Abstract
Protein aggregation is a hallmark of a large and diverse number of conformational diseases. Molecular chaperones of the Hsp40 family (Escherichia coli DnaJ homologs) recognize misfolded disease proteins and suppress the accumulation of toxic protein species. Type I Hsp40s are very potent at suppressing protein aggregation and facilitating the refolding of damaged proteins. Yet, the molecular mechanism for the recognition of nonnative polypeptides by Type I Hsp40s such as yeast Ydj1 is not clear. Here we computationally identify a unique motif that is selectively recognized by Ydj1p. The motif is characterized by the consensus sequence GX[LMQ]{P}X{P}{CIMPVW}, where [XY] denotes either X or Y and {XY} denotes neither X nor Y. We further verify the validity of the motif by site-directed mutagenesis and show that substrate binding by Ydj1 requires recognition of this motif. A yeast proteome screen revealed that many proteins contain more than one stretch of residues that contain the motif and are separated by varying numbers of amino acids. In light of our results, we propose a 2-site peptide-binding model and a plausible mechanism of peptide presentation by Ydj1p to the chaperones of the Hsp70 family. Based on our results, and given that Ydj1p and its human ortholog Hdj2 are functionally interchangeable, we hypothesize that our results can be extended to understanding human diseases.
Keywords: molecular chaperones, protein aggregation, protein misfolding, conformational diseases, peptide recognition
Molecular chaperones facilitate cellular protein metabolism by promoting protein folding (1, 2), suppression of protein aggregation, and protein degradation (3, 4). In addition, Hsp70s and Hsp40s also play a major role in suppressing the formation of toxic protein species that cause neurodegeneration (5). Enhancing the function of chaperones could therefore provide an avenue for the treatment of protein-misfolding diseases. Hsp70s are ubiquitous chaperones (6) whose cellular functions (7, 8) are specified through interaction with their cochaperones of the Hsp40 family (9, 10). These chaperones consist of an N-terminal nucleotide-binding domain (NBD) and a C-terminal substrate-binding domain (SBD) connected by a short linker peptide (11). ATP hydrolysis in the NBD of Hsp70 drives a cycle of conformational changes in its SBD, subsequently leading to binding and release of protein substrates (12). Recent structural studies have added significantly to our understanding of J cochaperone binding and regulation of Hsp70s (13). However, the mechanism by which Hsp40s bind and deliver substrates to Hsp70 remains a mystery.
The Hsp40 family (proteins with molecular weights of ≈40 kDa) is large and structurally and functionally diverse. Members of this family are grouped into 3 subtypes (types I, II, and III) (14) based on the degree of conservation of their domains with those of Escherichia coli DnaJ. Type I Hsp40 proteins are considered descendents of E. coli DnaJ (14). They are characterized by the presence of a J domain, a glycine-phenylalanine (G/F)-rich region, 2 zinc-finger-like motifs, and a conserved carboxyl-terminal domain (CTD). Type II Hsp40s are different from Type I in that they lack the zinc-finger-like motifs. Type III Hsp40s have only conserved J domains. Hsp40s also function as “molecular chaperones” (15–17) involved in various steps of protein maturation starting from biogenesis (2) and assembly (1), through translocation (18) to degradation (3, 4). Besides, these proteins contribute to other basic and complex functions such as prion propagation (19), amyloid plaque formation (20), cell-cycle regulation, and mitogenic signal transduction (21). Although Type I and Type II Hsp40s are shown to bind nonnative polypeptides, they contain unique protein modules for chaperone activity (22) and have different tertiary structures (23, 24). Thus, it is possible that different Hsp40s use unique mechanisms for binding and delivery of nonnative polypeptides to Hsp70.
The focus of our study is on substrate binding by Ydj1p, a chaperone and representative member of the Type I Hsp40 family of proteins in yeast Saccharomyces cerevisiae (14, 25, 26). Ydj1p is essential for normal cell growth and survival of yeast from heat stress and is involved in protein translocation across membrane, protein folding, and protein degradation. Ydj1p is shown to influence the assembly-state of endogenous yeast prions and it influences the aggregation of fragments of huntingtin that are expressed in yeast (27, 28). Ydj1 and its human homolog Hdj2 are functionally interchangeable, and hence studies on Ydj1p function will provide insights into Hdj2 function in human cells and neurons.
Ydj1p is made up of an N-terminal J domain located adjacent to a highly flexible G/F-rich region that is followed by a zinc-finger-like region (ZFLR) and conserved carboxyl-terminal domains I and II (CTDI and CTDII, respectively). Polypeptide-bound Ydj1p forms transient complexes with Hsp70s, and presents the nonnative polypeptides to Hsp70s for subsequent protein folding (29, 30). Structure-based mutagenesis studies indicate that a conserved hydrophobic pocket located on the peptide-binding fragment of Type I Hsp40s, that is also found in Type II Hsp40s, plays a critical role in its molecular chaperone activity by mediating interactions with its substrates (31, 32). In addition, there is also evidence that suggests that a conserved zinc-finger-like domain (33, 34) and a C-terminal farnesyl moiety participate in polypeptide binding and presentation by Type I Hsp40 (34, 35). The combined action of these different polypeptide-binding sites is proposed to confer the ability of Type I Hsp40s to bind a broad range of protein conformers (5, 27, 36, 37). However, the molecular mechanism for recognition of nonnative substrates by Type I Hsp40s such as Ydj1 remains obscure.
Here, we use computational and biochemical tools to investigate the sequence specificity and structural basis for substrate binding to the conserved hydrophobic depression in CTDI of the Type I Hsp40 Ydj1p. We analyze known Ydj1p substrates to identify patterns of amino acids that explain the mechanism of peptide binding by this chaperone. We find that interacting peptides follow a consensus given by G[LMQ]L{P}X{P}{CIPMVW}, where [XY] represents either X or Y and {XY} is negation of [XY]. We experimentally demonstrate that the consensus is critical for substrate binding by Ydj1p. We further explore the physiological relevance of this consensus by screening the yeast proteome computationally and find that proteins from different families display such consensus sequences. We experimentally validate the genuineness of the motif and find that binding peptides in the prion domain of the yeast prion Rnq1 by Ydj1p is drastically mitigated by mutations at a critical position in the motif.
Results
Recognition Motif: Sufficient Condition for Binding to Ydj1p.
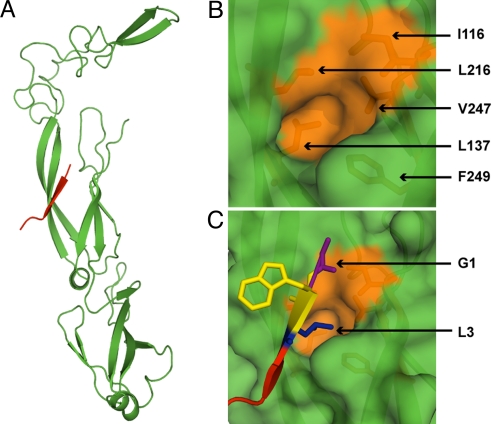
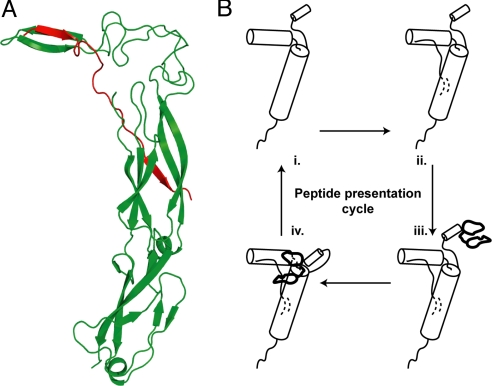
The crystal structure of a fragment of Ydj1—a peptide-binding domain in CTDI and a 7-aa-long peptide (GWLYEIS)—suggests that this region of Type I Hsp40s contains a 2-stranded antiparallel beta sheet connected by a short helix that forms a β-strand with polypeptide substrates (26, 38) (Fig. 1A). Binding is mediated by a hydrophobic pocket formed by residues I116, L137, L216, V247, and F249 of the peptide-binding fragment of Ydj1p (Fig. 1B), into which the side chain of the third residue on the heptapeptide is inserted (Fig. 1C).
Fig. 1.
Peptide-binding site on Ydj1. (A) The structure (PDB ID: 1NLT) of the peptide binding fragment of Ydj1p (green) in complex with a peptide substrate GWLYEIS (red). (B) Surface representation of the active pocket on Ydj1p. Residues forming the hydrophobic pocket are colored orange and shown in stick representation. The peptide fragment is removed for a clear view of the binding pocket. (C) Orientation of peptide substrate near the active site. The side chain of L3 fits the active pocket. Residue numbering on the peptide conforms to the description in Results.
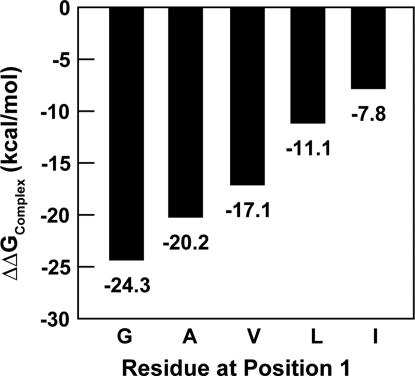
To understand sequence specificity of peptide-binding to this region of Ydj1p, we computationally analyzed the conformation of residues at each position on the peptide by using Medusa (39, 40), a suite of programs developed in-house (see Methods). Because the first residue of the peptide is in a hydrophobic environment (Fig. 1 B and C), we studied the effect of increasing hydrophobicity of this residue on binding. We observed that glycine is preferred energetically over other residues at the first position on the peptide (Fig. 2). We used a heptaglutamine peptide for this analysis for 2 reasons: (i) Because Ydj1p modulates polyglutamine aggregation and toxicity in vivo(5), use of a heptaglutamine peptide would be physiologically relevant; and (ii) the contribution of rotameric states of the side chains from different residues can be normalized. We ignored the residue at the second position of the peptide in our study because the side chain of any residue at that position points away from Ydj1p and, hence, does not contribute to total energy of binding (Fig. 1C). For computational ease, we used alanine at this position throughout our analyses. The residue at the third position docks its side chain into the binding pocket on Ydj1p (Fig. 1C). To arrive at the consensus, we computationally estimated the ΔΔGBinding (see Methods) of each heptapeptide from the entire Rnq prion domain. We found that the peptides with binding scores comparable (≥75%) to that of the wild-type peptide [wild-type defined as GWLYEIS that was cocrystallized with Ydj1 (PDB ID: 1NLT)], had either L, M, or Q in the third position. These results are in agreement with experimental binding studies recently reported by Summers et al. (34). Upon modifying the cutoff for comparison to 50% (or better), we observed an enrichment of N in the third position. Besides, a 50% cutoff also resulted in an enrichment of S and T in the third position, which is in agreement with the trends observed by Li and Sha using phage display library screens (38).
Fig. 2.
Computational mutation analysis of position 1. Effect of side-chain-length on binding: Increasing length of the side chain decreases binding affinity linearly.
To account for the remaining 4 positions on the peptide, we rationally designed the polypeptide backbone and assigned scores for the binding of all possible 7-mers starting with GAL to Ydj1p. We compared the scores of different complexes with that of the native peptide crystallized along with Ydj1p. The results from our analysis suggest that peptides binding Ydj1p follow a consensus given by G{P}[LMQ]{P}X{P}{CIMPVW}, where {XY} denotes any residue other than X and Y; [XY] denotes either X or Y.
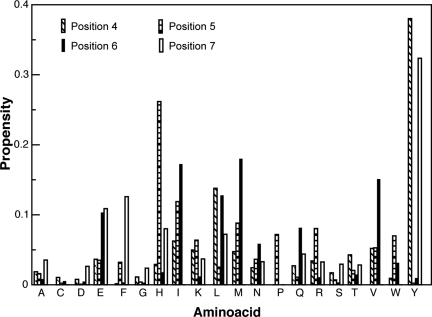
From the results reported thus far, it is clear that besides strand formation, the residues at positions 1 and 3 on the peptide mediate binding to Ydj1p. Furthermore, as beta-strand formation involves nonspecific backbone–backbone interactions, it follows that the residues at positions 1 and 3 confer specificity and increase stability of the peptide-bound form of Ydj1p. To estimate the contribution of other residues, we calculated the propensities of occurrence of each amino acid in each position on the peptide. For this analysis, we considered only those complexes with an estimated energy of binding (ΔΔG) ≥−30 kcal/mol (binding energy with native peptide: −32.69 kcal/mol). From our peptide design, we observed a striking enrichment of tyrosine at positions 4 and 7, that of histidine at position 5, and methionine at position 6 (Fig. 3 and Fig. S1). The position-specific propensity scores are reported in Table S1.
Fig. 3.
Propensity of amino acids at different positions on the heptapeptide. The bar chart shows propensities of occurrence of each amino acid at positions 4, 5, 6, and 7. The first 3 positions were fixed as GAL for this study (see Results).
Experimental Validation of the Identified Consensus.
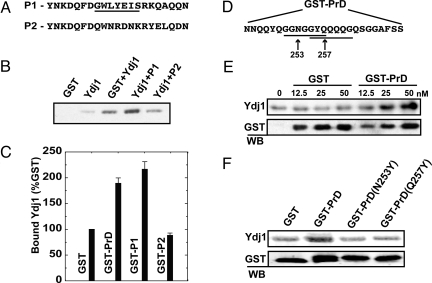
To validate the consensus sequence, we designed a peptide 21-aa long (Fig. 4A), containing exactly 1 heptapeptide representative of the consensus. We fused this peptide to GST and estimated direct binding to recombinant Ydj1p in vitro by GST pull-down. The GST-fusion peptide enhanced binding with Ydj1p ≈2-fold over background (Fig. 4 B lanes 3 and 4, and C). To establish the importance of the motif in mediating Ydj1p binding, we designed a scrambled peptide (Fig. 4A) that did not contain any representative binding motifs. We found that the scrambled peptide reduced Ydj1-binding down to background (Fig. 4 B lanes 3 and 5, and C). These results show that the identified consensus sequence mediates interaction between Ydj1p and its substrate polypeptide.
Fig. 4.
Experimental validation of Ydj1-binding motif. (A) (P1) 21-aa fragment designed with exactly 1 consensus-binding sequence (underlined). (P2) 21-aa scrambled peptide with no consensus-binding sequence. (B) The protein complex was isolated with glutathione-conjugated beads and resolved by SDS/PAGE. (C) P1 showed a 2-fold increase in binding compared with GST alone, whereas signals from binding to P2 remained at the background. (D) A 25-aa fragment from the Rnq1 prion domain (amino acids 245–269) contains 2 overlapping Ydj1-binding motifs (underlined). To demonstrate direct binding, this fragment was fused to GST and incubated with purified Ydj1. Similar experiments were conducted to demonstrate binding of the PrD peptide to Ydj1p. (E) The GST-PrD fusion was titrated with 50 nM Ydj1 to determine the concentration at which binding between GST-PrD and Ydj1 is saturated. Data points were generated by quantifying bound Ydj1 after Western immunoblotting and normalizing to background (no GST protein). (F) Point mutations were made in the third position of 2 putative Ydj1-binding motifs in GST-PrD. Ydj1 was incubated with GST alone, GST-PrD, or GST-PrD with these point-mutants at a 1:2 ratio to determine binding affinity.
Biological Relevance of the Motif.
To verify the biological relevance of the unique motif, we screened the yeast proteome for sequences that conform to the identified motif. The yeast proteome was obtained as FASTA sequences from the National Center for Biotechnology Information genome database (ftp://ftp.ncbi.nih.gov/genomes). We mined all of the yeast protein sequences for all possible sequence combinations spanned by the identified motif. Furthermore, we searched for repeats of the identified motif separated by n residues where n varies from 5 to 20. The hits obtained from the yeast proteome scan indicated that Hsp60s, Hsp70s, Hsp82s, Hsp90s and other family members; ATPases, GTPases and hydrolases; GTPase-activating proteins (GAPs) and guanine exchange factors (GEFs); and proteins involved in peptide synthesis, sorting, and trafficking express more than one stretch of residues following the consensus. In addition to the proteins mentioned above, we noted the presence of the recognition motif in certain yeast prions like Sup35 (41, 42), Ure2 (28, 43), and Rnq1 (34), which are known to interact with Ydj1p.
Experimental Validation of Physiologically Relevant Motifs.
To confirm that our consensus sequence represents a bona fide substrate-binding motif, we experimentally examined interaction between Ydj1p and a model substrate containing this motif. Ydj1p binds the Gln/Asn-rich prion domain (amino acids 153–405) from the yeast protein Rnq1 (34). This domain possesses several motifs that match the consensus sequence described above. Rather than mutate every potential binding site in the prion domain, we focused our analysis on a 25-aa peptide that Ydj1 recognizes in a peptide array from Rnq1 (Fig. 4D). We predicted that interaction between Ydj1 and this fragment would depend on one of the 2 binding-motifs in this peptide. To test this hypothesis, we fused the prion domain peptide to GST and assessed direct binding in vitro with recombinant Ydj1 by GST pull-down. The GST-prion fusion protein showed almost 2-fold enhancement in binding with Ydj1 over GST alone (Fig. 4C). Furthermore, this interaction saturates at higher GST-prion concentrations (Fig. 4E). To test the dependence of Ydj1-binding on these motifs, the third position in each motif was mutated to tyrosine. This residue should sterically hinder interactions with the hydrophobic peptide-binding pocket of Ydj1. Indeed, mutating the third position to tyrosine in either motif reduced Ydj1-binding down to background (Fig. 4F lanes 3 and 4). Thus, interaction between Ydj1 and this prion substrate is mediated at least in part through discrete motifs located throughout a Gln/Asn-rich domain.
Two-Site Peptide-Binding Model.
In an attempt to understand the role of Ydj1p in modulation of the assembly status of yeast prions, we analyzed a set of 34 sequences of peptides from the prion domains of yeast prions (Table S2) to see whether they carry the motif presented above. Each of these peptide sequences is nearly 25-aa long. We considered a window of 7 residues and slid the entire peptide over the active pocket of Ydj1p, one window at a time. Our aim was to identify stretches of residues on yeast prions that showed energetically favorable binding to the chaperone. For most of the yeast prion sequences, we observed that the most favorable binding patch on the peptide conforms to the novel motif. We did not observe the motif on some of the yeast prions, which indicates the scope for alternate binding mechanisms. Interestingly, in a set of a significant number of yeast prion sequences, we observed more than one such sequence competent of binding to Ydj1p. Typically, 2 such sequence motifs were separated by 8 to 11 residues. Furthermore, hits obtained from the yeast proteome screen had multiple motifs separated by varying number of amino acids. In light of the fact that the ZFLR of Ydj1p is vital in suppression of prion toxicity (34) and transfer of substrates for Hsp40 to Hsp70 (33), we propose a peptide-binding and presentation hypothesis discussed below and outlined in Fig. 5.
Fig. 5.
Two-site peptide-binding and presentation model. (A) A nonnative polypeptide is extended between the characterized peptide-binding domain and the ZFLR of Ydj1p. (B) The schematic shows the peptide-presentation cycle where a Ydj1p monomer (i) binds a polypeptide (ii), interacts with Hsp70 via its J-domain (iii), and eventually delivers the peptide to the Hsp70 (iv).
Discussion
The purpose of this study was to understand the mechanism of chaperone activity of the Type I Hsp40s, and the yeast chaperone Ydj1p was studied as a model. We used a combination of computational and experimental techniques to analyze the peptide binding characteristics of the chaperone and propose a unique motif characterized by the consensus sequence G{P}[LMQ]{P}X{P}{CIPMVW}. The presence of this motif in many prion sequences as well as in a foray of proteins in the yeast proteome suggests that it could be a sufficient condition to estimate, by looking at its sequence, whether a peptide binds Ydj1p. Our results can further be extrapolated to other members of the Hsp40 family and additional heat-shock proteins. Thus, our study lays a path to understanding the mechanism of action of molecular chaperones. Our methodology can further be extended to predict the sequence of a peptide of least-binding-energy to a given protein with a well-characterized active site.
We conducted a proteome-wide scan to look for the presence of the identified recognition motif in the yeast proteome. It being a short peptide, a possible argument would be that the probability of finding a consensus that is restrained at only 2 positions is very high. If proteins bearing this consensus are prone to binding by Ydj1, such sequences must be preserved by nature in the intrinsic folds of proteins and be exposed in their unfolded conformations [see Pelham (44) for supporting argument]. By using the dictionary of protein secondary structure (45), analyses of the extent-of-buriedness of residues forming the consensus show that proteins in general bear this consensus well within their folded forms, whereas those displaying a rather solvent-exposed stretch of residues are cochaperones and other proteins supporting chaperone activity of Ydj1p (such as HSP60, HSP70, GAPs, and GEFs).
Thus, binding of Ydj1 to the consensus motif, when exposed in protein folding intermediates, will assist in facilitation of folding. In addition, because Ydj1p is a homodimer, it is possible that one monomer in the dimer can bind a substrate protein, whereas the other monomer recognizes solvent-exposed recognition motifs in its partner chaperones. Dual recognition of nonnative proteins and partner chaperones may serve to increase the productivity of substrate transfer between chaperones and thereby increase protein folding efficiency.
Full-length proteins have many occurrences of the recognition motif, making it difficult to study the effect of mutagenesis of one such motif on binding to Ydj1p. We did not prefer to consider shorter peptides with 1 or 2 motifs from full-length proteins either, because such peptides could be conformationally biased toward their native folded forms. Hence, we chose to conduct mutagenesis on prion peptides to test our hypothesis. Moreover, it is relevant to work with amyloid-like protein aggregates as they show direct implication in many diseases. Our results clearly support the presence of a recognition motif that acts as a sufficient condition for peptide recognition by Ydj1p.
Given that Ydj1p forms dimers in solution, we hypothesized that the chaperone binds peptides with 2 sequence motifs by stretching the peptide between 2 monomers. However, we observed that Ydj1p monomers are equally competent in regulating prion propagation in yeast (34). Hence, dimer formation is a possibility but not a necessity for Ydj1p function. Furthermore, there is evidence that the zinc-finger-like domains in Ydj1p act in binding some peptides (33, 46). Recent small angle X-ray scattering studies by Ramos et al. (24) show that the space between 2 monomers in Ydj1p is not void, suggesting that it may not be implicated in docking HSP70s during peptide binding, as speculated earlier (47). Putting the above pieces together in light of our results, we propose the following mechanism of peptide presentation to HSP70s. Because the zinc-finger-like domains act in peptide binding, it is logical that this region cooperates with CTDI in polypeptide binding. Peptides containing 2 sequence motifs may bind Ydj1 by forming a psi-loop motif with the zinc-finger-like region and the peptide-binding site on CTDI (Fig. 5A), enabling the predominantly hydrophobic peptide fragment to be presented to HSP70 (Fig. 5B). This hypothesis fits the result that Ydj1p monomers are sufficient to regulate yeast prion propagation. Furthermore, it explains the involvement of zinc-finger-like regions in peptide binding and the presence of 2 or more recognition motifs in yeast prions. We believe that these speculations are good starting points to unravel the mechanisms of peptide binding and presentation by molecular chaperones of the HSP40 family.
Conclusions
Peptides display position-specific binding patterns to Ydj1p. A unique motif GX{P}[LMQ]X{P}XX{P}X{CIMPVW} was identified as a sufficient condition for peptide binding to Ydj1p. Yeast proteome screen revealed that the motif is biologically relevant and could be observed in many proteins involved in cochaperone activity, with Ydj1p and others involved in critical cellular events. We believe that our results can be extrapolated to human chaperones. Similar studies on the homologous human chaperone Hdj2 may give enough leads for understanding the molecular basis of conformational diseases.
Methods
Estimation of Binding Energy of Complexes.
We used Medusa (39, 40) to rationally design and estimate the binding free energy of a complex (ΔΔG). Proteins were modeled by using the United Atom model, which includes all heavy atoms and polar hydrogen atoms. The free energy of binding (ΔΔG) was computed by using
where ΔΔGBinding is the free energy of binding of a polypeptide to Ydj1p, ΔGComplex is the total energy of the protein-peptide complex, ΔGYdj1 is the energy of the protein without the peptide bound to it, and ΔGPolypeptide is the energy of the peptide in its free form. To model each mutant computationally, the native residues were substituted with the target residues and optimal packing was achieved by sampling the side-chain rotameric states. The lowest energy from multiple optimization runs was used to compute the stability of the mutant. The stability change upon mutation (ΔΔG) was obtained by subtracting the energy of the wild-type protein from that of the mutant. This methodology for estimating ΔΔGBinding was benchmarked by Yin et al. (48). A detailed description of how the energy terms are computed is given by Ding et al. (39). The units of ΔG values obtained from Medusa are kcal/mol.
Fixed-Backbone Custom-Redesign.
Due to the stochastic nature of the redesign algorithm described by Ding et al. (39), we performed 20 runs for each mutation and calculated the average total energy of the system. Such average ΔG values were used to compute the binding free energy by using the equation given in the previous section.
GST Fusion Proteins.
Recombinant GST-fusions were generated by PCR amplifying a 75-nt fragment (732–807) from Rnq1 with a 5′ BamHI cut site and short-flanking sequence encoding 3 alanines. A XhoI cut site and stop codon were fused to the 3′ end. This fragment was ligated in-frame into a pGEX5x-1 vector and confirmed by sequencing. Mutations were generated in this backbone by Quikchange mutagenesis (Stratagene). GST-fusion proteins were purified from E. coli with a 5-ml GSTrap FF affinity column (Amersham) by using the manufacturer's recommended protocol, and dialyzed into GST buffer (20 mM Hepes pH 7.4, 150 mM NaCl).
GST Pull-Down Assay.
GST-fusion proteins were incubated in GST buffer with recombinant Ydj1p purified as previously described (9). Sepharose beads conjugated with glutathione (Amersham Biosciences) were incubated in the reaction and washed 3 times with GST buffer. Beads were resuspended in sample buffer, boiled, and analyzed by SDS/PAGE followed by Western immunoblotting for Ydj1 and GST (Sigma).
Supplementary Material
Acknowledgments.
We thank Dr. Feng Ding for useful discussions and interesting suggestions. This work was supported in part by National Institutes of Health Grant R01 GM080742.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900746106/DCSupplemental.
References
- 1.Cheng MY, et al. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989;337:620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- 2.Craig EA, Gambill BD, Nelson RJ. Heat shock proteins: Molecular chaperones of protein biogenesis. Microbiol Rev. 1993;57:402–414. doi: 10.1128/mr.57.2.402-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 4.Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 5.Meriin AB, et al. Huntingtin toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James P, Pfund C, Craig EA. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 7.Bukau KT, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 8.Young JC, Barral JM, Hartl FU. More than folding: Localized functions of cytosolic chaperones. Trends Biochem Sci. 2003;28:541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Cyr DM, Langer T, Douglas MG. DnaJ-like proteins: Molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 10.Cyr DM, Lu X, Douglas MG. Regulation of HSP70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- 11.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the hsc70 chaperone. Mol Cell. 2005;20:513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayer MP, et al. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struc Biol. 2000;7:586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J, et al. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: Conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgopoulos C, Welch WJ. Role of the major heat shock proteins as molecular chaperones. Annu Rev Cell Biol. 1993;9:601–634. doi: 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- 16.Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 17.Michels AA, et al. Hsp70 and hsp40 chaperone activities in the cytoplasm and the nucleus of mammalian cells. J Biol Chem. 1997;272:33283–33289. doi: 10.1074/jbc.272.52.33283. [DOI] [PubMed] [Google Scholar]
- 18.Chirico WJ, Waters MG, Blobel G. 70k heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- 19.Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic HSP70 in Yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fonte V, et al. Interaction of intracellular beta amyloid peptide with chaperone proteins. Proc Natl Acad Sci USA. 2002;99:9439–9444. doi: 10.1073/pnas.152313999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helmbrecht K, Zeise E, Rensing L. Chaperones in cell cycle regulation and mitogenic signal transduction: A review. Cell Prolif. 2000;33:341–365. doi: 10.1046/j.1365-2184.2000.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan C-Y, Lee S, Ren H-Y, Cyr DM. Exchangeable chaperone modules contribute to specification of type I and type II hsp40 cellular function. Mol Biol Cell. 2004;15:761–773. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borges JC, Hannes F, Craievich AF, Ramos CHI. Low-resolution structural study of two human Hsp40 chaperones in solution. HJA1 from subfamily A and HJB4 from subfamily B, have different quaternary structures. J Biol Chem. 2005;280:13671–13681. doi: 10.1074/jbc.M408349200. [DOI] [PubMed] [Google Scholar]
- 24.Ramos CH, Oliveira CLP, Yang-Fan C, Torriani IL, Cyr DM. Conserved central domains control the quaternary structure of Type I and Type II HSP40 molecular chaperones. J Mol Biol. 2008;383:155–166. doi: 10.1016/j.jmb.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caplan AJ, Douglas MG. Characterization of YDJ1: A yeast homologue of the bacterial DnaJ protein. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Qian X, Sha B. The crystal structure of the yeast HSP40 Ydj1 complexed with its peptide substrate. Structure. 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the Yeast model. J Biol Chem. 2005;280:22809–22818. doi: 10.1074/jbc.M500390200. [DOI] [PubMed] [Google Scholar]
- 28.Lian H-Y, et al. Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J Biol Chem. 2007;282:11931–11940. doi: 10.1074/jbc.M606856200. [DOI] [PubMed] [Google Scholar]
- 29.Laufen T, et al. Mechanism of regulation of HSP70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan C-Y, Lee S, Cyr DM. Mechanisms for regulation of HSP70 function by HSP40. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Sha B. Structure-based mutagenesis studies of the peptide substrate binding fragment of type I heat-shock protein 40. Biochem J. 2005;386:453–460. doi: 10.1042/BJ20041050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S, Fan C-Y, Younger JM, Ren H-Y, Cyr DM. Identification of essential residues in the type II hsp40 Sis1 that function in polypeptide binding. J Biol Chem. 2002;277:21675–21682. doi: 10.1074/jbc.M111075200. [DOI] [PubMed] [Google Scholar]
- 33.Fan C-Y, Ren H-Y, Lee P, Caplan AJ, Cyr DM. The type I hsp40 zinc finger-like region is required for hsp70 to capture non-native polypeptides from Ydj1. J Biol Chem. 2005;280:695–702. doi: 10.1074/jbc.M410645200. [DOI] [PubMed] [Google Scholar]
- 34.Summers DW, Douglas PM, Ren HY, Cyr DM. The type I HSP40 YDJ1 utilizes a farnesyl moiety and zinc finger like-region to suppress prion toxicity. J Biol Chem. 2009;284:3628–3639. doi: 10.1074/jbc.M807369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flom GA, Lemieszek M, Fortunato EA, Johnson JL. Farnesylation of Ydj1 is required for in vivo interaction with hsp90 client proteins. Mol Biol Cell. 2008;19:5249–5258. doi: 10.1091/mbc.E08-04-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muchowski PJ, et al. HSP70 and HSP40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakahira H, Breuer P, Hayer-Hartl MK, Hartl FU. Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. Proc Natl Acad Sci USA. 2002;99:16412–16418. doi: 10.1073/pnas.182426899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Sha B. Peptide substrate identification for Yeast HSP40 Ydj1 by screening the phage display library. Biol Proced Online. 2004;6:204–208. doi: 10.1251/bpo90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Comput Biol. 2006;2 doi: 10.1371/journal.pcbi.0020085. 0725–0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15:1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 41.Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 2006;25:822–833. doi: 10.1038/sj.emboj.7600985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagriantsev SN, Gracheva EO, Richmond RE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19:2433–2443. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savistchenko J, Krzewska J, Fay N, Melki R. Molecular chaperones and the assembly of the prion URE2p in vitro. J Biol Chem. 2008;283:15732–15739. doi: 10.1074/jbc.M800728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelham HRB. Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell. 1986;46:959–961. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- 45.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 46.Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y, Li J, Jin Z, Fu Z, Sha BD. The crystal structure of the C-terminal fragment of yeast Hsp40 Ydj1 reveals novel dimerization motif for Hsp40. J Mol Biol. 2005;346:1005–1011. doi: 10.1016/j.jmb.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 48.Yin S, Ding F, Dokholyan NV. Eris: An automated estimator of protein stability. Nat Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.