Abstract
A vitamin B6 degradative pathway has recently been identified and characterized in Mesorhizobium loti MAFF303099. One of the enzymes on this pathway, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO), is a flavin-dependent enzyme and catalyzes the oxidative ring opening of 2-methyl-3-hydroxypyridine-5-carboxylic acid to form E-2-acetaminomethylene succinate. The gene for this enzyme has been cloned and the corresponding protein has been overexpressed in Escherichia coli and purified. The crystal structure of MHPCO has been solved to 2.1 Å using SAD phasing with and without the substrate MHPC bound. These crystal structures provide insight into the reaction mechanism and suggest roles for active site residues in the catalysis of a novel oxidative ring-opening reaction.
Two pathways for the degradation of pyridoxal-phosphate vitamers (vitamin B6), have been identified in soil microbes able to grow on vitamin B6 as the sole source of carbon and nitrogen (1). The first pathway, which converts vitamin B6 to succinic semialdehyde, occurs in Pseudomonas sp. MA-1 (Figure 1), while the second pathway has been identified in Pseudomonas IA and Arthrobacter Cr-7 and converts pyridoxine to 2-(hydroxymethyl)-4-oxobutanoate. The enzymes and intermediates for both pathways have been characterized (2–4). However, the corresponding genes remained unidentified until the recent discovery that the first pathway is also present in the symbiotic bacterium Mesorhizobium loti MAFF303099 (5). All the M. loti PLP catabolic genes have now been identified and the corresponding catabolic reactions have been characterized (6–12). One of the later enzymes on this pathway, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO), catalyzes a particularly interesting reaction in which 2-methyl-3-hydroxypyridine-5-carboxylic acid (MHPC) undergoes an oxidative ring opening to generate E-2-acetamidomethylene succinate, the first acyclic intermediate on the pathway (7 to 8 in Figure 1).
Figure 1.
The vitamin B6 degradative pathway found in M. loti. The genes are named as using the RhizoBase numbering system. 1, pyridoxine; 2, pyridoxal; 3, 4-pyridoxolactone; 4, 4-pyridoxic acid; 5, 5-formyl-2-methyl-3-hydroxypyridine-4-carboxylate; 6, 2-methyl-3-hydroxypyridine-4,5-dicarboxylate; 7, 2-methyl-3-hydroxypyridine-5-carboxylate; 8, 2-(acetamidomethylene)succinate; 9, succinic semialdehyde.
MHPCO (E.C. 1.14.12.4) has been classified as a flavin-dependent monooxygenase based on its sequence and biochemical similarity to other enzymes in this family, which includes the well-characterized p-hydroxybenzoate hydroxylase (PHBH) (13, 14). This class of enzymes utilize FAD and molecular oxygen to form a C(4a)-hydroperoxy-FAD intermediate which then donates one oxygen atom to the substrate (15). The second reaction catalyzed by MHPCO, the pyridine ring-opening reaction, is an unusual reaction for an enzyme of the monooxygenase family because most oxidative ring-opening reactions of aromatic compounds are catalyzed by iron-dependent dioxygenases. MHPCO was confirmed as a flavin-dependent monooxygenase, incorporating one oxygen atom from molecular oxygen and one oxygen atom from water to produce E-2-(acetamidomethylene)succinate (E-2AMS) the ring-opened product (16). Previous work has shown the Pseudomonas MA-1 MHPCO to crystallize and diffract to reasonable resolution, although the structure was not reported (17). We have determined the 2.1 Å resolution crystal structures of M. loti MHPCO and its complex with MHPC. These structures confirm the relationship of MHPCO to other members of the flavin monooxygenase family and provide insight into the catalytic mechanism.
MATERIALS AND METHODS
Materials
M9 minimal salts and CaCl2 were purchased from Sigma (St. Louis, MO). Ni-NTA resin came from Qiagen (Valencia, CA) and Acros Chemicals (Morris Plains, NJ) supplied the imidazole and l-selenomethionine. Dextrose, FeSO4, MgSO4, NaCl, Tris buffer, β-mercaptoethanol, chloramphenicol, ampicillin, and all natural amino acids were purchased from Fisher Scientific (Fairlawn, NJ). 100X MEM vitamin mix were from Invitrogen (Carlsbad, CA), as was the Gateway system used for DNA manipulations.
Cloning of M. loti MHPCO
Standard methods were used for DNA manipulations (18, 19). The M. loti MHPCO gene was amplified from genomic DNA (purified from ATCC strain # 700743) by PCR with the following primer pair: 5’-CAC CAT GGC CAA TGT AAA CAA AAC TCC-3’ and 5’-CTA CTG CGG CCA CGA GTA GAC ACG GCG CAG C-3’. The PCR product was purified and used in a topoisomerase-mediated reaction with pENTR-TEV-D-TOPO (Invitrogen) following the manufacturer’s instructions. Clones were screened by PCR and verified by sequencing. A correct clone was used in an LR recombination reaction with the plasmid pDESTF1, a Gateway-adapted vector based on the pET-system from Novagen that encodes an N-terminal 6xHis tag under the control of the T7lac promoter. Clones were again screened by restriction digestion. A correct clone was named pMl5332.XF1.
Overexpression and Purification of Selenomethionyl (SeMet) MHPCO E. coli
B834(DE3) was transformed first using the chaperone-encoding plasmid pGro7 (Takara Bio Inc.) and then using pMl5332.XF1. Cells were grown with shaking at 37 °C in minimal media supplemented with 20 mg/L of all amino acids except methionine, 50 mg/L l-selenomethionine, 1X MEM vitamin mix, 0.4% glucose, 2 mM MgSO4, 0.1 mM CaCl2, 25 mg/L FeSO4, 35 mg/L chloramphenicol, and 100 mg/L ampicillin. Chaperone proteins GroES and GroEL were induced at cell culture inoculation by addition of 2.5 g/L l-arabinose. When cells reached an OD600 of 0.7 the temperature was lowered to 15 °C and overexpression of MHPCO was induced with 0.5 mM isopropyl β-d-thiogalactopyranoside. After overnight growth, cells were pelleted by centrifugation at 4 °C for 15 min at 6,000 g and then stored at −20 °C until purification. Cells were lysed by sonication and the cell lysate was cleared by centrifugation at 40,000 g for 45 min at 4 °C. SeMet MHPCO was purified from the clarified cell lysate by nickel affinity chromatography. The clarified lysate was twice passed over the column equilibrated with Buffer A (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, and 3 mM β-mercaptoethanol (β-ME)) and 10 mM imidazole and the column was washed with this buffer for 20 column volumes. The column was then washed with Buffer A containing 35 mM imidazole for 15 column volumes. Buffer A and 250 mM imidazole were used to elute SeMet MHPCO from the nickel column and the protein was 80% pure by SDS-PAGE analysis (results not shown). Size exclusion chromatography (HiLoad 26/60 Superdex 200 pg, GE Healthcare) was used to further purify SeMet MHPCO to >95% homogeneity. One liter of culture produced 0.5 mg pure SeMet MHPCO. The protein was then buffer exchanged into 20 mM Tris (pH 7.7), 50 mM NaCl, and 1 mM DTT and concentrated to 8 mg/mL.
Enzymatic Synthesis of the Substrate
The enzymatic synthesis and purification of 2-methyl-3-hydroxypyridine-4,5-dicarboxylic acid (MHPD) has been previously described (10). MHPC was enzymatically synthesized from MHPD in a 10 mL reaction mixture containing 5 mM MHPD and 100 µM of MHPD decarboxylase in 100 mM sodium phosphate buffer (pH 8.0) incubated over night at room temperature. MHPC was purified by the HPLC (10) and upon lyophilization yielded a stable white solid. 1H NMR (300 MHz, D2O) δ 2.46 (s, 3H, CH3), 7.68 (s, 1H, C4-H), and 8.12 (s, 1H, C6-H). The purified MHPC was dissolved in 100 mM Tris (pH 7.7) to a final concentration of 100 mM and stored frozen at −20 °C until use.
Activity Assay for MHPCO
The recombinant His-tagged MHPCO was assayed by monitoring the disappearance of NADH absorbance at 340 nm as previously described (12).
Analytical Ultracentrifugation
Samples of MHPCO were prepared as described above and analyzed by analytical ultracentrifugation to determine the oligomeric state of the enzyme. To obtain MHPCO with cleaved His tag, the sample was dialyzed overnight into 50 mM Tris (pH 8.0), 150 mM NaCl, and 2 mM DTT at 4 °C. It was then incubated with TEV protease at 4 °C for 20 hours and then passed over a Ni-NTA column to remove uncleaved MHPCO and the TEV protease. The cleavage reaction was monitored by SDS-PAGE analysis. The protein samples, both with and without the His tag, were extensively dialyzed into 20 mM Tris (pH 8.0) 150 mM NaCl and then subjected to sedimentation velocity analytical ultracentrifugation on a ProteomeLab XL-I protein characterization system (Beckman Coulter). Data were analyzed using the programs SEDNTERP and DCDT+ (20).
Protein Crystallization
Initial crystallization conditions were identified using the hanging drop vapor diffusion method (Crystal Screens 1 and 2, Hampton Research; Wizard Screens 1 and 2, Emerald Biosystems) at room temperature. Hanging drops were set up by mixing 1 µL of reservoir solution with 1 µL of protein sample. Optimized crystallization conditions were 6–9% PEG8000 and 100 mM Tris (pH 8.1–8.5). Yellow plate-like crystals grew to 100–200 µm × 50–100 µm in five days. To obtain crystals complexed with the substrate MHPC, SeMet MHPCO crystals were transferred to a drop consisting of the reservoir solution and 10 mM MHPC then allowed to soak for 3–5 h. This crystallization condition, supplemented with 17% glycerol, was used as cryoprotectant and for crystals soaked with MHPC, 10 mM MHPC was also present in the cryoprotectant. After briefly soaking the crystals in the cryoprotectant, they were flash frozen by plunging in liquid nitrogen. Both SeMet MHPCO and the substrate complex crystals belong to the space group C2 with unit cell parameters a = 111.3 Å, b = 130.2 Å, c = 89.5 Å, and β = 122.6°, for unliganded MHPCO. The asymmetric unit contains two monomers with a Matthew’s number of 3.28 Å3/Da and solvent content of 60% (21).
X-Ray Data Collection and Processing
For structure determination, data were collected on a single SeMet MHPCO crystal at the Advanced Photon Source beamline 24-ID-E (Argonne National Laboratory) using a Quantum315 detector (Area Detector Systems Corporation). To maximize the anomalous signal, a single wavelength anomalous diffraction experiment was conducted at the peak f″ for selenium. Data were collected to 2.1 Å resolution using a 1° oscillation range for 330 frames. Data collection on a crystal containing SeMet MHPCO complexed with MHPC was done on the 24-ID-C beamline using a Quantum315 detector. Data were collected at 0.9795 Å over 180° using a 1° oscillation range. Datasets were indexed, integrated, and scaled using the HKL2000 suite of programs (22). The data collection statistics are summarized in Table 1.
Table 1.
Summary of Data Collection Statistics
| SeMet MHPCO | MHPCO + MHPC | |
|---|---|---|
| Beamline | APS NE-CAT 24-IDE | APS NE-CAT 24-IDC |
| Resolution (Å) | 2.10 | 2.10 |
| Wavelength (Å) | 0.97918 | 0.97949 |
| Space Group | C2 | C2 |
| a (Å) | 111.3 | 110.4 |
| b (Å) | 130.2 | 129.5 |
| c (Å) | 89.5 | 89.1 |
| β (°) | 122.6 | 122.6 |
| Reflections | 408805 | 208350 |
| Unique reflectionsa | 121606 (11380) b | 58150 (4574) |
| Average I/σ | 13.9 (3.2) | 12.6 (3.5) |
| Redundancy | 3.4 (2.5) | 3.6 (3.0) |
| Completeness (%) | 98.6 (91.8) | 94.4 (74.2) |
| Rsymc (%) | 12.0 (33.5) | 9.4 (21.3) |
Unique reflections include Bijvoet pairs.
Values in parentheses are for the highest resolution shell.
Rsym = ∑∑i|Ii−<I>|/ ∑<I>, where <I> is the mean intensity of the N reflections with intensities Ii and common indices h,k,l.
Structure Determination, Model Building, and Refinement
Se atom positions were located using data cut off at 2.5 Å resolution and the program hkl2map; 18 of a possible 20 Se atoms were located (23–25). These heavy atom positions were then used in CNS for phasing, density modification, and calculation of electron density maps (26). Further improvement to the maps and automated model building were performed using RESOLVE, which succeeded in modeling 377 residues with correct side chains and 196 residues as alanine out of 758 total residues in the asymmetric unit (27). Iterative rounds of model building in COOT and refinement in CNS successfully positioned residues 11–379 for both chains (28). An FAD molecule was then modeled into each chain, followed by addition of water molecules using CNS. Lastly, for the complex MHPC was modeled into MHPCO before a final round of refinement. The refinement statistics are summarized in Table 2. The geometry of MHPCO was verified using PROCHECK which identified one residue, Ala51, in each chain in the disallowed region (29). Figures were generated using PYMOL and ChemBioDraw (30).
Table 2.
Summary of Data Refinement Statistics
| SeMet MHPCO | MHPCO + MHPC | |
|---|---|---|
| Resolution (Å) | 50.00 – 2.10 | 50.00 – 2.10 |
| # of protein atoms | 5730 | 5730 |
| # of ligand atoms | 108 | 130 |
| # of water atoms | 744 | 483 |
| Reflections in working set | 59384 | 52927 |
| Reflections in test set | 3175 | 2867 |
| Rfactora (%) | 19.3 | 20.1 |
| Rfreeb (%) | 22.4 | 22.9 |
| Rmsd from ideals | ||
| Bonds (Å) | 0.0055 | 0.0061 |
| Angles (°) | 1.3 | 1.3 |
| Avg B factor (Å2) | 19.4 | 22.7 |
| Ramachandran Plot | ||
| Most favored (%) | 90.0 | 90.3 |
| Additionally allowed (%) | 9.7 | 9.1 |
| Generously allowed (%) | 0.0 | 0.3 |
| Disallowed (%) | 0.3 | 0.3 |
R factor = ∑hkl| |Fobs |−k|Fcal||/∑hkl|Fobs|where Fobs and Fcal are observed and calculated structure factors, respectively.
For Rfree the sum is extended over a subset of reflections (5%) excluded from all stages of refinement.
RESULTS
MHPCO Activity
The recombinant His-tagged MHPCO was purified using metal ion chromatography and was found to be active using the assay previously described (12). The activity of recombinant MHPCO with the His tag cleaved by digestion with TEV protease was comparable to the activity of the His-tagged MHPCO sample.
Structure of the MHPCO monomer
The structure of MHPCO was determined at 2.1 Å resolution using SAD phasing. The final model of the MHPCO monomer is shown in Figure 2 and contains residues 11–379 of the protein and one molecule of FAD. The final model of the ternary substrate complex also contains one molecule of MHPC. MHPCO shows strong structural similarity to other members of the flavin-dependent hydroxylase family. MHPCO comprises three domains exhibiting mixed α/β folds. The N-terminal domain has a five-stranded parallel β-sheet (β7↑β2↑β1↑β11↑β18↑) flanked on one side by a small, three stranded antiparallel β-sheet (β10↑β9↓β8↑) and seven α-helices arranged in two layers on the opposite side. This domain forms most of the interactions with the FAD molecule. The second domain consists of a six-stranded mostly antiparallel β-sheet with a strand order of β5↓β4↑β14↑β15↓β16↑β13↓ that is flanked on the solvent exposed side by three short α-helices and forms part of the substrate binding pocket on the other face. The third domain connects the first two domains and consists of α11 stretching the length of MHPCO and the two stranded antiparallel β-sheet β3↓β6↑. Domain 3 is largely solvent exposed on the back side of MHPCO.
Figure 2.
The monomeric structure of MHPCO. A. Stereoview ribbon diagram of MHPCO with FAD and MHPC bound and secondary structural elements labeled. The α-helices are shown in blue and the β-strands are shown in green with loops colored yellow. The bound ligands, FAD and MHPC, are shown in stick representation and colored red. B. Topology diagram of MHPCO with the same color scheme as 3A.
Oligomeric Structure
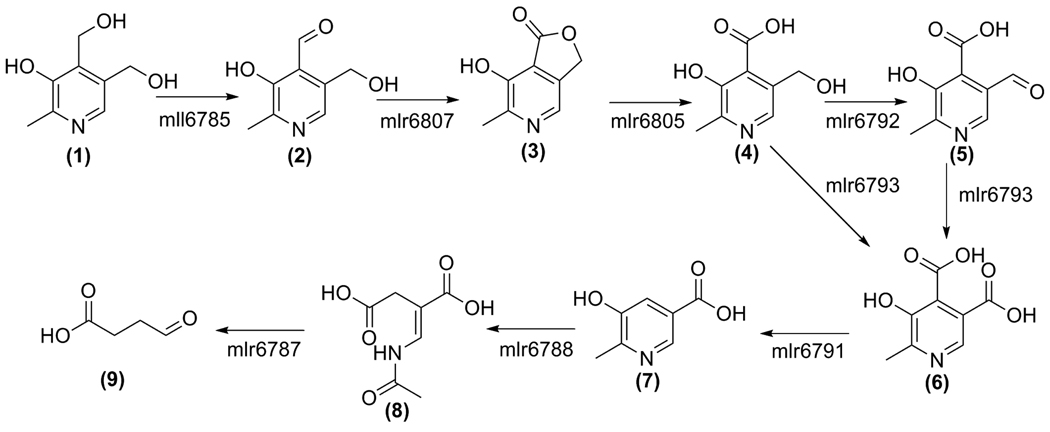
Two protomers are found within the asymmetric unit lying antiparallel to each other and the dimer spans approximately 75 Å, (Figure 3A). The two active sites are independent of each other, but face in the same direction and are located more than 40 Å apart. The total surface area for the dimer of MHPCO is 28750 Å2 and the total surface area buried is 1375 Å2 at the dimeric interface (31). The primary interactions occur through β12 to form an extended β-sheet consisting of β17↑β12↓β12*↑β17*↓ where strands marked with an asterisk are located in the opposing protomer. β12 is five residues long and consists of mostly hydrophilic residues, stretching from Glu170 to Val174. Five hydrogen bonds are formed between carbonyl oxygen atoms and nitrogen atoms from the protein backbone, stabilizing the dimer formation and ranging in length from 2.65 Å and 3.1 Å. A nitrogen atom of Arg172 is involved in an additional hydrogen bond to the carbonyl oxygen atom of the side chain of Asn170*. One salt bridge is also present at this interface, between Arg172 and Asp164*. These interactions are shown in Figure 3B. The oligomeric state of MHPCO was further investigated using analytical ultracentrifugation. To determine if the N-terminal 6X-His tag was responsible for dimer formation over tetramer formation, sedimentation velocity analytical ultracentrifugation experiments were performed on MHPCO samples with and without the His tag (32). These experiments showed that both sets of samples formed tetramers in solution with a molecular weight of roughly 160 kDa, as shown in Figures 3C and 3D.
Figure 3.
The dimeric structure of MHPCO. A. Ribbon diagram of the MHPCO dimer with FAD and MHPC bound and shown in red. The first protomer is shown in green and the opposing protomer is colored yellow. Hydrogen bonds and salt bridges are shown using dashed lines. B. Ball and stick representation of the dimeric interface at β12β12*. The color scheme is the same as Figure 4A. C. Overlay of sedimentation velocity analytical ultracentrifugation data with the fitted curve for a single species. The data points are shown as open blue circles and the best fit line is shown in red. The overlay for MHPCO with the N-terminal His tag attached. D. The overlay for MHPCO with the N-terminal His tag cleaved. The fits were prepared using the program DCDT+.
FAD Binding Site
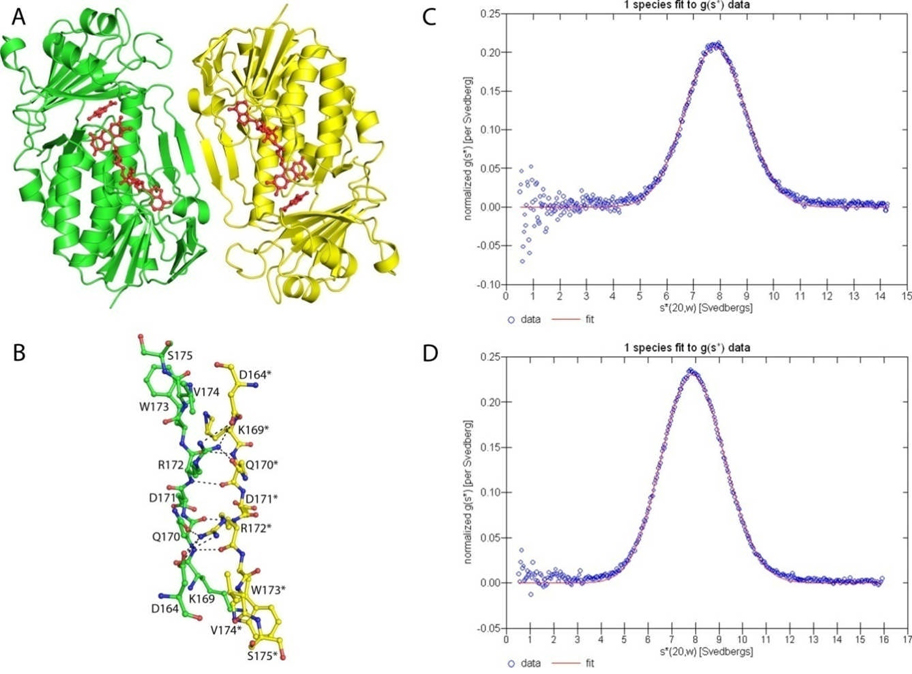
The FAD molecule binds within a long cleft roughly 30 Å long and 15 Å wide. FAD is tightly bound in a non-covalent fashion and copurified with the enzyme. Each protomer within the asymmetric unit contains one bound FAD molecule (Figure 4). The adenine base and ribose sugar are partially buried and surrounded by the hydrophilic residues Glu41, Lys42, Glu129, Asp156 and Lys161. N1 of the adenine base forms a hydrogen bond to the amide nitrogen of Ala130 and both N6 and N7 form water-mediated hydrogen bonds to Lys161. The ribose sugar is stabilized through water-mediated hydrogen bonds to the backbone carbonyl oxygen atom of Arg47. The 2′-hydroxyl group also forms a strong hydrogen bond (2.5 Å) with Glu41. The α-phosphate group is largely solvent exposed, while the β-phosphate group interacts with two water molecules and two backbone amide nitrogen atoms found at the start of helix α1. The isoalloxazine ring adopts an “IN” conformation for both structures and is positioned in a large pocket lined with hydrophobic and aromatic residues. The N1 atom and the carbonyl oxygen atom of C2 are hydrogen bonded to the amide nitrogen atom of Ala301 and Gly302, the first residues of helix α10. N3 is hydrogen bonded to the main chain carbonyl oxygen atom of Tyr54. N5 and the carbonyl oxygen of C4 form a water-mediated hydrogen bond to Tyr223. There are no interactions with the rest of the isoalloxazine ring.
Figure 4.
Stereoview diagram of FAD conformation found in MHPCO both with and without MHPC bound. Residues from MHPCO are shown in green using the ball and stick representation and the interactions with the protein indicated using black dashed lines. Water molecules are shown as red nonbonded spheres. FAD adopts the “IN” conformation and is shown with carbon atoms colored yellow.
Substrate Binding Site
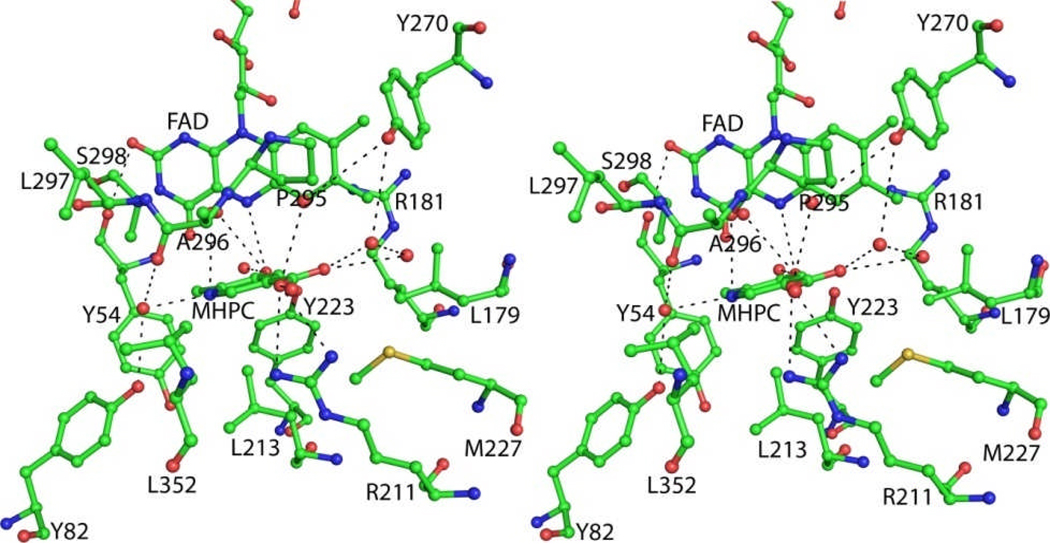
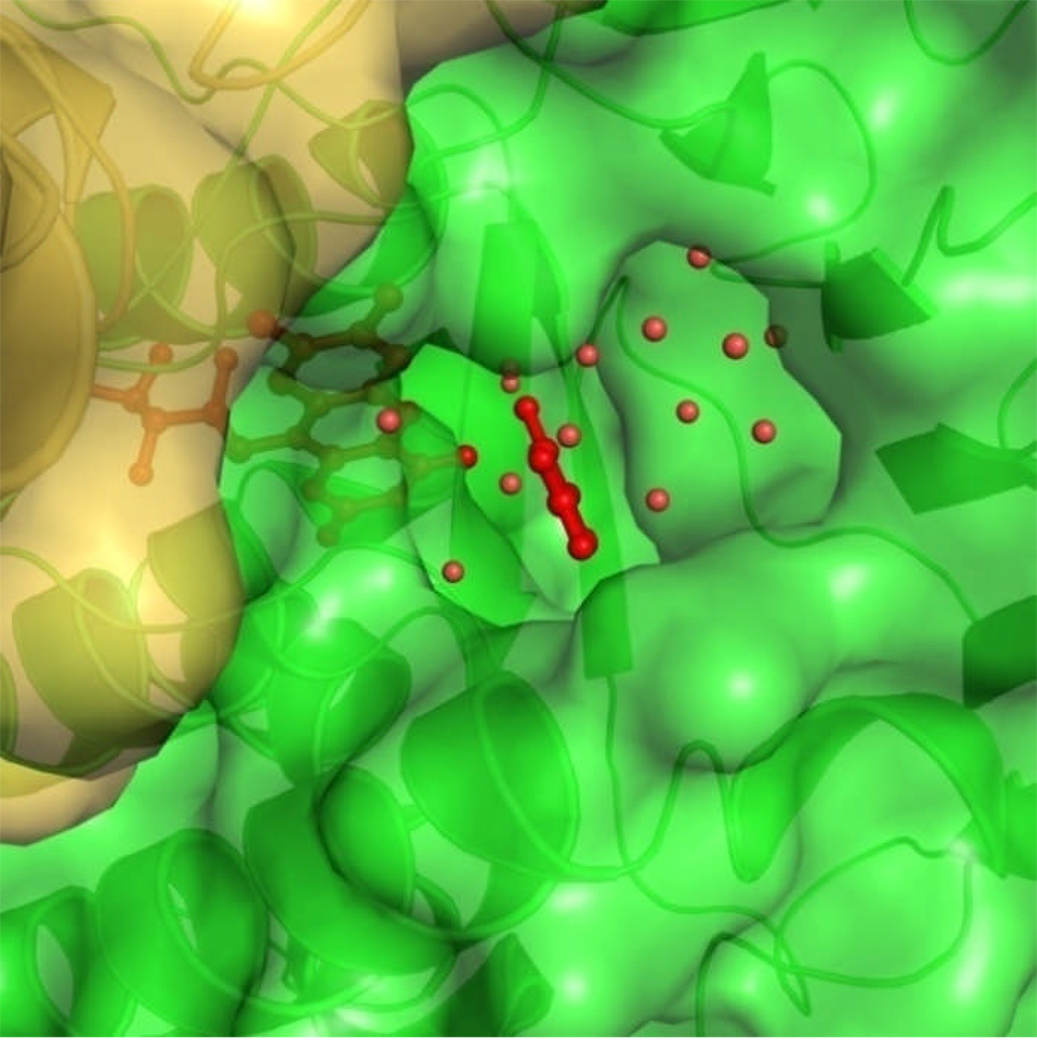
Soaking MHPCO crystals in a reservoir solution containing 10 mM MHPC for 3 h resulted in MHPC binding in each active site of the MHPCO dimer (Figure 5). The active site forms a pocket 15 Å by 10 Å by 12 Å and is much larger than the MHPC molecule. In addition to MHPC, nine water molecules are found in the binding pocket. A channel 7 Å wide lined with aromatic and hydrophobic residues points away from the FAD binding site leading from the back side of the dimer interface to the active site (Figure 6). This tunnel is roughly 17 Å long. MHPC binds beneath the loop connecting α9 to α10 and is stacked 5 Å above Tyr54. The C2 carbon atom of MHPC is 5.3 Å from the C4a atom of the isoalloxazine ring. MHPC has no direct interactions with the residues lining the active site; instead, all interactions are mediated through hydrogen bonds with water (Figure 5). The hydroxyl group is hydrogen bonded to Tyr223 through a water molecule (2.4 Å from the hydroxyl group to the water molecule and 2.7 Å to Tyr223) and N1 is coordinated to Tyr82 through another water molecule 2.6 Å from N1 and 3.0 Å from the hydroxyl group of Tyr82. Each oxygen atom of the carboxylate group is hydrogen bonded to a water molecule that is part of a water molecule network leading down the tunnel to the solvent. Arg211 is oriented 3.2 Å and 90° from the carboxylate group of MHPC and is positioned through a salt bridge with Asp201, but is poorly positioned to contribute a hydrogen bond to MHPC.
Figure 5.
Stereoview diagram of the active site of MHPCO with FAD and MHPC bound. Water molecules are shown as nonbonded red spheres. Hydrogen bonds between MHPC and water molecules and water molecules and the protein are shown as dashed lines.
Figure 6.
Space filling representation of the substrate tunnel leading to the binding pocket. FAD and MHPC are red and shown in ball and stick and the electrostatic surface of MHPCO is colored by protomer with green being the chain with FAD and MHPC bound and yellow being the opposing chain. Water molecules in the tunnel are shown as red spheres. The secondary structural elements are shown in cartoon representation.
DISCUSSION
Oligomeric State of MHPCO
Previous work on MHPCO in both Pseudomonas MA-1 and M. loti has identified MHPCO as a homotetramer using gel filtration and analytical ultracentrifugation. The stoichiometry of FAD in each tetramer has been less conclusively determined with different experiments indicating either two or four FAD molecules per tetramer (12–14, 33). The crystal structure of M. loti MHPCO suggests that each active site contains an FAD molecule, and sample preparation did not require excess flavin to be added during purification or crystallization. However, the MHPCO is a dimer in the crystal structures reported here. A dimer has only been reported for Pseudomonas MA-1 MHPCO apoenzyme where the FAD had been chemically removed from the active site (33). Activity assays indicate that samples of MlMHPCO with and without the His tag are equally active. It is still possible that the His tag disrupted the tetramer assembly as opposed to crystal packing. The His tag and the first ten residues of MHPCO are disordered in the crystal structures and Thr11 is solvent exposed at one end of the monomeric structure. It is conceivable that the His tag could have adopted a conformation that prevents the coupling of two dimers to form the tetramer. Examination of crystal packing shows some weak interactions between opposing dimers in adjacent asymmetric units. Arg47 hydrogen bonds to Ser44*, the carboxylate side chain of Asp123 forms hydrogen bonds to the amino side chain of Lys263*, and several water-mediated hydrogen bonds occur between the three stranded antiparallel β-sheet (β10↑β9↓β8) of one protomer and the loop region between α8 and β17 of the second protomer. These interactions could be remnants of a tetramer disrupted by the His tag or by crystal packing. To investigate the role the N-terminal His tag may have played in disrupting the tetramer, analytical ultracentrifugation was performed. The presence of the His tag does not appear to be sufficient to prevent tetramer formation in solution as shown in Figure 3C; however, it is possible that the His tag at the N-terminus could weaken the interactions occurring at the interface. In combination with crystallization conditions and crystal lattice packing, the tetramer may have been disrupted, resulting in the dimer observed in the crystal structures solved.
Comparison of MHPCO to Other Enzymes
Previous work has identified MHPCO as a member of the flavin aromatic monooxygenases on the basis of cofactor requirement, substrate structure, and sequence similarity (16, 34). Both BLAST and DALI searches were performed to determine which enzymes in this family are the closest structural relatives of MHPCO (35, 36). The BLAST search revealed that MHPCO is most closely related to flavin monooxygenases. The most similar proteins were hypothetical proteins, salicylate 1-monooxygenases, and FAD monooxygenases. These proteins had sequence identity with MHPCO of 22–25% and sequence similarity below 50%. Because of the nucleotide binding motif, the DALI search identified many enzymes as being structurally similar to MHPCO (Table 3). As expected, these enzymes are all flavin-dependent aromatic monooxygenases that use a flavin hydroperoxide intermediate to hydroxylate an aromatic ring (37–40). The overall fold of these enzymes is the same as that seen for MHPCO and first described for PHBH (41). The substrates for three of these enzymes, p-hydroxybenzoate hydroxylase (PHBH), m-hydroxybenzoate hydroxylase (MHBH), and 2,6-dihydroxypyridine hydroxylase (DHPH) are aromatic rings with a hydroxyl group ortho to the position being hydroxylated, as seen in MHPCO (42). The RMSD for each of these enzymes to MHPCO is less than 3 Å for at least 325 aligned residues with Z-scores of 26 or higher, and the structures are compared in Figure 7. Each of these enzymes bind FAD in a long narrow cleft stretching between two domains and bind very specifically to their respective substrates.
Table 3.
Enzymes Identified as Structurally Similar to MHPCO through DALI
| Protein | PDB ID | Z Score | RMSD | % Identical | # aligned residues |
|---|---|---|---|---|---|
| Pyocyanin biosynthetic enzyme, PhzS |
2RGJ | 35.1 | 2.3 | 25.7 | 325 |
| p-Hydroxybenzoate hydroxylase (PHBH) |
1D7L | 33.7 | 2.7 | 17.2 | 338 |
| 2,6-Dihydroxypyridine hydroxylase (DHPH) |
2VOU | 33.3 | 2.8 | 21.9 | 336 |
| Probable tryptophan hydroxylase, VioD |
3C4A | 32.6 | 2.8 | 20.1 | 329 |
| m-Hydroxybenzoate hydroxylase (MHBH) |
2DKI | 31.9 | 2.6 | 13.1 | 332 |
| Phenol hydroxylase | 1FOH | 31.9 | 2.8 | 14.0 | 341 |
| Rebeccamycin biosynthetic enzyme, RebC |
2R0G | 25.7 | 2.9 | 15.8 | 334 |
Figure 7.
Possible mechanism for the reaction catalyzed by MHPCO. The aromatic ring is opened through an electrocyclic mechanism after pyridine hydroxylation by the flavin hydroperoxide.
The mechanism of substrate binding within these enzymes appears to be different. Some enzymes, such as PhzS and MHBH, have a tunnel that provides access to the binding pocket without requiring a conformational change in the FAD (38, 43). Other structurally similar enzymes do not have such a channel. For example, DHPH could not be crystallized with substrate because the ligand would not bind. PHBH is believed to require the flavin to adopt multiple, different conformations over the cycle of reaction. The flavin is found in an “IN” conformation when p-hydroxybenzoate (p-OHB) is bound, an “OPEN” conformation before p-OHB binds, and an “OUT” conformation when undergoing reduction by NAD(P)H (40, 44, 45). RebC appears to have a helix that can undergo conformational change upon substrate binding (39). Examination of MHPCO suggests that MHPC most likely enters the active site through a tunnel (Figure 6) without inducing any large conformational changes of the protein. Tyr270, Trp173, Arg211, Met227, Asp177, and Arg350 line the channel and form the mouth of the active site. Despite the presence of a substrate tunnel in several of the flavin monooxygenases, in MHPCO the tunnel is in a different location compared to PhzS and MHBH. Moreover, the MHBH tunnel has two openings leading to the surface of the protein, but these are pointed in different directions. One opening of MHBH is suggested to be for molecular oxygen to enter the active site and the second, filled with water molecules in the structure, is postulated to be the tunnel for substrate entrance (38). The channel of MHPCO does not have two entrances and does not appear to be as wide as that of MHBH although it is also filled with water molecules.
The binding of FAD by MHPCO also shares many similarities with other flavin monooxygenases but has some significant differences. In all structures of flavin aromatic monooxygenases determined, the FAD binds lengthwise in a groove contained within one protomer. The adenine base in all structures is bound in a pocket between arginine and aspartate residues and forms hydrogen bonds to the amide nitrogen atom of the protein backbone. A conserved glutamate residue (Glu41) is pointed toward the hydroxyl groups of the ribose ring in all structures. The phosphate tail is coordinated by water molecules hydrogen bonded to the protein backbone and several residues along the FAD are conserved, including Gly18, Gly23, and Asp288. The isoalloxazine ring is held in the “IN” conformation when the substrate is bound to the active site. In some structures where ligand is not bound, the isoalloxazine ring has been found in the “OUT” conformation. The crystal structures of PhzS, RebC, and MHBH (PDB ID 2rgj, 2roc, and 2dkh respectively) show the flavin in the “OUT” conformation. In contrast to these enzymes, the FAD of MHPCO remains in the “IN” conformation and shows very little movement, regardless of the presence or absence of substrate (Figure 4). Using PHBH as a model, it has been previously proposed that NADH could bind parallel to the FAD molecule for transfer of the hydride to yield the reduced flavin (13). However, the binding site for NADH in MHPCO has not been definitively identified. Additionally, for FAD in MHPCO to adopt the “OUT” conformation seen in PhzS, the loop connecting β2 and β3 would have to move 3 to 4 Å to accommodate the isoalloxazine ring. The “OUT” conformation of FAD in PhzS is stabilized largely through π stacking with a tryptophan residue found on a loop that extends to the active site. This loop structure in MHPCO, between β16 and α7, is much shorter with only 8 residues instead of 16 residues, when compared to the PhzS structure and does not extend to cover the active site of MHPCO. The π stacking between the flavin in MHBH occurs with a tyrosine residue. The equivalent aromatic residue in MHPCO, Tyr270, is one residue shifted and is the most likely residue to provide π stacking to FAD if the “OUT” conformation were adopted. However, studies done by Chaiyen et al. using FAD analogues substituted at the 8-position showed that the effects on rate of reduction were due solely to the changes in redox potential and that the size of the substituent had no effect (46). This suggests that flavin movement may not play a role in FAD reduction for MHPCO and raises the possibility that FAD could remain in the “IN” conformation during reduction by NADH. The FAD of MHPCO may be reduced by NADH in a manner different than for other flavin aromatic monooxygenases.
Active Site of MHPCO
The active site of MHPCO is a large pocket lined mostly by hydrophobic and aromatic residues (Figure 5). These residues include Tyr54, Tyr82, Tyr223, Phe358, Trp203, Asp 203, and Arg211. The hydroxyl groups of the tyrosine residues point into the active site and the side chain of Arg211 is also positioned within the active site. These residues are hydrogen bonded to the many water molecules that are found within the active site in both structures. Comparisons of the active sites of flavin aromatic monooxygenases whose structures are available show some similarities. All structures have an absolutely conserved proline residue above the FAD isoalloxazine ring containing C7 and C8 (Pro295 in MHPCO). The other commonality is the presence of several aromatic residues at the back of the binding pocket. MHPCO has three tyrosine residues, two tryptophan residues, and two phenylalanine residues in the rear of the active site and none form any direct contacts with the substrate MHPC. PHBH (PDB ID 1pbe) and DHPH have a similar wall in the active site. p-Hydroxybenzoate (p-OHB), the substrate of PHBH, forms a hydrogen bond to a tyrosine hydroxyl group, which can deprotonate p-OHB for activation. The carboxylate group of p-OHB forms a salt bridge with an arginine side chain, helping correctly orient the substrate within the active site. The binding of MHPC within the active site of MHPCO is quite different. Unlike PHBH, MHPCO makes no direct interactions with its substrate (Figure 5). The 3-hydroxyl group of MHPC forms a water-mediated hydrogen bond to Tyr223 and the N1 atom is hydrogen bonded to Tyr82 through a water molecule. The carboxyl oxygen atoms are each hydrogen bonded to two water molecules that lead into the tunnel. Arg211 is one of the few charged residues found in the active site and it is poorly positioned to help stabilize MHPC. The surprising lack of direct interactions between MHPC and MHPCO and the large open cavity of the active site is unusual and differentiates MHPCO from other members of this enzyme family.
Mechanistic Implications of the MHPCO Structure
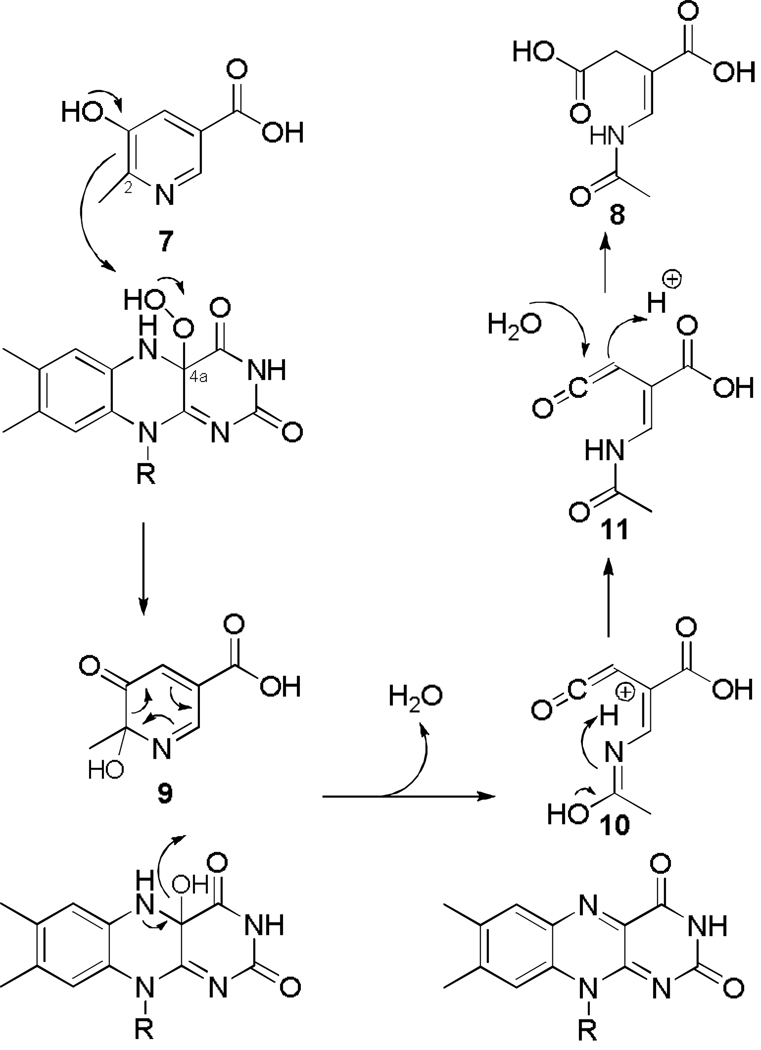
One possible mechanism for the oxidative ring opening of MHPC is shown in Figure 7. In this mechanism, the hydroxylation of MHPC first occurs by an electrophilic attack by the flavin hydroperoxide on C2 of the pyridine 7 to give 9, which could then undergo an electrocyclic ring opening to produce the ketene 10. Tautomerization to 11 followed by addition of water to the ketene would generate the product E-2AMS 8. Several other variations on this proposal for the conversion of 9 to 8 are possible. (34). This mechanism is consistent with all previous 18O2 and H2 18O labeling experiments, with substrate analogue studies, and with kinetics data (16, 33, 47, 48) and the current structure of MHPCO bound with MHPC seems to position the substrate optimally for this attack to occur.
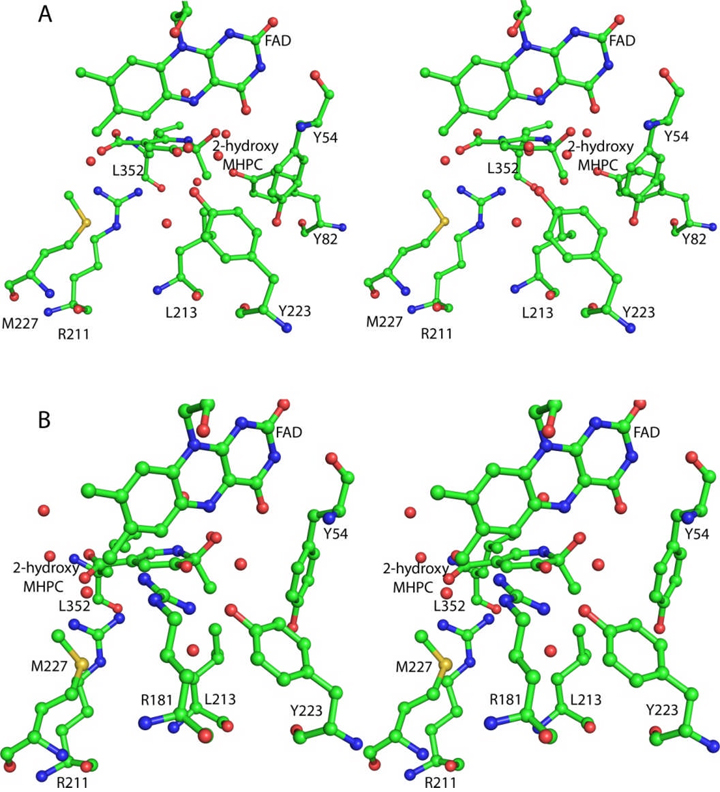
The structure of MHPCO complexed with the substrate MHPC 7 clarifies many of the details of the active site and provides further insight into the catalytic mechanism. The C2 of MHPC is located 5.3 Å from C4a of the isoalloxazine ring. This separation, in addition to the orientation of the planes of the pyridine and the isoalloxazine rings, gives a reasonable geometry for reaction of the distal oxygen atom of the flavin hydroperoxide with C2 of the substrate. The position of this oxygen is indicated by structurally conserved water molecules seen in both structures of MHPCO described here. Comparable substrate-C4a distances have been observed in other aromatic ring oxidizing flavin-dependent monooxygenases. Tyr223 and Tyr82 are likely catalytic residues (Figure 8A). Tyr223 could activate the water molecule located 2.7 Å from the substrate C3 oxygen and facilitate the conversion of 7 to 9. Tyr82 is hydrogen bonded to the water molecule located 2.6 Å from the pyridine nitrogen and may facilitate the conversion of 10 to 11. Arg181 is also a possible candidate for an active site base. In the crystal structure, this residue is oriented along the isoalloxazine ring but a conformational change of the side chain may position the guanidinium group within hydrogen bonding distance of the substrate C3 oxygen (modeled in Figure 8B). However, for Arg181 to adopt this conformation some conformational changes of the active site would have to occur to avoid close contacts between the isoalloxazine ring and the arginine. Removal of the hydroxyl group of Tyr223 (Y223F) or substitution of the guanidinium group of Arg181 with an amide group (R181Q) both render the enzyme inactive. Arg211 is located below the plane of the pyrimidine ring and its guanidinium group is facing the C5 carboxylate of MHPC. While poorly positioned to form a salt bridge in the enzyme substrate complex, conformational changes during the course of the reaction could increase the strength of this interaction.
Figure 8.
Stereoview diagrams of the active site with possible bases for the ring opening reaction. All residues and ligands are shown in ball and stick representation and colored according to atom type. Red nonbonded spheres are water molecules. The 2-hydroxy-MHPC intermediate 9 was manually modeled into the active site using the binding of MHPC as a guide. A. Tyr223 as the active site acid. Tyr223 activates a water molecule for attack at the C3 carbonyl and could provide stabilization of the intermediate. B. Arg181 as the active site base. Arg181 could protonate the carbonyl and stabilize the intermediate after attack of water at the carbonyl. Arg181 has adopted a different conformation than that seen in the crystal structure.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Yasunobu Ohkawa, National Institute of Agrobiological Sciences, Ibaraki, Japan for providing us with the M. loti/ MAFF303090 strain. The authors are grateful to the staff scientists at the APS NE-CAT beamlines for their assistance in data collection. We would also like to thank the Cornell University Protein Facility for providing the clones and assistance with the analytical ultracentrifugation experiments. Leslie Kinsland’s assistance during manuscript preparation is appreciated.
ABBREVIATIONS
- PLP
Pyridoxal 5′-phosphate
- MHPCO
2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
- MHPC
2-methyl-3-hydroxypyridine-5-carboxylic acid
- PHBH
p-Hydroxybenzoate hydroxylase
- FAD
flavin adenine dinucleotide
- E-2AMS
E-2-(acetamidomethylene)succinate
- SeMet
selenomethionyl
- β-ME
β-mercaptoethanol
- Tris
tris(hydroxymethyl)aminomethane base
- DTT
dithiothreitol
- MHPD
2-methyl-3-hydroxypyridine-4,5-dicarboxylic acid
- MHBH
3-hydroxybenzoate hydroxylase
- DHPH
2,6-dihydroxypyridine hydroxylase
- p-OHB
p-hydroxybenzoate
- 5-HN
5-hydroxynicotinic acid
Footnotes
This research was supported by a grant from the National Institutes of Health (GM069618). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
The Brookhaven Protein Data Bank codes for MHPCO and the MHPCO-MHPC complex are 3GMB and 3GMC respectively.
BIBLIOGRAPHY
- 1.Snell EE, Haskell BE. The Metabolism of Vitamin B6, In: Comprehensive Biochemistry. Vol. 21. New York: Elsevier/North Holland; 1971. [Google Scholar]
- 2.Burg RW, Rodwell VW, Snell EE. Bacterial oxidation of vitamin B6. II. Metabolites of pyridoxamine. J. Biol. Che. 1960;235:1164–1169. [PubMed] [Google Scholar]
- 3.Rodwell VW, Volcani BE, Ikawa M, Snell EE. Bacterial oxidation of vitamin B6. I. Isopyridoxal and 5-pyridoxic acid. J. Biol. Chem. 1958;233:1548–1554. [PubMed] [Google Scholar]
- 4.Ikawa M, Rodwell VW, Snell EE. Bacterial oxidation of vitamin B6. II. Structure of 260 compound. J. Biol. Chem. 1958;233:1555–1559. [PubMed] [Google Scholar]
- 5.Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Mochizuki Y, Nakayama S, Nakazaki N, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 6.Funami J, Yoshikane Y, Kobayashi H, Yokochi N, Yuan B, Iwasaki K, Ohnishi K, Yagi T. 4-Pyridoxolactonase from a symbiotic nitrogen-fixing bacterium Mesorhizobium loti: cloning, expression, and characterization. Biochim. Biophys. Acta. 2005;1753:234–239. doi: 10.1016/j.bbapap.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Ge F, Yokochi N, Yoshikane Y, Ohnishi K, Yagi T. Gene identification and characterization of the pyridoxine degradative enzyme 4-pyridoxic acid dehydrogenase from the nitrogen-fixing symbiotic bacterium Mesorhizobium loti MAFF303099. J. Biochem. 2008;143:603–609. doi: 10.1093/jb/mvn010. [DOI] [PubMed] [Google Scholar]
- 8.Mukherjee T, Hilmey DG, Begley TP. PLP catabolism: identification of the 2-(Acetamidomethylene)succinate hydrolase gene in Mesorhizobium loti MAFF303099. Biochemistry. 2008;47:6233–6241. doi: 10.1021/bi8004158. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee T, Kinsland C, Begley TP. PLP catabolism: identification of the 4-pyridoxic acid dehydrogenase gene in Mesorhizobium loti MAFF303099. Bioorg. Chem. 2007;35:458–464. doi: 10.1016/j.bioorg.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukherjee T, McCulloch KM, Ealick SE, Begley TP. Gene identification and structural characterization of the pyridoxal 5'-phosphate degradative protein 3-hydroxy-2-methylpyridine-4,5-dicarboxylate decarboxylase from Mesorhizobium loti MAFF303099. Biochemistry. 2007;46:13606–13615. doi: 10.1021/bi701439j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan B, Yokochi N, Yoshikane Y, Ohnishi K, Ge F, Yagi T. Gene identification and characterization of the pyridoxine degradative enzyme alpha-(N-acetylaminomethylene) succinic acid amidohydrolase from Mesorhizobium loti MAFF303099. J. Nutr. Sci. Vitaminol. (Tokyo) 2008;54:185–190. doi: 10.3177/jnsv.54.185. [DOI] [PubMed] [Google Scholar]
- 12.Yuan B, Yokochi N, Yoshikane Y, Ohnishi K, Yagi T. Molecular cloning, identification and characterization of 2-methyl-3-hydroxypyridine-5-carboxylic-acid-dioxygenase-coding gene from the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. J. Biosci. Bioeng. 2006;102:504–510. doi: 10.1263/jbb.102.504. [DOI] [PubMed] [Google Scholar]
- 13.Chaiyen P, Ballou DP, Massey V. Gene cloning, sequence analysis, and expression of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7233–7238. doi: 10.1073/pnas.94.14.7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sparrow LG, Ho PP, Sundaram TK, Zach D, Nyns EJ, Snell EE. The bacterial oxidation of vitamin B6. VII. Purification, properties, and mechanism of action of an oxygenase which cleaves the 3-hydroxypyridine ring. J. Biol. Chem. 1969;244:2590–2600. [PubMed] [Google Scholar]
- 15.van Berkel WJ, Kamerbeek NM, Fraaije MW. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 2006;124:670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 16.Chaiyen P, Brissette P, Ballou DP, Massey V. Unusual mechanism of oxygen atom transfer and product rearrangement in the catalytic reaction of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase. Biochemistry. 1997;36:8060–8070. doi: 10.1021/bi970089u. [DOI] [PubMed] [Google Scholar]
- 17.Oonanant W, Sucharitakul J, Yuvaniyama J, Chaiyen P. Crystallization and preliminary X-ray crystallographic analysis of 2-methyl-3-hydroxypyridine-5-carboxylic acid (MHPC) oxygenase from Pseudomonas sp. MA-1, Acta Crystallogr. Sect.F Struct. Biol. Cryst. Commun. 2005;61:312–314. doi: 10.1107/S1744309105004367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausubel FM, Brent F, editors. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1987. [Google Scholar]
- 19.Sambrook J, Fritsch GF, Maniatis T. Molecular Cloning: A Laboratory Guide. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 21.Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Cryst. 2004;37:843–844. [Google Scholar]
- 24.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallog. D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 25.Sheldrick GM. Macromolecular phasing with SHELXE. Z. Kristallog.r. 2002;217:644–650. [Google Scholar]
- 26.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 30.DeLano WL. The PyMOL Molecular Graphics Systems. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 31.Hasel W, Hendrickson TF, Still WC. A rapid approximation to the solvent-accessible surface areas of atoms. Tetrahedron Comp. Meth. 1988;1:103–116. [Google Scholar]
- 32.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishore GM, Snell EE. Interaction of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase with FAD, substrates, and analogues. Spectral and fluorescence investigations. J. Biol. Chem. 1981;256:4234–4240. [PubMed] [Google Scholar]
- 34.Chaiyen P, Brissette P, Ballou DP, Massey V. Reaction of 2-methyl-3-hydroxypyridine-5-carboxylic acid (MHPC) oxygenase with N-methyl-5-hydroxynicotinic acid: studies on the mode of binding, and protonation status of the substrate. Biochemistry. 1997;36:13856–13864. doi: 10.1021/bi9715122. [DOI] [PubMed] [Google Scholar]
- 35.Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Entsch B, van Berkel WJ. Structure and mechanism of para-hydroxybenzoate hydroxylase. FASEB J. 1995;9:476–483. doi: 10.1096/fasebj.9.7.7737455. [DOI] [PubMed] [Google Scholar]
- 38.Hiromoto T, Fujiwara S, Hosokawa K, Yamaguchi H. Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site. J. Mol. Biol. 2006;364:878–896. doi: 10.1016/j.jmb.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 39.Ryan KS, Howard-Jones AR, Hamill MJ, Elliott SJ, Walsh CT, Drennan CL. Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15311–15316. doi: 10.1073/pnas.0707190104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Treiber N, Schulz GE. Structure of 2,6-dihydroxypyridine 3-hydroxylase from a nicotine-degrading pathway. J. Mol. Biol. 2008;379:94–104. doi: 10.1016/j.jmb.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 41.Wierenga RK, de Jong RJ, Kalk KH, Hol WG, Drenth J. Crystal structure of p-hydroxybenzoate hydroxylase. J. Mol. Biol. 1979;131:55–73. doi: 10.1016/0022-2836(79)90301-2. [DOI] [PubMed] [Google Scholar]
- 42.Joosten V, van Berkel WJ. Flavoenzymes. Curr. Opin. Chem. Biol. 2007;11:195–202. doi: 10.1016/j.cbpa.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Greenhagen BT, Shi K, Robinson H, Gamage S, Bera AK, Ladner JE, Parsons JF. Crystal structure of the pyocyanin biosynthetic protein PhzS. Biochemistry. 2008;47:5281–5289. doi: 10.1021/bi702480t. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Ortiz-Maldonado M, Entsch B, Massey V, Ballou D, Gatti DL. Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase. Proc. Natl. Acad. Sci. U. S. A. 2002;99:608–613. doi: 10.1073/pnas.022640199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole LJ, Entsch B, Ortiz-Maldonado M, Ballou DP. Properties of p-hydroxybenzoate hydroxylase when stabilized in its open conformation. Biochemistry. 2005;44:14807–14817. doi: 10.1021/bi0512142. [DOI] [PubMed] [Google Scholar]
- 46.Chaiyen P, Sucharitakul J, Svasti J, Entsch B, Massey V, Ballou DP. Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase. Biochemistry. 2004;43:3933–3943. doi: 10.1021/bi035734d. [DOI] [PubMed] [Google Scholar]
- 47.Chaiyen P, Brissette P, Ballou DP, Massey V. Thermodynamics and reduction kinetics properties of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase. Biochemistry. 1997;36:2612–2621. doi: 10.1021/bi962325r. [DOI] [PubMed] [Google Scholar]
- 48.Kishore GM, Snell EE. Kinetic investigations on a flavoprotein oxygenase, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase. J Biol Chem. 1981;256:4228–4233. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.