Abstract
Paramecium bursaria Chlorella virus-1 is an icosahedrally shaped, 1,900-Å-diameter virus that infects unicellular eukaryotic green algae. A 5-fold symmetric, 3D reconstruction using cryoelectron microscopy images has now shown that the quasiicosahedral virus has a unique vertex, with a pocket on the inside and a spike structure on the outside of the capsid. The pocket might contain enzymes for use in the initial stages of infection. The unique vertex consists of virally coded proteins, some of which have been identified. Comparison of shape, size, and location of the spike with similar features in bacteriophages T4 and P22 suggests that the spike might be a cell-puncturing device. Similar asymmetric features may have been missed in previous analyses of many other viruses that had been assumed to be perfectly icosahedral.
Keywords: 5-fold averaging, cryoelectron microscopy reconstruction, Paramecium bursaria Chlorella virus-1, cell entry, specialized vertex protein
Paramecium bursaria Chlorella virus-1 (PBCV-1) belongs to the genus Chlorovirus of the Phycodnaviridae family. Together with iridoviruses, asfarviruses, and mimivirus, they belong to a group of nucleocytoplasmic large DNA viruses that may have evolved from a common ancestor, predating the divergence of prokaryotes and eukaryotes (1–4). PBCV-1 infects the unicellular eukaryotic green alga Chlorella NC64A by attaching rapidly and irreversibly to the algal cell wall (2, 5). Attachment appears to occur at one of the vertices and might involve hair-like appendages (3). After digestion of the cell wall by virion-packaged enzymes in the vicinity of the point of attachment, the internal viral membrane fuses with the host membrane. Viral DNA and some associated proteins then enter into the cell, leaving the empty capsid on the cell surface (3).
PBCV-1 has a multilayer structure, with an outer glycoprotein capsid that surrounds a lipid bilayer membrane and a dsDNA genome (4, 6). The 331-kbp PBCV-1 genome codes for 11 tRNAs and ≈365 putative proteins (7), of which more than 100 are present in the mature virion. Vp54, a glycoprotein and the major component of the capsid, represents about 40% by weight of the total protein content in the virus. The crystal structure of Vp54 showed that the subunit consists of two 8-stranded, antiparallel β-barrel, “jelly-roll” domains. The monomeric Vp54 forms a trimeric capsomer (a “major capsomer”), which has pseudo-6-fold symmetry, relating the jelly-roll structures (6).
Using cryoelectron microscopy (cryoEM) and assuming icosahedral symmetry (4), it had been shown that the virion has a diameter varying from 1,650 Å, measured along the 2-fold and 3-fold axes, and 1,900 Å, measured along the 5-fold axes. The PBCV-1 glycoprotein shell is composed of 20 triangular units, or “trisymmetrons,” and 12 pentagonal caps, or “pentasymmetrons,” at the 5-fold vertices (8). Each trisymmetron and pentasymmetron consists of a pseudohexagonal array of 66 and 30 trimeric capsomers, respectively. The triangulation number (9) (T) is 169d (h = 7, k = 8), where T represents the number of jelly rolls in the icosahedral asymmetric unit. The crystal structure of PBCV-1 Vp54 had been fitted into the 26-Å-resolution cryoEM density map (6, 10) to generate a pseudoatomic structure of the capsid. However, the 5 jelly rolls surrounding each pentameric vertex form a special capsomer in which, presumably, the monomers contain only 1 jelly roll. Thus, as in the adenovirus structure (11), the capsid protein at the virus vertices differs from the major capsid protein, Vp54.
Results and Discussion
PBCV-1 Has a Unique Vertex Topped by a Spike-like Structure.
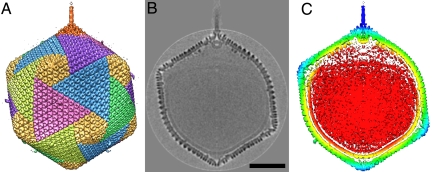
Unprocessed cryoEM images of PBCV-1 usually indicate that the virions have a dense external capsid surrounding a membrane-like feature that, in turn, surrounds a nucleocapsid core. In some images, there is a gap between the enveloped nucleocapsid core and the surrounding capsid at one of the icosahedral pentameric vertices (Fig. S1). A similar gap occurs in mimivirus particles, which also have a unique pentameric vertex (12). Previous reconstructions of PBCV-1 (4, 6) did not show a unique vertex, because of the icosahedral averaging process. Hence, the cryoEM images of PBCV-1 have now been reanalyzed by assuming only the presence of 5-fold symmetry about an axis passing through the center of one selected vertex (see Materials and Methods). The resulting reconstruction clearly showed the capsid with the previously described trisymmetrons and pentasymmetrons (Fig. 1A) and the associated lipid membrane (Fig. 1B). However, 1 of the 2 vertices on the enforced 5-fold axis had a 250-Å-long and 50-Å-wide cylindrical spike (Fig. 1C). This feature can be recognized in selected particles where the spike is in the plane of the micrograph (Fig. S1) (3, 13). The possibility that PBCV-1 might have a special vertex had been predicted earlier in a paper that describes the unique vertex of the dsDNA bacteriophage PRD1 (14).
Fig. 1.
The 5-fold-averaged cryoEM structure of PBCV-1 viewed down a quasi-2-fold axis. (A) Hexagonal arrays of major capsomers form trisymmetrons and pentasymmetrons (yellow). The unique vertex with its spike structure is at the top. Capsomers in neighboring trisymmetrons are related by a 60° rotation, giving rise to the boundary between trisymmetrons. (B) Central cross-section of the cryoEM density. (Scale bar: 500 Å.) (C) The same view as in B but colored radially, with red density being within 680 Å, yellow between 680 and 745 Å, green between 745 and 810 Å, light blue between 810 and 880 Å, and dark blue greater than 880 Å. Note the typical lipid low-density gap surrounding the red nucleocapsid density.
Recognition of the unique vertex of PBCV-1 had been missed for many years on account of assuming icosahedral symmetry in structural analyses (4, 10). After the discovery of a unique vertex for herpes simplex virus (HSV) (15) and mimivirus (12, 16), to the best of our knowledge, PBCV-1 is the third icosahedral, eukaryotic virus that has been recognized to have a unique vertex. It would therefore be possible that many more icosahedral eukaryotic viruses may require unique vertices for the successful execution of their life cycle. However, the complex structure of unique vertices as occurs in most tailed bacteriophages, PRD1, HSV, mimivirus, and now also PBCV-1 will be limited to larger viruses that have enough coding capacity to include genes for the portal and other proteins that are required to create a unique vertex.
The tails of bacteriophages are special assemblies that are used for the highly efficient infection of their hosts (17). Furthermore, bacteriophages provide enzymes for digesting bacterial cell walls, mechanical means for puncturing cell membranes, and a tube for delivering the DNA genome into the host cytoplasm. In addition, the special vertex may be required for genome packaging into preformed proheads, as in HSV (15) or in most bacteriophages (18, 19). Yet another function of special vertices can be to nucleate prohead assembly in some bacteriophages (20). However, whereas tailed bacteriophages use a proteinaceous tail tube, PRD1 appears to form a membranous tube that is about 650 Å long and 140 Å in diameter at the time of infection, probably for genome transfer (21, 22).
Some icosahedral viruses, including tailed bacteriophages, HSV (15, 23), mimivirus (12, 16), and PRD1 (21, 24), have a specialized pentameric vertex that is associated with genomic packaging and/or cell entry. Even the small icosahedral poliovirus has or develops a special pentameric vertex when delivering its genome to its host (25). Thus, it is seems reasonable to assume that the special vertex of PBCV-1 might also be associated with cell entry or infection. The PBCV-1 spike is too thin to be a tube, as is the case for most tailed bacteriophages and PRD1. However, the PBCV-1 spike has the same size and shape as the cell wall-puncturing device of phage T4 (26, 27) and of bacteriophage P22 (28). Thus, the spike of PBCV-1 might have a function similar to that of the T4 cell-puncturing device. Although there is no significant sequence similarity of special vertex proteins belonging to either T4 or PRD1 with any part of the PBCV-1 genome, that does not necessarily imply that there is no structural or functional similarity, because in general, protein tertiary structure changes much slower than the primary amino acid sequences. The PBCV-1 genome is known to enter the host, probably subsequent to fusion of the viral membrane and host plasma membrane while leaving the empty capsid outside the cell (7). By comparison, the function of the PBCV-1 spike therefore may be to puncture the cell wall, and perhaps the membrane, to initiate the fusion process. The density of the PBCV-1 cryoEM reconstruction shows a lower density linking the spike to the unique vertex, suggesting that the spike may be released during the infectious process, as is also the case for bacteriophage T4 (27).
The Unique Vertex Contains Special Capsomers.
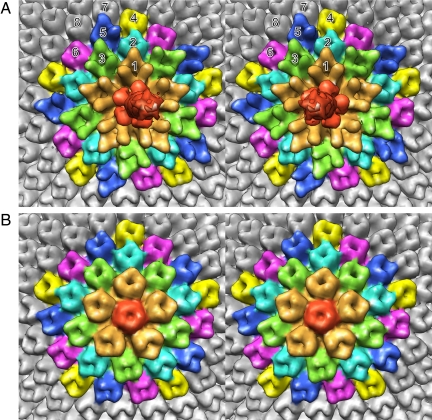
As mentioned above, the capsomers on 11 of the 12 vertices consist, presumably, of 5 single jelly-roll monomers. However, the pentasymmetron at the unique vertex (Fig. 2A) has a different density distribution for the pentameric capsomer than those at the 11 other vertices (Fig. 2B). The pentameric capsomer at the unique vertex binds the spike and is surrounded by 5 peripentonal capsomers. The peripentonal capsomers (no. 1 capsomers in Fig. 2A) have additional density on their surfaces compared with the major capsomers (Vp54 trimers; no. 7 and no. 8 capsomers in Fig. 2A). The 5 peripentonal capsomers at the unique vertex are surrounded by 10 capsomers (no. 2 and no. 3 capsomers in Fig. 2A) that are alternatively similar to the major capsomers and the peripentonal capsomers. These, in turn, are surrounded by 15 capsomers that form the edge of the pentasymmetron. Of these, 10 resemble the major capsomer (no. 4 and no. 6 capsomers in Fig. 2A), and the other 5 (no. 5 capsomers in Fig. 2A) are different (Table S1).
Fig. 2.
Stereodiagrams of the pentasymmetrons at (A) the unique vertex and (B) the 5-fold axis opposite the unique vertex. Note the difference in the density distribution of the pentameric capsomer (red) in A and B. The top end of the narrow spike can also be seen in A. Although all of the major capsomers (Vp54 trimers) surrounding the pentasymmetrons and in the pentasymmetron shown in B have very similar density distributions, this is not the case for the pentasymmetron at the special vertex shown in A. The peripentonal capsomers around the unique vertex are shown in gold and have an extended amino acid insertion. The 10 capsomers surrounding the peripentonal capsomers are colored blue and green. Whereas the green capsomers are similar to the peripentonal capsomers, the blue capsomers are similar to the major capsomers. The capsomers in the outer pentasymmetron ring are colored yellow, dark blue, and pink. The yellow and pink capsomers are similar in structure to the major capsid protein, whereas the dark blue capsomers have a slightly different density distribution.
Large insertions in jelly-roll structures occur invariably between the D and E and between the F and G β-strands in either the first or second jelly-roll fold within the capsid proteins of the nucleocytoplasmic large DNA viruses (Fig. S2). Thus, the additional density in the peripentonal capsomers might correspond to an insertion at one of these loops. BLAST analysis (29) of the 6 PBCV-1-encoded proteins with sequence similarity to Vp54 showed that only the gene product of ORF A622L is larger than Vp54 and has an insertion of 111 residues between β-strands D and E in the first jelly roll of the sequence. This number of amino acids occupies a predicted volume of ≈15,000 A3 (calculated by using the peptide property calculator of A. Chazan, available at www.basic.northwestern.edu/biotools/proteincalc.html), which agrees with the ≈15,500 Å3 extra density of the peripentonal capsomers in comparison with Vp54.
The Virus Has a Partially Empty Pocket.
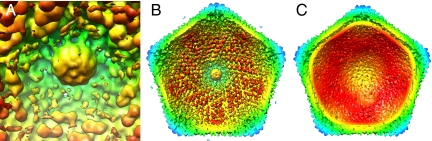
Within the virion, near the pentameric capsomer at the unique vertex, is an additional density (150 Å wide and 100 Å long) that is similar in size and location to the portal proteins in tailed phages, such as in phi29 (30), SPP1 (31), and P22 (Fig. 3A) (32, 33). The PBCV-1 portal-like protein is attached to the peripentonal capsomers and not to the special pentameric capsomer. The portal protein in bacteriophages is a dodecamer. If PBCV-1 also has a 12-fold symmetric portal, then the 5-fold averaging used in the cryoEM reconstruction would blur the boundaries between subunits. Although the role of the PBCV-1 portal-like protein is unclear, in tailed phages the portal protein is probably involved in both DNA packaging and DNA ejection (7).
Fig. 3.
View of features inside the PBCV-1 capsid. All panels are colored as in Fig. 1C. (A) The portal within the capsid at the unique vertex. (B) View of the unique vertex from the inside of the virus. Note the finger proteins at a slightly smaller radius appear red. No finger proteins are associated with the pentasymmetron at the unique vertex. (C) View of the vertex opposite the unique vertex. Note the lack of finger protein features and the low-density gap corresponding to the lipid bilayer membrane roughly in the yellow density.
As reported previously (4), an internal lipid bilayer envelopes the PBCV-1 nucleocapsid. In previous icosahedral reconstructions of PBCV-1, the lipid membrane follows the contour of the capsid. However, the present 5-fold averaged map shows that the lipid membrane departs from icosahedral symmetry in the region near the unique vertex, creating a partially empty pocket between the 5 icosahedral faces surrounding the unique vertex and the membrane (Fig. 1C). The pocket measures ≈325 Å along a radial direction at its widest point. The average density in the pocket is substantially less than that within the nucleocapsid. Cell wall-degrading enzymes are thought to be stored near the unique vertices of tailed bacteriophage (32–34). Thus, the pocket in PBCV-1 may function as a reservoir for enzymes and other proteins that might be excreted through the unique vertex to digest the cell wall of the chlorella cells in the initial stages of infection (3).
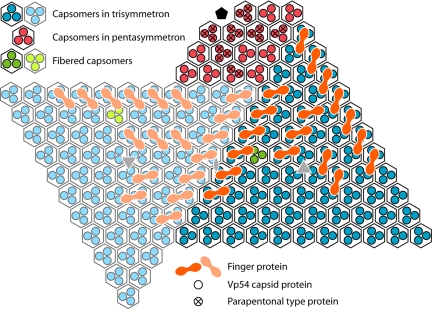
Apart from the general disordered distribution of density within the pocket, a set of well-ordered features line the inside of the viral capsid within the pocket. These features are similar to a class of minor capsid proteins (“finger proteins”) observed in the T = 147 Chilo iridescent virus (CIV) (35). The PBCV-1 finger proteins have 2 distinct globular domains (Fig. 3B): one is associated with the center of an external capsomer, and the other is associated with the gap between 3 neighboring capsomers (Fig. 4). However, in PBCV-1, unlike in CIV, the cryoEM data were only 5-fold-averaged, thereby making it possible to differentiate between finger proteins associated with trisymmetrons around the unique vertex (Fig. 3B) and the absence of finger proteins in the other trisymmetrons (Fig. 3C). It may be that finger proteins prevent the lipid membrane from attaching to the inside of the viral capsid in the vicinity of the unique vertex, thus facilitating the formation of the pocket that contains the molecular “soup” necessary for infection.
Fig. 4.
Diagrammatic organization of capsomers as seen from inside the virus viewed toward the unique vertex. Within each quasihexagonal capsomer, there are 3 Vp54 monomers, each represented by a dot (blue within one trisymmetron, light blue within the neighboring trisymmetron, and red within the pentasymmetron). Some of the capsomers are associated with minor capsid finger proteins, shown in orange in one trisymmetron and a lighter orange in the neighboring trisymmetron.
The Fibered Capsomers Have a Unique Symmetry.
A previously published PBCV-1 cryoEM reconstruction (36) showed that 1 capsomer per icosahedral asymmetric unit (3 per trisymmetron) has a protruding fiber. However, the 5-fold-averaged reconstruction reported here shows that none of the 20 trisymmetrons have all 3 symmetry-related fibers (Fig. S3). Furthermore, some trisymmetrons have an additional type of fibered capsomer (Fig. S4). These fibers might be involved in attaching and orienting the virus to the host during infection.
Conclusions
The major capsid proteins of PRD1, Sulfolobus turreted icosahedral virus, and PBCV-1, infecting prokaryotes, archaea, and eukaryotes, have a “double jelly-roll” fold, suggesting that at least as far as their capsids are concerned, they may have had a common precursor (6, 37–40). Similarly, many or most tailed dsDNA bacteriophages (41, 42) and HSVs (43) use the “HK97” fold for the structure of their major capsid protein, suggesting a common ancestry for these viruses. Yet, it is shown here that there might be some structural and functional relationship between the cell-puncturing device of bacteriophage T4 and the spike structure of PBCV-1, which could indicate perhaps convergent evolution for the development of a mechanical device for penetrating the external cell defenses for viruses that have quite different origins for their major capsid proteins.
The results also demonstrate that global enforcement of icosahedral symmetry in viruses can obscure vital information about the structural properties of virions. Indeed, transient slight deviation from icosahedral symmetry may be as important to the viral life cycle (23, 44) as precise icosahedral symmetry is for viral assembly. Larger viruses, such as PBCV-1, PRD1, HSV, mimivirus, or T4, have the capacity to code for special proteins associated with a special vertex, as is witnessed by the presence of genes that are homologues of the major capsid protein and might be required for spike or tail formation. However, smaller viruses that do not have specialized genes might also have special vertices (25) that are associated with the ends of the genome that are stored for exit when infection is initiated. Nevertheless, the presence of unique features required for cell entry might not be necessary for all viruses. Whereas some viruses, such as PBCV-1 and tailed bacteriophages, remain outside the cell on infection, inserting only their genome through the cell wall or plasma membrane, other viruses merely become unstable within the acid pH of endosomes. Thus, it is likely that many or even most virus structures reported to date might be reexamined to establish the presence of special nonsymmetric features, such as special vertices and attachment fibers.
Materials and Methods
PBCV-1 samples were prepared for cryoEM as described previously (4). Micrographs of frozen-hydrated samples were recorded on Kodak SO-163 film in CM200 and CM300 FEG transmission electron microscopes (Philips). Micrographs were digitized at 7-μm intervals, and the resulting data were interpolated to yield pixels corresponding to 8.0 Å at the specimen. The program RobEM (45) was used to select 5,149 particles from 228 micrographs and to determine their defocus levels by fitting the contrast-transfer functions to the Fourier transforms averaged incoherently over all particle images for each micrograph. The 3D reconstruction was computed by using images that had only been corrected for phase reversals. A similar procedure for producing a 5-fold average density map, described previously (44), was developed by integrating functions with the SPIDER program (46) and a modified version of XMIPP (47). The first stage of the procedure assumed icosahedral symmetry to determine the center position and relative orientation of each recorded particle. The second stage of the procedure assumed only 5-fold symmetry by selecting a unique orientation from the 12 icosahedrally related orientations with respect to an increasingly more asymmetric model as refinement proceeded. The test for selecting the unique orientation from the 12 possible orientations was based on comparing only the density in the vicinity of the unique vertex with each particle projection. Various limits were tried for defining the size and shape of the mask that defined the volume around the unique vertex. After 10 cycles, the choice of orientation of each boxed image had mostly converged. The final resolution of the cryoEM reconstruction was 22-Å resolution, determined as the point at which the Fourier shell correlation coefficient between 2 half-datasets fell below 0.5.
Figures were created by using the program CHIMERA (48).
Supplementary Material
Acknowledgments.
We are grateful to Susan Hafenstein, Ye Xiang, Anastasia Aksyuk, Ping Zhang, and Bärbel Kaufmann for helpful discussions. We thank Carol Greski for help in preparation of the manuscript. The work was supported by National Institutes of Health Grants AI11219 (to M.G.R.), GM32441 (to J.L.V.E.), and GM33050 (to T.S.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The EM density map reported in this paper has been deposited in the EM Databank, http://emdatabank.org (accession no. EMD-1597).
This article contains supporting information online at www.pnas.org/cgi/content/full/0904716106/DCSupplemental.
References
- 1.Dunigan DD, Fitzgerald LA, Van Etten JL. Phycodnaviruses: A peek at genetic diversity. Virus Res. 2006;117:119–132. doi: 10.1016/j.virusres.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 2.Meints RH, Lee K, Burbank DE, Van Etten JL. Infection of a Chlorella-like alga with the virus, PBCV-1: Ultrastructural studies. Virology. 1984;138:341–346. doi: 10.1016/0042-6822(84)90358-1. [DOI] [PubMed] [Google Scholar]
- 3.Van Etten JL, Lane LC, Meints RH. Viruses and viruslike particles of eukaryotic algae. Microbiol Rev. 1991;55:586–620. doi: 10.1128/mr.55.4.586-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan X, et al. Structure and assembly of large lipid-containing dsDNA viruses. Nat Struct Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meints RH, Burbank DE, Van Etten JL, Lamport DT. Properties of the Chlorella receptor for the virus PBCV-1. Virology. 1988;164:15–21. doi: 10.1016/0042-6822(88)90614-9. [DOI] [PubMed] [Google Scholar]
- 6.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamada T, Onimatsu H, Van Etten JL. Chlorella viruses. Adv Virus Res. 2006;66:293–336. doi: 10.1016/S0065-3527(06)66006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrigley NG. An electron microscope study of the structure of Sericesthis iridescent virus. J Gen Virol. 1969;5:123–134. doi: 10.1099/0022-1317-5-1-123. [DOI] [PubMed] [Google Scholar]
- 9.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 10.Simpson AA, Nandhagopal N, Van Etten JL, Rossmann MG. Structural analyses of Phycodnaviridae and Iridoviridae. Acta Crystallogr D. 2003;59:2053–2059. doi: 10.1107/s090744490302225x. [DOI] [PubMed] [Google Scholar]
- 11.Rux JJ, Kuser PR, Burnett RM. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution x-ray crystallographic, molecular modeling, and sequence-based methods. J Virol. 2003;77:9553–9566. doi: 10.1128/JVI.77.17.9553-9566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao C, et al. Structural studies of the giant mimivirus. PLoS Biol. 2009;7:958–966. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onimatsu H, Suganuma K, Uenoyama S, Yamada T. C-terminal repetitive motifs in Vp130 present at the unique vertex of the Chlorovirus capsid are essential for binding to the host Chlorella cell wall. Virology. 2006;353:433–442. doi: 10.1016/j.virol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Stromsten NJ, Bamford DH, Bamford JK. The unique vertex of bacterial virus PRD1 is connected to the viral internal membrane. J Virol. 2003;77:6314–6321. doi: 10.1128/JVI.77.11.6314-6321.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newcomb WW, et al. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75:10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zauberman N, et al. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008;6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Curr Opin Struct Biol. 2007;17:237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Moffitt JR, et al. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun S, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Morais MC, et al. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- 21.Bamford D, Mindich L. Structure of the lipid-containing bacteriophage PRD1: Disruption of wild-type and nonsense mutant phage particles with guanidine hydrochloride. J Virol. 1982;44:1031–1038. doi: 10.1128/jvi.44.3.1031-1038.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grahn AM, Daugelavicius R, Bamford DH. Sequential model of phage PRD1 DNA delivery: Active involvement of the viral membrane. Mol Microbiol. 2002;46:1199–1209. doi: 10.1046/j.1365-2958.2002.03250.x. [DOI] [PubMed] [Google Scholar]
- 23.Cardone G, et al. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361:426–434. doi: 10.1016/j.virol.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karhu NJ, Ziedaite G, Bamford DH, Bamford JK. Efficient DNA packaging of bacteriophage PRD1 requires the unique vertex protein P6. J Virol. 2007;81:2970–2979. doi: 10.1128/JVI.02211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bubeck D, Filman DJ, Hogle JM. Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat Struct Mol Biol. 2005;12:615–618. doi: 10.1038/nsmb955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanamaru S, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 27.Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG. The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol. 2004;14:171–180. doi: 10.1016/j.sbi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Olia AS, Casjens S, Cingolani G. Structure of phage P22 cell envelope-penetrating needle. Nat Struct Mol Biol. 2007;14:1221–1226. doi: 10.1038/nsmb1317. [DOI] [PubMed] [Google Scholar]
- 29.Madden TL, Tatusov RL, Zhang J. Applications of network BLAST server. Methods Enzymol. 1996;266:131–141. doi: 10.1016/s0076-6879(96)66011-x. [DOI] [PubMed] [Google Scholar]
- 30.Simpson AA, et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lebedev AA, et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang J, et al. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14:1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Lander GC, et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 34.Leiman PG, et al. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 35.Yan X, et al. The capsid proteins of a large, icosahedral dsDNA virus. J Mol Biol. 2009;385:1287–1299. doi: 10.1016/j.jmb.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Etten JL. Unusual life style of giant chlorella viruses. Annu Rev Genet. 2003;37:153–195. doi: 10.1146/annurev.genet.37.110801.143915. [DOI] [PubMed] [Google Scholar]
- 37.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 38.Benson SD, Bamford JK, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 39.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khayat R, et al. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci USA. 2005;102:18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morais MC, et al. Conservation of the capsid structure in tailed dsDNA bacteriophages: The pseudoatomic structure of phi29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 42.Wikoff WR, et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 43.Trus BL, et al. The herpes simplex virus procapsid: Structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 44.Hafenstein S, et al. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc Natl Acad Sci USA. 2007;104:6585–6589. doi: 10.1073/pnas.0701574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 46.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 47.Sorzano CO, et al. XMIPP: A new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.