Abstract
Recent studies have shown that a number of genes that are not mutated in various forms of muscular dystrophy may serve as surrogates to protect skeletal myofibers from injury. One such gene is Galgt2, which is also called cytotoxic T cell GalNAc transferase in mice. In this study, we show that Galgt2 overexpression reduces the development of dystrophic pathology in the skeletal muscles of mice lacking α sarcoglycan (Sgca), a mouse model for limb girdle muscular dystrophy 2D. Galgt2 transgenic Sgca−/− mice showed reduced levels of myofiber damage, as evidenced by i) normal levels of serum creatine kinase activity, ii) a lack of Evans blue dye uptake into myofibers, iii) normal levels of mouse locomotor activity, and iv) near normal percentages of myofibers with centrally located nuclei. In addition, the overexpression of Galgt2 in the early postnatal period using an adeno-associated virus gene therapy vector protected Sgca−/− myofibers from damage, as observed using histopathology measurements. Galgt2 transgenic Sgca−/− mice also had increased levels of glycosylation of α dystroglycan with the CT carbohydrate, but showed no up-regulation of β, γ, δ, or ε sarcoglycan. These data, coupled with results from our previous studies, show that Galgt2 has therapeutic effects in three distinct forms of muscular dystrophy and may, therefore, have a broad spectrum of therapeutic potential for the treatment of various myopathies.
Recent work from a number of laboratories has identified therapeutic targets in skeletal muscle that can, when up-regulated, diminish the severity of disease despite the fact that these molecules do not replace the genetic defect. Such targets cut a wide swath across aspects of muscle biology, ranging from inhibitors of apoptosis (eg, Bcl-21) to stimulators of muscle growth and regeneration (eg, myostatin inhibitors,2,3,4,5 Igf16) to inducers of, or members of, alternative transmembrane protein complexes (eg, utrophin,7,8,9 agrin,10,11,12 neuregulin,13 integrin α7B,14,15 sarcospan,16 ADAM12,17,18 and Galgt219,20,21). This last category contains a glycosyltransferase, Galgt2, which can alter the expression and properties of membrane proteins in skeletal muscle.19,22 Galgt2, also called the cytotoxic T cell (CT) GalNAc transferase by virtue of its original description in activated CD8+ T cells in mice,23,24 is a β1,4-N-acetylgalactosaminyltransferase that transfers β1,4GalNAc onto a relative small number of glycoproteins and at least one glycolipid to create the CT carbohydrate antigen (Neu5Ac [or Neu5Gc]α2,3[GalNAcβ1,4]Galβ1,4GlcNAcβ-R).23,25,26,27 In adult skeletal muscle, Galgt2 protein,28 and the CT carbohydrate it creates,29 become confined to the neuromuscular junction and are not present in extrasynaptic regions of the sarcolemmal membrane.
We have created transgenic mice that overexpress Galgt2 specifically in skeletal muscle to maintain extrasynaptic expression of the CT carbohydrate in adult animals.22 These mice show that the predominant glycoprotein glycosylated by Galgt2 is α dystroglycan,22 a member of the dystrophin-associated glycoprotein complex30,31,32 implicated in numerous forms of muscular dystrophy.33,34 Importantly, muscles in Galgt2 transgenic mice show ectopic extrasynaptic expression of synaptic proteins known to bind to dystroglycan, including utrophin, laminin α4, and laminin α5.22 In light of the changed distribution of these molecules, we have tested the therapeutic effects of Galgt2 transgene overexpression in two mouse models of muscular dystrophy–mdx mice, which lack dystrophin,35,36 and in dyW/dyW mice,21 which have reduced expression of laminin α2.37,38 In both instances, Galgt2 overexpression in skeletal muscle significantly reduced the development of pathology related to muscular dystrophy.19,20,21 We have also used gene therapy vectors to increase Galgt2 expression in skeletal muscles postnatally. These results also demonstrate efficacy in both the mdx and dyW/dyW models, but with fewer molecular and developmental changes than occur in Galgt2 transgenic animals.20,21 Here, we have analyzed the effects of Galgt2 overexpression in a third muscular dystrophy model–-mice lacking α sarcoglycan (Sgca), a model for limb girdle muscular dystrophy 2D (LGMD2D).39
Sarcoglycans are transmembrane proteins that comprise a portion of the dystrophin-associated glycoprotein complex in cardiac muscle, smooth muscle, and skeletal muscle.40,41,42 In skeletal muscle, the sarcoglycan complex is comprised of four proteins (α, β, γ, and δ sarcoglycan), and loss of any of proteins results in a form of Limb Girdle muscular dystrophy (LGMD2D, LGMD2E, LGMD2C, and LGMD2F, respectively). LGMD2C-2F are all autosomal recessive disorders.43,44,45,46,47,48,49,50 Loss of any one sarcoglycan due to genetic mutation leads to the concomitant loss of the other sarcoglycan proteins in skeletal muscle membrane50,51 (with exceptions52). Mouse models lacking each of these proteins has been made and these mimic important aspects of LGMD, including increased myofiber damage, necrosis, and regeneration, decreased lifespan, and variably increased cardiomyopathy.39,53,54,55,56,57 Cardiomyopathy is associated with loss of β, γ, and δ sarcoglycan, but not α sarcoglycan, in patients43 and in mice.53 This may result from alternative sarcoglycan complexes made in heart between β, γ, and δ sarcoglycan and ε sarcoglycan, a sarcoglycan with significant homology to α sarcoglycan.58,59 While null mouse models of sarcoglycan deficiency mimic human disease, knock-in mouse models of the most common LGMD2D missense mutation (R77C) do not, suggesting that mice have more robust quality control mechanisms for protein folding and/or protein stability than humans.60,61 The sarcoglycan complex also tightly associates with sarcospan, another transmembrane protein whose expression is reduced in their absence.39,62,63 Loss of sarcospan in mice does not result in muscular dystrophy,64 though overexpression in mdx muscles can reduce disease.16 Loss of sarcoglycans can also alter the stability of dystroglycan and dystrophin in skeletal muscle,39,55 though these proteins are still present on the sarcolemmal membrane.39,55,65 By contrast with sarcospan, loss of dystrophin (in the mdx mouse) results in muscular dystrophy,35,36,66 as does loss of dystroglycan specifically in skeletal muscle.67,68
LGMD2D is the most common sarcoglycanopathy.43,69 Age of disease onset can range from 3 to 40 years, usually presenting with weakness in the pelvic and girdle muscles, though truncal and distal muscles may also be affected. Disease severity can range from mild impairment with slow progression to severe disability with rapid deterioration.43,69 While there are treatments aimed at ameliorating symptoms of LGMD2D and related sarcoglycanopathies, including medications to manage cardiac symptoms and pain,69 there are currently no treatments that have been shown to slow disease progression in patients, though corticosteroids can have positive effects.70 Moreover, only a scattering of approaches have shown promise in animal models for these diseases.71,72,73,74,75,76,77 Here we provide a proof of principal demonstration that a surrogate gene approach can reduce muscular dystrophy in the Sgca−/− mouse model of LGMD2D, akin to what we have previously shown in mdx mice and dyW/dyW mice. This makes Galgt2 the first surrogate gene approach shown to prevent muscle pathology in these three forms of the disease.
Materials and Methods
Materials
Agarose-bound lectins (Wisteria floribunda agglutinin, WFA; and Wheat germ agglutinin, WGA) were purchased from EY Laboratories (San Mateo, CA). AAV1-CMV-Galgt2 was made and purified by Virapure (San Diego, CA). AAV8-CMV-Galgt2 was made by the Viral Vector Core at Nationwide Children’s Hospital. Monoclonal antibodies to dystrophin (Dy4/6D3), utrophin (DRP3/20C5), β-dystroglycan (43DAG1/8D5), α-sarcoglycan (Ad1/20A6), β-sarcoglycan (Sarc1/5B1), δ-sarcoglycan (δSarc3/12C1), and γ-sarcoglycan (35DAG/21B5) were obtained from Nova Castra (Newcastle On Tyne, UK). A rabbit polyclonal antiserum to ε sarcoglycan (H-67) and a mouse monoclonal antibody to Plectin 1 (10F6) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Polyclonal antibody to amino acids 20 to 33 of actin (Sigma, A5060) and a polyclonal antiserum to EHS laminin (Sigma, L9393) were obtained from Sigma (St. Louis, MO). Antibody to α-dystroglycan (IIH6) was obtained from Upstate Biotechnology (Lake Placid, NY). An affinity-purified rabbit anti-peptide antibody to mouse CT GalNAc transferase (Galgt2) was produced in our laboratory (CT6822), as was CT2,29 a monoclonal antibody that recognizes the CT carbohydrate.24 Secondary antibodies conjugated to horseradish peroxidase, fluorescein isothiocyanate, Cy3, or Cy2 were purchased from Jackson Immunochemicals (Seattle, WA).
Mice
Transgenic mice bearing the CT GalNAc transferase (Galgt2) specifically in skeletal muscles via the human skeletal α-actin promoter (CT)22 were described by us previously, as were Galgt2 transgenic mdx mice.19 Sgca−/− mice were originally made by Kevin Campbell and colleagues (HHMI, U. Iowa).39 For these experiments, we generated four genotypes: Sgca+/−, Sgca+/−CT, Sgca−/−, and Sgca−/−CT. All mice were maintained on a similarly mixed background, and all control animals were age-matched littermates.
Infection of Skeletal Muscles with AAV1-CMV-Galgt2 or AAV8-CMV-Galgt2
The tibialis anterior (TA) or gastrocnemius muscle on the left side of 4- to 7-day-old Sgca−/− mice was injected with 1 to 5 × 1010 vector genomes (vg) of AAV1-CMV-Galgt2 or AAV8-CMV-Galgt2. AAV8 used in these experiments was isolated in Rhesus macaque strain variant 74 (rh.74). Packaged AAV vectors were produced and purified using the triple transfection method in HEK293 cells and purified by iodixinol chromatography as previously described.78 In these vectors, the mouse CT GalNAc transferase gene (Galgt2) was expressed via the cytomegalovirus promoter (CMV). The gastrocnemius muscle of the left leg was injected with AAV vector in a volume of 50 μl of sterile PBS using a 0.3cc insulin syringe, whereas tibialis anterior muscle was injected with a 25-μl volume. Muscles were always injected near the midpoint of the belly of the muscle. Contralateral muscles (on the right side) were injected with the same volume of sterile PBS (mock-infected). Control infections were also done with AAV1-CMV-lacZ or AAV1-CMV-GFP to confirm that no changes came from nonspecific effects of AAV infection (not shown). After 4 to 6 weeks, mice were sacrificed and muscles dissected and snap-frozen in liquid nitrogen-cooled isopentane, as before.20,21
Serum Creatine Kinase Activity
Blood was collected from the tail vein and allowed to clot for 1 hour at 37°C. Clotted cells were centrifuged at 1500 × g for 3 minutes, and serum was collected and analyzed without freezing. Creatine kinase activity assays were done using an enzyme-coupled absorbance assay kit (CK-SL; Diagnostic Chemicals Limited; Charlottetown, PEI, Canada) following the manufacturer’s instructions, as previously described.19 Absorbance was measured at 340 nm every 30 seconds for 4 minutes at 25°C to calculate enzyme activity. All measurements were done in triplicate.
Histology
Muscles were dissected and snap-frozen in liquid nitrogen-cooled isopentane and sectioned at 8 to 10 μm on a cryostat. Sections were either stained with H&E (Sigma; St. Louis, MO), or immunostained with antibodies against the CT carbohydrate (CT2), α-, β-, δ-, ε-, and γ-sarcoglycans, α- and β-dystroglycan, dystrophin, or utrophin as previously described.22 For use of mouse monoclonal antibodies, a mouse Ab-on-mouse blocking reagent was used (Dako). Antibody used here for δ-sarcoglycan did not stain, but worked well on Western blots, while the antibody used for ε-sarcoglycan stained but was ineffective on Western blot. All other antibodies were used with both methods. Quantification of central nuclei and myofiber diameters was done as previously described.19,20,21 Briefly, cross-sections were cut from the midsection (belly) of the muscle at 8 to 10 μm and stained with H&E. For central nuclei measurements, all myofibers (ca. 500 to 1000/section) from each section were counted and the percentage of central nuclei averaged over eight sections per animal. For myofiber diameters, 50 myofibers were measured at random per section for at least five sections per animal, and the data were then averaged to generate the measures of myofiber diameter. Determinations of the presence of central nuclei versus CT carbohydrate overexpression were done at or near the midsection of infected skeletal muscles, again as previously described.20,21 Fifty myofibers were measured in each condition (expressing or non-expressing) per section for gastroc and TA and 3 to 4 sections were averaged per animal. All myofibers were counted in each section analyzed, and all data obtained was used in determinations of significance. Averages of central nuclei represent analysis of individual myofibers where n is always a single muscle from a unique animal, with the exception of AAV experiments, in which data for the AAV1-CMV-Galgt2 and mock-infected muscles were obtained from the same muscles. Identical time exposures were used for all comparisons of immunostaining. Addition of secondary reagents alone did not generate significant staining.
Lectin Precipitation and Immunoblotting
Immunoblotting of whole muscle lysates and of lectin precipitations were done as previously described.19,20,21 For lectin precipitations, we extracted identical amounts (50 mg) of minced gastrocnemius muscle from Galgt2 transgenic (CT) or non-transgenic Sgca+/− or Sgca−/− mice with Nonidet P-40 buffer (1% Nonidet P-40, 50 mmol/L Tris-HCl, pH 7.4, 150 mmol/L NaCl, 2 mmol/L EDTA, and 1:200 protease inhibitor cocktail; Sigma, St. Louis, MO). For whole cell lysates, we extracted a similar weight of muscle with SDS/urea buffer (2% SDS, 4 mol/L urea, 50 mmol/L Tris-HCl, pH 6.8, 2 mmol/L EDTA). Extractions were performed at 4°C with light shaking overnight. Protein amounts were measured using the Bio-Rad DC protein assay (Bio-Rad, Hercules, CA) versus standard curve made in the appropriate buffer. For lectin pull-downs, 150 μg of NP-40-extracted protein per sample was incubated with 50 μl WGA agarose or WFA agarose at 4°C overnight. Pellets were washed four times in excess volume of NP-40 buffer and boiled in SDS sample buffer. The entire precipitate for each genotype was then resolved by SDS-polyacrylamide gel electrophoresis (PAGE, 4% to 12% linear gradient gels, Invitrogen, CA, or standard 6% or 12% SDS-PAGE gels) and transferred to nitrocellulose membrane (Bio-Rad Laboratories, CA). Antibodies against the CT carbohydrate (CT2), Galgt2, α-, β-, δ-, and γ-sarcoglycans, α- and β-dystroglycan, dystrophin, utrophin, and actin were used in immunoblot analysis as previously described.19,20,21,28 For whole cell SDS-urea extracted lysates, extracted proteins were boiled in SDS gel running buffer and 40 μg of cell lysate loaded per lane for separation on SDS-PAGE gels, as above. Antibodies against Galgt2, α-, β-, δ-, and γ-sarcoglycans, α- and β-dystroglycan, dystrophin, utrophin, plectin 1, and actin were used in immunoblot analysis as previously described.19,22,28 Secondary antibody alone never generated any positive bands, with the exception of mouse secondary antibodies, which recognized a 50 to 55 kDa band not shown in the results in all genotypes (presumed to be endogenous IgG Fc receptor). Protein levels were compared by scanning densitometry of the appropriate protein bands on Western blots, as previously described.19 Data from four independent experiments were averaged to obtain measures of changed expression.
Measurement of Gene Expression by TaqMan
Total RNA was isolated using Trizol reagent (Invitrogen; Carlsbad, CA) from gastrocnemius, quadriceps, tibialis anterior, diaphragm, or triceps muscle samples stabilized in RNALater (Ambion; Austin, TX). RNA was further purified on a silica-gel-based membrane (RNeasy-Mini; Qiagen, Valencia, CA) and the integrity of RNA was determined by capillary electrophoresis using 6000 Nano LabChip kit on a Bioanalyzer 2100 (Agilent; Foster City, CA). RNA content was measured using an ND-1000 spectrophotometer (Nanodrop; Wilmington, DE). Only samples with no evidence of RNA degradation were used for TaqMan gene expression studies. A high capacity cDNA archive kit (Applied Biosystems; Foster City, CA) was used to reverse transcribe 3 μg of total RNA as per the manufacturer’s guidelines. Samples were subjected to real-time PCR in triplicate using TaqMan ABI 7500 sequence detection system (Applied Biosystems; Foster City, CA) with 18S ribosomal RNA (product no. 4308329, Applied Biosystems) as internal control. Primers and probe against CT GalNAc transferase (Galgt2) were custom-made and provided as a 20× reaction mix containing 18μmol/L each of primers (forward primer sequence: 5′-GATGTCCTGGAGAAAACCGAACT-3′; reverse primer sequence: 5′-GCAGCCTGAACTGGTAAGTATTCC-3′) and 5μmol/L of probe (probe sequence: 5′-CCGCCCACCACATCC-3′) (Applied Biosystems). Each 25 μl PCR reaction mix consisted of 1× primer-probe mix, 1× TaqMan Universal PCR master mix (product no. 4304437; Applied Biosystems). After an initial hold of 2 minutes at 50°C to allow activation of AmpErase and 10 minutes at 95°C to activate the AmpliTaq polymerase, the samples were cycled 40 times at 95°C for 15 seconds and 60°C for 1 minute. Gene expression was determined as relative changes by the 2−ΔΔCt method.79 Data are presented as fold difference normalized to 18S ribosomal RNA. All measures were done in triplicate for each data point.
Treadmill and Evans Blue Dye uptake
To measure exercise physiology, a treadmill experiment was performed on mice that were 3 months old. All strains of mice (Sgca+/−, Sgca+/−CT, Sgca−/−, and Sgca−/−CT) underwent a 10-minute run on a horizontal treadmill (Treadmill Simplex II, Columbus Instruments) at a speed of 10 m/min twice a week for 2 to 3 weeks. Time remaining on the treadmill, up to a total time of 10 minutes, was recorded. For Evans blue dye (EBD) uptake analysis, EBD was injected intraperitoneally (50 μg/g body weight in 100 μl of sterile PBS). Five hours later, mouse activity was normalized by subjecting them to 15 minutes of exercise on a treadmill at a constant speed of 15m/min, as before.39 Thirty-six hours after the injection, mice were sacrificed, and skeletal and heart muscles were snap-frozen in liquid nitrogen-cooled isopentane. Quantification of EBD-positive stained areas in sections of skeletal muscle was done using Olympus imaging program (Slidebook) and Olympus BX61 epifluorescence microscope to quantify the area of the muscle with spontaneous fluorescence (in the rhodamine channel) of EBD. Percentage area with dye uptake in skeletal muscle was calculated by dividing the area of positive EBD staining by the total area of the muscle section analyzed for at least eight sections per muscle.
Statistics
Determinations of significance for comparisons of transgenic animals were done using analysis of variance with pre- and posthoc Bonferroni analysis. Comparisons for AAV-infected muscles versus non-infected ones were done using a paired Student’s t-test with equal weighting between samples.
Results
Characterization of Galgt2 Transgenic (CT) α Sarcoglycan-Deficient (Sgca−/−) Mice
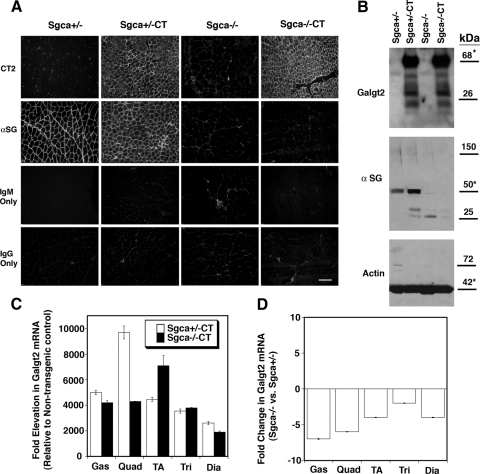
We first measured the level of Galgt2 transgene (CT) overexpression in Sgca+/−CT and Sgca−/−CT skeletal muscle compared with non-transgenic Sgca+/− and Sgca−/− controls (Figure 1). Sgca+/−CT and Sgca−/−CT muscles had equivalent levels of overexpression of CT carbohydrate, the product of Galgt2 activity,25,27 as determined by immunostaining (Figure 1A). They also had equivalent levels of overexpressed Galgt2 protein, as determined by immunoblotting of whole muscle SDS cell lysates (Figure 1B). For the gastrocnemius muscle (shown in Figure 1), Galgt2 protein was increased by 9.7 ± 1.7-fold in Sgca+/−CT versus Sgca+/− and by 9.3 ± 1.8-fold in Sgca−/−CT versus Sgca−/− (P < 0.001 for both comparisons versus non-transgenic littermates, n = 4). There was no change in Sgca−/− vs. Sgca+/− (1.1 ± 0.7). The fold-overexpression of Galgt2 mRNA was also consistent with our previous studies in wild-type, mdx, and dyW/dyW transgenic mice,19,20,21,22 though the amplitude of increased expression was higher (Figure 1C). Galgt2 mRNA levels were increased between 2000- and 10,000-fold in both Sgca+/−CT versus Sgca+/− and Sgca−/−CT versus Sgca−/− comparisons. Sgca−/− muscles, however, showed reduced Galgt2 mRNA expression compared with Sgca+/−, with decreases ranging from two- to sevenfold depending on the muscle (Figure 1D). This stands in contrast to previous mdx versus wild-type and dyW/dyW vs. dyW/+ comparisons, where Galgt2 mRNA expression was increased.21 The lower denominator in Sgca−/− muscle may explain the 0.5–1-log increase in Galgt2 mRNA expression in Sgca−/−CT compared with mdxCT and dyW/dyWCT muscles.21 As expected,39 immunostaining for α sarcoglycan was lost in both Sgca−/− and Sgca−/−CT muscles (Figure 1A), and α sarcoglycan protein was absent in Sgca−/− and Sgca−/−CT muscles by Western blot (Figure 1B). As we have previously reported, Galgt2 transgenic wild-type muscles have increased levels of sarcoglycan proteins.19 Consistent with this, Sgca+/−CT muscles had increased levels of α sarcoglycan relative to Sgca+/− (a 1.6- ± 0.9-fold increase, n = 4, Figure 1B). Control secondary antibodies for CT2 (mouse IgM only) or α sarcoglycan (mouse IgG only) yielded only minimal background (Figure 1A), and blots for actin showed equivalent levels of protein loading and transfer (Figure 1B).
Figure 1.
Creation of Galgt2 transgenic α sarcoglycan-deficient (Sgca−/−) mice. A: Galgt2 transgenic (CT) mice and non-transgenic littermates that were either heterozygous (Sgca+/−) or homozygous (Sgca−/−) for a deletion of α sarcoglycan were stained with a monoclonal antibody to α sarcoglycan or with CT2, an antibody that recognizes the CT carbohydrate. Control secondary antibody for CT2 (IgM only) and α sarcoglycan (IgG only) are also shown. Scale bar = 100 μm. B: 40 μg of whole muscle SDS protein lysate was blotted for Galgt2 protein and for α sarcoglycan protein. Actin was used as a control for protein loading and transfer. Asterisk indicates native molecular weight for the protein of interest. C: Sgca+/−CT and Sgca−/−CT muscles show similar increases in Galgt2 mRNA, relative to Sgca+/− and Sgca−/−, respectively, as measured by quantitative RT-PCR. D: Galgt2 mRNA was reduced in Sgca−/− muscles relative to Sgca+/−. Gas, gastrocnemius; Quad, quadriceps; TA, tibialis anterior; Tri, triceps; Dia, diaphragm. Errors are SD for 3 to 4 animals per condition (in C and D).
Analysis of Muscular Dystrophy in Galgt2 Transgenic (CT) Sgca−/− Animals
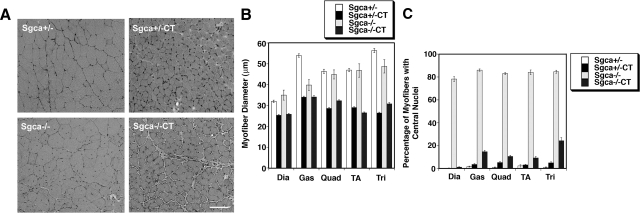
Having made Sgca+/−CT and Sgca−/−CT animals, we proceeded to determine whether Galgt2 transgenic Sgca−/− mice would reduced levels of muscle pathology (as we had shown previously in mdxCT19 and dyW/dyWCT21 mice). We began by analyzing the histopathology of individual skeletal muscles from Sgca+/−, Sgca+/−CT, Sgca−/−, and Sgca−/−CT mice (Figure 2, A–C). Sgca+/−CT and Sgca−/−CT muscles were reduced in size relative to their Sgca+/− and Sgca−/− littermates (Figure 2, A and B), a finding we had observed previously in all other Galgt2 transgenic strains we have studied. Reductions in myofiber diameter were highly significant in the diaphragm, gastrocnemius, quadriceps, tibialis anterior, and triceps for both Sgca+/−CT muscles (compared with Sgca+/−) and Sgca−/−CT muscles (compared with Sgca−/−) (P < 0.01 for all such comparisons, Figure 2B). The percentage reduction ranged from 20% to 50%, with the diaphragm being least affected. Diaphragm, gastrocnemius, and triceps muscles were all reduced as well for Sgca−/− muscles compared with Sgca+/− (P < 0.05 for all). As expected,39 Sgca−/− muscles also showed increased variability in myofiber diameter relative to Sgca+/−, which is likely due to the presence of smaller regenerating myofibers in these dystrophic muscles (Figure 2B). Sgca−/− muscles also had high percentages of myofibers with centrally located nuclei (ca. 80% for all muscles studied), again indicating the presence of muscular dystrophy (Figure 2C). This elevation was highly significant (P < 0.001) for all Sgca−/− vs. Sgca+/− comparisons. By contrast, Sgca−/−CT muscles, like mdxCT19 and dyW/dyWCT muscles,21 showed very reduced levels of myofibers with central nuclei (P < 0.001 for all Sgca−/−CT versus Sgca−/− comparisons). As with dyW/dyWCT muscles,21 this level was not quite reduced to wild-type (non-dystrophic) levels, but was nevertheless a highly significant in all muscles studied (Figure 2C).
Figure 2.
Characterization of muscle pathology in Galgt2 transgenic Sgca+/− and Sgca−/− mice. A: H&E staining of the gastrocnemius muscle of Galgt2 transgenic (CT) Sgca+/− and Sgca−/− mice and non-transgenic littermates. Scale bar =100 μm. B: Myofiber diameters were measured in the diaphragm (Dia), gastrocnemius (Gas), quadriceps (Quad), tibialis anterior (TA) and triceps (Tri) muscle of Galgt2 transgenic (CT) Sgca+/− and Sgca−/− mice and their non-transgenic, age-matched, littermates. CT muscles were reduced in size, regardless of Sgca genotype (P < 0.01 for all). C: Percentage of myofibers with central nuclei was greatly increased in Sgca−/− muscles (P < 0.001 for all versus Sgca+/−). Galgt2 transgene expression significantly lowered central nuclei in Sgca−/− muscles relative to Sgca−/− (P < 0.001 for all). Errors are SD from n = 3 to 4 animals per genotype (in B and C).
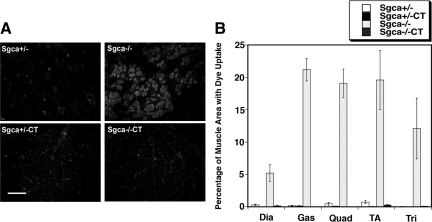
We next assessed resistance to membrane damage in vivo by measuring Evans blue dye uptake into myofibers. Only Sgca−/− muscles had increased dye uptake (Figure 3, A and B). Area of uptake ranged from 5% in the diaphragm to 20% in the gastrocnemius, quadriceps, and tibialis anterior (P < 0.05 for all Sgca−/− muscles versus Sgca+/−, Figure 3B). By contrast, no Sgca+/−, Sgca+/−CT, or Sgca−/−CT muscle exceeded 1% of total area with dye uptake (Figure 3B), and no significant uptake was observed in the heart (not shown). Thus, overexpression of Galgt2 prevented dye uptake in all Sgca−/− skeletal muscles studied, indicating prevention of muscle damage (P < 0.05 for all comparisons of Sgca−/−CT to Sgca−/−).
Figure 3.
Evans blue dye uptake is reduced in exercised Galgt2 transgenic Sgca−/−muscles. Galgt2 transgenic (CT) Sgca+/− and Sgca−/− mice were compared with their non-transgenic littermates for uptake of Evans blue dye. A: Dye uptake is increased in the quadriceps muscles in Sgca−/− animals and is not elevated in Sgca+/−, Sgca+/−CT, and Sgca−/−CT animals. Scale bar = 100 μm. B: Quantification of the percentage of total area with dye uptake in the diaphragm (Dia), gastrocnemius (Gas), quadriceps (Quad), tibialis anterior (TA) and the triceps (Tri) muscle. Dye uptake was reduced in all Sgca−/−CT muscles compared with Sgca−/− (P < 0.05 for all). Errors are SD for n = 3 to 4 animals per condition.
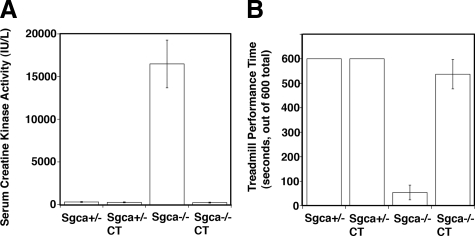
Overexpression of the Galgt2 was similarly effective in preventing other whole animal measures of muscular dystrophy in Sgca−/− animals (Figure 4). Sgca−/− mice had, on average, over a tenfold elevation in their serum creatine kinase activity, a measure of acute damage in skeletal muscles throughout the animal (Figure 4A). As with mdx mice,19,80 this measure is highly variable from mouse to mouse, nevertheless, serum CK activity levels in Sgca−/− mice were significantly elevated compared with Sgca+/− or Sgca+/−CT (P < 0.01 for both comparisons). Sgca−/−CT animals showed a significant decrease in serum CK activity relative to Sgca−/− (P < 0.05), reaching levels indistinguishable from Sgca+/− controls. Sgca−/− animals perform poorly on a constant treadmill measure of locomotor activity when compared with Sgca+/− littermates (Figure 4B). We therefore also assessed the ability of the four strains of mice to maintain walking on a treadmill of constant speed for a period of 10 minutes. All Sgca+/− and Sgca+/−CT mice studied were able to stay on the apparatus for the duration of the experiment. By contrast, Sgca−/− mice lasted, on average, only 1 minute, while Sgca−/−CT mice lasted 9 minutes. Thus, locomotor activity of Sgca−/−CT mice was significantly different from Sgca−/− (P < 0.001) and approached levels found in non-dystrophic Sgca+/− mice (Figure 4B). In summary, using histopathology measures of muscle regeneration (Figure 2), muscle membrane damage (Figure 3), and measures of muscular dystrophy in the whole animal (Figure 4), the overexpression of the Galgt2 transgene inhibited the development of muscular dystrophy in Sgca−/− mice.
Figure 4.
Whole animal measures of muscular dystrophy are reduced in Galgt2 transgenic Sgca−/− mice. Galgt2 transgenic (CT) and non-transgenic Sgca+/− and Sgca−/− mice were compared for levels of (A) serum creatine kinase activity and (B) locomotor activity. A: Serum creatine kinase activity was increased by an order of magnitude in Sgca−/− animals and this was significantly reduced, to wild-type levels, in Sgca−/−CT mice (P < 0.01 for Sgca−/− vs. Sgca+/− and P < 0.05 for Sgca−/−CT versus Sgca−/−). B: Sgca−/− mice had significantly reduced locomotor activity, and this too was significantly reversed in Sgca−/−CT mice (P < 0.001 for Sgca−/− vs. Sgca+/− and for Sgca−/−CT versus Sgca−/−). Errors are SEM for n = 11(Sgca+/−), 7 (Sgca+/−CT), 6 (Sgca−/−), or 17 (Sgca−/−CT) in (A) and n = 8 (Sgca+/− and Sgca−/−) or 7 (Sgca+/−CT and Sgca−/−CT) animals in (B).
Postnatal Overexpression of Galgt2 Reduces Muscular Dystrophy in Sgca−/− Muscle without Affecting Myofiber Size
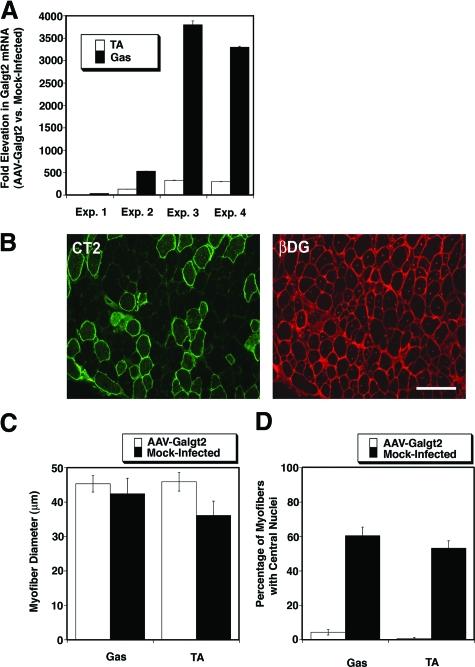
We infected Sgca−/− mice with AAV1-CMV-Galgt2 at 1 week of age in the tibialis anterior and gastrocnemius muscle and then compared infected and contralateral mock-infected muscles for Galgt2 overexpression at 5 weeks of age (Figure 5A). We performed four experiments where we dosed AAV-CMV-Galgt2 injections over a range from 1 × 1010 to 5 × 1010 vector genomes (vg). QRT-PCR showed a range of increased Galgt2 mRNA expression (Figure 5A). At the high end, this equaled levels measured in Galgt2 transgenic Sgca−/− mice (Figure 1C). Here, as before,20 the range of expression in the gene therapy experiments was wider than was seen in transgenic mice (10-fold to 3800-fold). To some extent this reflected altered percentages of myofibers that were infected (which ranged from 25% for lowest expressing muscles to 95% for the highest expressing ones). As before, increased expression was elevated by 0.5 to 1 log compared with similar experiments in mdx muscles.20 This may be due to a changed denominator in such measures between strains; Galgt2 levels in mdx are increased compared with wild-type,21 while levels in Sgca−/− were reduced (Figure 1D). Despite this larger range, myofibers within muscles with only 40-fold increased Galgt2 expression overexpressed the CT carbohydrate at robust levels and showed reduced to absent dystrophic pathology (Figure 5B). This was assessed by immunostaining for the CT carbohydrate and counterstaining with an antibody to β dystroglycan (Figure 5B), which reveals central nuclei due to non-specific binding of the goat anti-mouse secondary antibody.20 Similar results were obtained if we analyzed serial sections where staining for CT2 (to score overexpression) and hematoxylin and eosin (to score central nuclei) were done on subsequent sections (not shown).
Figure 5.
Postnatal overexpression of Galgt2 inhibits the development of muscle pathology in Sgca−/− mice. A: Sgca−/− muscles infected with AAV1-CMV-Galgt2 show a range of increased Galgt2 mRNA expression, as measured by QRT-PCR, in four separate experiments. TA, tibialis anterior; Gas, gastrocnemius. B: Infected muscles overexpressing Galgt2 were visualized by immunostaining with CT2, an antibody to the CT carbohydrate (green). Central nuclei were evident on counterstaining with an antibody to β dystroglycan (red). Scale bar =100 μm. C: Myofiber diameters were measured in Galgt2-overexpressing myofibers and compared with muscles not overexpressing transgene. D: Muscles overexpressing Galgt2 had significantly fewer myofibers with central nuclei than muscles not overexpressing transgene (P < 0.001 for comparison in Gastroc and TA). Errors are SD for n = 3 to 4 animals per condition in C and D. Errors in (A) are SEM for n = 9 measurements per condition.
Almost all AAV1-CMV-Galgt2-infected myofibers in all experiments did not display muscle pathology or centrally located nuclei. Sgca−/− myofibers overexpressing Galgt2 and Sgca−/− myofibers not overexpressing Galgt2 were compared for myofiber diameter (Figure 5C) and myofibers with centrally located nuclei (Figure 5D). As before,20,21 Galgt2 overexpression at this time point did not inhibit muscle growth; Galgt2-overexpressing myofibers were, on average, significantly larger than non-expressing myofibers in the gastrocnemius (and insignificantly larger the tibialis anterior). This is not a function of Galgt2 expression inducing muscle growth, but instead results from the presence of small regenerating myofibers in non-expressing muscle, which result from dystrophic processes in unprotected muscles. As in previous mdx and dyW/dyW experiments,20,21 postnatal overexpression of Galgt2 prevented histopathology associated with muscular dystrophy; Galgt2 overexpressing myofibers had normal (wild-type) levels of centrally located nuclei, while non-expressing myofibers had significantly elevated levels (P < 0.001 for gastroc and TA, Figure 5D). Allowing transgene overexpression to continue for longer time periods of time (up to 3 months) led to the maintenance of these normal CT-positive Sgca−/− myofibers (not shown), and this suggests that postnatal Galgt2 overexpression in fact protects skeletal muscles against the development of the dystrophic phenotype.
Glycosylation of α Dystroglycan in Skeletal Galgt2 Transgenic Sgca+/− and Sgca−/− Muscle
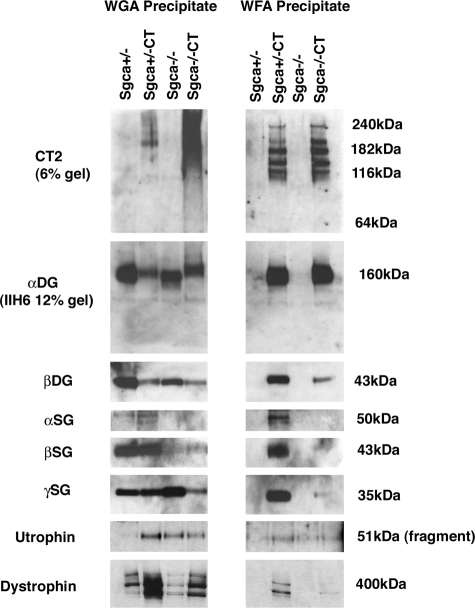
We had previously shown that α dystroglycan was the predominant glycoprotein glycosylated with the CT carbohydrate in Galgt2 transgenic skeletal muscle, including in transgenic mdx19 and dyW/dyW mice.21 This can be shown by differential lectin precipitation of α dystroglycan from non-denaturing muscle cell extracts; Wheat germ agglutinin (WGA) normally binds α dystroglycan by virtue of its dense concentration of sialic acid (and also GlcNAc), but WGA binds very poorly to the CT-glycosylated form of the protein.19,22 By contrast, Wisteria floribunda agglutinin (WFA) only binds CT-glycosylated α dystroglycan,19,22 which it does because WFA is a βGalNAc-binding lectin and β1,4GalNAc is an essential glycan defining the CT carbohydrate.81 α dystroglycan was precipitated equally well by WFA in Sgca+/−CT and Sgca−/−CT muscle, while it precipitated no α dystroglycan from Sgca+/− or Sgca−/− muscle. WGA, however, precipitated α dystroglycan from both Sgca+/− and Sgca−/− muscle (and also Sgca−/−CT), demonstrating that α dystroglycan was present in all four lysates in roughly equal amounts. Immunoblotting with antibody to the CT carbohydrate (CT2) identified multiple glycoforms of α dystroglycan containing this glycan in WFA precipitates, and these forms were equivalent in Sgca+/−CT and Sgca−/−CT muscle. These blots were analyzed using a low (6%) percentage SDS-PAGE gel to clarify the glycoforms present. Thus, loss of α sarcoglycan did not inhibit Galgt2 glycosylation of α dystroglycan with the CT carbohydrate. In general, similar amounts of β dystroglycan co-precipitated with α dystroglycan, to which it binds tightly.30,31 The one exception was Sgca−/−CT precipitates, where far more α dystroglycan was present than β dystroglycan (Figure 6). α, β, and γ sarcoglycan, utrophin, and dystrophin were all similarly co-precipitated by WFA in Sgca+/−CT muscle but were reduced or absent in Sgca−/−CT muscle (despite abundant α dystroglycan being present). This reduction in sarcoglycans may reflect their reduced expression in Sgca−/− muscle. Both utrophin and dystrophin were enriched in WGA and WFA precipitates of Sgca+/−CT muscle compared with Sgca+/− (and to a lesser degree in Sgca−/−CT compared with Sgca−/−). While dystrophin was precipitated as a full-length native protein of 427 kDa, utrophin was only precipitated as a proteolytic fragment (51 kDa) and not as a native (ca. 400 kDa) protein.
Figure 6.
Loss of α sarcoglycan does not affect glycosylation of α dystroglycan with the CT carbohydrate in Galgt2 transgenic mice. Gastrocnemius muscle was solubilized in non-ionic detergent and precipitated with Wheat germ agglutinin (WGA), a control lectin known to bind α dystroglycan, and Wisteria floribunda agglutinin, a βGalNAc-binding lectin that binds the CT-glycosylated form of α dystroglycan. α dystoglycan was glycosylated by Galgt2 such that it could be precipitated by WFA equally well in Sgca+/−CT and Sgca−/−CT muscle. In non-CT muscle, WGA bound as much α dystroglycan as WFA did in CT muscle. Precipitates resolved on a low percentage gel (6%) and blotted with CT2 show equivalent glycoforms of α dystroglycan precipitated by WFA in Sgca+/−CT and Sgca−/−CT muscle. β dystroglycan was co-precipitated with α dystroglycan in all muscles, but was relatively reduced in WFA precipitates from Sgca−/−CT, as were β and γ sarcoglycan, due to their reduced overall expression in these lysates (see Figure 7). Full-length dystrophin (427 kDa) and a utrophin protein fragment (51 kDa) were also enriched in CT muscles. Data are representative of three experiments with similar results.
Expression of Sarcoglycans, Dystroglycan, Dystrophin, and Utrophin in Galgt2 Transgenic Sgca+/− and Sgca−/− Muscle
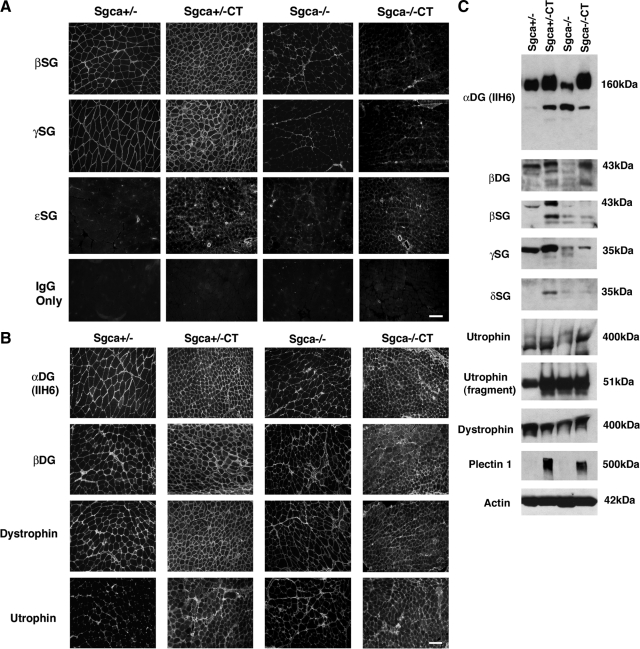
Last, we analyzed the expression of members of the dystrophin-associated glycoprotein complex, including dystrophin, utrophin, α dystroglycan, β dystroglycan, β sarcoglycan, γ sarcoglycan, δ sarcoglycan, and ε sarcoglycan by immunostaining (Figure 7, A and B), immunoblotting (Figure 7C), or both in Sgca+/−, Sgca+/−CT, Sgca−/−, and Sgca−/−CT muscle. Importantly, β, γ, and ε sarcoglycan were not increased in expression along myofibers in Sgca−/−CT muscle (Figure 7A), suggesting that the inhibition of muscular dystrophy seen in these muscles (Figure 2–4) did not come from compensation by an alternative sarcoglycan protein complex. Immunostaining of α dystroglycan, β dystroglycan, dystrophin, and utrophin was high in both Sgca+/−CT and Sgca−/−CT muscle. Staining showed modestly increased intracellular expression in Sgca−/−CT muscle compared to Sgca+/−CT. (Figure 7B). Analysis of these same proteins using immunoblotting of whole muscle SDS lysates showed that expression of β, γ, and δ sarcoglycan protein were not increased in Sgca−/−CT muscle compared with Sgca−/− (1.03 ± 0.19-fold, 0.98 ± 0.14 and 1.52 ± 0.47-fold, respectively, n = 4 for all), while the increase in utrophin and α dystroglycan found in Sgca+/−CT muscle (5.4 ± 1.4-fold and 1.22 ± 0.02-fold versus Sgca+/−) was still evident in Sgca−/−CT muscle (2.03 ± 0.27 and 1.3 ± 0.19-fold versus Sgca−/−) (Figure 7C). As previously found,39 Sgca−/− muscle had reduced levels of α-δ sarcoglycans and also α and β dystroglycan (compared with Sgca+/−) (Figures 1B and 7C). Last, we found a dramatic increase in Plectin 1, a protein that can bind both to cytoskeletal elements such as vimetin, desmin, and F-actin82,83,84 and to dystroglycan, dystrophin, and utrophin,85 in both Sgca+/−CT (20 ± 10-fold versus Sgca+/−) and Sgca−/−CT muscle (14.0 ± 3-fold versus Sgca−/−). Actin was used as a control for protein loading and transfer and showed equivalent signals (no more than 9% difference between strains) in all blotting experiments.
Figure 7.
β, γ, δ, and ε sarcoglycan are not overexpressed in Galgt2 transgenic Sgca−/− muscle. A: Immunostaining of gastrocnemius muscle in Galgt2 transgenic (CT) and non-transgenic Sgca+/− and Sgca−/− mice. β, γ, and ε sarcoglycan were not increased in expression along myofibers in Sgca−/−CT muscle, while (B) immunostaining of α dystroglycan, β dystroglycan, dystrophin, and utrophin was high in Sgca+/−CT and Sgca−/−CT muscle. Scale bar =100 μm (A and B). C: 40 μg of SDS whole muscle lysate from gastrocnemius is loaded per lane. β, γ and δ sarcoglycan protein levels were not increased in Sgca−/−CT muscle as they were in Sgca+/−CT muscle. Utrophin, α dystroglycan, and plectin 1, by contrast, were similarly increased in both Sgca−/−CT and Sgca+/−CT muscle. Actin is shown as a control for protein loading and transfer.
Discussion
We have provided proof of principal evidence that overexpression of the Galgt2 gene in skeletal muscles of α sarcoglycan-deficient (Sgca−/−) mice lessens the development of dystrophic skeletal muscle pathology. Overexpression of Galgt2 inhibits the increased incidence of myofibers with central nuclei, the increased uptake of Evans blue dye into myofibers, the increase in serum creatine kinase activity, and the decrease in locomotor activity that occur in Sgca−/− animals. Thus, by a variety of measures that pertain to muscle degeneration and regeneration, membrane permeability, and muscle activity, Galgt2 overexpression reduces dystrophic changes to near wild-type levels. Use of gene therapy techniques to overexpress Galgt2 in skeletal muscles of young adult Sgca−/− mice led to equivalently dramatic improvements in histopathology measures, but without impacting important developmental aspects such as muscle growth. These experiments suggest that Galgt2 should be considered as a therapeutic target for limb girdle muscular dystrophy 2D (LGMD2D).
The results we have shown here are very similar to our previous results showing reduced muscular dystrophy in Galgt2 transgenic mdx mice19,20 and dyW/dyW mice.21 As such, the experiments presented suggest that Galgt2 may have a broader impact on muscular dystrophies than other surrogate gene therapies previously tested. Many therapies that have been shown to be effective in one of these mouse muscular dystrophy models have not been as effective when tested in others. For example, Bcl2 overexpression ameliorates disease in dy/dy mice but not in mdx mice,1 ADAM12 overexpression is effective in mdx mice but not in dy/dy mice,18,86 myostatin inhibition is effective in boosting muscle mass in mdx mice but not in dy/dy or Sgca−/− mice,2,3,4,87,88 stimulation of calcineurin signaling is effective in mdx mice but not in dy/dy mice,89,90,91 and integrin α7B overexpression ameliorates disease in mdxutrn−/− mice but not in Scgd−/− mice.14,92 By contrast, proteosome inhibitors appear to promote membrane localization of sarcoglycan complexes in Sgca-deficient cells93 and have also been shown to be therapeutic in mdx mice.94,95 Perhaps the most promising surrogate gene therapy thus far for LGMD2D has been transgenic overexpression of ε sarcoglycan, which appears to inhibit muscular dystrophy for many months and increase expression of β-δ sarcoglycan on the muscle membrane.96 However, endogenous ε sarcoglycan, which other groups have reported is present in skeletal muscle,39,59 does not seem to inhibit the disease process or maintain elevated β-δ sarcoglycan levels in α sarcoglycan-deficient mice,39,59 even though it clearly can associate with these subunits in Sgca−/− muscle.97 In our study, we found no increase in ε sarcoglycan or in β, γ, or δ sarcoglycan to suggest that alternative sarcoglycan complexes were responsible for the therapeutic effects of the Galgt2 transgene.
While all of these models show defects in a member of the dystrophin-associated glycoprotein complex or laminin α2, which one presumes are all linked together in a functional complex, many of these differences in therapeutic efficacy could be nevertheless be attributed to fundamental differences in the disease models. For example, dyW/dyW mice have very impaired muscle regeneration, while mdx mice do not.18,98,99 Similarly, mdx muscles show decreased resistance to injury in eccentric contraction paradigms, while Sgcg−/− muscles, which also lack Sgca, do not.100 Thus, there are distinct differences in these disease models, despite the fact that they all possess deletions in proteins that work, at least to some extent, in concert. As such, differences in therapeutic efficacy of different surrogate genes may also reflect underlying mechanistic differences that occur in different forms of muscular dystrophy. Galgt2 appears to overcome such differences in a manner that many other therapies do not.
The fundamental questions for Galgt2, going forward, are: 1. How does altering membrane glycosylation reduce muscular dystrophy in these different disease forms and 2. How can these effects be translated into a therapy? As to question 1, it is certainly beneficial from a mechanistic standpoint that glycosylation, by its very nature, affects multiple protein targets and multiple protein functions. While Galgt2 overexpression in skeletal muscle predominantly leads to increased CT-glycosylation of only several molecules, in particular α dystroglycan22,28,101 and an as yet defined glycolipid,21 these glycosylation changes could impact multiple biological systems that would be beneficial in these diseases. It is clear from the seminal work of Ervasti, Campbell and colleagues that the glycans on α dystroglycan are required for the binding of extracellular matrix proteins, including laminins, agrin, perlecan, and also neurexins.30,31,102 Preliminary studies from our lab suggest that CT-glycosylation of α dystroglycan increases extracellular matrix binding. As such, CT-glycosylation of α dystroglycan may tighten extracellular matrix-membrane adhesion via the dystrophin-associated glycoprotein complex. The lectin precipitation studies shown here suggest a tight association of dystrophin and perhaps also utrophin with dystroglycan in CT transgenic muscles, which could be consistent with such a notion. Similarly, the increased expression of Plectin1, which could serve to link cytoskeletal-associated proteins including dystrophin and utrophin with membrane proteins such as dystroglycan,85 may facilitate these interactions. Regardless of the complexities of the mechanism involved, this study, coupled with studies in mdx19,20 and in dyW/dyW mice,21 suggest that elevation of Galgt2 expression in skeletal muscles would be therapeutic in these diseases.
How to answer question 2 is of primary importance going forwards. We have undertaken two strategies in this regard. First, we have developed a gene therapy approach to overexpress the Galgt2 cDNA using adeno-associated virus. This approach works to prevent dystrophic changes in infected muscles in Sgca−/−, mdx,20 and dyW/dyW mice.21 Future work will entail developing methods to allow systemic delivery of such gene therapy vectors using the human Galgt2 cDNA driven by muscle- or muscle/heart-specific promoters. A second approach is to use pharmacology. Galgt2 gene expression is very low in skeletal muscle,21 consistent with the synaptic distribution of the protein22,28 and its carbohydrate product.29 Identification of drugs that would increase extrasynaptic expression in muscle, therefore, is another approach to stimulate the therapeutic effects of Galgt2 overexpression.
Acknowledgments
We thank Jerry Mendell (Center for Gene Therapy, Nationwide Children’s Hospital) for comments on the manuscript and K. Reed Clark (Center for Gene Therapy, Nationwide Children’s Hospital) for AAV Viral Vector production.
Footnotes
Address reprint requests to Paul T. Martin, Center for Gene Therapy, The Research Institute at Nationwide Children’s Hospital, Departments of Pediatrics and of Physiology and Cell Biology, Ohio State University College of Medicine, 700 Children’s Drive, Columbus, OH 43205. E-mail: MartinPT@pediatrics.ohio-state.edu.
Supported by grants from the National Institutes of Health (AR050202 and AR049722) to PTM.
No conflicts of interest to declare.
References
- Dominov JA, Kravetz AJ, Ardelt M, Kostek CA, Beermann ML, Miller JB. Muscle-specific BCL2 expression ameliorates muscle disease in laminin {alpha}2-deficient, but not in dystrophin-deficient, mice. Hum Mol Genet. 2005;14:1029–1040. doi: 10.1093/hmg/ddi095. [DOI] [PubMed] [Google Scholar]
- Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LA, Ahima RS, Khurana TS. Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002;420:418–421. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- Wagner KR, McPherron AC, Winik N, Lee SJ. Loss of myostatin attenuates severity of muscular dystrophy in mdx mice. Ann Neurol. 2002;52:832–836. doi: 10.1002/ana.10385. [DOI] [PubMed] [Google Scholar]
- Bogdanovich S, Perkins KJ, Krag TO, Whittemore LA, Khurana TS. Myostatin propeptide-mediated amelioration of dystrophic pathophysiology. FASEB J. 2005;19:543–549. doi: 10.1096/fj.04-2796com. [DOI] [PubMed] [Google Scholar]
- Haidet AM, Rizo L, Handy C, Umapathi P, Eagle A, Shilling C, Boue D, Martin PT, Sahenk Z, Mendell JR, Kaspar BK. Long-term enhancement of skeletal muscle mass and strength by single gene administration of myostatin inhibitors. Proc Natl Acad Sci USA: 2008;105:4318–4322. doi: 10.1073/pnas.0709144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abmayr S, Gregorevic P, Allen JM, Chamberlain JS. Phenotypic improvement of dystrophic muscles by rAAV/microdystrophin vectors is augmented by Igf1 codelivery. Mol Ther. 2005;12:441–450. doi: 10.1016/j.ymthe.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Deconinck N, Tinsley J, De Backer F, Fisher R, Kahn D, Phelps S, Davies K, Gillis JM. Expression of truncated utrophin leads to major functional improvements in dystrophin-deficient muscles of mice. Nat Med. 1997;3:1216–1221. doi: 10.1038/nm1197-1216. [DOI] [PubMed] [Google Scholar]
- Deconinck AE, Rafael JA, Skinner JA, Brown SC, Potter AC, Metzinger L, Watt DJ, Dickson JG, Tinsley JM, Davies KE. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90:717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis JM, Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Barzaghi P, Lin S, Ruegg MA. Overexpression of mini-agrin in skeletal muscle increases muscle integrity and regenerative capacity in laminin-alpha2-deficient mice. FASEB J. 2005;19:934–942. doi: 10.1096/fj.04-3376com. [DOI] [PubMed] [Google Scholar]
- Moll J, Barzaghi P, Lin S, Bezakova G, Lochmuller H, Engvall E, Muller U, Ruegg MA. An agrin minigene rescues dystrophic symptoms in a mouse model for congenital muscular dystrophy. Nature. 2001;413:302–307. doi: 10.1038/35095054. [DOI] [PubMed] [Google Scholar]
- Qiao C, Li J, Zhu T, Draviam R, Watkins S, Ye X, Chen C, Li J, Xiao X. Amelioration of laminin-alpha2-deficient congenital muscular dystrophy by somatic gene transfer of miniagrin. Proc Natl Acad Sci USA. 2005;102:11999–12004. doi: 10.1073/pnas.0502137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krag TO, Bogdanovich S, Jensen CJ, Fischer MD, Hansen-Schwartz J, Javazon EH, Flake AW, Edvinsson L, Khurana TS. Heregulin ameliorates the dystrophic phenotype in mdx mice. Proc Natl Acad Sci USA. 2004;101:13856–13860. doi: 10.1073/pnas.0405972101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkin DJ, Wallace GQ, Nicol KJ, Kaufman DJ, Kaufman SJ. Enhanced expression of the alpha 7 beta 1 integrin reduces muscular dystrophy and restores viability in dystrophic mice. J Cell Biol. 2001;152:1207–1218. doi: 10.1083/jcb.152.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkin DJ, Wallace GQ, Milner DJ, Chaney EJ, Mulligan JA, Kaufman SJ. Transgenic expression of {alpha}7{beta}1 integrin maintains muscle integrity, increases regenerative capacity, promotes hypertrophy, and reduces cardiomyopathy in dystrophic mice. Am J Pathol. 2005;166:253–263. doi: 10.1016/s0002-9440(10)62249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter AK, Marshall JL, Crosbie RH. Sarcospan reduces dystrophic pathology: stabilization of the utrophin-glycoprotein complex. J Cell Biol. 2008;183:419–427. doi: 10.1083/jcb.200808027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadaszadeh B, Albrechtsen R, Guo LT, Zaik M, Kawaguchi N, Borup RH, Kronqvist P, Schroder HD, Davies KE, Voit T, Nielsen FC, Engvall E, Wewer UM. Compensation for dystrophin-deficiency: aDAM12 overexpression in skeletal muscle results in increased alpha 7 integrin, utrophin and associated glycoproteins. Hum Mol Genet. 2003;12:2467–2479. doi: 10.1093/hmg/ddg264. [DOI] [PubMed] [Google Scholar]
- Kronqvist P, Kawaguchi N, Albrechtsen R, Xu X, Schroder HD, Moghadaszadeh B, Nielsen FC, Frohlich C, Engvall E, Wewer UM. ADAM12 alleviates the skeletal muscle pathology in mdx dystrophic mice. Am J Pathol. 2002;161:1535–1540. doi: 10.1016/S0002-9440(10)64431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HH, Jayasinha V, Xia B, Hoyte K, Martin PT. Overexpression of the cytotoxic T cell GalNAc transferase in skeletal muscle inhibits muscular dystrophy in mdx mice, Proc Natl Acad Sci USA. 2002;99:5616–5621. doi: 10.1073/pnas.082613599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Camboni M, Martin PT. Postnatal overexpression of the CT GalNAc transferase inhibits muscular dystrophy in mdx mice without altering muscle growth or neuromuscular development: evidence for a utrophin-independent mechanism. Neuromuscul Disord. 2007;17:209–220. doi: 10.1016/j.nmd.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Chandrasekharan K, Yoon JH, Camboni M, Martin PT. Overexpression of the cytotoxic T cell (CT) carbohydrate inhibits muscular dystrophy in the dyW mouse model of congenital muscular dystrophy 1A. Am J Pathol. 2007;171:181–199. doi: 10.2353/ajpath.2007.060927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Hoyte K, Kammesheidt A, Deerinck T, Ellisman M, Martin PT. Overexpression of the CT GalNAc transferase in skeletal muscle alters myofiber growth, neuromuscular structure, and laminin expression. Dev Biol. 2002;242:58–73. doi: 10.1006/dbio.2001.0530. [DOI] [PubMed] [Google Scholar]
- Lefrancois L, Bevan MJ. Novel antigenic determinants of the T200 glycoprotein expressed preferentially by activated cytotoxic T lymphocytes. J Immunol. 1985;135:374–383. [PubMed] [Google Scholar]
- Conzelmann A, Lefrancois L. Monoclonal antibodies specific for T cell-associated carbohydrate determinants react with human blood group antigens CAD and SDA. J Exp Med. 1988;167:119–131. doi: 10.1084/jem.167.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann A, Kornfeld S. A murine cytotoxic T lymphocyte cell line resistant to Vicia villosa lectin is deficient in UDP-GalNAc:beta-galactose beta 1,4-N-acetylgalactosaminyltransferase. J Biol Chem. 1984;259:12536–12542. [PubMed] [Google Scholar]
- Lefrancois L, Bevan MJ. Functional modifications of cytotoxic T-lymphocyte T200 glycoprotein recognized by monoclonal antibodies. Nature. 1985;314:449–452. doi: 10.1038/314449a0. [DOI] [PubMed] [Google Scholar]
- Smith PL, Lowe JB. Molecular cloning of a murine N-acetylgalactosamine transferase cDNA that determines expression of the T lymphocyte-specific CT oligosaccharide differentiation antigen. J Biol Chem. 1994;269:15162–15171. [PubMed] [Google Scholar]
- Hoyte K, Kang C, Martin PT. Definition of pre- and postsynaptic forms of the CT carbohydrate antigen at the neuromuscular junction: ubiquitous expression of the CT antigens and the CT GalNAc transferase in mouse tissues. Brain Res Mol Brain Res. 2002;109:146–160. doi: 10.1016/s0169-328x(02)00551-x. [DOI] [PubMed] [Google Scholar]
- Martin PT, Scott LJ, Porter BE, Sanes JR. Distinct structures and functions of related pre- and postsynaptic carbohydrates at the mammalian neuromuscular junction. Mol Cell Neurosci. 1999;13:105–118. doi: 10.1006/mcne.1999.0737. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- Martin PT. Mechanisms of Disease: congenital muscular dystrophies-glycosylation takes center stage. Nat Clin Pract Neurol. 2006;2:222–230. doi: 10.1038/ncpneuro0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PT. The dystroglycanopathies: the new disorders of O-linked glycosylation. Semin Pediatr Neurol. 2005;12:152–158. doi: 10.1016/j.spen.2005.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Xu H, Christmas P, Wu XR, Wewer UM, Engvall E. Defective muscle basement membrane and lack of M-laminin in the dystrophic dy/dy mouse. Proc Natl Acad Sci USA. 1994;91:5572–5576. doi: 10.1073/pnas.91.12.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wu XR, Wewer UM, Engvall E. Murine muscular dystrophy caused by a mutation in the laminin alpha 2 (Lama2) gene. Nat Genet. 1994;8:297–302. doi: 10.1038/ng1194-297. [DOI] [PubMed] [Google Scholar]
- Duclos F, Straub V, Moore SA, Venzke DP, Hrstka RF, Crosbie RH, Durbeej M, Lebakken CS, Ettinger AJ, van der Meulen J, Holt KH, Lim LE, Sanes JR, Davidson BL, Faulkner JA, Williamson R, Campbell KP. Progressive muscular dystrophy in alpha-sarcoglycan-deficient mice. J Cell Biol. 1998;142:1461–1471. doi: 10.1083/jcb.142.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack AA, Groh ME, McNally EM. Sarcoglycans in muscular dystrophy. Microsc Res Tech. 2000;48:167–180. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<167::AID-JEMT5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Ozawa E, Mizuno Y, Hagiwara Y, Sasaoka T, Yoshida M. Molecular and cell biology of the sarcoglycan complex. Muscle Nerve. 2005;32:563–576. doi: 10.1002/mus.20349. [DOI] [PubMed] [Google Scholar]
- Lim LE, Campbell KP. The sarcoglycan complex in limb-girdle muscular dystrophy. Curr Opin Neurol. 1998;11:443–452. doi: 10.1097/00019052-199810000-00006. [DOI] [PubMed] [Google Scholar]
- Guglieri M, Straub V, Bushby K, Lochmuller H. Limb-girdle muscular dystrophies. Curr Opin Neurol. 2008;21:576–584. doi: 10.1097/WCO.0b013e32830efdc2. [DOI] [PubMed] [Google Scholar]
- Roberds SL, Leturcq F, Allamand V, Piccolo F, Jeanpierre M, Anderson RD, Lim LE, Lee JC, Tome FM, Romero NB, Fardeau M, Beckmann JS, Kaplan J-C, Campbell KP. Missense mutations in the adhalin gene linked to autosomal recessive muscular dystrophy. Cell. 1994;78:625–633. doi: 10.1016/0092-8674(94)90527-4. [DOI] [PubMed] [Google Scholar]
- Lim LE, Duclos F, Broux O, Bourg N, Sunada Y, Allamand V, Meyer J, Richard I, Moomaw C, Slaughter C, Tomé FMS, Fardeau M, Jackson CE, Beckmann JS, Campbell KP. Beta-sarcoglycan: characterization and role in limb-girdle muscular dystrophy linked to 4q12. Nat Genet. 1995;11:257–265. doi: 10.1038/ng1195-257. [DOI] [PubMed] [Google Scholar]
- Bonnemann CG, Modi R, Noguchi S, Mizuno Y, Yoshida M, Gussoni E, McNally EM, Duggan DJ, Angelini C, Hoffman EP. Beta-sarcoglycan (A3b) mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat Genet. 1995;11:266–273. doi: 10.1038/ng1195-266. [DOI] [PubMed] [Google Scholar]
- Noguchi S, McNally EM, Ben Othmane K, Hagiwara Y, Mizuno Y, Yoshida M, Yamamoto H, Bonnemann CG, Gussoni E, Denton PH, Kyriakides T, Middleton L, Hentati F, Ben Hamida M, Nonaka I, Vance JM, Kunkel LM, Ozawa E. Mutations in the dystrophin-associated protein gamma-sarcoglycan in chromosome 13 muscular dystrophy. Science. 1995;270:819–822. doi: 10.1126/science.270.5237.819. [DOI] [PubMed] [Google Scholar]
- Nigro V, de Sa Moreira E, Piluso G, Vainzof M, Belsito A, Politano L, Puca AA, Passos-Bueno MR, Zatz M. Autosomal recessive limb-girdle muscular dystrophy. LGMD2F, is caused by a mutation in the delta-sarcoglycan gene. Nat Genet. 1996;14:195–198. doi: 10.1038/ng1096-195. [DOI] [PubMed] [Google Scholar]
- Jung D, Duclos F, Apostol B, Straub V, Lee JC, Allamand V, Venzke DP, Sunada Y, Moomaw CR, Leveille CJ, Slaughter CA, Crawford TO, McPherson JD, Campbell KP. Characterization of delta-sarcoglycan, a novel component of the oligomeric sarcoglycan complex involved in limb-girdle muscular dystrophy. J Biol Chem. 1996;271:32321–32329. doi: 10.1074/jbc.271.50.32321. [DOI] [PubMed] [Google Scholar]
- Piccolo F, Roberds SL, Jeanpierre M, Leturcq F, Azibi K, Beldjord C, Carrie A, Recan D, Chaouch M, Reghis A, El Kerch F, Sefiani A, Voit T, Merlini L, Collin H, Eymard B, Beckmann JS, Romero NB, Tomé FMS, Fardeau M, Campbell KP, Kaplan J-C. Primary adhalinopathy: a common cause of autosomal recessive muscular dystrophy of variable severity. Nat Genet. 1995;10:243–245. doi: 10.1038/ng0695-243. [DOI] [PubMed] [Google Scholar]
- Vainzof M, Passos-Bueno MR, Canovas M, Moreira ES, Pavanello RC, Marie SK, Anderson LV, Bonnemann CG, McNally EM, Nigro V, Kunkel LM, Zatz M. The sarcoglycan complex in the six autosomal recessive limb-girdle muscular dystrophies. Hum Mol Genet. 1996;5:1963–1969. doi: 10.1093/hmg/5.12.1963. [DOI] [PubMed] [Google Scholar]
- Gouveia TL, Kossugue PM, Paim JF, Zatz M, Anderson LV, Nigro V, Vainzof M. A new evidence for the maintenance of the sarcoglycan complex in muscle sarcolemma in spite of the primary absence of delta-SG protein. J Mol Med. 2007;85:415–420. doi: 10.1007/s00109-007-0163-8. [DOI] [PubMed] [Google Scholar]
- Durbeej M, Campbell KP. Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Curr Opin Genet Dev. 2002;12:349–361. doi: 10.1016/s0959-437x(02)00309-x. [DOI] [PubMed] [Google Scholar]
- Hack AA, Ly CT, Jiang F, Clendenin CJ, Sigrist KS, Wollmann RL, McNally EM. Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J Cell Biol. 1998;142:1279–1287. doi: 10.1083/jcb.142.5.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araishi K, Sasaoka T, Imamura M, Noguchi S, Hama H, Wakabayashi E, Yoshida M, Hori T, Ozawa E. Loss of the sarcoglycan complex and sarcospan leads to muscular dystrophy in beta-sarcoglycan-deficient mice. Hum Mol Genet. 1999;8:1589–1598. doi: 10.1093/hmg/8.9.1589. [DOI] [PubMed] [Google Scholar]
- Durbeej M, Cohn RD, Hrstka RF, Moore SA, Allamand V, Davidson BL, Williamson RA, Campbell KP. Disruption of the beta-sarcoglycan gene reveals pathogenetic complexity of limb-girdle muscular dystrophy type 2E. Mol Cell. 2000;5:141–151. doi: 10.1016/s1097-2765(00)80410-4. [DOI] [PubMed] [Google Scholar]
- Coral-Vazquez R, Cohn RD, Moore SA, Hill JA, Weiss RM, Davisson RL, Straub V, Barresi R, Bansal D, Hrstka RF, Williamson R, Campbell KP. Disruption of the sarcoglycan-sarcospan complex in vascular smooth muscle: a novel mechanism for cardiomyopathy and muscular dystrophy. Cell. 1999;98:465–474. doi: 10.1016/s0092-8674(00)81975-3. [DOI] [PubMed] [Google Scholar]
- Ettinger AJ, Feng G, Sanes JR. epsilon-Sarcoglycan, a broadly expressed homologue of the gene mutated in limb-girdle muscular dystrophy 2D. J Biol Chem. 1997;272:32534–32538. doi: 10.1074/jbc.272.51.32534. [DOI] [PubMed] [Google Scholar]
- Straub V, Ettinger AJ, Durbeej M, Venzke DP, Cutshall S, Sanes JR, Campbell KP. epsilon-sarcoglycan replaces alpha-sarcoglycan in smooth muscle to form a unique dystrophin-glycoprotein complex. J Biol Chem. 1999;274:27989–27996. doi: 10.1074/jbc.274.39.27989. [DOI] [PubMed] [Google Scholar]
- Kobuke K, Piccolo F, Garringer KW, Moore SA, Sweezer E, Yang B, Campbell KP. A common disease-associated missense mutation in alpha-sarcoglycan fails to cause muscular dystrophy in mice. Hum Mol Genet. 2008;17:1201–1213. doi: 10.1093/hmg/ddn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli M, Gicquel E, Barrault L, Soheili T, Malissen M, Malissen B, Vincent-Lacaze N, Perez N, Udd B, Danos O, Richard I. Mannosidase I inhibition rescues the human alpha-sarcoglycan R77C recurrent mutation. Hum Mol Genet. 2008;17:1214–1221. doi: 10.1093/hmg/ddn029. [DOI] [PubMed] [Google Scholar]
- Crosbie RH, Lim LE, Moore SA, Hirano M, Hays AP, Maybaum SW, Collin H, Dovico SA, Stolle CA, Fardeau M, Tome FM, Campbell KP. Molecular and genetic characterization of sarcospan: insights into sarcoglycan-sarcospan interactions. Hum Mol Genet. 2000;9:2019–2027. doi: 10.1093/hmg/9.13.2019. [DOI] [PubMed] [Google Scholar]
- Crosbie RH, Lebakken CS, Holt KH, Venzke DP, Straub V, Lee JC, Grady RM, Chamberlain JS, Sanes JR, Campbell KP. Membrane targeting and stabilization of sarcospan is mediated by the sarcoglycan subcomplex. J Cell Biol. 1999;145:153–165. doi: 10.1083/jcb.145.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebakken CS, Venzke DP, Hrstka RF, Consolino CM, Faulkner JA, Williamson RA, Campbell KP. Sarcospan-deficient mice maintain normal muscle function. Mol Cell Biol. 2000;20:1669–1677. doi: 10.1128/mcb.20.5.1669-1677.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack AA, Lam MY, Cordier L, Shoturma DI, Ly CT, Hadhazy MA, Hadhazy MR, Sweeney HL, McNally EM. Differential requirement for individual sarcoglycans and dystrophin in the assembly and function of the dystrophin-glycoprotein complex. J Cell Sci. 2000;113 (Pt 14):2535–2544. doi: 10.1242/jcs.113.14.2535. [DOI] [PubMed] [Google Scholar]
- Head SI, Williams DA, Stephenson DG. Abnormalities in structure and function of limb skeletal muscle fibres of dystrophic mdx mice. Proc Biol Sci. 1992;248:163–169. doi: 10.1098/rspb.1992.0058. [DOI] [PubMed] [Google Scholar]
- Cohn RD, Henry MD, Michele DE, Barresi R, Saito F, Moore SA, Flanagan JD, Skwarchuk MW, Robbins ME, Mendell JR, Williamson RA, Campbell KP. Disruption of DAG1 in differentiated skeletal muscle reveals a role for dystroglycan in muscle regeneration. Cell. 2002;110:639–648. doi: 10.1016/s0092-8674(02)00907-8. [DOI] [PubMed] [Google Scholar]
- Cote PD, Moukhles H, Lindenbaum M, Carbonetto S. Chimaeric mice deficient in dystroglycans develop muscular dystrophy and have disrupted myoneural synapses. Nat Genet. 1999;23:338–342. doi: 10.1038/15519. [DOI] [PubMed] [Google Scholar]
- Straub V, Bushby K. The childhood limb-girdle muscular dystrophies. Semin Pediatr Neurol. 2006;13:104–114. doi: 10.1016/j.spen.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Connolly AM, Pestronk A, Mehta S, Al-Lozi M. Primary alpha-sarcoglycan deficiency responsive to immunosuppression over three years. Muscle Nerve. 1998;21:1549–1553. doi: 10.1002/(sici)1097-4598(199811)21:11<1549::aid-mus30>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Daniele N, Richard I, Bartoli M. Ins and outs of therapy in limb girdle muscular dystrophies. Int J Biochem Cell Biol. 2007;39:1608–1624. doi: 10.1016/j.biocel.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Allamand V, Donahue KM, Straub V, Davisson RL, Davidson BL, Campbell KP. Early adenovirus-mediated gene transfer effectively prevents muscular dystrophy in alpha-sarcoglycan-deficient mice. Gene Ther. 2000;7:1385–1391. doi: 10.1038/sj.gt.3301247. [DOI] [PubMed] [Google Scholar]
- Dressman D, Araishi K, Imamura M, Sasaoka T, Liu LA, Engvall E, Hoffman EP. Delivery of alpha- and beta-sarcoglycan by recombinant adeno-associated virus: efficient rescue of muscle, but differential toxicity. Hum Gene Ther. 2002;13:1631–1646. doi: 10.1089/10430340260201725. [DOI] [PubMed] [Google Scholar]
- Rodino-Klapac LR, Lee JS, Mulligan RC, Clark KR, Mendell JR. Lack of toxicity of alpha-sarcoglycan overexpression supports clinical gene transfer trial in LGMD2D. Neurology. 2008;71:240–247. doi: 10.1212/01.wnl.0000306309.85301.e2. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Ampong BN, Ohshima S, Shin JH, Nakai H, Imamura M, Miyagoe-Suzuki Y, Okada T, Takeda S. Recombinant adeno-associated virus type 8-mediated extensive therapeutic gene delivery into skeletal muscle of alpha-sarcoglycan-deficient mice. Hum Gene Ther. 2008;19:719–730. doi: 10.1089/hum.2007.184. [DOI] [PubMed] [Google Scholar]
- Pacak CA, Walter GA, Gaidosh G, Bryant N, Lewis MA, Germain S, Mah CS, Campbell KP, Byrne BJ. Long-term skeletal muscle protection after gene transfer in a mouse model of LGMD-2D. Mol Ther. 2007;15:1775–1781. doi: 10.1038/sj.mt.6300246. [DOI] [PubMed] [Google Scholar]
- Sampaolesi M, Torrente Y, Innocenzi A, Tonlorenzi R, D'Antona G, Pellegrino MA, Barresi R, Bresolin N, De Angelis MG, Campbell KP, Bottinelli R, Cossu G. Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301:487–492. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Jayasinha V, Nguyen HH, Xia B, Kammesheidt A, Hoyte K, Martin PT. Inhibition of dystroglycan cleavage causes muscular dystrophy in transgenic mice. Neuromuscul Disord. 2003;13:365–375. doi: 10.1016/s0960-8966(03)00040-3. [DOI] [PubMed] [Google Scholar]
- Martin PT. Glycobiology of the neuromuscular junction. J Neurocytol. 2003;32:915–929. doi: 10.1023/B:NEUR.0000020632.41508.83. [DOI] [PubMed] [Google Scholar]
- Hijikata T, Murakami T, Ishikawa H, Yorifuji H. Plectin tethers desmin intermediate filaments onto subsarcolemmal dense plaques containing dystrophin and vinculin. Histochem Cell Biol. 2003;119:109–123. doi: 10.1007/s00418-003-0496-5. [DOI] [PubMed] [Google Scholar]
- Litjens SH, Wilhelmsen K, de Pereda JM, Perrakis A, Sonnenberg A. Modeling and experimental validation of the binary complex of the plectin actin-binding domain and the first pair of fibronectin type III (FNIII) domains of the beta4 integrin. J Biol Chem. 2005;280:22270–22277. doi: 10.1074/jbc.M411818200. [DOI] [PubMed] [Google Scholar]
- Garcia-Alvarez B, Bobkov A, Sonnenberg A, de Pereda JM. Structural and functional analysis of the actin binding domain of plectin suggests alternative mechanisms for binding to F-actin and integrin beta4. Structure. 2003;11:615–625. doi: 10.1016/s0969-2126(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Rezniczek GA, Konieczny P, Nikolic B, Reipert S, Schneller D, Abrahamsberg C, Davies KE, Winder SJ, Wiche G. Plectin 1f scaffolding at the sarcolemma of dystrophic (mdx) muscle fibers through multiple interactions with beta-dystroglycan. J Cell Biol. 2007;176:965–977. doi: 10.1083/jcb.200604179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LT, Shelton GD, Wewer UM, Engvall E. ADAM12 overexpression does not improve outcome in mice with laminin alpha2-deficient muscular dystrophy. Neuromuscul Disord. 2005;15:786–789. doi: 10.1016/j.nmd.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Li ZF, Shelton GD, Engvall E. Elimination of myostatin does not combat muscular dystrophy in dy mice but increases postnatal lethality. Am J Pathol. 2005;166:491–497. doi: 10.1016/S0002-9440(10)62271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli M, Poupiot J, Vulin A, Fougerousse F, Arandel L, Daniele N, Roudaut C, Noulet F, Garcia L, Danos O, Richard I. AAV-mediated delivery of a mutated myostatin propeptide ameliorates calpain 3 but not alpha-sarcoglycan deficiency. Gene Ther. 2007;14:733–740. doi: 10.1038/sj.gt.3302928. [DOI] [PubMed] [Google Scholar]
- Chakkalakal JV, Harrison MA, Carbonetto S, Chin E, Michel RN, Jasmin BJ. Stimulation of calcineurin signaling attenuates the dystrophic pathology in mdx mice. Hum Mol Genet. 2004;13:379–388. doi: 10.1093/hmg/ddh037. [DOI] [PubMed] [Google Scholar]
- Parsons SA, Millay DP, Sargent MA, Naya FJ, McNally EM, Sweeney HL, Molkentin JD. Genetic disruption of calcineurin improves skeletal muscle pathology and cardiac disease in a mouse model of limb-girdle muscular dystrophy. J Biol Chem. 2007;282:10068–10078. doi: 10.1074/jbc.M609368200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal JV, Michel SA, Chin ER, Michel RN, Jasmin BJ. Targeted inhibition of Ca2+/calmodulin signaling exacerbates the dystrophic phenotype in mdx mouse muscle. Hum Mol Genet. 2006;15:1423–1435. doi: 10.1093/hmg/ddl065. [DOI] [PubMed] [Google Scholar]
- Milner DJ, Kaufman SJ. Alpha7beta1 integrin does not alleviate disease in a mouse model of limb girdle muscular dystrophy type 2F. Am J Pathol. 2007;170:609–619. doi: 10.2353/ajpath.2007.060686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaldello S, D'Angelo S, Franzoso S, Fanin M, Angelini C, Betto R, Sandona D. Inhibition of proteasome activity promotes the correct localization of disease-causing alpha-sarcoglycan mutants in HEK-293 cells constitutively expressing beta-, gamma-, and delta-sarcoglycan. Am J Pathol. 2008;173:170–181. doi: 10.2353/ajpath.2008.071146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assereto S, Stringara S, Sotgia F, Bonuccelli G, Broccolini A, Pedemonte M, Traverso M, Biancheri R, Zara F, Bruno C, Lisanti MP, Minetti C. Pharmacological rescue of the dystrophin-glycoprotein complex in Duchenne and Becker skeletal muscle explants by proteasome inhibitor treatment. Am J Physiol Cell Physiol. 2006;290:C577–C582. doi: 10.1152/ajpcell.00434.2005. [DOI] [PubMed] [Google Scholar]
- Bonuccelli G, Sotgia F, Schubert W, Park DS, Frank PG, Woodman SE, Insabato L, Cammer M, Minetti C, Lisanti MP. Proteasome inhibitor (MG-132) treatment of mdx mice rescues the expression and membrane localization of dystrophin and dystrophin-associated proteins. Am J Pathol. 2003;163:1663–1675. doi: 10.1016/S0002-9440(10)63523-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura M, Mochizuki Y, Engvall E, Takeda S. Epsilon-sarcoglycan compensates for lack of alpha-sarcoglycan in a mouse model of limb-girdle muscular dystrophy. Hum Mol Genet. 2005;14:775–783. doi: 10.1093/hmg/ddi072. [DOI] [PubMed] [Google Scholar]
- Liu LA, Engvall E. Sarcoglycan isoforms in skeletal muscle. J Biol Chem. 1999;274:38171–38176. doi: 10.1074/jbc.274.53.38171. [DOI] [PubMed] [Google Scholar]
- Kuang W, Xu H, Vilquin JT, Engvall E. Activation of the lama2 gene in muscle regeneration: abortive regeneration in laminin alpha2-deficiency. Lab Invest. 1999;79:1601–1613. [PubMed] [Google Scholar]
- De la Porte S, Morin S, Koenig J. Characteristics of skeletal muscle in mdx mutant mice. Int Rev Cytol. 1999;191:99–148. doi: 10.1016/s0074-7696(08)60158-8. [DOI] [PubMed] [Google Scholar]
- Hack AA, Cordier L, Shoturma DI, Lam MY, Sweeney HL, McNally EM. Muscle degeneration without mechanical injury in sarcoglycan deficiency. Proc Natl Acad Sci USA. 1999;96:10723–10728. doi: 10.1073/pnas.96.19.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Martin PT. Modulation of agrin binding and activity by the CT and related carbohydrate antigens. Mol Cell Neurosci. 2002;19:539–551. doi: 10.1006/mcne.2001.1095. [DOI] [PubMed] [Google Scholar]
- Michele DE, Barresi R, Kanagawa M, Saito F, Cohn RD, Satz JS, Dollar J, Nishino I, Kelley RI, Somer H, Straub V, Mathews KD, Moore SA, Campbell KP. Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature. 2002;418:417–422. doi: 10.1038/nature00837. [DOI] [PubMed] [Google Scholar]