Abstract
Long-chain acyl-acyl carrier proteins (acyl-ACP) are established biochemical regulators of bacterial type II fatty acid synthases due to their ability to feedback-inhibit the early steps in the biosynthetic pathway. In Streptococcus pneumoniae, the expression of the fatty acid synthase (fab) genes is controlled by a helix-turn-helix transcriptional repressor called FabT. A screen of pathway intermediates identified acyl-ACP as a ligand that increased the affinity of FabT for DNA. FabT bound to a wide range of acyl-ACP chain lengths in the absence of DNA, but only the long-chain acyl-ACPs increase the affinity of FabT for DNA. FabT affinity for DNA increased with increasing acyl-ACP chain length with cis-vaccenoyl-ACP being the most effective ligand. Thus, FabT is a new ACP-interacting partner that acts as a transcriptional rheostat to fine tune the expression of the fab genes based on the demand for fatty acids.
Fatty acid biosynthesis is a vital facet of bacterial physiology that is carried out by a set of discrete enzymes collectively known as the as the type II fatty acid synthase system (FAS-II)3 (for reviews, see Refs. 1 and 2). Fatty acid synthesis is an energy intensive process, and the rate of fatty acid production is tightly regulated to ensure that the supply of membrane phospholipid exactly matches the demand (1, 2). The intermediates in FAS-II are carried on ACP, a 9-kDa, four-helix protein that carries the fatty acid as a thioester attached to its 4′-phosphopantetheine prosthetic group (3). ACP is one of the most interactive proteins in biology, binding to a large number of intracellular proteins in a variety of biochemical pathways (4). The long-chain acyl-ACP end products of FAS-II (primarily 16- and 18-carbon fatty acids) are established biochemical regulators of the rate of FAS-II in Escherichia coli. These acyl-ACPs inhibit the initiation of FAS-II, through their feedback regulation of FabH (5), and limit the rate of FAS-II through their inhibition of acetyl-CoA carboxylase (6). The genetic regulation of FAS-II in E. coli is focused on two genes, fabA and fabB. Transcription of the fabA and fabB genes is governed by two transcriptional regulators: FadR, the activator; and FabR, the repressor (for reviews, see Refs. 2, 7, and 8). FadR was discovered as a repressor of the β-oxidation regulon (9, 10) and subsequently discovered to also be an activator of fabA and fabB transcription (11–13). The ligands that control FadR DNA binding are acyl-CoAs that are ≥12 carbons long (14–20). FabR was identified as a protein that bound to a palindrome located in the fabB and fabA promoters (21), where it functions as a transcriptional repressor based on the increase in fabA and fabB mRNA in a fabR deletion mutant (22). A regulatory ligand for FabR has not been identified.
The genetic regulation of FAS-II in Gram-positive bacteria is different and more diverse. Like E. coli, the Bacillus subtilis fatty acid biosynthetic genes are scattered throughout the chromosome, and this organism has a transcription factor, called FapR, that functions as a global regulator of the fab genes, with the notable exception of acetyl-CoA carboxylase (8, 23). FapR is a repressor, and deletion of fapR results in significant up-regulation of the target FAS-II genes, leading to membrane compositional alterations and a cold-sensitive phenotype. Malonyl-CoA releases FapR from its DNA binding site (24, 25). In contrast, the human pathogen, S. pneumoniae, has a distinctive clustering of the fab genes within the genome coupled with an atypical mechanism for unsaturated fatty acid biosynthesis (26) and enoyl-ACP reduction (26, 27). The second gene in the fab cluster, fabT (fatty acid biosynthesis transcriptional regulator), encodes a helix-turn-helix DNA-binding protein belonging to the MarR superfamily of transcriptional regulators that binds to a sequence-specific DNA palindrome located within the two promoters that control fab gene expression (28). A knock-out of the fabT gene leads to an increase in the expression of all FAS-II genes with the exception of fabM (28).
Although the role of FabT as a repressor of fatty acid biosynthesis gene expression is clear from the gene knock-out experiments, the increased expression of FabT does not decrease the expression of the FAS-II genes (28), suggesting the presence of a limiting intracellular regulatory ligand that positively controls FabT association with its DNA binding elements. This study identifies this activating ligand as long-chain acyl-ACP, which increases the affinity of FabT for its sequence-specific DNA binding sites. Thus, the acyl-ACP end products of fatty acid synthesis act as a feedback regulators of FAS-II gene expression.
EXPERIMENTAL PROCEDURES
Materials
Sources of supplies were: Avanti Polar Lipids Inc., oleoyl-lysophosphatidic acid, palmitoyl-lysophosphatidic acid, oleoyl-CoA, palmitoyl-CoA; TriLink BioTechnologies, guanosine-3′,5′-bisdiphosphate (ppGpp); Fluka, 4-hydroxy-6-methyl-2-pyrone; Sigma, sodium salicylate and malonyl-CoA; Invitrogen, Novex® 6% DNA retardation gel, Novex Tris-borate-EDTA Hi-Density sample buffer. Acylphosphate (29), acyl-ACPs (30), and anti-ACP IgG (31) were prepared following procedures in the literature. Expression and purification of the amino-terminal His-tagged FabT was performed as described (28). The removal of the His tag did not alter FabT DNA binding properties, so for most of the experiments, the His-tagged version of FabT was used.
Preparation of DNA Probe for Electrophoretic Mobility Shift Assay
The two FabT binding sites of the fab gene cluster are located in the promoter region of the fabT and fabK genes (28). The DNA fragment used in the gel shift assay contained the palindrome sequence from the fabK promoter (AGTTTGACTGTCAAATT) in the center and was extended on both sides to obtain a long probe containing 279 bp of the FabT palindrome (LP). The use of this probe, rather than a shorter oligonucleotide, was required for our experiments to counteract the charge on the highly positive FabT (pI = 9.4) to give rise to a FabT·DNA complex with a net negative charge that would migrate into a Novex 6% DNA retardation gel (pH 8.3). FabT binding was specific for the palindrome sequence identified previously within LP (28) based on the ability of an unlabeled oligonucleotide probe containing the palindrome sequence to prevent binding of FabT to LP. A scrambled oligonucleotide had no effect. LP was prepared starting with the double-stranded 35-bp fragment from the fabK promoter, 5′-TTAGGTAATAGTTTGACTGTCAAATTATGGTGAAA-3′, containing the underlined palindrome sequence in the middle; this fragment was generated from two custom-synthesized complementary single-stranded oligonucleotides. The resulting blunt end DNA was ligated into pCRII-Blunt TOPO (Invitrogen) to yield plasmid pAJ050, which served as template DNA to amplify LP using PCR primers 5′-CAGGAAACAGCTATGACCATGATTACG-3′ and 5′-GTAAAACGACGGCCAGTGAATTG-3′. Following purification, LP was 32P-labeled using a DNA 5′ end-labeling kit (Promega). We also used a DNA probe constructed from the related palindrome that binds FabT located in the fabT-fabH-acpP promoter (GTTTTGATTGTAAAAGT), and in all cases, obtained the same results, although FabT bound this palindrome less tightly than the more perfect palindrome located in the fabK promoter.
Electrophoretic Mobility Shift Assay
FabT DNA binding was assessed by gel mobility shift assays using Novex 6% DNA retardation gel (Invitrogen) and 32P-labeled LP. In a representative assay, 10 pm 32P-labeled LP, 60 nm FabT, and 0–1500 nm acyl-ACP were mixed in a buffer containing 0.5× Novex Hi-Density Tris-borate-EDTA sample buffer (Invitrogen) and 50 mm NaCl; the mixture was incubated at 25 °C for 5 min, and then 15 μl (3000 dpm) was loaded per lane. The gel was exposed to a phosphor screen and was scanned with a Typhoon 9600 PhosphorImager (GE Healthcare).
Gel Filtration Chromatography
Interaction between FabT and acyl-ACP was assessed based on the elution profile of [1-14C]18:1Δ9-ACP in the presence of 0, 5, or 12 μm FabT. The 250-μl mixtures of [1-14C]18:1Δ9-ACP (18,000 dpm) and FabT in 25 mm HEPES, pH 7.4, and 400 mm NaCl were incubated at 25 °C for 10 min and applied to a SuperdexTM 10/300 GL column (Amersham Biosciences) by an AKTATM fast protein liquid chromatography (GE Healthcare) and eluted in the same buffer collecting 200-μl fractions in a 96-well microtiter plate. The radioactivity in 100-μl aliquots from each fraction was determined using liquid scintillation counting.
AlphaScreen Binding Assay
The AlphaScreen assay is a bead-based technology developed for assessing protein-protein interactions in a homogeneous microplate format (32, 33). We developed a binding assay in a 384-well microplate format using this technology to monitor interaction between FabT and acyl-ACP, which employed the nickel chelate histidine detection kit (PerkinElmer Life Sciences), amino-terminal His-tagged FabT, and biotinylated acyl-ACPs. An EZ-Link® Sulfo-NHS-LC biotinylation kit (Pierce) was used to label ACPs. The AlphaScreen assay is a sensitive method to reliably determine relative binding affinities, although the high avidity of the assay leads to an overestimate of the strength of the interaction when compared with other techniques (34). Reagents were diluted with buffer containing 25 mm HEPES at pH 7.4, 400 mm LiCl, and 1 mg/ml bovine serum albumin as follows: FabT, to a stock concentration of 200 nm; acyl-ACPs, serial dilution between 104 and 1.7 nm; streptavidin-coated donor beads and nickel chelate acceptor beads, to 0.1 mg/ml. In a representative experiment, 7.5 μl of biotinylated acyl-ACP was mixed with 7.5 μl of His-tagged FabT, 5 μl of donor beads, and 5 μl of acceptor beads followed by incubation in the dark at 25 °C for 1 h and data collection on a Fusion-Alpha plate reader (Packard). Each assay point was determined in triplicate. Background was obtained by eliminating FabT from the well mixture, and the resulting signal to background ratio was ∼600:1. There was no signal generated when His-tagged DkgB (35) was substituted for FabT as a nonspecific protein control. Substituting His-tagged DgkB for FabT did not generate a signal.
Data Analyses
Binding isotherms were analyzed using Prism 5 (GraphPad) statistical software to calculate the apparent dissociation constants. The triplicate data points were corrected for background counts obtained in a control assay without FabT, and the data points between 0 and 15 nm were fitted to a single site binding hyperbola, Y = Ymax*[ACP]/(Kd + [ACP]), that yielded the dissociation constant and the maximum interaction between binding partners. Positive cooperativity was observed for the dependence of FabT binding to malonyl- and holo-ACP on ACP concentration; thus, dissociation constants were determined by fitting the data to the Y = Ymax*[ACP]h/(Kdh + [ACP]h) equation. Data are reported as value ± standard error.
RESULTS
Long-chain Fatty Acyl-ACPs Activate FabT DNA Binding
FabT functions as a transcriptional repressor based on the up-regulation of the 12-gene fab cluster in a S. pneumonia strain lacking FabT (28). However, increased expression of FabT using a multicopy plasmid did not suppress expression of the fatty acid biosynthetic genes in the wild-type strain (28), suggesting that FabT repression of the fab genes was positively controlled by an intracellular ligand that regulates the affinity of FabT for its DNA binding sites. To test this hypothesis, we developed an electrophoretic mobility shift assay to screen candidate intracellular metabolites related to fatty acid synthesis for their ability to promote the interaction between FabT and it sequence-specific DNA binding site. Although the FabT repressor will bind to its cognate DNA palindrome if enough protein is added to the assay (28), our strategy was to detect interactions that increase the affinity of FabT for DNA; therefore, the amount of FabT protein used in the screen was below that required to observe DNA binding in the absence of an effector. Most of the metabolites tested, including malonyl-CoA, had no effect on FabT DNA binding (Fig. 1A). However, long-chain acyl-ACP end products of fatty acid synthesis significantly increased FabT DNA binding (Fig. 1A). FabT binding to LP in the presence of acyl-ACP was not detected in assays spiked with the oligonucleotide containing the palindrome previously identified to bind to FabT (28) (not shown). 16:0-ACP and 18:1Δ11-ACP were selected for the screen because 16:0 (22%) and 18:1Δ11 (43%) are two abundant fatty acid chains found in S. pneumoniae phospholipids (28). Also, 18:1Δ9-CoA slightly increased FabT DNA binding (Fig. 1A). Although the difference in the migration of the DNA shifted with acyl-ACP and acyl-CoA was small, the band shifted with the acyl-CoA consistently ran farther into the gel than the band shifted with acyl-ACP (Fig. 1A). We next tested a series of acyl-ACP chain lengths to determine the acyl-ACP chain length dependence for FabT DNA binding (Fig. 1B). 18:1Δ11-ACP was the most potent activator of FabT binding, and the ability of acyl-ACP to promote FabT-DNA interaction fell as the chain length decreased, illustrating that the two end products of the pathway were the most potent regulators of FabT.
FIGURE 1.
Long-chain acyl-ACP induces FabT DNA binding activity. A, identification of the specific ligands that promote FabT DNA binding using the gel electrophoretic mobility shift assay described under “Experimental Procedures.” Each lane contained 60 nm purified FabT, 1.5 μm of the indicated compounds, and 10 pm 32P-labeled DNA fragment (LP). LPA, lysophosphatidic acid; TAL, 4-hydroxy-6-methyl-2-pyrone. B, acyl-ACP chain length specificity in the activation of FabT DNA binding determined under the same assay conditions. holo-, holo-ACP; malonyl-, malonyl-ACP.
The observation that 18:1Δ9-CoA increased FabT binding to DNA was curious because there is no evidence that these fatty acid metabolites exist in S. pneumoniae. There are no predicted acyl-CoA synthetases found in the genome, and there are no β-oxidation enzymes present either, a major route for acyl-CoA metabolism in bacteria such as E. coli. Furthermore, the PlsY and PlsC acyltransferases of S. pneumoniae do not recognize acyl-CoA thioesters and exclusively utilize acyl-ACP substrates, ruling out a role for acyl-CoA in membrane lipid synthesis in this organism (29). Therefore, we compared the concentration dependence of acyl-ACP and acyl-CoA to promote FabT DNA binding (Fig. 2). 18:1Δ11-ACP was more effective in enhancing FabT DNA binding than 16:0-ACP, although FabT binding was saturable in both cases. FabT DNA binding promoted by 18:1Δ9-CoA occurred at a much lower affinity and failed to saturate. 16:0-CoA was even less effective. These data show that 18:1Δ11-ACP, corresponding to the most abundant fatty acid in S. pneumoniae membranes, was the most effective FabT ligand. We attribute the activity of 16:0-CoA and 18:1Δ9-CoA to it acting as a weak binding analog of acyl-ACP. It is widely appreciated that acyl-CoA and N-acetyl-cystamine thioesters act as ACP analogs that function in FAS-II enzyme systems at much lower affinities than their corresponding acyl-ACPs (36–38).
FIGURE 2.
Long-chain acyl-CoAs are lower affinity analogs of acyl-ACP. Electrophoretic mobility shift assays were performed using 60 nm purified FabT, 10 pm 32P-labeled LP, and increasing concentrations as follows: A, 18:1Δ11-ACP; B, 16:0-ACP; C, 18:1Δ9-CoA; and D, 16:0-CoA. Ligand concentrations, expressed as nm, are indicated above the lanes.
Interaction between Acyl-ACP and FabT
We found no evidence for the transfer of the labeled fatty acid to FabT from [14C]16:0-ACP or that FabT caused the hydrolysis of acyl-ACP, arguing against the acylation of FabT or the metabolism of acyl-ACP by FabT (not shown). These data suggested that the DNA complex observed in the gel retardation assays was a FabT·acyl-ACP·DNA ternary complex. This idea was confirmed using anti-ACP IgG to signal the presence of ACP in the complex revealed by a “supershift” the FabT·acyl-ACP·[32P]DNA complex in the presence of anti-ACP IgG (Fig. 3A). Next, we determined whether the FabT·acyl-ACP complex formed in the absence of DNA. Gel filtration chromatography was used to separate acyl-ACP from FabT·acyl-ACP complexes (Fig. 3B). FabT alone eluted at 15.8 ml, which was slower than the FabT·acyl-ACP complex eluting at 16.5 ml but faster than the elution of acyl-ACP at 15.5 ml. These data clearly show the formation of a FabT·acyl-ACP complex in the absence of DNA. A sensitive, direct binding AlphaScreen assay was used to determine the rank order of FabT binding by different acyl-ACP species (Fig. 3C). This assay yielded the same apparent dissociation constants for both long-chain and medium-chain ACPs and higher apparent dissociation constants for malonyl-ACP and ACP. These data show that although the presence of a fatty acid chain on acyl-ACP increases the affinity of FabT for DNA, all acyl-ACP species bind with approximately the same apparent affinity. These data, taken together with the information in Fig. 1B, lead to the conclusion that the interaction between FabT and acyl-ACP is primarily mediated by FabT-ACP protein interactions and that FabT monitors the entire acyl-ACP pool in vivo. However, the length of the fatty acid chain is critical to triggering the conformational change that converts FabT from a low to a high affinity DNA-binding protein. Thus, FabT functions in much the same way as DesT, a transcription factor from Pseudomonas aeruginosa that binds to all acyl-CoA species equally, but only those with a double bond induce the conformational change necessary for DNA binding (39).
FIGURE 3.
Acyl-ACPs bind to FabT in the absence of DNA. A, electrophoretic mobility shift assays were performed using 60 nm FabT and 10 pm 32P-labeled LP as described under “Experimental Procedures.” When present, 18:1Δ11-ACP was 188 nm, and purified anti-ACP IgG antibody (31) was 0.1 μg. B, gel filtration chromatography of samples containing 6 μm [1-14C]18:1Δ9-ACP either alone or in the presence of 12 μm FabT using a Superdex 10/300 GL column (Amersham Biosciences) as described under “Experimental Procedures.” The same results were obtained using [1-14C]16:0-ACP. C, FabT binding to various acyl-ACPs determined by an AlphaScreen assay. Triplicate values are plotted as mean ± standard errors (smaller than the points). The apparent dissociation constants were calculated from the binding isotherms: 18:1Δ11-ACP, 0.5 ± 0.1 nm; 16:0-ACP, 0.5 ± 0.1 nm; 14:0-ACP, 0.5 ± 0.2 nm; 10:0-ACP, 0.7 ± 0.3 nm; malonyl-ACP, 2.0 ± 0.2 nm; and ACP, 4.5 ± 0.2 nm.
DISCUSSION
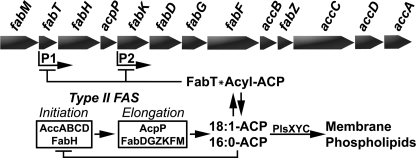
Our discovery reveals an important new level of FAS-II regulation by acyl-ACP (Fig. 4). Acyl-ACP binds to the FabT repressor and significantly increases its affinity for its binding sites in the two promoters in the fab gene cluster. The potency of acyl-ACP increases with increasing chain length, and the most effective acyl-ACP, 18:1Δ11-ACP (Fig. 2), corresponds to the most abundant fatty acid (48%) found in S. pneumoniae membrane phospholipids (28). These observations lead to a model where the end products of FAS-II act as feedback inhibitors of the transcription of the genes encoding the enzymes in the biosynthetic pathway (Fig. 4). This regulatory circuit coupled with the genetic organization of the FAS-II gene cluster ensures that transcription of the entire fab gene set is coordinately linked to the level of acyl-ACP precursors to membrane phospholipids. This genetic regulatory loop is complemented by the biochemical regulation of FAS-II by acyl-ACPs through their ability to inhibit two key enzymes involved in the initiation and maintenance of the pathway, FabH (5) and acetyl-CoA carboxylase (6) (Fig. 4). Both of these enzymes are negatively regulated by acyl-ACP with the longer chain lengths being the most potent effectors. Finally, the regulatory roles for acyl-ACP end products of FAS-II depicted in Fig. 4 points to coordination of membrane phospholipid synthesis with other macromolecular processes occurring at the acyltransferase step.
FIGURE 4.
Role of long-chain acyl-ACP in the biochemical and genetic regulation of FAS-II. Membrane phospholipid fatty acids are produced by FAS-II, which is conceptually divided into initiation and elongation components. The initiation enzymes, β-ketoacyl-ACP synthase III (FabH), which catalyzes the first step in the pathway, and acetyl-CoA carboxylase (AccABCD), which supplies malonyl-CoA for each condensation step, control the number of fatty acids produced and the rate of elongation. The elongation cycles of fatty acid biosynthesis process the nascent fatty acid chain into the end products of the pathway, which in S. pneumoniae consist primarily of 16:0 and 18:1Δ11 acyl-ACP. These acyl-ACPs are used by the PlsXYC acyltransferase system (29) to generate phosphatidic acid, which is the common intermediate used in the formation of membrane phospholipids. Long-chain acyl-ACPs biochemically regulate FAS-II by their feedback inhibition of FabH (5) and acetyl-CoA carboxylase (6). This study extends the role of acyl-ACP to the transcriptional regulation of FAS-II via their binding to FabT, which represses the expression of the FAS-II genes by binding of the FabT-acyl-ACP complex to the two promoters (P1 and P2) that control expression of the FAS-II gene cluster (28).
The most thoroughly studied FAS-II transcriptional regulator is FapR (23–25). This protein acts as a repressor to coordinately control the dispersed genes of FAS-II in B. subtilis. In this case, malonyl-CoA is the ligand that releases FapR from DNA. Thus, the FabT and FapR systems appear quite different. FabT participates in a defined feedback loop (Fig. 4), whereas FapR operates by a less well characterized feed-forward regulatory loop. Acetyl-CoA carboxylase is perhaps a regulatory target for ligands that coordinate membrane biogenesis with cell growth in B. subtilis, but the details of how acetyl-CoA carboxylase is regulated and how a feed-forward system could coordinate the rate of FAS-II with the demand for acyl-ACP remain to be defined. Another important transcriptional repressor of the type II system is FabR (22), but nothing is known about its regulatory ligand. A continued effort to understand these regulatory interactions is warranted due to their importance in maintaining bacterial membrane lipid homeostasis and the potential to exploit these control systems for the development of novel antibacterial therapeutics.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34496, Cancer Center (CORE) Support Grant CA21765, and the American Lebanese Syrian Associated Charities.
- FAS-II
- dissociated type II fatty acid synthase
- ACP
- acyl carrier protein
- 18:1Δ11
- cis-vaccenic acid
- 18:1Δ9
- oleic acid
- 16:0
- palmitic acid
- fab genes
- general term referring to all the genes encoding the enzymes of FAS-II.
REFERENCES
- 1.Zhang Y. M., Rock C. O. ( 2008) Nat. Rev. Microbiol. 6, 222– 233 [DOI] [PubMed] [Google Scholar]
- 2.Cronan J. E., Jr., Rock C. O. ( 2008) in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology( Böck I., Curtis R., 3rd, Kaper J. B., Karp P. D., Neidhardt F. C., Nyström T., Slauch J. M., Squires C. L., Ussery D. eds) ASM Press, Washington, DC [Google Scholar]
- 3.Rock C. O., Jackowski S. ( 2002) Biochem. Biophys. Res. Commun. 292, 1155– 1166 [DOI] [PubMed] [Google Scholar]
- 4.Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. ( 2005) Nature 433, 531– 537 [DOI] [PubMed] [Google Scholar]
- 5.Heath R. J., Rock C. O. ( 1996) J. Biol. Chem. 271, 10996– 11000 [DOI] [PubMed] [Google Scholar]
- 6.Davis M. S., Cronan J. E., Jr. ( 2001) J. Bacteriol. 183, 1499– 1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y. M., Rock C. O. ( 2009) J. Lipid Res. 50, 5115– 5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schujman G. E., de Mendoza D. ( 2005) Curr. Opin. Microbiol. 8, 149– 153 [DOI] [PubMed] [Google Scholar]
- 9.Overath P., Pauli G., Schairer H. U. ( 1969) Eur. J. Biochem. 7, 559– 574 [PubMed] [Google Scholar]
- 10.DiRusso C. C., Nunn W. D. ( 1985) J. Bacteriol. 161, 583– 588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry M. F., Cronan J. E., Jr.( 1991) J. Mol. Biol. 222, 843– 849 [DOI] [PubMed] [Google Scholar]
- 12.Henry M. F., Cronan J. E., Jr.( 1992) Cell 70, 671– 679 [DOI] [PubMed] [Google Scholar]
- 13.Campbell J. W., Cronan J. E., Jr.( 2001) J. Bacteriol. 183, 5982– 5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiRusso C. C., Heimert T. L., Metzger A. K. ( 1992) J. Biol. Chem. 267, 8685– 8691 [PubMed] [Google Scholar]
- 15.Raman N., DiRusso C. C. ( 1995) J. Biol. Chem. 270, 1092– 1097 [DOI] [PubMed] [Google Scholar]
- 16.DiRusso C. C., Tsvetnitsky V., Højrup P., Knudsen J. ( 1998) J. Biol. Chem. 273, 33652– 33659 [DOI] [PubMed] [Google Scholar]
- 17.Cronan J. E., Jr.( 1997) J. Bacteriol. 179, 1819– 1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Aalten D. M., DiRusso C. C., Knudsen J., Wierenga R. K. ( 2000) EMBO J. 19, 5167– 5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y., Heath R. J., Li Z., Rock C. O., White S. W. ( 2001) J. Biol. Chem. 276, 17373– 17379 [DOI] [PubMed] [Google Scholar]
- 20.van Aalten D. M., DiRusso C. C., Knudsen J. ( 2001) EMBO J. 20, 2041– 2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCue L., Thompson W., Carmack C., Ryan M. P., Liu J. S., Derbyshire V., Lawrence C. E. ( 2001) Nucleic Acids Res. 29, 774– 782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y. M., Marrakchi H., Rock C. O. ( 2002) J. Biol. Chem. 277, 15558– 15565 [DOI] [PubMed] [Google Scholar]
- 23.Schujman G. E., Paoletti L., Grossman A. D., de Mendoza D. ( 2003) Dev. Cell 4, 663– 672 [DOI] [PubMed] [Google Scholar]
- 24.Schujman G. E., Guerin M., Buschiazzo A., Schaeffer F., Llarrull L. I., Reh G., Vila A. J., Alzari P. M., de Mendoza D. ( 2006) EMBO J. 25, 4074– 4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schujman G. E., Altabe S., de Mendoza D. ( 2008) Mol. Microbiol. 68, 987– 996 [DOI] [PubMed] [Google Scholar]
- 26.Marrakchi H., Choi K. H., Rock C. O. ( 2002) J. Biol. Chem. 277, 44809– 44816 [DOI] [PubMed] [Google Scholar]
- 27.Heath R. J., Rock C. O. ( 2000) Nature 406, 145– 146 [DOI] [PubMed] [Google Scholar]
- 28.Lu Y. J., Rock C. O. ( 2006) Mol. Microbiol. 59, 551– 566 [DOI] [PubMed] [Google Scholar]
- 29.Lu Y. J., Zhang Y. M., Grimes K. D., Qi J., Lee R. E., Rock C. O. ( 2006) Mol. Cell 23, 765– 772 [DOI] [PubMed] [Google Scholar]
- 30.Rock C. O., Garwin J. L. ( 1979) J. Biol. Chem. 254, 7123– 7128 [PubMed] [Google Scholar]
- 31.Jackowski S., Rock C. O. ( 1983) J. Biol. Chem. 258, 15186– 15191 [PubMed] [Google Scholar]
- 32.Lishanski A., Kurn N., Ullman E. F. ( 2000) Nucleic Acids Res. 28, E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ullman E. F., Kirakossian H., Singh S., Wu Z. P., Irvin B. R., Pease J. S., Switchenko A. C., Irvine J. D., Dafforn A., Skold C. N., Wagner D. B. ( 1994) Proc. Natl. Acad. Sci. U. S. A. 91, 5426– 5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazar G. A., Dang W., Karki S., Vafa O., Peng J. S., Hyun L., Chan C., Chung H. S., Eivazi A., Yoder S. C., Vielmetter J., Carmichael D. F., Hayes R. J., Dahiyat B. I. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 4005– 4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jerga A., Miller D. J., White S. W., Rock C. O. ( 2009) J. Biol. Chem. 284, 7246– 7254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helmkamp G. M., Jr., Brock D. J., Bloch K. ( 1968) J. Biol. Chem. 243, 3229– 3231 [PubMed] [Google Scholar]
- 37.Alberts A. W., Bell R. M., Vagelos P. R. ( 1972) J. Biol. Chem. 247, 3190– 3198 [PubMed] [Google Scholar]
- 38.Heath R. J., Rubin J. R., Holland D. R., Zhang E., Snow M. E., Rock C. O. ( 1999) J. Biol. Chem. 274, 11110– 11114 [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y. M., Zhu K., Frank M. W., Rock C. O. ( 2007) Mol. Microbiol. 66, 622– 632 [DOI] [PubMed] [Google Scholar]