Abstract
The extreme pathological diversity of non-Hodgkin’s lymphomas has made their accurate histological assessment difficult. New diagnostics and treatment modalities are urgently needed for these lymphomas, particularly in drug development for cancer-specific targets. Previously, we showed that a subset of B cell lymphoma, diffuse large B cell lymphoma, may be characterized by two major, orthogonal axes of gene expression: one set of transcripts that is differentially expressed between resting and proliferating, nonmalignant cells (ie, a “proliferative signature”) and another set that is expressed only in proliferating malignant cells (ie, a “cancer signature”). A differential proteomic analysis of B cell proliferative states, similar to previous transcriptional profiling analyses, holds great promise either to reveal novel factors that participate in lymphomagenesis or to define biomarkers of onset or progression. Here, we use a murine model of diffuse large B cell lymphoma to conduct unbiased two-dimensional gel electrophoresis and mass spectrometry-based comparative proteomic analyses of malignant proliferating B cells and tissue-matched, normal resting, or normal proliferating cells. We show that the expression patterns of particular proteins or isoforms across these states fall into eight specific trends that provide a framework to identify malignancy-associated biomarkers and potential drug targets, a signature proteome. Our results support the central hypothesis that clusters of proteins of known function represent a panel of expression markers uniquely associated with malignancy and not normal proliferation.
Lymphomas, which include non-Hodgkin’s lymphoma, are the fifth most common type of diagnosed cancer in males, the sixth most common type in females and the fifth most common cause of cancer mortality in the United States.1 Current standard chemotherapy/antibody-directed immunotherapy regimens are effective in only 40% of cases.2 Non-Hodgkin’s lymphoma is more common than Hodgkin’s lymphoma, with 16,000 new cases diagnosed annually. Moreover, patients with aggressive forms of non-Hodgkin’s lymphoma, such as diffuse large B cell lymphoma (DLBCL) with poor scores on the revised International Prognostic Index, have a four year overall survival of 55% when treated with the current standard of care.3 More effective early diagnostic measures are urgently needed, such as sensitive biomarker assays that exploit proteomic signatures potentially unique to aggressive lymphomas.
Major progress in lymphoma diagnosis and prognosis has been built on recent advances in genome-wide transcriptional profiling methods.4,5 For example, it has been possible to resolve transcriptional signatures of B cells of different origins and proliferative states (eg, normal germinal center B cells, mitogenically activated B cells, tonsillar B cells, and resting peripheral B cells, all exhibiting unique and resolvable transcriptional signatures), as well as clinical samples of DLBCL malignancies of greater or lesser aggressiveness that are otherwise indistinguishable histologically.5,6,7 However, there remains a major challenge in the development of useful biomarkers: the identification of a small, relevant set of functionally important targets from the vast sums of genomic, proteomic, metabolomic or transcriptomic information of uncertain etiological significance that can be gathered from diseased cells or tissues. Biomarkers that display robust prognostic significance or confer insight into disease mechanism are difficult to discern, even by the most sophisticated statistical analyses,6 within immense data sets derived from analyses of “normal” and diseased tissues.7
We have established that one reasonable approach to simplifying the complexity of this task in the context of discovery of biomarkers and potential drug targets in malignancies is to subtract out the largest possible set of normal tissue-derived background signals from diseased tissue signals: in the case of B cell lymphomas, to subtract out signals from all of the nonmalignant B cell proliferative states (such as resting B cells and normal proliferating B cells) from those of malignant B cells, thus enriching for potential disease biomarker signals of functional importance. As a model system for DLBCL, we used our previously developed transgenic murine model based on Eμ-driven, B cell-restricted, constitutive expression of the double bromodomain protein Brd2 (Tg).8 This DLBCL model is well characterized, stable and exhibits monoclonal expansion of only mature B cells; it offers a highly reproducible system for the determination of genome-wide and proteome-wide biomarkers for lymphoma expansion and progression.8 The splenic lymphoma’s genome-wide transcriptional signature is most similar to the “activated B cell-like” form of human DLBCL,8,9 the more aggressive subtype, which is associated with a worse prognosis.3,4,5,6 Previously, we compared genome-wide transcriptional expression profiles of independently arising Tg splenic lymphomas with those of nonmalignant proliferating or normal resting primary splenic B cells from a syngeneic, inbred strain of mice, which controlled as much as possible for variation between individuals and stromal microenvironments. Principal component analysis10 of these signals identified two distinct axes of differential gene expression. One group of genes, which we called the “proliferation signature,” was differentially expressed between normal resting primary B cells and mitogenically stimulated B cells. Another group of genes, which we called the “cancer signature,” was differentially expressed along an orthogonal axis of gene expression unrelated to normal proliferation. This axis included genes specific to lymphomagenesis, progression and survival, several human orthologs of which are implicated in human lymphomagenesis. Furthermore, we identified seven statistically distinct clusters of gene expression that agreed with cellular function and pathology.9 Thus, we were able to use normal B cell counterparts in a subtractive fashion to identify genes of functional importance for the malignant B cell that are uniquely relevant to cancer.
Although transcriptional profiling has matured as a diagnostic and prognostic research tool in pathology11 and has been widely used to characterize malignancies as diverse as renal cell carcinoma,12 prostate cancer,13 metastatic bone cancer14 and breast cancer,15 protein-based biomarkers may have potentially greater diagnostic power than genetic or transcriptional profiles because they may reflect disease-related alterations to tissue that are invisible to gene-based analyses.16 Protein expression and activity are subject to additional layers of metabolic regulation over and above the regulation of gene expression.17 Moreover, disease states are often associated with deregulation of protein expression or function, which may include significant posttranslational modification, truncation, mislocalization within the cell and other variations.18 Many cancers, in particular, show deregulated activity of growth factor receptor tyrosine kinases, which in turn alter protein phosphorylation patterns within the cell, and which genome-wide transcriptional analysis is unable to capture. Furthermore, although protein biomarkers may originally be identified through tissue- or proximal fluid-based analyses, it is likely that they may be detectable as species shed from the site of disease into blood or other body fluids, which may be obtained noninvasively. Thus, protein biomarkers potentially represent a powerful source of disease diagnostics. Proteomic analysis, therefore, can play an important, non-redundant role in the identification and characterization of potential biomarkers and targets of cancer intervention.19
Several studies have used advanced proteomics methods and statistical analyses to attempt to distinguish normal and cancer specimens for lung,20 ovarian,21 and breast cancer.22 Based on our transcriptome results, we framed a similar hypothesis: nonmalignant proliferating B cells and malignant proliferating B cells will share the induction of important proliferation-associated proteins in comparison with non-proliferating controls; however, malignant cells will show additional changes in protein expression, including posttranslational modification, that are unique to their malignant state, over and above those changes associated with normal, mitogen-stimulated proliferation alone. Here, we have investigated this hypothesis, using a differential proteomic approach within our model of subtractive profiling of B cell lymphomas to develop a framework to evaluate potential protein biomarkers that are malignancy-associated. Specifically, we have conducted differential proteomics23 using subfractionated protein nuclear lysate from resting and proliferating normal B cells and proliferating malignant B cells. We identified the major protein species that differ between these states and show that distinct patterns of expression define a potentially functionally relevant, malignancy-associated protein expression pattern.
Materials and Methods
Preparation of Murine Normal and Malignant B Cell Extracts
Mouse splenic B cells were isolated by magnetic cell sorting-based magnetic bead separation with anti-CD43 negative selection, as previously described.9 Normal resting B cells were stimulated in vitro with 30 μg/ml Salmonella typhimurium lipopolysaccharide for 48 hours. Unless otherwise described, all reagents were from Sigma (St. Louis, MO). Percoll (Amersham Biosciences, Piscataway, NJ) gradient centrifugation was used to enrich for stimulated B cells, while removing cellular debris, apoptotic cells and resting B cells. Percoll gradients of 50%, 60%, 66%, and 70% were prepared from an ice-cold Percoll solution supplemented with Hanks’ balanced salt solution (Invitrogen/GIBCO-BRL, Grand Island, NY). The Eμ-BRD2-driven large B cell lymphoma, adoptively transferred between mice, provided a constant source of lymphoma cells for the study.8 After sublethal irradiation (6 Gy), mice were inoculated by intraperitoneal injection of 107 cells. Mice were maintained on antibiotic water (trimethoprim and sulfamethoxazole). Malignant splenic B cells were purified and cell extractions performed as previously described.24 Tumor cells were obtained from female FVB mice that were 6 to 8 weeks old and normal cells were obtained from syngeneic age-matched female controls housed under the same conditions. Animals were handled humanely in accordance with Federal and institutional requirements; this study was conducted with Boston University Institutional Animal Care and Use Committee authorization.
Two-Dimensional Polyacrylamide Gel Electrophoresis Fractionation of Cell Extracts and Gel Image Analysis
Two-dimensional (2D) polyacrylamide gel electrophoresis (PAGE) separation defined 2D reference maps. B cell-soluble nuclear extracts were prepared and subjected to size exclusion chromatography with Bio-Gel P-60 gel (Bio-Rad). This step enriched for proteins with a molecular mass >60 kd, which significantly improved 2D gel electrophoresis and resulted in more accurate mass spectrometry protein identifications. Proteins were acetone-precipitated and solubilized in DeStreak rehydration solution (Amersham Biosciences). First-dimension isoelectric focusing was performed overnight on a Protean IEF cell (Bio-Rad, Richmond, CA) with pI 3–10 immobilized pH gradients, in accordance with the manufacturer’s recommendations, in the presence of 1 mmol/L dithiothreitol. In-gel reduction and alkylation was performed with dithiothreitol and iodoacetamide before running second dimension electrophoresis. Second dimension sodium dodecyl sulfate-PAGE was performed on 10% Tris-HCl gels or on 4 to 12% gradient bis-Tris gels. Gels were washed in water, followed by colloidal Coomassie staining overnight (or until protein spots were sufficiently visualized). Alternatively, gel replicates were fixed and stained with Plus One silver stain (GE HealthCare, Piscataway, NJ).
In-Gel Digestion Methods
Resolved, stained and quantitated protein spots of interest were excised, destained, washed extensively, and digested with trypsin, as previously described.25 Gel plugs were destained three times with 100 mmol/L ammonium bicarbonate (pH 8.8)/50% acetonitrile (Fisher Scientific, Pittsburgh, PA), and subjected to four rounds of washing with alternating solutions of 100 mmol/L ammonium bicarbonate (pH 8.8), 100 mmol/L ammonium bicarbonate (pH 8.8)/50% acetonitrile and 100% acetonitrile. Trypsin (Trypsin Gold, Promega, Madison, WI) digestion was conducted at 37°C overnight after swelling the gel plugs in digestion solutions containing a 1:10 enzyme-substrate (w/w) ratio (estimating substrate quantity by relative staining intensity of original spot) and 50 mmol/L ammonium bicarbonate (pH 8.8)/5% acetonitrile. Peptides were extracted from the gel pieces twice with alternating solutions of 20 mmol/L ammonium bicarbonate (pH 8.8), 1% trifluoroacetic acid/50% acetonitrile and 100% acetonitrile. Extracts were pooled and subjected to desalting with microreversed phase chromatography (ZipTips, Millipore, Bedford, MA).
Mass Spectrometric Data Acquisition
Matrix-assisted laser desorption ionization/time of flight mass spectrometry (MS) of purified peptides was conducted with a Bruker Reflex IV mass spectrometer (Bruker Daltonics, Billerica, MA) using the matrix, 2,5-dihydroxybenzoic acid, and an AnchorChip target (Bruker Daltonics). Mass spectra were internally recalibrated to within 50 ppm mass accuracy using known peptide ions and peak lists were generated using the software MoverZ (Genomic Solutions, Ann Arbor, MI).
Data Analysis
Gel images were captured with a VersaDoc3000 (Bio-Rad) and were processed with Proteomeweaver (Bio-Rad), for generation of aligned and warped gel overlay images for graphical representation. In-depth gel image analyses was performed with SameSpots (Nonlinear Dynamics, Durham, NC), for cross-experiment gel alignment, feature identification, feature volume quantification, expression, principal component analysis and hierarchical clustering analyses. Peak lists were submitted for peptide mass fingerprint database search with Mascot (Matrix Science, Boston, MA). The databases used were SwissProt version 50.3 and NCBInr 20060718, limiting the searches to Mus musculus entries, tryptic peptides with up to one missed cleavage, carbamidomethylation of cysteines, variable oxidation of methionines, and an error tolerance within 80 ppm. Under these searching parameters, Mascot scores of >53 for the SwissProt database and scores of >63 for the NCBInr database corresponded to statistically significant (P ≤ 0.05) protein assignments. Protein functional assignment, Gene Ontology (GO) term annotation and graphical rendering were accomplished with software written in-house that mines protein GO annotations from the public repository available through the http://uniprot.org website.
Results
To define biomarkers associated with lymphoid malignancies more effectively, we previously used a Tg model for DLBCL to demonstrate that B cell gene expression may be characterized by both a proliferation signature and an orthogonal cancer signature.9 Consequently, we hypothesize here that on subtraction of proteomic signatures of normal resting and nonmalignant proliferating B cells from the proteome of malignant B cells, we will be able to categorize a set of unique protein biomarkers that define proliferating malignant B cells, distinct from nonmalignant proliferating cells. We have explored this hypothesis by undertaking a 2D-PAGE and mass spectrometric-based proteomic analysis of our Tg DLBCL, in relation to syngeneic, normal B cells, both resting and proliferating.
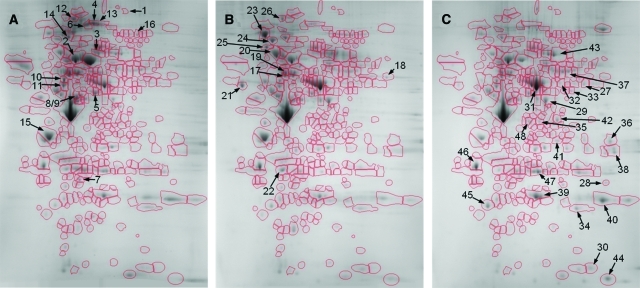
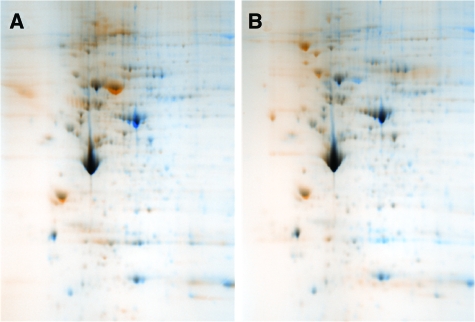
Two-dimensional proteomic reference maps were produced for each of the three B cell proliferative states: normal resting B cells (Figure 1A), nonmalignant proliferating B cells (Figure 1B) and malignant B cells (Figure 1C). Differential image analysis allowed the comparison of the 2D reference map for the lymphoma state both to that of the resting state and to that of the nonmalignant proliferating state (Figure 2, A and B). Above the background of similarity, particular protein spot features were seen to change robustly between the three states.
Figure 1.
Example of 2D reference maps of resting, proliferating, and lymphoma B cell proteomes. Nuclear extract was isolated from spontaneous Tg B cell lymphomas, syngeneic resting splenic B cells, and B cells mitogenically stimulated to proliferate in culture. Lysate was subjected to desalting by size exclusion chromatography and then to 2D gel electrophoresis over the pI range of 3–10 and the molecular mass range of approximately 10 to 250 kd, followed by Coomassie staining. Gel images were warped, aligned and analyzed for changes in protein expression allowing for equivalent spot features defined across all data sets. A: Resting B cells. B: Normal, mitogenically stimulated, proliferating B cells. C: Proliferating lymphoma B cells. The gel region of approximately pI 4–9 (from left to right) and molecular mass 250 to 15 kd (from top to bottom) are shown, with spot features outlined and numbered according to the protein assignments reported in Table 1.
Figure 2.
Comparative proteomic analysis of B cell proliferative states. Differential image analysis was conducted using warped and aligned 2D proteome reference maps for lymphoma B cells in comparison with normal resting and mitogenically stimulated, proliferating B cells. Overlapping areas of protein expression appear black while non-overlapping areas that are unique to each state are shown in color. A: Lymphoma B cells (blue) versus normal resting B cells (orange). B: Lymphoma B cells (blue) versus normal, mitogenically stimulated, proliferating B cells (orange).
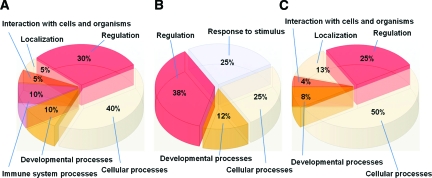
We identified major proteins that underwent dramatic expression changes between B cell proliferative states through the use of in-gel trypsin digestion, matrix-assisted laser desorption ionization/time of flight MS and peptide mass fingerprint analyses (Table 1). Relative expression levels (as indicated in Table 1) of this set of proteins across all of the states showed that they could be assigned to three distinct groups: those that were up-regulated in the resting state with respect to the other two states (Table 1, first grouping), those that were up-regulated in the nonmalignant proliferative state with respect to the other two states (Table 1, second grouping), and those that were up-regulated in the lymphoma state with respect to the other two states (Table 1, third grouping). GO functional assignments (within the GO supercategory of Biological Process) highlight functional differences between the major up-regulated proteins in each B cell proliferative state (Figure 3, A–C).
Table 1.
Assignments for the Major Proteins That Differ among B Cell Proliferative States, as Determined by Mass Spectrometry and Mass Fingerprint Analyses
| Protein no. | Protein name | Accession | Score | Expect | R | P | L |
|---|---|---|---|---|---|---|---|
| Normal resting | |||||||
| 1 | Eukaryotic translation initiation factor 3 | IF3A_MOUSE | 82 | 7.80E-05 | + | − | − |
| 2 | Heat shock cognate 71 kd protein | HSP7C_MOUSE | 263 | 6.20E-23 | +++ | + | + |
| (Heat shock 70 kd protein 8) | |||||||
| 3 | Ig μ chain C region | MUC_MOUSE | 109 | 1.60E-07 | ++ | + | − |
| 4 | Leukemia inhibitory factor precursor (LIF) | LIF_MOUSE | 59 | 1.40E-02 | + | v− | − |
| 5 | Heterogeneous nuclear ribonucleoprotein H (hnRNP H) | HNRH1_MOUSE | 157 | 2.50E-12 | +++ | ++ | + |
| 6 | Matrin-3 | MATR3_MOUSE | 71 | 1.00E-03 | +++ | − | + |
| 7 | Prohibitin (B cell receptor-associated protein 32) | PHB_MOUSE | 193 | 6.20E-16 | + | − | − |
| 8 + 9 | Protein DEK + Actin-like protein 6a | DEK_MOUSE+ACL6A_MOUSE | 113 | 5.70E-08 | +++ | + | ++ |
| 10 | Vimentin | VIME_MOUSE | 272 | 7.80E-24 | +++ | ++ | ++ |
| 11 | Tubulin α6 | TBA2_MOUSE | 147 | 2.50E-11 | ++ | + | + |
| 12 | Splicing factor 3b | 74191506* | 156 | 2.70E-11 | ++ | − | − |
| 13 | Heterogeneous nuclear ribonucleoprotein U | 17390825* | 131 | 8.50E-09 | +++ | + | + |
| 14 | Transitional endoplasmic reticulum ATPase 1 | TERA_MOUSE | 120 | 1.20E-08 | + | + | − |
| 15 | Nucleophosmin (NPM) | NPM_MOUSE | 108 | 2.00E-07 | ++ | ++ | + |
| 16 | Elongation factor 2 (EF-2) | EF2_MOUSE | 140 | 1.20E-10 | + | + | − |
| Normal, mitogenically stimulated, proliferating | |||||||
| 17 | Splicing factor 3A | SF3A3_MOUSE | 62 | 8.80E-03 | + | +++ | + |
| 18 | Pyruvate kinase isozyme M2 | KPYM_MOUSE | 130 | 1.20E-09 | − | + | − |
| 19 | Heterogeneous nuclear ribonucleoprotein K | HNRPK_MOUSE | 164 | 4.90E-13 | − | ++ | + |
| 20 | 78 kd glucose-regulated protein precursor (GRP 78) | GRP78_MOUSE | 256 | 3.10E-22 | + | +++ | − |
| 21 | Calreticulin | CALR_MOUSE | 118 | 2.00E-08 | − | + | − |
| 22 | Chloride intracellular channel protein 1 | CLIC1_MOUSE | 158 | 2.00E-12 | − | + | − |
| 23 | Endoplasmin (Heat shock protein 90 kd β member 1) | ENPL_MOUSE | 257 | 2.50E-22 | + | ++ | − |
| 24 | Hsp 90- β (HSP 84) | HS90B_MOUSE | 151 | 9.90E-12 | + | ++ | − |
| 25 | Hematopoietic cell specific Lyn substrate 1 | HCLS1_MOUSE | 142 | 7.80E-11 | − | + | − |
| 26 | 30 hypoxia up-regulated 1 homologue | 74192146* | 151 | 8.50E-11 | − | +++ | + |
| Proliferating lymphoma | |||||||
| 27 | Adenylyl cyclase-associated protein 1 (CAP 1) | CAP1_MOUSE | 93 | 6.80E-06 | − | − | + |
| 28 | Aldose reductase | ALDR_MOUSE | 63 | 6.80E-03 | − | − | + |
| 29 | Alpha enolase | ENOA_MOUSE | 252 | 7.80E-22 | − | + | +++ |
| 30 | Cofilin 1 | COF1_MOUSE | 62 | 8.40E-03 | − | − | + |
| 31 | Coronin 1A | COR1A_MOUSE | 213 | 6.20E-18 | + | + | ++ |
| 32 | Dihydrolipoyl dehydrogenase, mitochondrial | DLDH_MOUSE | 87 | 2.80E-05 | − | − | + |
| 33 | Glutamate dehydrogenase 1, mitochondrial | DHE3_MOUSE | 63 | 5.40E-03 | − | − | + |
| 34 | Ran GTP-binding nuclear protein | RAN_MOUSE | 123 | 6.20E-09 | − | − | + |
| 35 | Heterogeneous nuclear ribonucleoprotein A/B | ROAA_MOUSE | 65 | 3.80E-03 | − | − | + |
| 36 | Heterogeneous nuclear (hnRNP A3) ribonucleoprotein A3 | ROA3_MOUSE | 101 | 9.90E-07 | − | − | + |
| 37 | Heterogeneous nuclear (hnRNP L) ribonucleoprotein L | HNRPL_MOUSE | 89 | 1.70E-05 | − | + | +++ |
| 38 | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1) | ROA2_MOUSE | 127 | 2.50E-09 | − | − | + |
| 39 | High mobility group protein B1 | HMGB1_MOUSE | 61 | 1.10E-02 | − | − | + |
| 40 | High mobility group protein B2 | HMGB2_MOUSE | 91 | 1.00E-05 | − | − | + |
| 41 | LIM and SH3 domain protein 1 | LASP1_MOUSE | 65 | 3.90E-03 | − | − | + |
| 42 | Macrophage capping protein (Myc basic motif homolog 1) | CAPG_MOUSE | 83 | 6.20E-05 | − | − | + |
| 43 | Moesin | MOES_MOUSE | 244 | 4.90E-21 | − | + | +++ |
| 44 | Peptidyl-prolyl cis-trans isomerase A (PPIA) | PPIA_MOUSE | 96 | 3.00E-06 | − | − | + |
| 45 | Rho GDP-dissociation inhibitor 2 (Rho GDI 2) | GDIS_MOUSE | 108 | 2.00E-07 | + | + | +++ |
| 46 | Tropomyosin α−3 chain (Tropomyosin-3) | TPM3_MOUSE | 117 | 2.50E-08 | + | ++ | +++ |
| 47 | UTP-glucose-1-phosphate uridylyltransferase 2 | UGPA2_MOUSE | 76 | 2.50E-04 | ++ | + | +++ |
| 48 | Serpin b1a | 74354376* | 141 | 8.50E-10 | − | + | +++ |
Major protein spots that differed in staining intensity among the three states were excised, destained, digested with trypsin, and subjected to matrix-assisted laser desorption ionization/time of flight MS and peptide mass fingerprint analyses, as described in Materials and Methods. Protein numbers correspond to spot features as labeled in Figure 1. Protein names and database accession numbers are derived from the SwissProt database, except where marked with an asterisk, in which case they are from the NCBInr database. Mascot scores and corresponding expect values indicate the high degree of significance for each assignment. Protein expression levels, normalized to total protein, are shown qualitatively (as verified across staining types and replicate gels) across B cell proliferative states. −, +, ++, and +++ indicate 0–25%, 25–50%, 50–75%, and 75–100% normalized expression levels, respectively. R, normal resting B cells; P, normal mitogenically stimulated proliferating B cells; L, malignant proliferating lymphoma B cells.
Figure 3.
Biological Process GO functional annotation of proteins dramatically up-regulated in each B cell proliferative state. Proteins assigned by matrix-assisted laser desorption ionization/time of flight MS and peptide mass fingerprint were annotated with GO functional tags and graphed to show the relative contributions of each functional assignment within the protein groups. Shown is the GO supercategory of Biological Process. A: Proteins up-regulated in normal resting B cells. B: Proteins up-regulated in normal, mitogenically stimulated, proliferating B cells. C: Proteins up-regulated in proliferating lymphoma B cells.
Clustering of spot features into groups of relevant expression trends was performed to create a useful framework for the improved identification of potential biomarkers. We conducted differential analysis on the individual gel spot features for each of the B cell proliferative states. Raw data were rendered as vectors in three dimensions, comprising the X, Y, Z coordinates of resting state intensity, proliferating state intensity and lymphoma state intensity. Relative expression differences between states for each feature are depicted (Figure 4), calculated by determining the components of each vector that were perpendicular to a vector of equivalent expression across all states.
Figure 4.
Expression difference profiling of protein spots between proliferative states. Gel spot feature intensity was quantified for each of the B cell proliferative states (Figure 1). Relative differences in expression levels between proliferative states were calculated and are shown plotted on a difference vector diagram, in which each spot feature is represented as a point with coordinates in the three intensity difference axes of resting, proliferating and lymphoma (reflecting its expression level difference for each state). The distance of the points from the origin (the point representing equivalent expression across all of the states) toward one set of axes or another quantitatively reflects the expression level bias of the protein spots they represent toward one or another state.
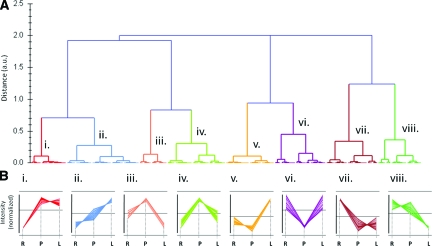
These relative expression differences were then subjected to principal component analysis and hierarchical clustering. Clusters were grouped according to expression changes, as shown in Figure 5. At a relatedness distance metric of 0.5 out of 2.0 on the dendrogram (Figure 5A), granularity became sufficient that functionally relevant expression trends were apparent in the clusters (Figure 5B; members of each expression cluster that were identified are shown in Table 2). Clusters i and vii contain proteins (or protein isoforms) the behavior of which is similar between the normal proliferative and lymphoma states, but different from the resting state (ie, a proliferation-associated signature), wherein i contains proteins that are up-regulated with respect to resting, while vii contains proteins that are down-regulated with respect to resting. Clusters iii, iv, and vi contain proteins the expression behavior of which is similar between the resting and the lymphoma, but different from the normal proliferative state, wherein iii and iv contain proteins that are down-regulated with respect to the proliferative state, while vi contains proteins that are up-regulated with respect to the proliferative state. Clusters ii, v, and viii contain proteins the expression behavior of which for the lymphoma state is different from both the resting and the nonmalignant proliferative states. This may be regarded as a malignancy-associated protein expression pattern. Cluster viii contains proteins that are down-regulated in lymphoma B cells by comparison with the other two states. Clusters ii and v contain proteins that are up-regulated in lymphoma cells with respect to the other states. Thus, the expression clusters ii, v, and viii represent a source of malignancy-associated biomarkers. The significance of the result lies in the identification of proteins of known function that are uniquely associated with malignancy, and not with normal proliferation.
Figure 5.
Hierarchical clustering of protein expression profiles across B cell proliferative states. Expression profiles of gel spot features were subjected to unsupervised clustering into expression trend groups. A: Dendrogram delineating the relatedness clusters of the expression profiles of the major sets of protein spots that varied between B cell proliferative states. Left axis, relatedness distance metric (in arbitrary units). Clusters at relatedness distance below the arbitrary value of 0.5 are labeled i–viii. B: Expression trends displayed by each cluster i-viii from A. Normalized spot intensities are graphed for each member of the cluster. R, normal resting B cells; P, normal mitogenically stimulated proliferating B cells; L, malignant proliferating lymphoma B cells. The average value for each cluster is shown in each graph by a horizontal line. Clusters i and vii represent a proliferative signature. Clusters ii, v, and viii represent potential malignancy-associated biomarkers.
Table 2.
Proteins Grouped into Eight Co-Expression Clusters Associated with B Cell Proliferative States
| Cluster i | Cluster ii | Cluster iii | Cluster iv |
|---|---|---|---|
| Coronin 1A (isoforms) | hnRNP A2/B1 (isoforms) | Hematopoietic cell-specific | Calreticulin |
| Dihydrolipoyl dehydrogenase | Macrophage capping protein | Lyn substrate 1 | Endoplasmin |
| Alpha enolase | Rho GDI 2 | Hsp 90b | Moesin (isoform) |
| Pyruvate kinase M2 | Coronin 1A (isoform) | Moesin (isoform) | Coronin 1A isoforms |
| LIM and SH3 domain 1 | EF-2 (isoforms) | EF-2 (isoforms) | |
| Cofilin-1 | GRP-78 | ||
| Glutamate dehydrogenase | Coronin 1A (isoform) | ||
| hnRNP L (isoforms) | |||
| High mobility group protein B1 | |||
| Tropomyosin α3 | |||
| Peptidyl-prolyl cis-trans | |||
| isomerase A (isoform) | |||
| Cluster v
|
Cluster vi
|
Cluster vii
|
Cluster viii
|
| Aldose reductase | UTP-glucose-1-phosphate | hnRNP U | Chloride intracellular |
| hnRNP A3 (isoform) | uridylyltransferase 2 (isoform) | Splicing factor 3b | channel protein 1 |
| Serpin b1a protein | High mobility group protein B2 | homologue | Prohibitin |
| hnRNP L (isoform) | Matrin-3 | Ig μ C region | Nucleophosmin |
| Adenylyl cyclase-associated | hnRNP A/B | DEK protein | Unnamed protein product |
| protein 1 | hnRNP H1 (isoform) | Actin-like protein 6A | similar to 30 hypoxia |
| Ran | Hsp 71 | Eukaryotic translation | Up-regulated-1 |
| initiation factor 3 | hnRNP H1 (isoform) | ||
| Vimentin | EF-2 (isoform) | ||
| Peptidyl-prolyl cis-trans | Transitional ER ATPase 1 | ||
| isomerase A (isoform) | hnRNP K | ||
| Tubulin α6 | |||
| hnRNP H1 (isoform) | |||
| Splicing factor 3A |
Many of the spot features included in the expression trend groups shown in Figure 5, or their isoforms were assigned by matrix-assisted laser desorption ionization/time of flight MS and peptide mass fingerprint analyses. Listed are the classifications of the assigned proteins or their isoforms into the expression trend groups i–viii.
Discussion
Previous genome-wide transcriptional analyses of DLBCL, originally pioneered by Staudt, Shipp, and others,4,5,6,7 were able to link disease severity, specific pathological characteristics and prognosis to transcriptional signature. We used normal resting and mitogenically stimulated proliferating B cells in a well-controlled model system to extend this approach by defining axes of gene expression that are related to normal proliferation or malignant proliferation, but not both, which enabled a malignancy-associated transcriptional signature to be deduced.9 From these studies, the hypothesis followed that a limited and reproducible set of malignancy-associated protein biomarkers could be identified in particular lymphoid malignancies; most importantly, these proteins would be different from the vast majority of proteins associated with normal cellular states, including normal proliferation.
Specifically, here we sought to define a simplified set of potential biomarkers of lymphoid malignancy, exploiting our Tg model to analyze the proteomic signatures of DLBCL, while subtracting out the signatures of both proliferating and resting normal B cells. Using 2D-PAGE, MS and peptide mass fingerprint analyses, we identified highly expressed protein species that were differentially expressed in each of the three B cell proliferative states. More detailed expression profiling and clustering analysis elucidated common expression trends that can be used for classifying potential malignancy-associated biomarkers and distinguishing them from markers that are shared with normal resting or normal proliferating cells. We defined specific clusters of protein isoform expression that appeared to constitute lymphoma-associated markers, because they were specifically differentially up-regulated in lymphoma, and were different from clusters that constituted merely a proliferative signature, which was nonspecific for lymphoma. We also defined a set of potential markers that are down-regulated in lymphoma with respect to both resting and normal proliferating cells, which might reflect tumor suppressor-like properties of the proteins.
DLBCL-Associated Proteome
In particular, we found that several proteins of interest were more highly expressed in resting B cells than in lymphoma cells, including factors that are involved in cell cycle regulation, such as leukemia inhibitory factor (Lif), which is an autocrine/paracrine B cell regulator that mediates B cell arrest through the Ras and Jak/Stat pathways,26 prohibitin, a tumor suppressor and cell cycle regulator that plays role in growth regulation of fibroblasts,27,28 which our group has previously shown is an important negative regulator of E2Fs,29 and matrin-3 (p130), a cell cycle regulator that mediates cell cycle arrest by p130 and regulates Cdk2.30 We also identified heat shock cognate 71 kd protein (Hsp70), which is a chaperone protein associated with growth inhibition in lymphoid cells31 and was more highly expressed in resting B cells than in lymphoma cells. This resting B cell group included proteins that are important for chromatin remodeling, such as actin-like protein 6A (Baf53a), which forms a repressor complex with Rb, a histone deacetylase that inhibits transcription of certain E2F target genes such as cyclin E, cyclin A, and CDC232 and suppresses p53-dependent transcription32; DEK, a chromatin remodeling protein and proto-oncogene product that is phosphorylated by Cdk233,34; and heterogeneous nuclear ribonucleoprotein U (SAF-A; scaffold attachment factor A), a chromatin remodeling factor together with p300 (and likely p/CAF), which primes sites for transcriptional activation and regulates transcription through scaffold/matrix attachment regions (S/MARs), the chromatin regions that bind the nuclear matrix.35 We also identified mRNA splicing factors, heterogeneous nuclear ribonucleoprotein H (hnRNP H), and splicing factor 3B subunit 2 (SAP 145). Additionally, in resting B cells we detected elevated levels of vimentin, a B cell intermediate filament protein important for cell structure, that is reduced in B cells as they mature,36 consistent with the observation that some lymphoma lines lack vimentin altogether. Immunoglobulin μ-chain constant region, membrane-bound and secreted forms, and tubulin α-6 chain were also present in this group.
Other proteins were present in relatively equal amounts in resting and normal proliferating B cells, a group that defines normal B cell biology, including the transitional endoplasmic reticulum ATPase, p97 (valosin-containing protein), which is a molecular chaperone37 with cell cycle control properties that is responsible for degradation of cyclin E, is involved in the transcriptional activation of NFκB, and is expressed at high levels in rapidly dividing cells.37 Valosin is up-regulated after B cell stimulation and is required for normal cell growth control.37 This normal B cell group also included nucleophosmin, which binds the tumor suppressors p53 and p19arf and is essential for ribogenesis, cell proliferation and survival after DNA damage, but is often mutated in human lymphoma and leukemia, and is associated with induction of proliferation in B cells38,39; and elongation factor 2, which promotes the GTP-dependent translocation of the nascent protein chain from the A-site to the P-site of the ribosome.40
In normal proliferating B cells, as expected, we identified factors that are important for cell cycle control such as calreticulin, which is a calcium-dependent regulator of p53 transcription41; endoplasmin (polymorphic tumor rejection antigen 1, tumor rejection antigen gp96), which is necessary for the Raf-1-MEK-MAPK signaling pathway; heat shock protein Hsp 90-β (tumor-specific transplantation 84-kd antigen), also necessary for Raf-1-MEK-MAPK signaling42; hematopoietic lineage cell-specific protein (HS1), which is required for B cell activation and B cell antigen receptor-mediated signaling43; and adenylyl cyclase-associated protein 1 (CAP-1), which is a multifunctional protein that activates Ras by posttranslational modification, directly regulates filament dynamics, and has been implicated in a number of complex developmental and morphological processes.44 We identified proteins with roles in translation:78 kd glucose-regulated protein Hspa5 (Bip), which is required for B cell maturation into antibody secreting cells,45 and splicing factor 3A subunit 3 (SAP 61). We identified proteins involved in energy metabolism, such as pyruvate kinase isozyme M2, which controls glycolysis necessary for cell proliferation.46 We found that normal proliferating B cells also overexpress a large group of transcription-related proteins, including heterogeneous nuclear ribonucleoprotein K, which assembles either transcriptional repressors or activators on DNA, accounting for the observations that K protein can either increase or decrease rates of transcription and that K protein is required for a p53 mediated cell cycle checkpoint.47,48
The malignancy-associated proteome identified several up-regulated proteins known not to be up-regulated during normal proliferation. The proteins up-regulated in lymphoma cells include proteins that are important for energy metabolism, such as aldose reductase, which has been shown to be up-regulated in many cancers, specifically liver and colon cancer, and indirectly mediates the expression of COX-2 and production of PGE49,50; α-enolase, a glycolytic enzyme that has been implicated in the differentiation of lymphoma51; dihydrolipoyl dehydrogenase, a mitochondrial enzyme that is a component of the glycine cleavage system and the α-ketoacid dehydrogenase complexes associated with the pyruvate dehydrogenase complex; glutamate dehydrogenase 1, a mitochondrial enzyme that supplements energy metabolism in mitogenically stimulated B cells and lymphoid cells; and UTP-glucose-1-phosphate uridylyltransferase 2. The lymphoma-associated group also included up-regulated isoforms of proteins of importance for the cell cycle such as coronin-1A (p57), which is a tumor suppressor gene and is known to be involved in lymphoid malignancies52; the GTP-binding nuclear protein Ran, which directly regulates hepatocarcinoma-up-regulated protein, which interacts with several mitotic spindle assembly factors53; peptidyl-prolyl cis-trans isomerase A (cyclophilin), which is overexpressed in many cancers54 and has been found to stimulate cell proliferation through CD147 and activation of ERK1/2 and p38 MAPKs54; and rho GDP-dissociation inhibitor 2, which has been implicated as a tumor suppressor gene.55 The malignancy-associated signature also included proteins that are important for cell structure, such as cofilin-1 (non-muscle isoform), a lim-K1 substrate that has been shown by gene array to be unregulated in metastases56,57; LIM and SH3 domain protein 1 (Lasp-1), which plays a role in cell motility by changing cell structure and is implicated in breast cancer58; macrophage capping protein (myc basic motif homolog 1) (CapG), which shows increased expression in certain cancers and appears to be a tumor promoter59,60; and membrane-organizing extension spike protein (moesin), which is involved in connecting major cytoskeletal structures to the plasma membrane and in cell motility. Last, the malignancy-associated group included numerous heterogeneous nuclear ribonucleoproteins, which have a variety of biological functions, have many implications in cancer, and have been suggested as potential cancer biomarkers, including heterogeneous nuclear ribonucleoprotein A/B, which interacts with the OPN promoter, decreasing OPN promoter activity and mRNA levels,61 which correlates with metastatic behavior, motility, and invasion in breast cancer.62 This ribonucleoprotein is also involved in telomere biogenesis and is important for B cell proliferation; its up-regulation has been found to be consistent with cancer and cell proliferation.61 Also identified were the heterogeneous nuclear ribonucleoproteins A3, L, and A2/B1, which have been shown to be unregulated in many cancers and play a role in mRNA splicing and cell cycle control,61,63 along with high mobility group (HMG) proteins B1 and B2,64 which are oncogenes and cause highly aggressive lymphoid malignancy in mice,65 are required for c-myc function, are overexpressed in human leukemia, and bind chromatin as architectural regulators of transcription.64
A Generalizable Approach to Cancer-Associated Proteomes
Proteomic and transcriptional profiling tools have the potential to reveal the signatures of cancer. Combining proteomic and transcriptional signatures of the same murine B cell malignancies yields largely orthogonal sets of biomarkers, consistent with the experience of others.18,66,67 We found that proteomic signatures correlated with differentially expressed genes9 for heat shock protein 8, immunoglobulin heavy chain, vimentin, splicing factor 3b and heterogeneous nuclear ribonucleoprotein U in normal, resting cells. Proteomic identifications agreed with transcriptome for pyruvate kinase isozyme M2 in normal proliferating cells and for Ran GTP-binding nuclear protein, high mobility group protein B1, LIM, and SH3 domain protein 1 and serpin b1a in proliferating malignant cells. Discordance between proteomic profiling and gene expression profiling datasets derived from the same animal model will likely resolve as both technologies advance. Within each methodological approach, robust correlations can be obtained between clinical variables and transcriptional signatures, such as the DLBCL analyses already discussed.5 However, we concur that transcriptional data alone must be used with great caution in drug discovery efforts, because transcriptional readouts are often of greater distance from biological function than are proteomic, phosphoproteomic, or metabolomic readouts.16,17,18,66 These methods of profiling are complementary and non-redundant.16 Although gene expression profiling provided information on the differential expression of a vastly larger number of features (22,690 transcriptional probe sets9), proteomic analysis arguably provides a more direct view of the cell’s architecture and protein machinery that constitute the normal and disease-altered cell states. Thus, the cancer-specific proteome offers a unique and highly useful set of biomarkers for disease detection and differential diagnosis.
Multiple biomarkers that constitute a signature of a disease state ultimately can have much stronger diagnostic and predictive power among a heterogeneous population than any single biomarker alone.4,5,6 Thus, a constellation of protein markers, each associated with a specific subtype of lymphoma, would provide a biomarker set for the accurate detection and determination of the malignancy status of a patient, and when correlated with International Prognostic Index markers, would establish criteria for prognostic classification. Although we have shown that several proteins that we have identified through MS-based proteomic methodologies appear to constitute a malignancy-associated signature, the usefulness of these proteins as potential biomarkers of lymphoma remains to be qualified, verified and validated in this and other models, as well as in human DLBCL cell lines and patient samples.68 Similarly, although these state-specific markers hold great promise for our understanding of lymphomagenesis, maintenance, and progression, functional hypothesis testing will be required to reveal their role in the pathogenesis of DLBCL.
Nevertheless, we demonstrate the potential utility of comparing the proteome of malignant B cells with normal resting and proliferating counterparts. Subtraction of non-malignancy signals effectively simplifies proteomic biomarker discovery and analysis. This approach represents a paradigm ideally applied in personalized medicine and diagnosis of B cell malignancies; a patient’s unique lymphoma-associated protein signature may thus be resolved from the background profile of normal B cell proliferative proteins. Significant progress toward this goal in genomic terms has been reported recently for the genetic signature of M1 subtype acute myeloid leukemia compared with normal skin from the same patient.69 Given the importance of phosphorylation cascades elicited by abnormal growth factor receptor activity and cytokine signal transduction in lymphoid and myeloid cancers, the malignancy-associated phosphoproteome is clearly of crucial importance to deduce for individual patients as well. Biomarker discovery of this type should aid personalized diagnosis and allow individualized tracking of chemotherapeutic efficacy and improved, earlier detection of relapse. Notably, it may be extended to biomarker discovery strategies in many other types of lymphoid and myeloid malignancy, including cancers of the macrophage, monocyte, neutrophil, and granulocyte lineages, as well as endothelial cells.
Acknowledgments
We thank Arthur Lambert, Harold Longe, Ronald Myint, Andrew Rankin, Anupama Sinha, and other members of the Cancer Research Center, Boston University School of Medicine, for invaluable assistance with experiments and helpful criticism; and we thank Vivek Bhatia, Yang Su, Weiwei Tong, and James West of the Cardiovascular Proteomics Center, Boston University School of Medicine, for development of bioinformatics tools for data analysis.
Footnotes
Address reprint requests to Gerald V. Denis, Ph.D., Cancer Research Center, Boston University School of Medicine, Room K520, 72 East Concord Street, Boston, MA 02118. E-mail: gdenis@bu.edu.
Supported in part by grant RSG-05-072-01 (to G.V.D.) from the American Cancer Society; grants CA128006 (to G.V.D.), CA84193 (to D.V.F.), P41-RR10888 (to C.E.C.), and S10-RR15942 (to C.E.C.) from the National Institutes of Health; and NIH-NHLBI contract N01-HV-28178 (to C.E.C.). This project was supported by the American Cancer Society’s Betty Lea Stone Research Fellowship (to P.B.R.) and ΑΩΑ Carolyn L. Kuckein Student Research Fellowship (to P.B.R.).
P.R. and D.H. contributed equally to this work.
References
- Edwards BK, Brown ML, Wingo PA, Howe HL, Ward E, Ries LAG, Schrag D, Jamison PM, Jemal A, Wu XC, Friedman C, Harlan L, Warren J, Anderson RN, Pickle LW. Annual report to the nation on the status of cancer, 1975–2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst. 2005;97:1407–1427. doi: 10.1093/jnci/dji289. [DOI] [PubMed] [Google Scholar]
- Ng AK. Diffuse large B cell lymphoma. Semin Radiat Oncol. 2007;17:169–175. doi: 10.1016/j.semradonc.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Illidge T, Tolan S. Current treatment approaches for diffuse large B cell lymphoma. Leukemia Lymphoma. 2008;49:663–676. doi: 10.1080/10428190701882187. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Staudt LM. Genomic-scale gene expression profiling of normal and malignant immune cells. Curr Opin Immunol. 2000;12:219–225. doi: 10.1016/s0952-7915(99)00078-3. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen M, Davis RE, C Ma, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large-B cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR. Diffuse large B cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- Staudt LM. Gene expression profiling of lymphoid malignancies. Annu Rev Med. 2002;53:303–318. doi: 10.1146/annurev.med.53.082901.103941. [DOI] [PubMed] [Google Scholar]
- Greenwald R, Tumang JR, Sinha A, Currier N, Cardiff RD, Rothstein TL, Faller DV, Denis GV. Eμ-BRD2 transgenic mice develop B cell lymphoma and leukemia. Blood. 2004;103:1475–1484. doi: 10.1182/blood-2003-06-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg M, Sinha S, Faller DV, Denis GV. Tumor-specific and proliferation-specific gene expression typifies murine transgenic B cell lymphomagenesis. J Biol Chem. 2007;282:4803–4811. doi: 10.1074/jbc.M605870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson K. On lines and planes of closest fit to systems of points in space. Philosoph Mag. 1901;2:559–572. [Google Scholar]
- Pollack JR. A perspective on DNA microarrays in pathology research and practice. Am J Pathol. 2007;171:375–385. doi: 10.2353/ajpath.2007.070342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Rogers CG, Cooper JT, Ditlev JA, Maatman TJ, Yang X, Furge KA, Teh BT. Gene expression profiling of renal cell carcinoma. Clin Cancer Res. 2004;10:6315S–6321S. doi: 10.1158/1078-0432.CCR-050002. [DOI] [PubMed] [Google Scholar]
- Duggan BJ, McKnight JJ, Williamson KE, Loughrey M, O'Rourke D, Hamilton PW, Johnston SR, Schulman CC, Zlotta AR. The need to embrace molecular profiling of tumor cells in prostate and bladder cancer. Clin Cancer Res. 2003;9:1240–1247. [PubMed] [Google Scholar]
- Marguiles AG, Klimberg VS, Bhattacharrya S, Gaddy D, Suva LJ. Genomics and proteomics of bone cancer. Clin Cancer Res. 2006;12:6217s–6221s. doi: 10.1158/1078-0432.CCR-06-1070. [DOI] [PubMed] [Google Scholar]
- Bhati R, Patterson C, Livasy CA, Fan C, Ketelsen D, Hu Z, Reynolds E, Tanner C, Moore DT, Gabrielli F, Perou CM, Klauber-DeMore N. Molecular characterization of human breast tumor vascular cells. Am J Pathol. 2008;172:1381–1390. doi: 10.2353/ajpath.2008.070988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamoon AS, Kohn EC, Azad NS. The ongoing evolution of proteomics in malignancy. Drug Discov Today. 2007;12:700–708. doi: 10.1016/j.drudis.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Steuer R. on the analysis and interpretation of correlations in metabolomic data. Brief Bioinform. 2006;7:151–158. doi: 10.1093/bib/bbl009. [DOI] [PubMed] [Google Scholar]
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Chen R, Pan S, Brentnall TA, Aebersold R. Proteomic profiling of pancreatic cancer for biomarker discovery. Mol Cell Proteom. 2005;4:523–533. doi: 10.1074/mcp.R500004-MCP200. [DOI] [PubMed] [Google Scholar]
- Alfonso P, Catala M, Rico-Morales ML, Durante-Rodriguez G, Moro-Rodriguez E, Fernandez-Garcia H, Escribano JM, Alvarez-Fernandez E, Garcia-Poblete E. Proteomic analysis of lung biopsies: differential protein expression profile between peritumoral and tumoral tissue. Proteomics. 2004;4:442–447. doi: 10.1002/pmic.200300647. [DOI] [PubMed] [Google Scholar]
- Alexe G, Alexe S, Liotta LA, Petricoin E, Reiss M, Hammer PL. Ovarian cancer detection by logical analysis of proteomic data. Proteomics. 2004;4:766–783. doi: 10.1002/pmic.200300574. [DOI] [PubMed] [Google Scholar]
- Neubauer H, Fehm T, Schutz C, Speer R, Solomayer E, Schrattenholz A, Cahill MA, Kurek R. Proteomic expression profiling of breast cancer. Recent Results Cancer Res. 2007;176:89–120. doi: 10.1007/978-3-540-46091-6_9. [DOI] [PubMed] [Google Scholar]
- Yates JR., 3rd Database searching using mass spectrometry data. Electrophoresis. 1998;19:893–900. doi: 10.1002/elps.1150190604. [DOI] [PubMed] [Google Scholar]
- Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res. 2006;5:502–511. doi: 10.1021/pr050430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Park JI, Strock CJ, Ball DW, Nelkin BD. The Ras/Raf/MEK/Extracellular signal-regulated kinase pathway induces autocrine-paracrine growth inhibition via the leukemia inhibitory factor/JAK/STAT pathway. Mol Cell Biol. 2003;23:543–554. doi: 10.1128/MCB.23.2.543-554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupe ER, Liu XT, Kiehlbauch JL, McClung JK, Dell'Orco RT. The 3′ untranslated region of prohibitin and cellular immortalization. Exp Cell Res. 1996;224:128–135. doi: 10.1006/excr.1996.0120. [DOI] [PubMed] [Google Scholar]
- McClung JK, Jupe ER, Liu XT, Dell'Orco RT. Prohibitin: potential role in senescence Development, and tumor suppression. Exp Gerontol. 1995;30:99–124. doi: 10.1016/0531-5565(94)00069-7. [DOI] [PubMed] [Google Scholar]
- Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene. 1999;18:3501–3510. doi: 10.1038/sj.onc.1202684. [DOI] [PubMed] [Google Scholar]
- Soeiro I, Mohamedali A, Romanska HM, Lea NC, Child ES, Glassford J, Orr SJ, Roberts C, Naresh KN, Lalani EN, Mann DJ, Watson RJ, Thomas NSB, Lam EWF. P27KIP1 and p130 cooperate to regulate hematopoietic cell proliferation in vivo. Mol Cell Biol. 2006;26:6170–6184. doi: 10.1128/MCB.02182-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DD, Caron AW, Bourget L, Meriin AB, Sherman MY, Morimoto RI, Massie B. The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Mol Cell Biol. 2000;20:7146–7159. doi: 10.1128/mcb.20.19.7146-7159.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lee JY, Chang SH, Kang MJ, Kwon H. Effects of Ser2 and Tyr6 mutants of BAF53 on cell growth and p53-dependent transcription. Mol Cells. 2005;19:289–293. [PubMed] [Google Scholar]
- Kappes F, Scholten I, Richter N, Gruss C, Waldmann T. Functional domains of the ubiquitous chromatin protein DEK. Mol Cell Biol. 2004;24:6000–6010. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitwala KV, Adams K, Markovitz DM. YY1 and NF-Y binding sites regulate the transcriptional activity of the dek and dek-can promoter. Oncogene. 2002;21:8862–8870. doi: 10.1038/sj.onc.1206041. [DOI] [PubMed] [Google Scholar]
- Martens JHA, Verlaan M, Kalkhoven E, Dorsman JC, Zantema A. Scaffold/Matrix attachment region elements interact with a p300-scaffold attachment factor A complex and are bound by acetylated nucleosomes. Mol Cell Biol. 2002;22:2598–2606. doi: 10.1128/MCB.22.8.2598-2606.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller P, Momburg F, Hofmann WJ, Matthaei-Maurer DU. Lack of vimentin occurring during intrafollicular stages of B cell development characterizes follicular center cell lymphomas. Blood. 1988;71:1033–1038. [PubMed] [Google Scholar]
- Wang Q, Song C, Li CH. Molecular perspectives on p97-VCP progress in understanding its structure and diverse biological functions. J Struct Biol. 2003;146:44–57. doi: 10.1016/j.jsb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Colombo E, Bonetti P, Lazzerini Denchi E, Martinelli P, Zamponi R, Marine JC, Helin K, Falini B, Pelicci PG. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol Cell Biol. 2005;25:8874–8886. doi: 10.1128/MCB.25.20.8874-8886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisendi S, Pandolfi PP. NPM mutations in acute myelogenous leukemia. N Engl J Med. 2005;352:291–292. doi: 10.1056/NEJMe048337. [DOI] [PubMed] [Google Scholar]
- Feurstein N, Mond JJ. “Numatrin,” a nuclear matrix protein associated with induction of proliferation in B lymphocytes. J Biol Chem. 1987;262:11389–11397. [PubMed] [Google Scholar]
- Mesaeli N, Phillipson C. Impaired p53 expression, function, and nuclear localization in calreticulin-deficient cells. Mol Biol Cell. 2004;15:1862–1870. doi: 10.1091/mbc.E03-04-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikakis N, Lin JH, Grammatikakis, Tsichlis PN, Cochran BH. p50cdc37 acting in concert with Hsp90 is required for RAF-1 function. Mol Cell Biol. 1999;19:1661–1672. doi: 10.1128/mcb.19.3.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanashi Y, Okada M, Semba T, Yamori T, Umemori H, Tsunasawa S, Toyoshima K, Kitamura D, Watanabe T, Yamamoto T. Identification of HS1 protein as a major substrate of protein-tyrosine kinase(s) upon B cell antigen receptor-mediated signaling. Proc Natl Acad Sci USA. 1993;90:3631–3635. doi: 10.1073/pnas.90.8.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara C, Shinkai M, Kariya KI, Yamawaski-Kataoka Y, Hu CD, Masuda T, Kataoka T. Association of elongation factor 1α and ribosomal protein L3 with the proline-rich region of yeast adenylyl cyclase-associated protein CAP. Biochem Biophys Res Commun. 1997;232:503–507. doi: 10.1006/bbrc.1997.6326. [DOI] [PubMed] [Google Scholar]
- Gass JN, Gifford NM, Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277:49047–49054. doi: 10.1074/jbc.M205011200. [DOI] [PubMed] [Google Scholar]
- Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- Moumen A, Masterson P, O'Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Bomsztyk K, Seuningen IV, Suzuki H, Denisenko O, Ostrowski J. Diverse molecular interactions of the hnRNP K protein. FEBS Lett. 1997;403:113–115. doi: 10.1016/s0014-5793(97)00041-0. [DOI] [PubMed] [Google Scholar]
- Saraswat M, Mrudula T, Kumar PU, Suneetha A, Rao TS, Srinivasulu M, Reddy B. Overexpression of aldose reductase in human cancer tissues. Med Sci Monitor. 2006;12:525–529. [PubMed] [Google Scholar]
- Tammali R, Ramana KV, Singhal SS, Awasthi S, Srivastava SK. Aldose reductase regulates growth factor-induced cyclooxygenase-2 expression and prostaglandin E2 production in human colon cancer cells. Cancer Res. 2006;66:9705–9713. doi: 10.1158/0008-5472.CAN-06-2105. [DOI] [PubMed] [Google Scholar]
- Mohammad RM, Hamdan MY, al-Katib A. Induced expression of alpha-enolase in differentiated diffuse large cell lymphoma. Enzyme Protein. 1994–95;48:37–44. doi: 10.1159/000474967. [DOI] [PubMed] [Google Scholar]
- Li Y, Nagai H, Ohno T, Yuge M, Hatano S, Ito E, Mori N, Saito H, Kinoshita T. Aberrant DNA methylation of p57KIP2 gene in the promoter region in lymphoid malignancies of B cell phenotype. Blood. 2002;100:2582–2577. doi: 10.1182/blood-2001-11-0026. [DOI] [PubMed] [Google Scholar]
- Sanderson HS, Clarke PR. Cell Biology: ran, mitosis and the cancer connection. Current Biol. 2006;16:R466–R468. doi: 10.1016/j.cub.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Li M, Zhai Q, Bharadwaj U, Wang H, Li F, Fisher WE, Chen C, Yao Q. Cyclophilin A is overexpressed in human pancreatic cancer cells and stimulates cell proliferation through CD147. Cancer. 2006;106:2284–2294. doi: 10.1002/cncr.21862. [DOI] [PubMed] [Google Scholar]
- Gildea JJ, Seraj MJ, Oxford G, Harding Ma, Hampton GM, Moskaluk CA, Frierson HF, Conaway MR, Theodorescu D. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer. Cancer Res. 2002;62:6418–6423. [PubMed] [Google Scholar]
- Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick A, Condeelis JS. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Park ZY, Lin D, Brahmbhatt AA, Rio MC, Yates JR, Klemke RL. Regulation of cell migration and survival by focal adhesion targeting of LASP-1. J Cell Biol. 2004;165:421–432. doi: 10.1083/jcb.200311045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corte VD, Impe KV, Bruyneel E, Boucherie C, Mareel M, Vandekerckhove J, Gettemans J. Increased importin-β-dependent nuclear import of the actin modulating protein CapG promotes cell invasion. J Cell Sci. 2004;117:5283–5292. doi: 10.1242/jcs.01410. [DOI] [PubMed] [Google Scholar]
- Watari A, Takaki K, Higashiyama, Li Y, Satomi Y, Takao T, Tanemura A, Yamaguchi Y, Katayama I, Shimakage M, Miyashiro I, Takami K, Kodama K, Yutsudo M. Suppression of tumorigenicity, but not anchorage independence, of human cancer cells by new candidate tumor suppressor gene CapG. Oncogene. 2005;25:7373–7380. doi: 10.1038/sj.onc.1209732. [DOI] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS. hnRNP complexes: composition, structure and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Gao C, Mi Z, Guo H, Wei J, Wai P, Kuo PC. A transcriptional repressor of osteopontin expression in the 4T1 murine breast cancer cell line. Biochem Biophys Res Commun. 2004;321:1010–1016. doi: 10.1016/j.bbrc.2004.07.063. [DOI] [PubMed] [Google Scholar]
- Yan-Sanders Y, Hammons GJ, Lyn-Cook BD. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett. 2002;183:215–220. doi: 10.1016/s0304-3835(02)00168-4. [DOI] [PubMed] [Google Scholar]
- Xu Y, Sumter TF, Bhattacharya R, Tesfaye A, Fuchs EJ, Wood LJ, Huso DL, Resar LM. The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia. Cancer Res. 2004;64:3371–3375. doi: 10.1158/0008-5472.CAN-04-0044. [DOI] [PubMed] [Google Scholar]
- Krynetski EY, Krynetskaia NF, Bianchi ME, Evans WE. A nuclear protein complex containing high mobility group proteins B1 and B2, heat shock cognate protein 70. ERp60, and glyceraldehyde-3-phosphate dehydrogenase is involved in the cytotoxic response to DNA modified by incorporation of anticancer nucleoside analogues. Cancer Res. 2003;63:100–106. [PubMed] [Google Scholar]
- Phizicky E, Bastiaens PI, Zhu H, Snyder M, Fields S. Protein analysis on a proteomic scale. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- Hanash S. Disease proteomics. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nature Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- Timothy JL, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, Cook L, Abbott R, Larson DE, Koboldt DC, Pohl C, Smith S, Hawkins A, Abbott S, Locke D, Hillier LW, Miner T, Fulton L, Magrini V, Wylie T, Glasscock J, Conyers J, Sander N, Shi X, Osborne JR, Minx P, Gordon D, Chinwalla A, Zhao Y, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson M, Baty J, Ivanovich J, Heath S, Shannon WD, Nagarajan R, Walter MJ, Link DC, Graubert TA, DiPersio JF, Wilson RK. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]