Abstract
The prevailing paradigm for G protein-coupled receptors is that each receptor is narrowly tuned to its ligand and closely related agonists. An outstanding problem is whether this paradigm applies to olfactory receptor (ORs), which is the largest gene family in the genome, in which each of 1,000 different G protein-coupled receptors is believed to interact with a range of different odor molecules from the many thousands that comprise “odor space.” Insights into how these interactions occur are essential for understanding the sense of smell. Key questions are: (i) Is there a binding pocket? (ii) Which amino acid residues in the binding pocket contribute to peak affinities? (iii) How do affinities change with changes in agonist structure? To approach these questions, we have combined single-cell PCR results [Malnic, B., Hirono, J., Sato, T. & Buck, L. B. (1999) Cell 96, 713–723] and well-established molecular dynamics methods to model the structure of a specific OR (OR S25) and its interactions with 24 odor compounds. This receptor structure not only points to a likely odor-binding site but also independently predicts the two compounds that experimentally best activate OR S25. The results provide a mechanistic model for olfactory transduction at the molecular level and show how the basic G protein-coupled receptor template is adapted for encoding the enormous odor space. This combined approach can significantly enhance the identification of ligands for the many members of the OR family and also may shed light on other protein families that exhibit broad specificities, such as chemokine receptors and P450 oxidases.
Olfactory (odor) receptors (ORs) in the mammalian olfaction system exhibit a combinatorial response to odorant molecules (1). A single odor elicits response from multiple receptors and a single receptor also responds to multiple odorants, so every odorant has been thought to have a unique combination of responses from several receptors. This endows a discriminatory power to the mammalian olfactory system that could discriminate thousands of odors. The mechanisms by which the olfactory system accomplishes its multitude tasks are not clear. However, it is known that each olfactory neuron expresses only one receptor. Odor detection is mediated by ≈1,000 ORs that are G protein-coupled membrane-bound proteins. Malnic et al. (1) recently reported the differential responses of individual mouse OR neurons to 24 organic odor compounds (linear alcohols, acids, diacids, and bromoacids with four to nine carbons) by using Ca2+-imaging techniques, followed by single-cell reverse transcription–PCR to determine the sequence of the responsive OR. These clean single-cell experimental results (1) lead to the compelling question “what is the molecular basis of odor recognition?” Such questions can be answered only with the atomic level model of these ORs. No structural information is available for ORs. Also for any member of the membrane protein family, the insolubility of membrane proteins and the difficulty in crystallizing membrane proteins makes it harder to obtain structural information. In this work, we have derived an atomic level structural model for the mammalian OR S25 sequenced by Malnic et al. (1) and also identified the potential binding site for simple aliphatic alcohol and acid odorants to this receptor. The order of binding energies correlate well with the experimental recognition profiles and the binding site predictions also correlate well with the speculations.
Modeling Techniques
Prediction of the Structure of ORs.
ORs are seven helical transmembrane G protein-coupled receptors. We have derived the atomic model for OR S25 by using a combination of hydrophobicity profile prediction methods (2) and large-scale coarse grain molecular dynamics (MD) methods (3–8) with proper description of differential solvent environment.
Prediction of helical regions by using hydrophobicity profiles and optimization.
The transmembrane helices were identified on the basis of hydrophobicity by the multisequence profile method of Donnelly (2), implemented in perscan. A window size of 21 residues was used. For validation, the analysis was done on 21 rat ORs reported by Singer et al. (9). Sequences were aligned by the iterative profile alignment utility of whatif (10). The sequence for the receptor S25 (1) was used to build canonical right-handed α-helices. The structure of these helices were optimized by using the Newton-Euler Inverse Mass Operator torsional MD method (5–7) that scales linearly with the number of torsional degrees of freedom.
Helix assembly.
The orientation of each helical axis was built by using the bovine rhodopsin 7.5-Å electron density map (11). Helical z-coordinates were set such that the midpoint of each helical axis was positioned in the same z-plane of the assembly. The packing of the helix bundle was further optimized by using rigid body dynamics with proper description of the membrane bilayers surrounding the receptor that we found critical for proper packing. We used the DREIDING force field (12) with polar group charges derived from charge equilibration to simulate the lipids. The performance of this combination of charges and parameters was evaluated through a series of constant temperature and pressure MD simulations of crystals. The systems 1,2-dilauroyl-dl-phosphatidyl ethanolamine acetic acid, disodium β-glycerophosphate hydrate, and l-α-glycerol phosphorylcholine were chosen for simulation to evaluate the performance of the force field and atomic charges progressively, from a simple polar head group to a crystal lipid bilayer. We compared our results with experimental data and other simulation results available in the literature. Our choice of charges and force field gives densities and cell parameters with less than 4% error from the experimentally determined parameters. Thus, we have used the combination DREIDING force field and charges calculated with charge equilibration method (13) for representing lipids. The helix bundle surrounded by lipid bilayers was optimized by using rigid body dynamics with DREIDING force field and charmm22 (14) charges for the protein. The rigid body dynamics was done for 150 ps by which equilibration was attained.
Optimization of the full atomic model.
After the rigid body dynamics, loops were added to the helices by using whatif (10) software. After the addition of loops, we performed a full-atom minimization of the complete model with a barrel of lipid surrounding the protein, followed by dynamics optimization of the structure by using the massively parallel simulation program (MPSim) (8). MPSim includes state-of-the-art MD techniques such as: (i) the cell multipole method (3, 4) for fast and accurate calculation of nonbond forces; (ii) fast torsional dynamic methods such as Newton-Euler Inverse Mass Operator (5–7) and Hierarchical Newton-Euler Inverse Mass Operator (15); (iii) continuum solvation techniques such as the Poisson-Boltzmann description (16) and surface-generalized Born model (17) that account for solvation in biological systems. Thus to optimize the solution structure, we performed mixed mode dynamics by using the following descriptions. The helices and loops in the protein were modeled with the Newton-Euler Inverse Mass Operator torsional MD, the lipids were treated as rigid bodies, and the counterions Na+ and Cl− as free Cartesian atoms. Constant temperature dynamics using the Hoover algorithm was performed for 50 ps with time steps of 1 and 5 fs. The outside of the lipid layer was simulated with surface-generalized Born model continuum solvent description. A low dielectric constant of 60.0 was used to simulate the low dielectric region surrounding the membrane.
Control simulations.
The protocol described above was first tested on bacteriorhodopsin (BRDP), a membrane protein for which the crystal structure has been fitted with fair accuracy in the transmembrane region of the protein. We started from the sequence of bacteriorhodopsin and used no knowledge of the crystal structure to build the complete model with the above protocol. The membrane was represented by bilayers of diphosphatidyl glycerophosphate that is the lipid present in the purple membrane from Halobacterium halobium. Although the sequence homology between BRDP and the ORs is not high (less than 30%), they share the same tertiary motif common to α-helical transmembrane proteins: a 7-helix barrel. The overall rms deviation in coordinates of Cα atoms from the crystal structure (18) for the final model is 5.98 Å for all 221 aa. The overall rms deviation in coordinates for the residues in the membrane region is 3.29 Å whereas that for the loops is 8.57 Å. Thus, this modeling procedure gives a very reasonable structure as compared with the crystal structure for a known membrane protein.
Modeling of S25 OR.
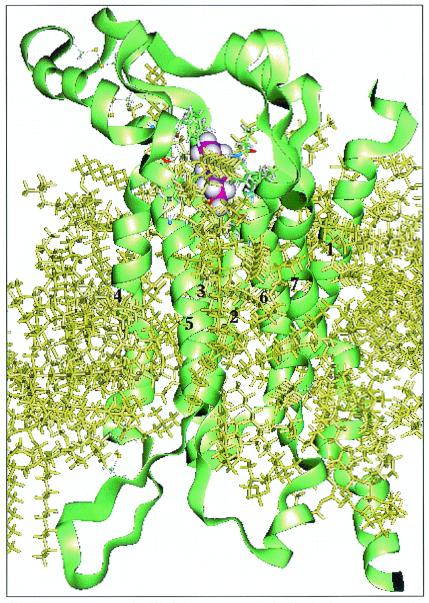
The sequence for OR S25 was taken from Malnic et al. (1). The membrane was simulated by using explicit lipid bilayers of dilauroylphosphatidyl choline. The choice of lipid in the OR case is supported by experimental indications (19, 20) that the membrane surrounding the ORs in vivo can be satisfactorily simulated by using a single-component lipid system of dilauroylphosphatidyl choline. The final atomic level model of S25 shown in Fig. 1 was used further in docking studies.
Figure 1.
Predicted structure for mouse OR S25 with predicted binding site for hexanol (purple). The membrane is represented by a barrel of dilauroylphosphatidyl choline bilayers (yellow) surrounding the TMs 1–7. The disulfide bonds were assigned between Cys-107 to Cys-209, Cys-132 to Cys-192, Cys-199 to Cys-219, and Cys-157 to Cys-171.
Docking Studies.
We have studied the binding of aliphatic alcohols and acids to OR S25 without any previous knowledge of the binding site. The list of ligands studied are those for which data on odor response preferences for several mouse OR was recorded recently (1). The list of 24 odorants includes aliphatic alcohols, carboxylic acids, dicarboxylic acids, and bromocarboxylic acids containing 4–9 carbon atoms. Among the odorants in that list, S25 responds positively to hexanol and heptanol only.
Each ligand was built in the extended conformation. The starting conformations were optimized by minimization of the potential energy by using the conjugate gradient method with DREIDING force field (12) and Gasteiger (21) charges. The minimized conformations were used as starting conformations for docking. The acids were considered in their protonated forms for docking because the pH range in the human nasal mucus is between 6 and 7 in normal individuals (22). The solvation energies for the ligands were calculated by using Poisson-Boltzmann continuum solvent model with the program jaguar (ref. 23; http://www.schrodinger.com). The solvation energies of the acids were calculated for the deprotonated species, because they are the dominant form in solution.
hier-dock protocol.
ORs interact with molecules of widely different structures and are therefore expected to exhibit high structural diversity in the ligand-binding region. Hypervariable regions in ORs have been identified in transmembrane domains (TMs) 3–5 (24–27) and are supposed to be involved in odorant binding. Nevertheless, the exact location of the binding site is not known. Therefore, we have derived a hierarchical docking protocol that could efficiently scan the whole receptor for all possible sites without bias from structural information. This was accomplished by combining the dock 4.0 program (28) with fine grain MD techniques. A complete scanning of all possible docking regions for S25 was done with the alcohol series because the odor response data available for S25 indicates preference for hexanol and heptanol (1), with no response reported for acids. Once the most probable binding site was identified, the complete alcohol and acid lists were optimized for calculating the binding energies. The protocol used for docking consists of the following steps:
- 1.
Mapping possible binding regions. The negative image of the receptor molecular surface, as defined by Connolly's method (29), was filled with a set of overlapping spheres. A probe of 1.4-Å radius was used to generate a 5 dots/Å molecular surface. Sphere clusters were generated for the whole receptor by using the program sphgen (30).
- 2.
Defining regions for docking. The sphere-filled volume representing the empty space inside the receptor was divided into five overlapping regions. They cover the extracellular portion of the receptor, as well as 2/3 of the inside of the helical barrel. Regions expected to be in contact with the membrane or involved in binding with the G protein were excluded from docking.
- 3.
Generating docked conformations of the receptor-ligand complexes. Orientations of the ligands into the receptor were generated by using the program dock 4.0 (28). We used flexible docking with torsion minimization of ligands, a nondistance-dependent dielectric constant of one, and a cutoff of 10 Å for energy evaluation. The configurations were ranked by using energy scoring. The best 10–30 configurations for each ligand in each possible binding region were used as input for the next step (annealing MD). We generated 110–120 configurations for each ligand in a total of 700 configurations covering the receptor space available for docking.
- 4.
Performing annealing MD for the complexes. Further optimization of ligand conformation in each binding region was done by using an annealing MD technique. The annealing MD also leads to a better scoring function by using a full atom force field and solvation effects. (Nonetheless, annealing MD does not allow for a long-range orientation search for which step 3 is necessary.) All configurations generated in step 3 were used in annealing MD simulations performed in 10 cycles of 1 ps from 50 to 600 K, by using the DREIDING force field (12), a nondistance-dependent dielectric constant of one, and a nonbond list cutoff of 9 Å. The best conformers from annealing were submitted to energy minimization.
- 5.
Selecting the best configuration and probable binding site. The conformations that have the lowest energy scores were selected. These exhibit a preferential region for binding.
- 6.
Redocking into the binding site. To obtain a comparative score for all ligands in the most possible binding site, we chose a 10 × 5 × 5-Å box enclosing the best configurations for butanol to heptanol. Steps 3–5 were then repeated for the alcohol and acid series.
- 7.
Cross-evaluating configuration energies by using perturbation techniques. The lowest energy configurations among the alcohols were used as template to build other members of the alcohol series. These complexes then were submitted to annealing MD. This was done to ensure that every ligand was evaluated in the same orientation starting from the best configuration of others.
- 8.
Ranking ligand affinities by using binding energies. The binding energies for the best complexes are calculated as the difference in the ligand energy in the receptor and in solution. The binding energies corresponding to different ligands then can be compared and ordered. The ligands for which the receptor–ligand complex has more favorable binding energies will have higher affinities to the receptor.
Results and Discussion
Identification of the Most Probable Binding Site.
A recent site-directed mutant (31) and several independent computational studies (32–35) have pointed to an odor-binding pocket composed of residues from TMs 3–7. We first tested this hypothesis by allowing the six test odors in the alcohol series to search the entire potential binding space; only parts of the structure in contact with membrane or facing the intracellular side were restricted from this analysis. Our docking protocol was applied for the alcohol series in each one of the docking regions chosen independently. The analysis of the best configurations by energy for each alcohol in each docking region clearly shows a preference for a particular region in the receptor. This preferred binding region is the most probable binding site for alcohols in this receptor. The best configurations for each ligand selected by energy criteria defined the most probable binding site. We cross-validated the results by each odorant in the preferred configuration for the other ligands.
Binding Affinity Ordering.
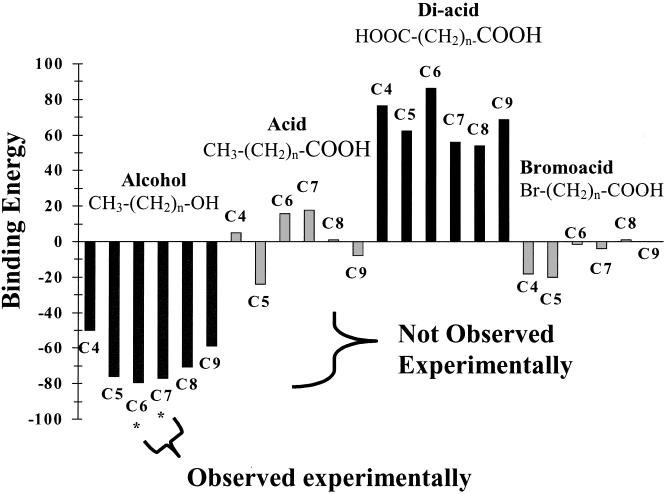
Having located the likely odor-binding pocket and found that the location is consistent with several previous lines of evidence, we proceeded to test how well the OR S25 model predicted relative odor affinities. We applied the docking protocol to the 24 ligands listed in Fig. 2 and predicted the ligand-receptor affinities, taking into account the ligand desolvation. The tests were run blindly, without reference to previous experimental results. The binding energies, shown in Fig. 2, correlated well with the experimental observations (1). Thus, hexanol and heptanol, the two compounds predicted by the OR S25 structure to have the highest binding energies, were the only two compounds that elicited measurable responses in the experiments. An important feature of the model is that it predicts affinities for other, less avidly bound compounds that may activate the receptor but are below the experimental detection threshold. For example, the structural model predicts that pentanol would have the third best binding energy, only 1.3 kcal/mol less favorable than heptanol. Because the responses observed for hexanol and heptanol were near threshold, binding studies of pentanol at higher concentrations or in other assay systems may show a response and would test our predicted energetics. It will be important for studies of expression systems to test carefully for these possible near-threshold responses, which may indicate broader ligand affinities than obtained thus far and which may be important for normal olfactory detection and perception.
Figure 2.
Calculated binding energies for 24 odorants docked to the mouse OR S25. The binding energies were calculated as the difference between the energy of the ligand in protein and in solution. The solvation corrections were calculated by using the Poisson–Boltzmann continuum model for the solvent (22). Binding energy bars are shaded according to the chemical classes indicated above them. The letter “C” followed by a number indicates the number of carbon atoms. Hexanol and heptanol (marked with *) are the only two compounds of the 24 found experimentally to elicit responses (1). These compounds are correctly predicted by our model as having the most favorable binding energies.
Residues Predicted as Directly Involved in Binding of Odorants.
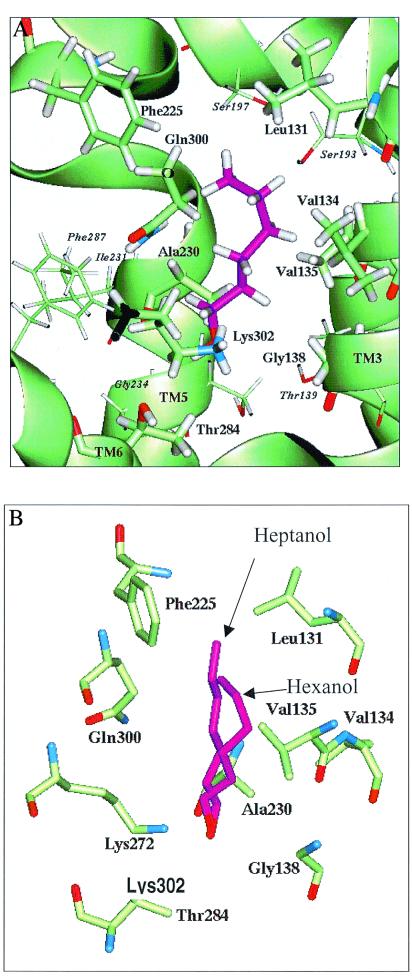
Fig. 3 shows the predicted binding pocket for the preferred compounds hexanol and heptanol. The pocket is situated between TMs 3–7, ≈10 Å deep from the extracellular surface. This is similar to the epinephrine-binding pocket of the β-adrenergic receptor (36). Docking results show that TMs 3, 5, and 6 have residues directly involved in binding. A closer look at our model allows us to infer that TM4 may have an important role in binding as it packs against TM3 and TM5 and therefore can alter their relative position if key residues of TM4 are mutated. TMs 3, 4, and 5 have been implicated in binding (25–27, 35). Fig. 3A shows 15 residues that constitute the hexanol-heptanol binding site. These residues are variable in the sequence alignment of ORs (1), consistent with their involvement in differential odor binding for different OR subtypes. Lys-302, which hydrogen bonds to the hydroxyl moiety, was critical for alcohol binding by OR S25. This presence of a critical Lys on TM7 is reminiscent of the closely related rhodopsin, where Lys-296 (TM7) binds the retinal chromophore (25). Substitutions in this residue could switch receptor specificity toward other functional groups. Other receptors we have reported had Val, Ser, and Ile residues at this position (1). Hydrophobic residues Phe-225, Leu-131, Val-134, Val-135, and Ala-230 formed Van der Waals contacts with the ligand, accounting for the specificity of the OR S25 model for 6–7 carbon compounds. Hydrophobic substitutions in these residues would be expected to modulate the preferred carbon length. In particular, the model predicts that substitutions of Val for Phe-225 and Val for Leu-131 would be expected to create more space in the pocket and shift its specificity toward larger ligands. Substitutions of Phe for Leu-131 would be predicted to have the opposite effect. Polar residues Thr-284 and Gln-300 were also in close contact with the ligand but did not appear to contribute any hydrogen bonding specificity. These residues could be important for interactions of other compounds with OR S25. This study thus identifies critical residues likely to contribute to the differential affinities of a specific OR for different odor ligands. Site-directed mutagenesis of these residues followed by functional assays will test the present observations and constrain future models. The agreement between the experimental and computational results suggests that the combination of functional assays and OR structure prediction will make it possible to identify potential odors for other ORs and propose site-directed mutants that increase selectivity for particular odor compounds. We suggest that this provides an efficient system for obtaining such knowledge of the atomic-level binding requirements for ORs, which can enhance the development of biosensors for the fragrance and food industries, industrial and environmental safety, and explosives and narcotics detection. The insights gathered also may apply to other protein families that display broad affinities for a range of compounds, such as the chemokine receptors and P450 oxidases, which have important implications for HIV, immunology, pharmacology, and toxicology.
Figure 3.
Predicted recognition site for hexanol and heptanol in mouse OR S25. Residues within 3 Å of the ligand (hexanol in purple) are displayed as thicker with labels in bold font. Residues within 3–5 Å of the ligand have labels in italics. Lys-302 forms a weak hydrogen bond to the hydroxyl group of the alcohols. Phe-225 and Leu-131 seem to limit the chain length suitable to the binding site. TM 3–7 have residues directly involved in binding. (A) Longitudinal view. (B) Detail view. Hydrogen atoms were suppressed.
Conclusions
We have developed a modeling and a docking protocol that can be combined to predict the binding site(s) of ORs from their sequence. We applied these protocols to the amino acid sequence of the mouse OR S25. Besides the sequence, the only additional information required for modeling are sequence alignment of other ORs and the bovine rhodopsin electron density map. By using binding energies that take into account desolvation of the ligands, we can qualitatively reproduce the odor response preferences reported for OR S25 (1). Among a set of 24 species of linear alcohols, acids, diacids, and bromoacids containing 4–9 carbon atoms, we successfully eliminated the acid series that are not recognized experimentally. Within the alcohol series, those with higher calculated binding affinities to the receptor are the ones with positive odor preference response (hexanol and heptanol). Some residues in TM 3, 5, and 6 are predicted as directly involved in binding of odor molecules, whereas TM4 and the four disulfide bridges in the model of OR S25 are implicated in the correct assembling of the recognition site.
We believe that the results of this work bring us one step forward in the use of theoretical methodologies for the design of biosensors for defense, perfume, or food industry applications. By applying the protocols presented here, it is possible to elaborate mutation studies aimed in the design of ORs for specific odorants or suggest potential odorants for natural-occurring ORs that can be tested experimentally. The knowledge of the binding requirements for ORs at an atomic level is also important for the development of artificial olfaction sensors.
Acknowledgments
We thank the Army Research Office (Multidisciplinary University Research Initiative, Dr. Robert Campbell) for support of this collaborative effort. The facilities of the Materials and Process Simulation Center used in this project are supported also by the Department of Energy (ASCI ASAP), National Science Foundation (CHE and MRI), Army Research Office, Chevron Corp., MMM, Beckman Institute, Seiko-Epson, Exxon, Dow Chemical, Avery-Dennison Corp., National Aeronautics and Space Administration, National Institutes of Health HD (W.A.G.), Asahi Chemical, and BP Chemical. This work is also supported by National Institute on Deafness and Other Communication Disorders (G.M.S.); National Institute on Aging, National Aeronautics and Space Administration, and National Institute of Mental Health (Human Brain Project, G.M.S.); Yale Medical Scientist Training Program (M.S.S.); and National Institute of Mental Health (IAIMS, M.S.S.).
Abbreviations
- OR
olfactory receptor
- MD
molecular dynamics
- TM
transmembrane domain
References
- 1.Malnic B, Hirono J, Sato T, Buck L B. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 2.Donnelly D. Biochem Soc Trans. 1993;21:36–39. doi: 10.1042/bst0210036. [DOI] [PubMed] [Google Scholar]
- 3.Ding H Q, Karasawa N, Goddard W A., III J Chem Phys. 1992;97:4309–4315. [Google Scholar]
- 4.Ding H Q, Karasawa N, Goddard W A., III Chem Phys Lett. 1992;196:6–10. [Google Scholar]
- 5.Jain A, Vaidehi N, Rodriguez G. J Comp Physiol. 1993;106:258–268. [Google Scholar]
- 6.Mathiowetz A M, Jain A, Karasawa N, Goddard W A., III Proteins. 1994;20:227–247. doi: 10.1002/prot.340200304. [DOI] [PubMed] [Google Scholar]
- 7.Vaidehi N, Jain A, Goddard W A., III J Phys Chem. 1996;100:10508–10517. [Google Scholar]
- 8.Lim K-T, Brunett S, Iotov M, McClurg R B, Vaidehi N, Dasgupta S, Taylor S, Goddard W A., III J Comput Chem. 1997;18:501–521. [Google Scholar]
- 9.Singer M S, Weisinger-Lewin Y, Lancet D, Shepherd G M. Recept Channels. 1995;4:141–147. [PubMed] [Google Scholar]
- 10.Vriend G. J Mol Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 11.Schertler G F X. Eye. 1998;12:504–510. doi: 10.1038/eye.1998.138. [DOI] [PubMed] [Google Scholar]
- 12.Mayo S L, Olafson B D, Goddard W A., III J Phys Chem. 1990;94:8897–8909. [Google Scholar]
- 13.Rappé A K, Goddard W A., III J Phys Chem. 1991;95:3358–3363. [Google Scholar]
- 14.MacKerell A D, Bashford D, Bellott M, Dunbrack R L, Evanseck J D, Field M J, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 15.Vaidehi N, Goddard W A., III J Phys Chem. 2000;104:2375–2383. [Google Scholar]
- 16.Tannor D J, Marten B, Murphy R, Friesner R A, Sitkoff D, Nicholls A, Ringnalda M, Goddard W A, III, Honig B. J Am Chem Soc. 1994;116:11875–11882. [Google Scholar]
- 17.Ghosh A, Rapp C S, Friesner R A. J Phys Chem B. 1998;102:10983–10990. [Google Scholar]
- 18.Grigorieff N, Ceska T A, Downing K H, Baldwin J M, Henderson R. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 19.Gimenez C. Rev Neurol (Paris) 1998;26:232–239. [PubMed] [Google Scholar]
- 20.Kiefer H, Krieger J, Olszewski J D, von Heijne G, Prestwich G D, Breer H. Biochemistry. 1996;35:16077–16084. doi: 10.1021/bi9612069. [DOI] [PubMed] [Google Scholar]
- 21.Gasteiger J, Marsili M. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 22.Sachdeva A, Sachdeva O P, Gulati S P, Kakkar V. Indian J Med Res B. 1993;98:265–268. [PubMed] [Google Scholar]
- 23.Schrodinger Inc. jaguar. Portland, OR: Schrodinger Inc.; 2000. , Version 4.0. [Google Scholar]
- 24.Pilpel Y, Lancet D. Protein Sci. 1999;8:969–977. doi: 10.1110/ps.8.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singer M S, Weisinger-Lewin Y, Lancet D, Shepherd G M. Recept Channels. 1995;4:141–147. [PubMed] [Google Scholar]
- 26.Mombaerts P. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- 27.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 28.Ewing T A, Kuntz I D. J Comput Chem. 1997;18:1175–1189. [Google Scholar]
- 29.Connolly M L. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 30.Kuntz I D, Blaney J M, Oatley S J, Langridge R, Ferrin T E. J Mol Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 31.Krautwurst D, Yau K W, Reed R R. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 32.Singer, M. S. (2000) Chem. Senses, in press. [DOI] [PubMed]
- 33.Poincelot R P, Millar P G, Kimbel R L, Jr, Abrahamson E W. Biochemistry. 1970;9:1809–1816. doi: 10.1021/bi00810a021. [DOI] [PubMed] [Google Scholar]
- 34.Singer M S, Shepherd G M. NeuroReport. 1994;5:1297–1300. doi: 10.1097/00001756-199406020-00036. [DOI] [PubMed] [Google Scholar]
- 35.Pilpel Y, Lancet D. Protein Sci. 1999;8:969–977. doi: 10.1110/ps.8.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strader C D, Sigal I S, Dixon R A F. FASEB J. 1989;3:1825–1832. doi: 10.1096/fasebj.3.7.2541037. [DOI] [PubMed] [Google Scholar]