Abstract
Rapid and effective differentiation between normal and cancer cells is an important challenge for the diagnosis and treatment of tumors. Here, we describe an array-based system for identification of normal and cancer cells based on a “chemical nose/tongue” approach that exploits subtle changes in the physicochemical nature of different cell surfaces. Their differential interactions with functionalized nanoparticles are transduced through displacement of a multivalent polymer fluorophore that is quenched when bound to the particle and fluorescent after release. Using this sensing strategy we can rapidly (minutes/seconds) and effectively distinguish (i) different cell types; (ii) normal, cancerous and metastatic human breast cells; and (iii) isogenic normal, cancerous and metastatic murine epithelial cell lines.
Keywords: fluorescence, gold nanoparticle, sensor, conjugated polymer
Each cell type has unique molecular signatures that distinguish between healthy and diseased tissues (1). In the case of cancers, the distinctions between normal vs. tumor and benign vs. metastatic cells are often subtle. The identification of cellular signatures for early cancer cell detection is a major hurdle for cancer therapy; the earlier these signatures can be established, the more effectively they can be treated (2). Cancerous cells are differentiated from noncancerous ones on the basis of intracellular or extracellular (cell surface) biomarkers. Detection methods based on specific recognition of intracellular biomarkers (e.g., DNA/RNA/Proteins) require previous knowledge of specific mutations in DNA/RNA (3) or changes in the regulation of protein expression inside the cells. Similarly, detection methods based on specific recognition of extracellular (cell surface) biomarkers such as histopathology (4), bioimaging (5), antibody arrays require prior knowledge of biomarkers on cell surfaces. Observation of overexpressed antigens (6) on tumor cells using antibody-based platforms have been explored using ELISA (7), surface plasmon resonance (8, 9), nanoparticles (10–13), microcantilevers (14), carbon nanotubes (15, 16), and expression microarrays (17). Antibody arrays provide an effective but complex approach for cancer detection, diagnosis and prognosis (18), however, there is no single marker or a combination of biomarkers that has sufficient sensitivity and specificity to differentiate between normal, cancerous, and metastatic cell types (19). Here, we describe a detection system that is based on selective noncovalent interactions between cell surface components and nanoparticle-based sensor elements that does not require any previous knowledge of intracellular or extracellular biomarkers.
The cell membrane surface consists primarily of a thin layer of amphipathic phospholipids, carbohydrates and many integral membrane proteins. The amount and types of which differ between species and according to function of cells (20, 21). This results into distinct cell membrane composition in different cell types. Therefore, one can predict, however, that there will be physicochemical (i.e., charge, hydrophobicity etc.) differences between cell types and between healthy and cancerous cells. Such physicochemical differences could potentially be detected by an array-based “chemical nose” approach that relies on selective interactions between multiple reporter elements and the target cell.
In the chemical nose approach, an array of different sensors is used where every element in the sensor array responds to a number of different chemicals or analytes (22). A distinct pattern of responses produced from a set of sensors in the array provide a fingerprint that allows classification and identification of the analyte (23). The collection of sensors should contain chemical diversity to respond to largest possible cross-section of analytes. The specific interactions involved between the reporter elements and the analyte are noncovalent and reversible. This approach provides an alternative to “lock–key” specific recognition (24) and has been used to detect metal ions (25), volatile agents (26), aromatic amines (27), amino acids (28, 29), and carbohydrates (25). In recent research we have demonstrated that the displacement of fluorescent polymers from differentially functionalized gold nanoparticles with concomitant restoration of fluorescence provides an effective array-based method for the identification of proteins (30). More recently, we have shown that this methodology can be used to differentiate between bacterial species and even between different strains of the same species (31). We report here a particle-polymer array that distinguishes between healthy, cancerous and metastatic human breast cells, and differentiates isogenic healthy and transformed cells.
Results and Discussion
Our detection system is based on conjugates between 3 structurally related cationic gold nanoparticles (NP1–NP3, Fig. 1A and Fig. S1) and the poly(para-phenyleneethynylene) (PPE) polymer PPE-CO2 featuring charge multivalency (32) and molecular wire properties (33) (Fig. 1A). In these noncovalent conjugates, the nanoparticle quenches the fluorescence of the polymer. The interactions between nanoparticles and anionic polymers are noncovalent, and predominantly electrostatic. When mammalian cells were incubated with these nanoparticle-polymer complexes, there is competitive binding between nanoparticle-polymer complexes and cell types (Fig. 1B). Because of their cationic surface, nanoparticles are expected to interact with phospholipids, membrane proteins and carbohydrates of the cell surface through both electrostatic and hydrophobic interactions. These interactions are responsible for displacement of the fluorophore polymer from the nanoparticle-polymer complexes generating a fluorescence response. The nanoparticles are expected to possess different affinities for dissimilar cell surfaces depending on cell membrane composition and surface of nanoparticles. Selective displacement of the polymer from the particle by the cell surface regenerates fluorescence, transducing the binding event in a “turn on” fashion.
Fig. 1.
Molecular structures of nanoparticles and polymers, and schematic of fluorophore displacement cell detection array. (A) Molecular structures of the cationic gold nanoparticles (NP1-NP3) and the fluorescent polymer (PPECO2). (B) Displacement of quenched fluorescent polymer (dark green strips, fluorescence off; light green strips, fluorescence on) by cell (in blue) with concomitant restoration of fluorescence.
The complex stability constants (KS) and association stoichiometries (n) for the nparticle-polymer dyads were obtained through nonlinear least-squares curve-fitting analysis (34). Complex stabilities vary within 1 order of magnitude (ΔΔG ≈ 4.5 kJ·mol−1), and the binding stoichiometry ranges from 2.5 for NP2 to 0.9 for NP3 (Fig. S2). After determining the saturation point for fluorescence quenching (Fig. S3), the appropriate stoichiometries of particle and polymer were mixed in 5 mM phosphate buffer (pH = 7.4) to yield nanoparticle-PPECO2 complexes with a final concentration of polymer of 100 nM and of nanoparticles 10–40 nM. The complexes of PPECO2 and NP1-3 were then incubated with different cell types to determine changes in fluorescence intensities. We observed increases and decreases in fluorescence intensities depending on the cell type and the nature of nanoparticle-polymer complexes. Increased fluorescence intensities are due to the displacement of the PPECO2 polymer from the NP-PPECO2 complexes by cell surfaces (Fig. 1B), whereas decreases in the fluorescence intensities are due to the quenching of the residual PPECO2 fluorescence by the cell surfaces. These differences in the fluorescence patterns depend on the cell type and are reproducible. We have performed array-based s sensing using 9 gold nanoparticles that possess different head groups and interact differently with polymers (Fig. S3a). We studied their interactions with the different cell types listed in Table 1, focusing on which particle set can best differentiate between different particles. (see below). From studies, we have observed the maximum differentiation grouping using 3 nanoparticles NP1-NP3, as established through jackknifed analysis (Fig. S3b).
Table 1.
Origin and nature of the normal, cancerous and metastatic cell lines used in this study.
| Cell line | Liver | HepG2 | Cancerous |
|---|---|---|---|
| Human | Cervix | HeLa | Cancerous |
| Testis | NT2 | Cancerous | |
| Breast | MCF10A | Normal immortalized | |
| MCF-7 | Cancerous | ||
| MDA-MB-231 | Metastatic | ||
| Mouse | BALB/c mice (breast) | CDBgeo | Normal immortalized |
| TD | Cancerous | ||
| V14 | Metastatic |
Detection of Differences in Cell Types.
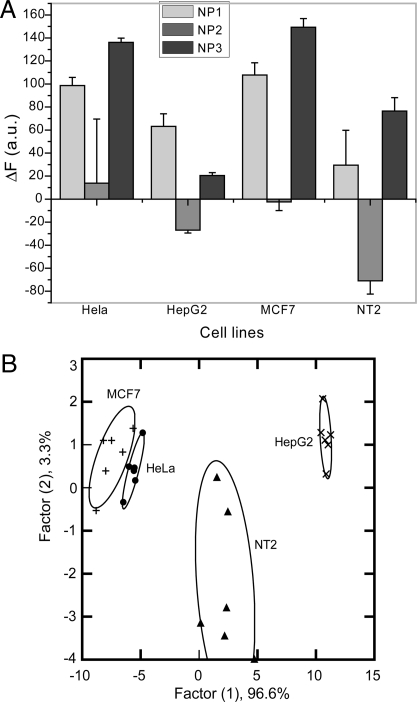
As an initial test of our method we used 4 different types of human cancer cells: HeLa (Cervical), HepG2 (Liver), NT2 (Testis) and MCF-7 (Breast). Fig. 2A presents the change in the fluorescence response for the nanoparticle-polymer supramolecular complexes upon addition of the different cancer cell types. Linear Discriminant Analysis (LDA) was used to statistically characterize the fluorescence changes. This analysis reduced the size of the training matrix (3 nanoparticles × 4 cell types × 6 replicates) and transformed them into canonical factors that are linear combinations of the response patterns (3 factors × 4 cell types × 6 replicates). The 2 canonical factors contain 96.6% and 3.3% of the variation, respectively as shown in Fig. 2B. In this plot, each point represents the response pattern for a single cell type to the NP-PPECO2 sensor array. In the canonical fluorescence response patterns, the different cell types are clustered into 4 nonoverlapping groups (95% level confidence ellipses) (Fig. 2B) with standard deviation of <5%. These initial results validate our ability to differentiate cancer cell types phenotypically based on their surface properties.
Fig. 2.
Detection of human cancerous cell lines. (A) Change in fluorescence intensities (F − F0) for 4 different cancer cell lines HeLa (Cervical), MCF7 (Breast), HepG2 (Liver) and NT2 (Testes) using nanoparticle-polymer supramolecular complexes. Each value is average of 6 parallel measurements. (B) Canonical score plot for the two factors of simplified fluorescence response patterns obtained with NP–PPECO2 assembly arrays against different mammalian cell types. The canonical scores were calculated by LDA for the identification of 4 cell lines.
Detection of Normal/Cancerous and Metastatic Cells.
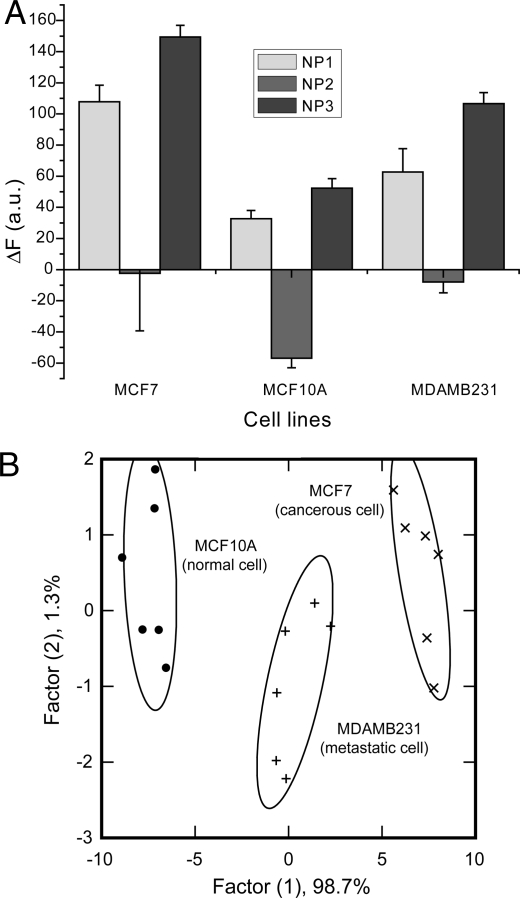
An important issue in cancer therapy is assessing whether tissue/cells are healthy, or either benign or metastatic tumors. We chose 3 different human breast cell lines to test our sensor array in this application: MCF10A a normal breast cell line, MCF7 a cancerous but nonmetastatic cell line, whereas MDA-MB-231 is a metastatic cancer cell line. The 3 cell lines show differential fluorescence patterns (Fig. 3A); LDA of their response indicates a 100% accuracy of detection (Fig. 3B).
Fig. 3.
Detection of normal, cancerous and metastatic human breast cells. (A) Change in fluorescence intensities (F − F0) for 3 breast cell lines of different nature MCF10A (normal), MCF-7 (cancer) and MDA-MB231 (metastatic) using nanoparticle-polymer supramolecular complexes. Each value is average of 6 parallel measurements. (B) Canonical score plot for the first two factors of simplified fluorescence response patterns obtained with NP–PPECO2 assembly arrays against different mammalian cell types.
Detection of Isogenic Cell Types.
The above studies suggest that we can differentiate normal, cancerous and metastatic cell types with our sensor array. Each of the 3 cell lines, however, came from different individuals. To provide a test bed where individual-to-individual variation is not present, we used 3 isogenic cell lines, CDBgeo, TD, and V14 cells. Due to their high genetic similarity, isogenic cells are expected to present a particularly stringent test for detection assays. Each of these isogenic cells was developed from BALB/c mice, and therefore possesses the same genotypic background. CDBgeo cells were prepared by retroviral infection with a marker gene encoding the fusion of β-galactosidase and neomycin resistance. These cells exhibit normal outgrowths when transplanted into mammary fat pads (35). The TD cells were prepared by treating CDBgeo-cells with 10 ng/mL TGF-β for 14 days. Withdrawal for 5 passages resulted in a persistent epithelial to mesenchymal transformation: Tumorogenic growth resulted when transplanted. The V14 cell line was established from a primary mammary tumor arising in BALB/c-Trp-53+/− mice. The cells lack p53 protein and form aggressive tumors that are locally invasive in mice (36). Fig. 4A presents the change in fluorescence intensities of 3 isogenic cell types toward nanoparticle-polymer complexes. The differential response indicates that these supramolecular complexes can effectively differentiate isogenic cell types. LDA classifies the cell types into 3 distinct clusters with 2 canonical factors containing 83.0% and 17.0% of the variation, with 100% identification accuracy among these isogenic cell types (Fig. 4B). Taken together, these studies indicate that our method rapidly and effectively differentiates cell lines based on cell type and disease state.
Fig. 4.
Detection of Isogenic cell types. (A) Change in fluorescence intensities (F − F0) for 3 cell lines of same genotype CDBgeo, TD cell and V14 using nanoparticle-polymer supramolecular complexes. Each value is average of 6 parallel measurements. (B) Canonical score plot for the first two factors of simplified fluorescence response patterns obtained with NP–PPECO2 assembly arrays against different mammalian cell types.
The efficacy of our approach indicates that there are distinct phenotypic differences in the physicochemical properties of cells. One question that arises is whether there is a response that is generally indicative of whether a cell is normal or cancerous. Metaanalysis of our studies using LDA indicates that normal epithelial cell lines CDBgeo and MCF10A were overlapping (Fig. 5a) even though both of these cell lines were isolated from mouse and human respectively. Likewise the metastatic murine (V14) and human MDMBA-231 metastatic cell lines were clustered, indicating a potential correlation between cell surface properties and disease states of cells.
Fig. 5.
Detection of normal and cancerous cell lines. (A) Changes in fluorescence intensities (F − F0) of noncancerous and cancerous cell types using nanoparticle-polymer supramolecular complexes. Each value is average of 6 parallel measurements. (B) Canonical score plot for the first two factors of simplified fluorescence response patterns obtained with NP–PPECO2 assembly arrays against different normal and cancerous cell types. Further differentiation was observed in the third dimension, allowing discrimination each of the species.
In summary, we have developed a rapid and effective array-based approach to differentiate between normal and cancerous cell lines. Significantly, full differentiation was achieved using only 3 nanoparticle-polymer dyads, indicating that a simple sensor array has ample diagnostic capacity when exposed to mammalian cells. These systems have the potential to help us understand the physical changes that occur on the surfaces of cells in various disease states. Taken together, “nose” based sensor systems are a fundamentally new way of looking into biodiagnostic, biophysical and surface science processes involving cell surfaces.
Materials and Methods
Nanoparticles (25, 37, 38) (SI Text, Scheme S1, and Fig. S1) and polymers (39) were synthesized as reported previously. All of the cells except MCF10A, CDBgeo, TD and V14 were grown in DMEM media supplemented with 10% FBS and 1% antibiotics in T75 flasks. NT2 cell line was obtained from R. T. Zoeller (Department or Biology, University of Massachusetts, Amherst, MA). CDBgeo, TD and V14 cells were grown in DMEM-F12 media supplemented with 2% ABS, 25 mM Hepes, 10 μg/mL insulin, 5 ng/mL EGF, 15 μg/mL gentamycin. Cells were washed with DPBS buffer, trypsinized with 1× trypsin and collected in the DMEM media. Fluorescence titration experiments determined the complexation between nanoparticles and PPECO2. Fluorescence intensity changes at 465 nm were recorded with an excitation wavelength of 430 nm. Polymer and stoichiometric amounts of NP1–NP3, as determined by the fluorescence titration study were diluted with phosphate buffer (5 mm, pH 7.4) to solutions with a final polymer concentration of 100 nM. Each solution (200 μL) was placed into a well on the micro plate. After incubation for 30 min, the fluorescence intensity at 465 nm was recorded with an excitation wavelength of 430 nm. Next, 100 μL of cell suspension (20,000 cells) was added to each well. After incubation for another 30 min, the fluorescence intensity at 465 nm was measured again. The fluorescence intensity before addition of the cells was subtracted from that obtained after addition of the cells to record the overall fluorescence response (DI) (Tables S1–S3). This process was completed for all cell lines to generate 6 replicates of each that was subjected to a classical linear discriminant analysis (LDA) using SYSTAT (version 11.0). Each cell line possesses a unique fluorescence response data with the NP-PPECO2 complex array, because cell interaction with the NP-PPECO2 complex array depends on the cell surface characteristic. Therefore, for each cell, we tested its fluorescence responses against 3 NP-PPECO2 adduct 6 times, generating (i) 3 × 6 × 4 matrix for 4 different cancer cell lines HeLa (Cervical), MCF7 (Breast), HepG2 (Liver) and NT2 (Testes) for Fig. 2, (ii) 3 × 6 × 3 matrix for 3 breast cell lines of different nature MCF10A (normal), MCF-7 (cancer) and MDA-MB231 (metastatic) for Fig. 3, and (iii) 3 × 6 × 3 matrix for 3 cell lines of same genotype CDBgeo, TD cell and V14 for Fig. 4. The raw data obtained were subjected to Linear Discriminant Analysis (LDA) (40, 41) to maximize the ratio between-class variance to the within-class variance, thus differentiate the fluorescence response patterns of the NP-PPECO2 system the cell targets. This analysis reduced the size of the training matrix and transformed them into canonical factors that are linear combination of the fluorescence response patterns (i) 2 factors × 6 replicates × 4 cell, (ii) 2 factors × 6 replicates × 3 cells, and (iii) 2 factors × 6 replicates × 3 cells, respectively. The canonical factors contain different percentage of variation and two of them were plotted in 2D as shown in Figs. 1B and 4B. In a blind experiment, the rates of fluorescence patterns of new case were first converted to canonical scores using discriminate functions established on training samples. Then, Mahalanobis distances [the distance of a case to the centroid of a group in a multidimensional space, in the current case it is 2-dimensional (42, 43)] of the new case to the centroid of respective groups (normal or cancerous or metastatic cells) of training samples were calculated. The new case was assigned to the group with shortest Mahalanobis distance. This processing protocol was performed on the SYSTAT 11 program, allowing the assignment of cells to specific groups.
Supplementary Material
Acknowledgments.
We thank Professor R. Thomas Zoeller for providing the NT2 cell lines. This work was supported by the National Science Foundation Center for Hierarchical Manufacturing at the University of Massachusetts Nanoscale Science and Engineering Center and National Science Foundation Grant DMI-0531171; and National Institutes of Health Grant GM077173; Allergy and Infectious Grant AI073425; Grants R01-CA095164 and R01-CA105452 (to D.J.J.); and Department of Energy Grant DE-FG02-04ER46141 (to U.H.F.B., R.L.P., I.-B.K. and V.M.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900975106/DCSupplemental.
References
- 1.Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001;2:698–704. doi: 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- 2.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 3.Jen J, Wu L, Sidransky D. An overview on the isolation and analysis of circulating tumor DNA in plasma and serum. Ann N Y Acad Sci. 2000;906:8–12. doi: 10.1111/j.1749-6632.2000.tb06581.x. [DOI] [PubMed] [Google Scholar]
- 4.Skvara H, Teban L, Fiebiger M, Binder M, Kittler H. Limitations of dermoscopy in the recognition of melanoma. Arch Dermatol. 2005;141:155–160. doi: 10.1001/archderm.141.2.155. [DOI] [PubMed] [Google Scholar]
- 5.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 6.Borrebaeck C. Antibody microarray-based oncoproteomics. Expert Opin Biol Ther. 2006;6:838. doi: 10.1517/14712598.6.8.833. [DOI] [PubMed] [Google Scholar]
- 7.Ward AM, Catto JWF, Hamdy FC. Prostate specific antigen: Biology, biochemistry and available commercial assays. Ann Clin Biochem. 2001;38:633–651. doi: 10.1258/0004563011901055. [DOI] [PubMed] [Google Scholar]
- 8.Chou SF, Hsu WL, Hwang JM, Chen CY. Development of an immunosensor for human ferritin, a nonspecific tumor marker, based on surface plasmon resonance. Biosens Bioelectron. 2004;19:999–1005. doi: 10.1016/j.bios.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Alivisatos P. The use of nanocrystals in biological detection. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 10.Soukka T, et al. Supersensitive time-resolved immunofluorometric assay of free prostate-specific antigen with nanoparticle label technology. Clin Chem. 2001;47:1269–1278. [PubMed] [Google Scholar]
- 11.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 12.Wu G, et al. Bioassay of prostate-specific antigen (PSA) using microcantilevers. Nat Biotechnol. 2001;19:856–860. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- 13.Chen RJ, et al. Noncovalent functionalization of carbon nanotubes for highly specific electronic biosensors. Proc Natl Acad Sci USA. 2003;100:4984–4989. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen RJ, et al. An investigation of the mechanisms of electronic sensing of protein adsorption on carbon nanotube devices. J Am Chem Soc. 2004;126:1563–1568. doi: 10.1021/ja038702m. [DOI] [PubMed] [Google Scholar]
- 15.Wang WU, Chen C, Lin KH, Fang Y, Lieber CM. Label-free detection of small-molecule-protein interactions by using nanowire nanosensors. Proc Natl Acad Sci USA. 2005;102:3208–3212. doi: 10.1073/pnas.0406368102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Y, Wei Q, Park H, Lieber CM. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 17.Campagnolo C, et al. Real-Time, label-free monitoring of tumor antigen and serum antibody interactions. J Biochem Biophys Methods. 2004;61:283–298. doi: 10.1016/j.jbbm.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Wingren C, Borrebaeck CA. Antibody microarray analysis of directly labelled complex proteomes. Curr Opin Biotech. 2008;18:55–61. doi: 10.1016/j.copbio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Carbayo M. Antibody arrays: Technical considerations and clinical applications in cancer. Clin Chem. 2006;52:1651–1659. doi: 10.1373/clinchem.2005.059592. [DOI] [PubMed] [Google Scholar]
- 20.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 21.Alberts B, Johnson A, Lewis J. Molecular Biology of the Cell. 4th Ed. New York: Garland Publishing; 2002. [Google Scholar]
- 22.Lavigne JL, Anslyn EV. Sensing a paradigm shift in the field of molecular recognition: From selective to differential receptors. Angew Chem Int Ed. 2001;40:3118–3130. doi: 10.1002/1521-3773(20010903)40:17<3118::AID-ANIE3118>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 23.Albert KJ, et al. Cross-reactive chemical sensor arrays. Chem Rev. 2000;100:2595–2626. doi: 10.1021/cr980102w. [DOI] [PubMed] [Google Scholar]
- 24.Wright AT, Anslyn EV. Differential receptor arrays and assays for solution-based molecular recognition. Chem Soc Rev. 2006;35:14–28. doi: 10.1039/b505518k. [DOI] [PubMed] [Google Scholar]
- 25.Lee JW, Lee JS, Chang YT. Colorimetric identification of carbohydrates by a pH indicator/pH change inducer ensemble. Angew Chem Int Ed. 2006;45:6485–6487. doi: 10.1002/anie.200602055. [DOI] [PubMed] [Google Scholar]
- 26.Rakow NA, Suslick KS. A colorimetric sensor array for odour visualization. Nature. 200;406:710–713. doi: 10.1038/35021028. [DOI] [PubMed] [Google Scholar]
- 27.Greene NT, Shimizu KD. Colorimetric molecularly imprinted polymer sensor array using dye displacement. J Am Chem Soc. 2005;127:5695–5700. doi: 10.1021/ja0468022. [DOI] [PubMed] [Google Scholar]
- 28.Folmer-Andersen JF, Kitamura M, Anslyn EV. Pattern-based discrimination of enantiomeric and structurally similar amino acids: An optical mimic of the mammalian taste response. J Am Chem Soc. 2006;128:5652–5653. doi: 10.1021/ja061313i. [DOI] [PubMed] [Google Scholar]
- 29.Buryak A, Severin K. A chemosensor array for the colorimetric identification of 20 natural amino acids. J Am Chem Soc. 2005;127:3700–3701. doi: 10.1021/ja042363v. [DOI] [PubMed] [Google Scholar]
- 30.You CC, et al. Detection and identification of proteins using nanoparticle-fluorescent polymer “chemical nose” sensors. Nat Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 31.Phillips RL, Miranda OR, You CC, Rotello VM, Bunz UH. Rapid and efficient identification of bacteria using gold-nanoparticle-poly(para-phenyleneethynylene) constructs. Angew Chem Int Ed. 2008;47:2590–2594. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 32.Kim IB, Erdogan B, Wilson JN, Bunz UHF. Sugar-poly(para-phenylene ethynylene) conjugates as sensory materials: Efficient quenching by Hg2+ and Pb2+ Chem Eur J. 2004;10:6247–6254. doi: 10.1002/chem.200400788. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Q, Swager TM. Fluorescent chemosensors based on energy migration in conjugated polymers: The molecular wire approach to increased sensitivity. J Am Chem Soc. 1995;117:12593–12602. [Google Scholar]
- 34.Phillips RL, et al. Gold nanoparticle-PPE constructs as biomolecular material mimics: Understanding the electrostatic and hydrophobic interactions. Soft Matter. 2009;5:607–612. [Google Scholar]
- 35.Deugnier MA, et al. Isolation of mouse mammary epithelial progenitor cells with basal characteristics from the Comma-Dbeta cell line. Dev Biol. 2006;293:414–425. doi: 10.1016/j.ydbio.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Blackburn AC, et al. Loss of heterozygosity occurs via mitotic recombination in Trp53+/− mice and associates with mammary tumor susceptibility of the BALB/c strain. Cancer Res. 2004;64:5140–5147. doi: 10.1158/0008-5472.CAN-03-3435. [DOI] [PubMed] [Google Scholar]
- 37.Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. Synthesis of thiol-derivatised gold nanoparticles in a two-phase liquid–liquid system. J Chem Soc Chem Commun. 1994:801–802. [Google Scholar]
- 38.Hostetler MJ, Templeton AC, Murray RW. Dynamics of place-exchange reactions on monolayer-protected gold cluster molecules. Langmuir. 1999;15:3782–3789. [Google Scholar]
- 39.Kim IB, Dunkhorst A, Gilbert J, Bunz Sensing of lead ions by a carboxylate-substituted PPE: Multivalency effects. Macromolecules. 2005;38:4560–4562. [Google Scholar]
- 40.Engelman L. Systat 11.0, Statistics. Chicago: SYSTAT Software; 2004. pp. i301–i358. [Google Scholar]
- 41.Jurs PC, Bakken GA, McClelland HE. Computational methods for the analysis of chemical sensor array data from volatile analytes. Chem Rev. 2000;100:2649–2678. doi: 10.1021/cr9800964. [DOI] [PubMed] [Google Scholar]
- 42.Mahalanobis PC. On the generalised distance in statistics. Proc Natl Inst Sci India. 1936;2:49–55. [Google Scholar]
- 43.Gnanadesikan R, Kettenring JR. Robust estimates, residuals, and outlier detection with multiresponse data. Biometrics. 1972;28:81–124. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.