlying the data presented in Fig. 4A; Y. Chun and R. Otsuka for assistance with experiments; F. Solomon for advice on NAD+ hydrolysis assays; A. Bicknell for microscopy advice; C. Sütterlin, A. Conzelmann, J. Rine, R. Schekman, W. S. Lo, S. L. Berger, M. Grunstein, F. Winston, and V. Cheung for reagents and advice; C.S. Chang, R. P. Darst, J. B. DuRose, S. N. Garcia, S. J. Jacobson, J. T. Kadonaga, E. M. Scott, F. Solomon, and J. M. Wilson for comments on the manuscript; and the entire Pillus lab for their contributions and suggestions during the course of this work. This work was initiated with support of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 10879.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900809106/DCSupplemental.
References
- 1.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 2.Brachmann CB, et al. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 3.Buck SW, Gallo CM, Smith JS. Diversity in the Sir2 family of protein deacetylases. J Leukoc Biol. 2004;75:939–950. doi: 10.1189/jlb.0903424. [DOI] [PubMed] [Google Scholar]
- 4.Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang B, Miller A, Kirchmaier AL. HST3/HST4-dependent deacetylation of lysine 56 of histone H3 in silent chromatin. Mol Biol Cell. 2008;19:4993–5005. doi: 10.1091/mbc.E08-05-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang B, Kirchmaier AL. Bypassing the catalytic activity of SIR2 for SIR protein spreading in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:5287–5297. doi: 10.1091/mbc.E06-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou CC, Li YC, Gartenberg MR. Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing. Mol Cell. 2008;31:650–659. doi: 10.1016/j.molcel.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 9.Nuoffer C, Jeno P, Conzelmann A, Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae Gas1 protein to the plasma membrane. Mol Cell Biol. 1991;11:27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popolo L, Vai M. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim Biophys Acta. 1999;1426:385–400. doi: 10.1016/s0304-4165(98)00138-x. [DOI] [PubMed] [Google Scholar]
- 11.Popolo L, et al. Physiological analysis of mutants indicates involvement of the Saccharomyces cerevisiae GPI-anchored protein gp115 in morphogenesis and cell separation. J Bacteriol. 1993;175:1879–1885. doi: 10.1128/jb.175.7.1879-1885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragni E, et al. GAS2 and GAS4, a pair of developmentally regulated genes required for spore wall assembly in Saccharomyces cerevisiae. Eukaryot Cell. 2007;6:302–316. doi: 10.1128/EC.00321-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suter B, et al. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics. 2004;167:579–591. doi: 10.1534/genetics.103.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell L, et al. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol Cell Biol. 2008;28:2244–2256. doi: 10.1128/MCB.01653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lafon A, Chang CS, Scott EM, Jacobson SJ, Pillus L. MYST opportunities for growth control: Yeast genes illuminate human cancer gene functions. Oncogene. 2007;26:5373–5384. doi: 10.1038/sj.onc.1210606. [DOI] [PubMed] [Google Scholar]
- 17.Clarke AS, Samal E, Pillus L. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol Biol Cell. 2006;17:1744–1757. doi: 10.1091/mbc.E05-07-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 19.Fritze CE, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renauld H, et al. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 21.Mrsa V, Klebl F, Tanner W. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-beta-1,3-glucanase. J Bacteriol. 1993;175:2102–2106. doi: 10.1128/jb.175.7.2102-2106.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagorce A, et al. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J Biol Chem. 2003;278:20345–20357. doi: 10.1074/jbc.M211604200. [DOI] [PubMed] [Google Scholar]
- 23.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 24.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 25.Garcia SN. Ph.D. dissertaion. La Jolla, CA: University of California at San Diego; 2003. [Google Scholar]
- 26.Boxem M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carotti C, et al. Characterization of recombinant forms of the yeast Gas1 protein and identification of residues essential for glucanosyltransferase activity and folding. Eur J Biochem. 2004;271:3635–3645. doi: 10.1111/j.1432-1033.2004.04297.x. [DOI] [PubMed] [Google Scholar]
- 28.Papaleo E, Fantucci P, Vai M, De Gioia L. Three-dimensional structure of the catalytic domain of the yeast beta-(1,3)-glucan transferase Gas1: A molecular modeling investigation. J Mol Model. 2006;12:237–248. doi: 10.1007/s00894-005-0025-7. [DOI] [PubMed] [Google Scholar]
- 29.Cid VJ, et al. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Meikle PJ, Bonig I, Hoogenraad NJ, Clarke AE, Stone BA. The location of (1→3)-β-glucans in the walls of pollen tubes of Nicotiana alata using a (1→3)-β-glucan-specific monoclonal antibody. Planta. 1991;185:1–8. doi: 10.1007/BF00194507. [DOI] [PubMed] [Google Scholar]
- 32.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Connelly JJ, Wang CL, Sternglanz R. Importance of the Sir3 N terminus and its acetylation for yeast transcriptional silencing. Genetics. 2004;168:547–551. doi: 10.1534/genetics.104.028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stone EM, Pillus L. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J Cell Biol. 1996;135:571–583. doi: 10.1083/jcb.135.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray A, et al. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat Genet. 2003;33:522–526. doi: 10.1038/ng1132. [DOI] [PubMed] [Google Scholar]
- 36.Ai W, Bertram PG, Tsang CK, Chan TF, Zheng XF. Regulation of subtelomeric silencing during stress response. Mol Cell. 2002;10:1295–1305. doi: 10.1016/s1097-2765(02)00695-0. [DOI] [PubMed] [Google Scholar]
- 37.Peng W, Togawa C, Zhang K, Kurdistani SK. Regulators of cellular levels of histone acetylation in Saccharomyces cerevisiae. Genetics. 2008;179:277–289. doi: 10.1534/genetics.107.085068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levy-Wilson B. Glycosylation, ADP-ribosylation, and methylation of Tetrahymena histones. Biochemistry. 1983;22:484–489. doi: 10.1021/bi00271a035. [DOI] [PubMed] [Google Scholar]
- 39.Reeves R, Chang D, Chung SC. Carbohydrate modifications of the high mobility group proteins. Proc Natl Acad Sci USA. 1981;78:6704–6708. doi: 10.1073/pnas.78.11.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 41.Jackson SP, Tjian R. O-glycosylation of eukaryotic transcription factors: Implications for mechanisms of transcriptional regulation. Cell. 1988;55:125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 42.Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 43.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2005. [Google Scholar]
- 44.Darst RP, Garcia SN, Koch MR, Pillus L. Slx5 promotes transcriptional silencing and is required for robust growth in the absence of Sir2. Mol Cell Biol. 2008;28:1361–1372. doi: 10.1128/MCB.01291-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith JS, Brachmann CB, Pillus L, Boeke JD. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 48.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.