Abstract
New experiences can trigger changes in gene expression in the brain. To understand this phenomenon better, we studied zebra finches hearing playbacks of birdsong. Earlier research had shown that initial playbacks of a novel song transiently increase the ZENK (ZIF-268, EGR1, NGFIA, KROX-24) mRNA in the auditory forebrain, but the response selectively habituates after repetition of the stimulus. Here, using DNA microarray analysis, we show that novel song exposure induces rapid changes in thousands of RNAs, with even more RNAs decreasing than increasing. Habituation training leads to the emergence of a different gene expression profile a day later, accompanied by loss of essentially all of the rapid “novel” molecular responses. The novel molecular profile is characterized by increases in genes involved in transcription and RNA processing and decreases in ion channels and putative noncoding RNAs. The “habituated” profile is dominated by changes in genes for mitochondrial proteins. A parallel proteomic analysis [2-dimensional difference gel electrophoresis (2D-DIGE) and sequencing by mass spectrometry] also detected changes in mitochondrial proteins, and direct enzyme assay demonstrated changes in both complexes I and IV in the habituated state. Thus a natural experience, in this case hearing the sound of birdsong, can lead to major shifts in energetics and macromolecular metabolism in higher centers in the brain.
Keywords: habituation, microarray, mitochondria, proteomic, songbird
The genome was once regarded as a passive agent in brain function, directing brain development but having little role in the moment-to-moment operation of the mature brain. Through the widespread application of technologies for measuring gene expression in experimental model systems, however, it now appears that changing social conditions, perceptual experience, and behavioral activity can all result in rapid changes in the expression of specific genes in the brain (1, 2). The challenge ahead is to understand the links between gene expression and natural experience, behavior, and cognition.
A striking example for study is the phenomenon of song response habituation in songbirds (3). In one of the first demonstrations of brain gene activation in response to a natural experience, Mello and colleagues (4) showed that the sound of another bird singing triggers a transient increase in expression of the ZENK gene (ZIF-268, EGR1, NGFIA, KROX-24) in an auditory/associative region of the songbird forebrain. This was not simply a sensory or an auditory response, as its magnitude showed clear categorical discrimination (conspecific song > heterospecific song > noise > pure tones). Further research showed that the same song stimulus would either activate ZENK expression or not, depending on the recent history of exposure to the stimulus and its context. After the stimulus was repeated for a few hours the ZENK RNA declined back to its initial starting level (5, 6). Subsequent presentations no longer elicited a response, but presentation of a different song (6) or even of the same song in a different spatial or visual context (7) activated the response once again. Stimulus-specific ZENK response habituation requires the initial activation of the ERK signaling pathway (8). Habituation is not simply the loss of all neural responses to the auditory stimulus, as neurons in the auditory forebrain still fire in response to the familiar song (albeit at a somewhat lower rate compared to when the song is first novel) (9–11). These changing neural responses may be related to shifts in how the singer is perceived in the context of territorial and colonial social life (3, 12, 13).
Studies of song response habituation have focused on a single gene, ZENK. However, other genes are likely to be involved as well, as studies in other systems have indicated that experience can affect the expression of many genes in the brain (e.g., refs. 14–16). Moreover, ZENK itself encodes a transcription factor protein that regulates expression of other target genes. What happens after the ZENK response itself has habituated? Do all other molecular responses to the song also habituate? Is habituation marked by a change in the basal complement of RNAs and proteins? Or is ZENK gene habituation countered by a new and different molecular response to the now-familiar song? Functional characterization of the broader profile of gene responses at different stages in the development of habituation might provide insight into the underlying cellular logic of the process as a whole.
With new resources available for comprehensive analysis of zebra finch genes and proteins (17), we now address these questions experimentally. To increase the power and depth of our analysis, in our primary experiment we measured both RNAs (via microarray hybridizations) and proteins (via 2-dimensional gel electrophoresis followed by mass-spectroscopy-based sequencing of individual spots). Recognizing the current limits of gene characterization and annotation in the zebra finch, we developed a statistical analysis that focused more on broad functional themes associated with groups of genes as opposed to the detailed characterization of individual gene products. Our analysis led to a specific functional prediction that song habituation is accompanied by slow changes in energy metabolism in the auditory forebrain, a prediction we then validated by direct enzyme assay. The results suggest that experience leads to large discrete shifts in the organization of regulatory networks that manage energy flow in the brain.
Results
Design of Combined Microarray and Proteomic Analysis.
Our initial experiment compared the auditory forebrain under 3 conditions: hearing a song for the first time (“novel”), hearing the same song again a day after development of habituation (“habituated”), and hearing no song (“silence”). We focused on an operationally defined brain region we call the auditory lobule (AL), which is easy to dissect quickly as a unit (18) and is enriched for auditory response areas including caudomedial nidopallium and mesopallium (NCM and CMM). Using this dissection, previous biochemical studies have demonstrated changes in ERK enzyme activation (18), caspase-3 release (19), and ZENK mRNA levels (8) in response to song playbacks, all of which habituate after song repetition. The AL is considered functionally analogous to auditory and association cortex and includes a diversity of cell types including neurons, glia, and endothelial cells. Molecular signaling processes in this region are essential for successful juvenile song tutoring (20).
Immediately after the final stimulus period, half of the birds in each group were euthanized for dissection of AL followed by RNA extraction. This time point captures the peak of the ZENK mRNA change as defined previously (5, 6). The other half of each group was euthanized 1 h later (90 min after stimulus onset), followed by protein extraction from dissected AL. This time point represents the peak of the ZENK protein response (21). This design is summarized schematically in Fig. 1. Video analysis (Fig. 2) confirmed the difference between the novel and habituated groups in their behavioral responses to the song as described in previous studies (8, 13).
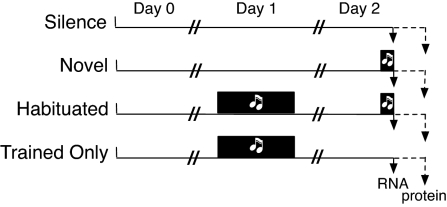
Fig. 1.
Experimental treatment groups. Following a balanced design, each bird was placed alone in a sound isolation chamber on the afternoon of day 0. On day 1, birds in the habituated and trained-only groups heard repeated playback of the song stimulus for 3 h. On day 2, birds in the novel and habituated groups heard the song stimulus for 30 min. The initial combined microarray and proteomic analysis focused just on the first 3 groups (silence, novel, and habituated). A subsequent microarray experiment compared a separate trained-only group matched to a new habituated group collected at the same time. New cohorts representing all 4 groups were then prepared and assayed for mitochondrial enzyme activities.
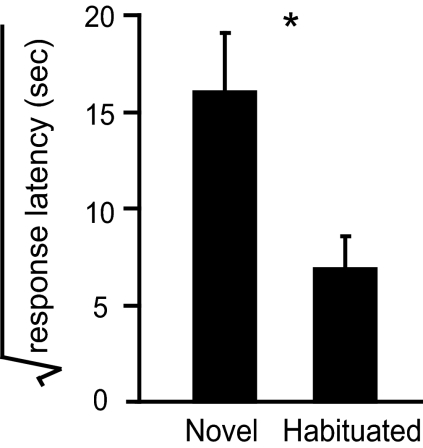
Fig. 2.
Behavioral validation of treatment groups. The birds from novel and habituated groups used in the combined microarray/proteomics experiment (n = 24) were videotaped during the test song presentation period and their responses to the onset of the song stimulus were scored as described (13) (SI). Upon onset of song playback, zebra finches arrest other behaviors and sit silently for periods of seconds to many minutes as though listening intently to the song. The duration from first song onset until the resumption of other visible activities is termed “response latency.” The data were square-root transformed to approximate normality. The birds in the habituated group had significantly shorter response latencies than those in the novel group (*, P < 0.01, Student's t test). Error bars represent SEM.
Microarray Analysis Distinguishes Each Group.
For RNA analysis we used the microarray resources of the Songbird Neurogenomics Initiative to measure the RNAs that hybridize to ≈18,000 cDNA probes (see Table 4 in ref. 17). These are “nonredundant” in that their sequences are nonoverlapping, although we have estimated that 15−35% may derive from genes in common (e.g., different splice forms or different segments of long transcripts). By mixed model ANOVA, 4,341 probes reported a significant difference across the 3 groups (false discovery rate, FDR < 0.05). t tests were then used to identify the RNAs significantly regulated in the novel or the habituated group compared to the silence group, and these were then classified according to the direction of change relative to the silence group (Fig. 3A) [see also supporting information (SI) Table S1].
Fig. 3.
Distinct gene sets define 4 major response patterns. (A) Venn diagram of differential expression. After pairwise comparisons of habituated and novel groups against the silence group, significant probes (Student's t test, FDR P < 0.05) were classified according to direction of change (up ↑ or down ↓) relative to the silence group. Probes present in multiple classes are indicated by overlaps, with the number of probes specific to each distinct subdivision indicated (see also Table S1). (B) Principal component analysis (PCA) of biological samples, using all probes on each array; axes represent the top 3 principal components (P1, P2, P3), which capture 54% of the overall variance. (C) PCA using only probes significant by ANOVA at FDR P < 0.05 (n = 4,341); the top 3 principal components capture 71% of the overall variance. Star, habituated group; circle, novel group; square, silence group; each symbol represents 1 bird. Note that birds naturally group according to treatment.
A total of 616 RNAs showed significant differences in the novel group compared to the silence group, with 153 RNAs increasing and an even larger number (463) decreasing. Moreover, most of these responses appeared to habituate, as 399 of these RNAs (65%) were not significantly different in the habituated versus silence comparison. Surprisingly, however, the habituated group was even more distinct from the silence group with changes in 2,923 additional RNAs not observed in the novel–silence comparison. In total, 3,140 probes (17% of the array) showed significant changes in the habituated group relative to the silence group (Fig. 3A). Principal components analysis (PCA) independently showed that the 3 treatment groups have distinguishable transcriptional profiles, with the habituated group even more readily separated than the novel and silence groups (Fig. 3 B and C). We corroborated our general conclusions with in situ hybridization measurements for a random subset of probes in a new set of birds (Fig. S1A).
Changes in Abundant Proteins Also Distinguish the Groups.
The protein extracts from each bird were analyzed using the 2-dimensional difference gel electrophoresis (2D-DIGE) approach (22), where samples from 2 birds were labeled with different dyes and run on the same gel along with a third common reference sample. In total, 9 gels were thus used to analyze the 18 birds in the analysis. A total of 1,394 spots were resolved across all 3 treatment groups (see examples in Fig. S2). A total of 113 (8% of detected proteins) varied by treatment, using a liberal significance criterion (ANOVA P < 0.1) to maximize the pool size. By Student's t test against the silence group, 93 spots displayed differential abundance in the novel group (37 up, 56 down, with changes from −1.76-fold to +1.59-fold), and 93 spots were different in the habituated group (41 up, 52 down, with changes from −1.72-fold to +1.79-fold). Only 17 spots were common to both novel and habituated groups (9 up in both, 8 down in both). Thus by protein composition and RNA content, the habituated group had a molecular profile that was quite different from that of both the silence and the novel groups. Although the 2D-DIGE analysis has considerably less statistical power than the microarray analysis, both approaches indicate that exposure to novel song initiates a wide array of molecular changes in the AL. Most of these initial changes habituate after song repetition, yet habituation leads to a new and different set of molecular changes evident a day later.
The Habituated Profile Is a Slow Response to Habituation Training.
Two different explanations could account for the appearance of the new molecular changes in the habituated treatment group. One possibility is that they developed slowly after the initial training experience and are independent of any stimulus presentation on the final testing day. Alternatively, they could represent a new and different molecular response to the final presentation of the now-familiar stimulus. To distinguish between these alternatives, we conducted an additional microarray comparison between 2 new groups of birds (n = 6 each group), using the same analysis procedures as above. One group was equivalent to the previous habituated group. The other (trained only) received the same initial training exposures as the habituated group, but then heard only silence on the following day before being killed. In the statistical comparison, no RNAs were different between the 2 groups at our standard significance threshold (FDR P = 0.05). Hence the habituation-specific RNA profile appears to be a stable consequence of the previous days of training and not a newly acquired response to acute presentation of the now-familiar song.
Functional Signatures from Microarray Analysis.
These results show that hearing a song playback can trigger changes in thousands of gene products in the AL, some increasing but at least as many decreasing. Moreover, the results reveal the existence of distinct molecular states in the tissue, one representing the resting state in silence, another the state reached shortly after initial song exposure, and a third in place a day after song habituation training. To tease out the larger functional significance of these molecular differences, we applied a statistical analysis (23) based on the Gene Ontology (GO) annotation terms associated with each expression profile. GO annotations were derived primarily from the annotations of apparent chicken orthologs as described (17). Overall, 72% of the regulated sequences had some degree of annotation. The remainder represent ones that are either unique to zebra finch or do not align with annotated protein-coding regions in other genomes (Table S1). The actual gene content of the zebra finch genome is still under analysis as part of the genome sequencing project (http://www.ncbi.nlm.nih.gov/genome/guide/finch/); hence the gene identifications and annotations must still be considered provisional. However, any robust enrichment of GO annotation terms in a particular gene expression group would be strong evidence of a probable function associated with that group, regardless of the details of the individual gene identities. Indeed, we found that each one of the 4 main RNA expression patterns (the circles in Fig. 3A) has a unique and almost nonoverlapping set of functional associations (Table 1). The analysis is presented in detail in SI (Fig. S3 and Table S1, Table S2, Table S3, and Table S4). Here we summarize the major conclusions that emerged.
Table 1.
Results of functional genomic analysis of the major gene expression sets (Fig. 3A)
| No. | GO term | NU | ND | HU | HD |
|---|---|---|---|---|---|
| 18 | Transcription factor activity | 0.01 | |||
| 27 | Carbohydrate metabolic process | 0.00 | |||
| 32 | Alcohol metabolic process | 0.00 | |||
| 50 | RNA metabolic process | 0.03 | |||
| 59 | Nuclease activity | 0.00 | |||
| 24 | Lipid biosynthetic process | 0.02 | 0.03 | ||
| 68 | Protein amino acid dephosphorylation | 0.03 | |||
| 64 | Notch signaling pathway | 0.00 | |||
| 38 | Ion channel activity | 0.00 | |||
| 69 | GTPase activity | 0.03 | |||
| 35 | Protein transport | 0.05 | |||
| 57 | Nucleotidyltransferase activity | 0.01 | |||
| 42 | Peptidase activity | 0.02 | |||
| 54 | Cytoplasmic membrane-bound vesicle | 0.04 | |||
| 28 | Electron transport | 0.00 | |||
| 29 | Oxidative phosphorylation | 0.00 | |||
| 30 | Macromolecular complex assembly | 0.01 | |||
| 40 | Receptor activity | 0.00 | |||
| 48 | Translation | 0.00 | |||
| 53 | Mitochondrion | 0.00 | |||
| 55 | Ribosome | 0.00 | |||
| 60 | NADH dehydrogenase activity | 0.00 | |||
| 61 | Cytochrome c oxidase activity | 0.00 | |||
| 62 | Ubiquinol–cytochrome c reductase activity | 0.00 | |||
| 65 | I-kappaB kinase/NF-kappaB cascade | 0.01 | |||
| 66 | Hydrogen ion transporter activity | 0.00 |
Onto-Express software (22) was used to derive FDR-corrected P values for the GO terms associated with the genes in each set (Fig. S3, Table S2, and Table S3), as shown in the 4 right columns. NU = Novel Up; ND = Novel Down; HU = Habituated Up; HD = Habituated Down. Only significant values are shown (<0.05). Four GO terms were significantly associated with novel or habituated groups but represented probes moving in opposite directions (labels 68, 57, 42, and 54); the up and down columns are combined for them. The ″No.″ column links to the number key for the nodes in Fig. S3, Table S2, and Table S3.
Novel Song Induces Transcription Factor RNAs.
The set of RNAs that are elevated in the novel compared to the silence group (“novel-up”) is dominated by ones encoding transcription factors or otherwise involved in RNA metabolism. Of the 10 RNAs that show the greatest magnitude of increase relative to silence, 5 encode known transcription factors, including ZENK itself (Table 2). In total on the array as a whole, 154 probes map onto the “transcription factor activity” GO category (GO ID: 0003700) and ≈10% of these (16 probes representing 12 transcripts) are included in the novel-up group, a significant overrepresentation (FDR P < 0.05) not observed in the other gene sets. Genes involved in glycolysis (carbohydrate and alcohol metabolism) are also upregulated specifically following novel song exposure (Table 1 and Table S3).
Table 2.
Top 10 RNAs showing the greatest relative increase in the novel group compared to the silence group (FDR P < 0.05, Fig. 3A)
| Clone ID | Gene ID | TF? | FDR P | Fold |
|---|---|---|---|---|
| SB03033A2C05.f1 | NR4A3 | ● | 3E-04 | 2.89 |
| SB02015A2B02.f1 | FOSL2 | ● | 7E-05 | 2.69 |
| SB03001B2H03.f1 | FOS | ● | 6E-05 | 2.56 |
| zf ZENK | EGR1 | ● | 8E-04 | 2.53 |
| SB03024B2C08.f1 | ASMTL | 0.017 | 1.99 | |
| SB010024000A07 | NA | 8E-05 | 1.84 | |
| SB03022A1C12.f1 | SFXN | 0.007 | 1.73 | |
| SB010019000G06 | NA | 0.017 | 1.68 | |
| SB010025000F05 | NR4A2 | ● | 0.005 | 1.64 |
| SB03050A1H11.f1 | MDGA2 | 0.005 | 1.62 |
Novel Song Suppresses Putative Noncoding and Ion Channel RNAs.
The RNAs that are decreased in the novel group versus silence (“novel-down”) are remarkable both for their number (463 probes) and for their relative obscurity: only 42% have annotated orthologs versus 75% for the other gene sets (Table S1). Of the 50 RNAs that show the greatest decline following song onset, only a single one has a definable protein-coding ortholog in the chicken. In a preliminary analysis we found that ≈60% of these map to introns in the chicken genome assembly, suggesting the possibility that some represent noncoding RNAs (see Discussion). Among the minority of novel-down genes that bear functional protein annotation, there is significant overrepresentation of ion channel genes (Table 1). These include genes encoding calcium channels (the T-type but not the L-type voltage-dependent channel and the ryanodine receptor), the voltage-gated potassium channel and 2 of the 4 subtypes of sodium channel, and the AMPA (but not NMDA) glutamate receptor (Table S3).
Altered Energetics and Metabolism in the Habituated Group.
The RNAs that are increased in the habituated versus silence comparison (“habituated-up”) comprise a large and diverse set of annotations, with significant enrichment for terms suggesting involvement of cytoskeleton-based intracellular transport (Table 1 and Table S3). A different pair of strong themes dominates the “habituated-down” gene set: electron transport and translation. Genes encoding proteins in all 5 of the complexes in the mitochondrial electron transport chain are downregulated in the habituated treatment group relative to the silence group (Table S4). No probe for any of these proteins is significantly regulated in the novel treatment group relative to the silence group. For the most part, the regulated mitochondrial components do not represent the core enzymatic subunits responsible for electron transport activity in the complexes, but are subunits that stabilize assembly or regulate complex activity. The theme of translation and ribosome assembly is represented by another fraction of the habituated-down gene set, as half of the 85 probes on the array for ribosomal protein genes (encoded on both nuclear and mitochondrial genomes) are downregulated.
Proteomic Analysis Also Implicates Mitochondria and Cellular Energetics.
To assess the identities and function of the proteins that changed in the 2D-DIGE experiment, we ran additional preparative gels and attempted to identify them by position and then purify and sequence them by MS/MS (22). The resulting sequences were then compared to known proteins and we obtained significant matches (MASCOT score >50) for 24 spots, representing 18 distinct protein sequences (Table S5). Twelve are mitochondrial proteins and 2 others are cytosolic proteins that also participate in cellular biosynthesis and energetics; we used immunoblotting to confirm the regulation of ATP synthase in an independent assay (Fig. S4).
Functional Validation by Mitochondrial Assay.
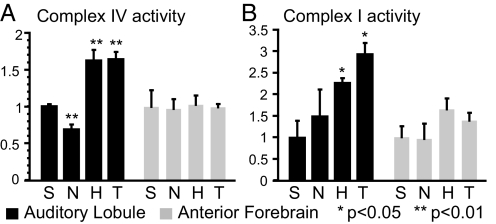
As both the microarray and proteomic components of our integrated experiment indicated major changes in molecules associated with mitochondrial function, we performed a direct biochemical analysis of 2 electron transport chain activities in a new set of birds exposed to the 3 original treatment conditions (silence, novel, and habituated). In this experiment we also included the fourth “trained-only” group (described above). We measured significant increases in both complex I and complex IV activities in the AL of birds in the habituated treatment group relative to either the silence or the novel group (Fig. 4). These effects appear specific to the auditory lobule as they were not observed in the extracts from the anterior forebrain. The habituated and trained-only groups were indistinguishable, indicating that training experience on one day leads to metabolic changes in the tissue that are expressed a day later independent of further stimulus presentations.
Fig. 4.
Mitochondrial complex activity analysis. Activities were measured (SI) in extracts from auditory lobule and anterior forebrain of birds in 4 treatment groups (n = 3 birds in each group): S, silence; N, novel; H, habituated; T, trained. All activities were normalized against the mean activity of the silence group within the same brain region. (A) complex IV activity; (B) complex I activity. ANOVA followed by post hoc test: **, P < 0.01; *, P < 0.05.
Discussion
Previous studies established that birdsong playbacks trigger a set of behavioral and neurophysiological responses in the songbird auditory forebrain, which habituate after stimulus repetition (reviewed in ref. 3). Habituation is one of the most basic forms of learning and memory, yet the mechanisms of habituation are not understood even in simple systems. Overtly it appears as a decrement or loss of a response, yet the specificity of the change implies the gain of some discriminative capacity. Here, using a combination of molecular and biochemical assays, we show that the experience of hearing birdsong deeply engages mechanisms at the genomic level and that habituation involves more than just a loss of these initial responses: indeed, it is marked by the acquisition of a new and distinct molecular profile. The progression from silence to novel to habituated appears to involve literally thousands of gene products and is associated with the emergence after a day of measurable changes in energy pathways in the auditory forebrain.
A major message from our primary microarray analysis is the large number of RNA changes associated with the seemingly simple experience of hearing birdsong. We confirmed the acute activation of familiar “immediate early genes” by novel birdsong (Table 2), but found that 20% of detectable transcripts (4,341 of 18,246 different probes) ultimately changed their expression in the 24 h following song exposure (considering both the novel and habituated treatment groups). This is an enormous increase over the number of gene products previously identified as “activity regulated” in the songbird brain, although it is not entirely unexpected on the basis of recent studies in rodents (24–27). In the zebra finch, using a smaller microarray and extensive in situ hybridization analyses, Wada et al. described 33 genes that are “singing regulated” in other parts of the brain responsible for song production (15). Seventeen of these are also present on our list of genes regulated by song listening in the auditory forebrain at a false discovery rate of 5%; 4 others do not appear to be represented on our array and the remaining 12 showed only nonsignificant trends in our experiment (Table S6). Further study is needed to determine how gene response patterns may differ by brain region and type of perceptual or motor activity. We have confidence in our overall analysis as we used a carefully qualified large microarray specifically designed to measure genes expressed in the zebra finch brain (17), participated in a centralized pipeline for execution and analysis that has been independently validated in related studies (28–30), executed a counterbalanced design with 3 treatment groups where the groups shared the same environment, controlling for circadian effects that confound interpretation of some other reports, applied a sensitive 2-stage mixed linear model for statistical analysis, and corroborated the overall accuracy of our microarray measurements with independent in situ hybridization measurements (SI). Moreover, the discrete composition of our 4 distinct gene response sets (Fig. 3A, Table 1) is an extremely improbable outcome if the sets were simply selected by chance.
Our matched proteomic analysis gave us a much narrower picture in part because it sampled only a slice of the most abundant proteins in the cell—so, for example, we detected no transcription factors by this means. Nevertheless, the proteomic data are important in showing that the effects of perceptual experience are not limited to subtle regulatory pathways or small cellular compartments but can leave an imprint on some of the major protein systems in the brain. Pinaud et al. (31) also recently reported a somewhat similar proteomic analysis of the acute response to song playbacks, sampling a different set of time points. They identified and sequenced 21 spots (representing 14 proteins) that changed out of ≈2,000 present on their gels—but only a single one is common between their dataset and ours (ATP5A) and the direction of change is different (up 1 h after song onset in their experiment, down 90 min after onset in ours). We suggest the lack of overlap probably reflects the fact that each study sampled only a small subset of the large number of proteins actually changing and also sampled different time points after the song stimulus. As has been commonly observed now in other comparisons of microarray and proteomic datasets (reviewed in refs. 32 and 33), we also saw only a weak correlation between the levels of proteins and their corresponding RNAs. Many factors may account for this, including the multiplicity of protein isoforms and differences in the synthesis, stability, and turnover of individual proteins versus their RNAs. Importantly, however, our RNA and protein analyses both indicated changes in composition of mitochondria and we confirmed a major functional effect (increased electron transport activity a day after training) directly by enzyme assay.
Given the large number of RNAs regulated in our novel and habituated groups, a major emphasis in our analysis was to dissect the structure of this population. To do this we combined statistical analyses of both expression pattern and GO-derived functional associations. We observed 4 major expression patterns in our dataset and found that each pattern involves a distinct and discrete set of RNAs, with relatively little overlap. Even more striking, each of the 4 groups has a unique functional annotation (Table 1). We must be cautious in interpretation given the preliminary and incomplete nature of zebra finch genome annotation. Nevertheless the results are worth some consideration. The gene set that increases immediately after novel song (novel-up) is enriched for familiar immediate early genes that encode transcription factors, like EGR1, FOS, NR4A2, and NR4A3. This fits with the common idea that exposure to novelty triggers a cascading program of gene regulation (14, 34) or a “genomic action potential” (1). However, we found that after novel song exposure, declining RNAs (novel-down) actually outnumbered the increasing ones, and by a large margin (463 vs. 153). Comparatively little attention has been given to understanding the mechanism and function of experience-dependent gene suppression in the brain, despite prior evidence for it in the literature (24–26, 35–37). Here we detected enrichment for ion channel protein genes in the novel-down set, including channels for calcium, potassium, and sodium and the AMPA glutamate receptor. We speculate that suppression of these channels may represent a homeostatic response to a surge in signaling activity (9, 10) associated with initial exposure to novelty. We also detected an unusually large population of downregulated RNAs that map to putative introns and may represent noncoding RNAs (ncRNAs). Appreciation is exploding for the potential role of ncRNAs in many aspects of biological regulation (38). Further study is warranted to determine if fluxes in particular ncRNAs are associated with different functional states in the brain.
A day after habituation training, presentation of the same song no longer induced most of the novel RNA changes. Of the 153 genes induced by novel song, only 64 were also significantly upregulated in the habituated–silence comparison (Fig. 4). Interestingly, among these were ZENK (EGR1) and FOS—they were at higher levels in the habituated than in the silence group, although at still higher levels in the novel group (Table S6). Thus complete “habituation to zero” did not occur for these genes in this experiment. These genes may respond somewhat to any kind of neural activity, regardless of its novelty or significance. Alternatively, their RNAs may have been induced by the novel stimulus and not yet returned to baseline a day later. The results of our second experiment favor the latter interpretation as there was no significant difference between habituated and trained-only groups. However, in the second experiment when the probes are simply rank ordered according to P value, EGR1, FOS, and NR4A3 are on the top of the list hovering at an FDR threshold of 30–40%, with slightly more signal in the habituated than in the trained group—suggesting that these genes may retain some persisting but small responsiveness to the familiar stimulus. Together these observations suggest an underlying response structure for some genes where novel song > habituated song > silence (after training) > silence (untrained).
The habituated condition, however, was marked by much more than just the suppression of novel responses. Indeed, it displayed an entirely new set of molecular differences from the silence group. In the microarray experiment, 3,140 probes had altered expression in the habituated group relative to the silence group, ≈5 times more than the number of novel-responding probes, and <10% of these overlap with the set of novel responders. To resolve whether these differences represented a new and different response to a now-familiar stimulus or a direct but delayed consequence of the previous day's training, we conducted an additional microarray comparison of birds hearing a replay of the habituated song with birds who had been trained but then heard only silence on testing day (trained only). We also incorporated this fourth trained-only group in the design of our mitochondrial enzyme activity analysis and one of our in situ validation studies (Fig. S1B). By all 3 assays we found no significant difference between habituated and trained-only groups (although see previous paragraph for discussion of trends). Hence we conclude that the training experience leads to a set of changes that are not immediately evident (in the novel treatment condition) but develop over the following day. These slowly developing changes may represent the “late” genes often hypothesized in learning and memory theory (1, 14, 34).
Although the set of genes regulated in the habituated condition is large and complex, one major focus is on gene products associated with mitochondrial energetics. We found that the majority of nuclear genes encoding regulatory components of the mitochondrial electron transport chain were decreased in expression in the habituated group (Table S4), and decreases were also observed at the protein level (Table S5). Somewhat counterintuitively, we measured an increase in enzyme electron transport chain activity by biochemical assay. One possible explanation is that the targeted regulatory components normally attenuate the electron transport pathway in brain mitochondria. Each electron transport complex is assembled from numerous regulatory and catalytic proteins, and net activity can be modulated by various posttranslational processes (39–41). It will be interesting to carry this analysis to the cellular and subcellular levels—Is mitochondrial function changing throughout the tissue or is it perhaps localized, e.g., to synapses? Recent studies using metabolic imaging have led to the concept of an important but still mysterious “intrinsic activity” in the brain—that is, the robust metabolic activity that proceeds in the absence of any acute stimulus (42). Might the ongoing metabolic changes we see a day after a training experience represent an aspect of this intrinsic activity?
Our experiments measured habituation at several different levels, including the behavioral, the transcriptional, the proteomic, and the metabolic. We see correlated changes at all of these levels but we cannot yet presume cause and effect. However, it should be possible to manipulate these processes to study their interrelationships. For example, we have recently shown that transient pharmacological interference with the ERK pathway blocks the ZENK response to novel song and also blocks the emergence of ZENK habituation a day after training exposures (8). Is ERK activation also necessary for other aspects of the genomic response to novelty? Are the initial responses to novelty necessary for the development of the habituated profile? If transcriptional habituation is blocked, does that also block the metabolic changes observed after song habituation training?
In this research we used the zebra finch as an experimental subject. The zebra finch is uniquely valuable for the insights it offers into mechanisms of neural plasticity, vocal communication, and social behavior. Sequencing of the zebra finch genome is nearing completion, and so the datasets produced here will be an important reference point for the ongoing analysis and annotation of genome function in this species. An important focus for the future will be to understand how these broad molecular and functional states relate to the episodic flow of natural experience. For example, what happens when an individual in the habituated state encounters a new stimulus that is both novel and significant? How do sequential experiences interact within the state chemistry of the brain? How do these molecular states map onto the complex cellular anatomy of the brain? The zebra finch will be a powerful model for probing these fundamental questions about genes, brains, and behavior.
Materials and Methods
All experiments were performed under protocols approved by the University of Illinois Laboratory Animal Care Advisory Committee. We used adult male zebra finches (Taeniopygia guttata) that were bred and raised in an aviary at the Beckman Institute animal facility. For the initial combined analysis of RNA (by microarray) and proteins (by 2D-DIGE), we prepared 3 treatment groups in a single counterbalanced design (Fig. 1: silence, novel, and habituated groups). On the afternoon of day 0 of the experiment, each bird (n = 36) was placed alone in a sound chamber to normalize recent experience. The next afternoon (day 1), for birds in the habituated group, a single song stimulus was played repeatedly over 3 h from a speaker in the chamber, following the established protocol of ZENK response habituation (8, 13). On day 2, the song stimulus was played for 30 min to both the novel and the habituated groups (but not to the silence groups). For all song exposures we used the same stimulus, song “zf101” recorded in a different aviary, which was presented once every 10 sec and accompanied by videotaping of behavioral responses, as described (8). The AL was dissected as described (18). Each RNA sample (from a single bird) was analyzed on a separate microarray using a universal reference design, as Community Collaboration 21 under the Songbird Neurogenomics Initiative (17). Main effects of treatment were calculated using 2-stage mixed linear modeling (43), with the threshold for significance set at an FDR-corrected P value of 5% (44). AL proteins from each bird were extracted, fluorescently labeled, and separated on 2D gels using the DIGE technique (2 samples plus a third reference on each gel). Statistical analyses of gel images were performed using DeCyder. Selected spots were sequenced by LC-MS/MS using MASCOT to match to Chordata sequences. A subsequent microarray experiment compared a second habituated group collected in parallel with a trained-only group. All of the raw microarray data are available in a MIAME-compliant format.
For mitochondrial enzyme assays, all 4 groups were prepared in parallel. Additional methods are given in SI, including descriptions of validation studies using in situ hybridization and immunoblotting and the protocols for measuring mitochondrial complex I and complex IV enzyme activities.
Supplementary Material
Acknowledgments.
We thank Gene Robinson and Wes Warren for helpful comments on the manuscript and Sarah London for comments and additional help preparing some of the SI Text. This work was supported by Public Health Service grants R01 NS051820, R01 NS045264, and P30 DA018310.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812998106/DCSupplemental.
References
- 1.Clayton DF. The genomic action potential. Neurobiol Learn Mem. 2000;74:185–216. doi: 10.1006/nlme.2000.3967. [DOI] [PubMed] [Google Scholar]
- 2.Robinson GE, Fernald RD, Clayton DF. Genes and social behavior. Science. 2008;322(5903):896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong S, Clayton DF. Habituation in songbirds. Neurobiol Learn Mem. 2008;92:183–188. doi: 10.1016/j.nlm.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mello CV, Vicario DS, Clayton DF. Song presentation induces gene expression in the songbird forebrain. Proc Natl Acad Sci USA. 1992;89:6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mello CV, Clayton DF. Song-induced ZENK gene expression in auditory pathways of songbird brain and its relation to the song control system. J Neurosci. 1994;14:6652–6666. doi: 10.1523/JNEUROSCI.14-11-06652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mello CV, Nottebohm F, Clayton DF. Repeated exposure to one song leads to a rapid and persistent decline in an immediate early gene's response to that song in zebra finch telencephalon. J Neurosci. 1995;15:6919–6925. doi: 10.1523/JNEUROSCI.15-10-06919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruse AA, Stripling R, Clayton DF. Context-specific habituation of the zenk gene response to song in adult zebra finches. Neurobiol Learn Mem. 2004;82:99–108. doi: 10.1016/j.nlm.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Dong S, Clayton DF. Partial dissociation of molecular and behavioral measures of song habituation in adult zebra finches. Genes Brain Behav. 2008;7:802–809. doi: 10.1111/j.1601-183X.2008.00423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stripling R, Volman SF, Clayton DF. Response modulation in the zebra finch caudal neostriatum: relationship to nuclear gene regulation. J Neurosci. 1997;17:3883–3893. doi: 10.1523/JNEUROSCI.17-10-03883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew SJ, Mello C, Nottebohm F, Jarvis E, Vicario DS. Decrements in auditory responses to a repeated conspecific song are long-lasting and require two periods of protein synthesis in the songbird forebrain. Proc Natl Acad Sci USA. 1995;92:3406–3410. doi: 10.1073/pnas.92.8.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chew SJ, Vicario DS, Nottebohm F. A large-capacity memory system that recognizes calls and songs of individual birds. Proc Natl Acad Sci USA. 1996;93:1950–1955. doi: 10.1073/pnas.93.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woolley SC, Doupe AJ. Social context-induced song variation affects female behavior and gene expression. PLoS Biol. 2008;6(3):e62. doi: 10.1371/journal.pbio.0060062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stripling R, Milewski L, Kruse AA, Clayton DF. Rapidly learned song-discrimination without behavioral reinforcement in adult male zebra finches (Taeniopygia guttata) Neurobiol Learn Mem. 2003;79(1):41–50. doi: 10.1016/s1074-7427(02)00005-9. [DOI] [PubMed] [Google Scholar]
- 14.Miyashita T, Kubik S, Lewandowski G, Guzowski JF. Networks of neurons, networks of genes: An integrated view of memory consolidation. Neurobiol Learn Mem. 2008;89(3):269–284. doi: 10.1016/j.nlm.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wada K, et al. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc Natl Acad Sci USA. 2006;103(41):15212–15217. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302(5643):296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- 17.Replogle K, et al. The Songbird Neurogenomics (SoNG) Initiative: Community-based tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng HY, Clayton DF. Activation and habituation of extracellular signal-regulated kinase phosphorylation in zebra finch auditory forebrain during song presentation. J Neurosci. 2004;24(34):7503–7513. doi: 10.1523/JNEUROSCI.1405-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huesmann G, Clayton DF. Dynamic role of postsynaptic caspase-3 and BIRC4 in zebra finch song response habituation. Neuron. 2006;52:1061–1072. doi: 10.1016/j.neuron.2006.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.London SE, Clayton DF. Functional identification of sensory mechanisms required for developmental song learning. Nat Neurosci. 2008;11(5):579–586. doi: 10.1038/nn.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mello CV, Ribeiro S. Zenk protein regulation by song in the brain of songbirds. J Comp Neurol. 1998;393:426–438. doi: 10.1002/(sici)1096-9861(19980420)393:4<426::aid-cne3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Zhou G, et al. 2D differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Mol Cell Proteomics. 2002;1(2):117–124. doi: 10.1074/mcp.m100015-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79(2):266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 24.O'Sullivan NC, et al. Temporal change in gene expression in the rat dentate gyrus following passive avoidance learning. J Neurochem. 2007;101(4):1085–1098. doi: 10.1111/j.1471-4159.2006.04418.x. [DOI] [PubMed] [Google Scholar]
- 25.Havik B, et al. Synaptic activity-induced global gene expression patterns in the dentate gyrus of adult behaving rats: Induction of immunity-linked genes. Neuroscience. 2007;148(4):925–936. doi: 10.1016/j.neuroscience.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 26.Park CS, Gong R, Stuart J, Tang SJ. Molecular network and chromosomal clustering of genes involved in synaptic plasticity in the hippocampus. J Biol Chem. 2006;281(40):30195–30211. doi: 10.1074/jbc.M605876200. [DOI] [PubMed] [Google Scholar]
- 27.Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41(1):35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 28.Lovell PV, Clayton DF, Replogle KL, Mello CV. Birdsong “transcriptomics”: Neurochemical specializations of the oscine song system. PLoS ONE. 2008;3(10):e3440. doi: 10.1371/journal.pone.0003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.London SE, Dong S, Replogle K, Clayton DF. Developmental shifts in gene expression in the auditory forebrain during the sensitive period for song learning. Dev Neurobiol. 2009;69(7):437–450. doi: 10.1002/dneu.20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomaszycki ML, et al. Sexual differentiation of the zebra finch song system: Potential roles for sex chromosome genes. BMC Neurosci. 2009;10:24. doi: 10.1186/1471-2202-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinaud R, Osorio C, Alzate O, Jarvis ED. Profiling of experience-regulated proteins in the songbird auditory forebrain using quantitative proteomics. Eur J Neurosci. 2008;27(6):1409–1422. doi: 10.1111/j.1460-9568.2008.06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers S, et al. Investigating the correspondence between transcriptomic and proteomic expression profiles using coupled cluster models. Bioinformatics. 2008;24(24):2894–2900. doi: 10.1093/bioinformatics/btn553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie L, Wu G, Culley DE, Scholten JC, Zhang W. Integrative analysis of transcriptomic and proteomic data: Challenges, solutions and applications. Crit Rev Biotechnol. 2007;27(2):63–75. doi: 10.1080/07388550701334212. [DOI] [PubMed] [Google Scholar]
- 34.Tischmeyer W, Grimm R. Activation of immediate early genes and memory formation. Cell Mol Life Sci. 1999;55:564–574. doi: 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavallaro S, D'Agata V, Manickam P, Dufour F, Alkon DL. Memory-specific temporal profiles of gene expression in the hippocampus. Proc Natl Acad Sci USA. 2002;99(25):16279–16284. doi: 10.1073/pnas.242597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'Agata V, Cavallaro S. Hippocampal gene expression profiles in passive avoidance conditioning. Eur J Neurosci. 2003;18(10):2835–2841. doi: 10.1111/j.1460-9568.2003.03025.x. [DOI] [PubMed] [Google Scholar]
- 37.Levenson JM, et al. A bioinformatics analysis of memory consolidation reveals involvement of the transcription factor c-rel. J Neurosci. 2004;24(16):3933–3943. doi: 10.1523/JNEUROSCI.5646-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2(12):919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 39.Fattoretti P, Bertoni-Freddari C, Giorgetti B, Balietti M. Increased mitochondrial and nuclear gene expression of cytochrome oxidase subunits I and IV in neuronal aging. Ann N Y Acad Sci. 2004;1030:303–309. doi: 10.1196/annals.1329.038. [DOI] [PubMed] [Google Scholar]
- 40.Verwer RW, et al. Decreased hippocampal metabolic activity in Alzheimer patients is not reflected in the immunoreactivity of cytochrome oxidase subunits. Exp Neurol. 2000;163(2):440–451. doi: 10.1006/exnr.2000.7385. [DOI] [PubMed] [Google Scholar]
- 41.Miyazaki T, Neff L, Tanaka S, Horne WC, Baron R. Regulation of cytochrome c oxidase activity by c-Src in osteoclasts. J Cell Biol. 2003;160(5):709–718. doi: 10.1083/jcb.200209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raichle ME. Neuroscience. The brain's dark energy. Science. 2006;314(5803):1249–1250. [PubMed] [Google Scholar]
- 43.Wolfinger R, et al. Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol. 2001;8(6):625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- 44.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.