Abstract
To elucidate the minimal lipopolysaccharide (LPS) structure needed for the viability of Escherichia coli, suppressor-free strains lacking either the 3-deoxy-d-manno-oct-2-ulosonic acid transferase waaA gene or derivatives of the heptosyltransferase I waaC deletion with lack of one or all late acyltransferases (lpxL/M/P) and/or various outer membrane biogenesis factors were constructed. Δ(waaC lpxL lpxM lpxP) and waaA mutants exhibited highly attenuated growth, whereas simultaneous deletion of waaC and surA was lethal. Analyses of LPS of suppressor-free waaA mutants grown at 21 °C, besides showing accumulation of free lipid IVA precursor, also revealed the presence of its pentaacylated and hexaacylated derivatives, indicating in vivo late acylation can occur without Kdo. In contrast, LPS of Δ(waaC lpxL lpxM lpxP) strains showed primarily Kdo2-lipid IVA, indicating that these minimal LPS structures are sufficient to support growth of E. coli under slow-growth conditions at 21/23 °C. These lipid IVA derivatives could be modified biosynthetically by phosphoethanolamine, but not by 4-amino-4-deoxy-l-arabinose, indicating export defects of such minimal LPS. ΔwaaA and Δ(waaC lpxL lpxM lpxP) exhibited cell-division defects with a decrease in the levels of FtsZ and OMP-folding factor PpiD. These mutations led to strong constitutive additive induction of envelope responsive CpxR/A and σE signal transduction pathways. Δ(lpxL lpxM lpxP) mutant, with intact waaC, synthesized tetraacylated lipid A and constitutively incorporated a third Kdo in growth medium inducing synthesis of P-EtN and l-Ara4N. Overexpression of msbA restored growth of Δ(lpxL lpxM lpxP) under fast-growing conditions, but only partially that of the Δ(waaC lpxL lpxM lpxP) mutant. This suppression could be alleviated by overexpression of certain mutant msbA alleles or the single-copy chromosomal MsbA-498V variant in the vicinity of Walker-box II.
Lipopolysacharides (LPS)4 are the major amphiphilic constituents of the outer leaflet of the outer membrane (OM) of Gram-negative bacteria, including Escherichia coli. LPS share a common architecture composed of a membrane-anchored phosphorylated and acylated β(1→6)-linked GlcN disaccharide, termed lipid A, to which a carbohydrate moiety of varying size is attached (1, 2). The latter may be divided into a lipid A proximal core oligosaccharide and, in smooth-type bacteria, a distal O-antigen. LPS always contain 3-deoxy-α-d-manno-oct-2-ulosonic acid (Kdo) linked to the lipid A.
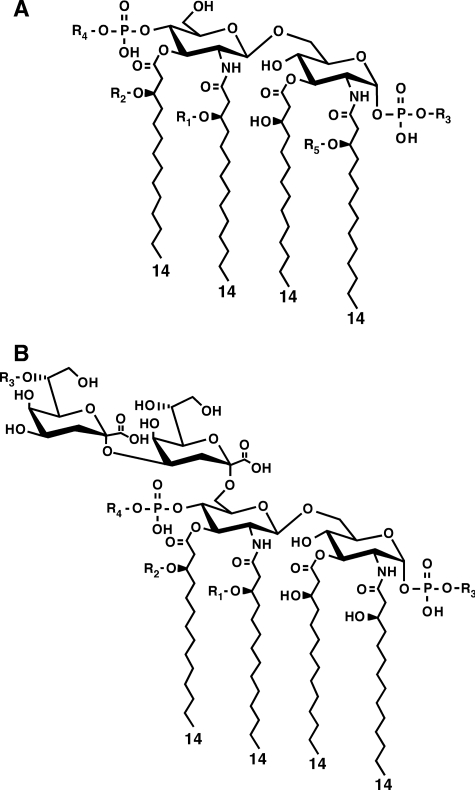
The physiological importance of the Kdo/lipid A region is reflected by its specific position within the pathway of LPS biosynthesis. In E. coli K-12, a bisphosphorylated lipid A precursor molecule with two amide and two ester-bound (R)-3-hydroxymyristate residues (lipid IVA) is synthesized from UDP-GlcNAc, following 6 distinct enzyme reactions (1). This intermediate serves as an acceptor for the Kdo transferase (WaaA), which transfers two Kdo residues from CMP-Kdo to yield an α(2→4)-linked Kdo disaccharide-attached α(2→6) to the non-reducing GlcN residue of lipid IVA (3). The latter reaction product, termed Kdo2-lipid IVA, comprises a key intermediate of LPS biosynthesis that acts 2-fold as a specific substrate: (i) for glycosyltransferases catalyzing further steps of the core oligosaccharide biosynthesis (4) and (ii) for acyltransferases that complete the lipid A moiety by the transfer of 2 additional fatty acids to the (R)-3-hydroxyl groups of both acyl chains, which are directly bound to position 2′ and 3′ of the non-reducing GlcN residue (1). Three acyltransferases, encoded by paralogous genes, have been described in E. coli K-12, which catalyze the latter enzyme reactions using acyl carrier protein-activated fatty acids as co-substrates (5–10). At ambient temperatures, a lauroyl residue is first transferred by LpxL (6) to the OH group of the amide-bound (R)-3-hydroxymyristate residue at position 2′. This catalytic step is partially replaced at low temperature (12 °C) by LpxP, which transfers palmitoleate to the same position in ∼80% of the LPS molecules (7). The free OH group of the ester-bound (R)-3-hydroxymyristate residue at position 3′ within both pentaacylated intermediates is then myristoylated by LpxM to give a hexaacylated lipid A moiety (Fig. 3) (5).
FIGURE 3.
Chemical structure of tetraacylated lipid IVA precursor (A) and Kdo2-lipid IVA (B). R1 represents C12:0 or C16:1; R2, C14:0; R3 and R4 are under LPS-modifying conditions P-EtN and l-Ara4N, respectively, and R5, C16:0.
Consistent with the essentiality of LPS in E. coli, all the genes, whose products are required for committed steps of biosynthesis of lipid IVA and subsequent transfer of Kdo to it, are essential (1, 2). However, individually neither the subsequent steps of addition of the secondary lauroyl and myristoyl residues to the distal glucosoamine unit by LpxL and LpxM to synthesize hexaacylated lipid A nor the later glycosylation of hexaacylated Kdo2-lipid A is essential for viability of bacteria like E. coli K-12 under defined growth conditions (8). Although Re mutants that possess LPS with only hexaacylated Kdo2-lipid A or mutants that synthesize complete LPS core with only lipid IVA are viable, they are impaired in several growth properties, including constitutive induction of RpoE signal transduction in Re mutants (8, 11–13). A triple null mutant, which lacks all 3 late acyltransferases, is viable but only in slow-growth conditions in accordance with lipid IVA being a poor substrate of the lipid A transporter MsbA (8). Mutants impaired in the synthesis of Kdo, which synthesize only lipid IVA lacking any glycosylation, can be constructed, but they require additional suppressor mutations either in msbA, or the yhjD gene (14, 15). Strains that potentially can only synthesize Kdo2-lipid IVA have not been reported up to now. Thus, suppressor-free minimal LPS structures that can support growth of E. coli K-12 bacteria known up to now have genetic compositions of Δ(lpxL lpxM lpxP) or Re mutants.
We describe the construction and characterization of suppressor-free ΔwaaA and Δ(waaC lpxL lpxM lpxP) mutants, synthesizing either free lipid IVA derivatives or Kdo2-lipid IVA LPS, respectively. Analyses of lipid A of ΔwaaA also revealed the presence of free penta- and hexaacylated lipid A derivatives, arising due to incorporation of secondary acyl chains. Such suppressor-free strains could be constructed only in slow-growth conditions at lower temperatures. Growth of Δ(waaC lpxL lpxM lpxP) could be restored by extragenic chromosomal MsbA-D498V suppressor mutation or by the overexpression of the msbA wild-type gene product. The LPS of Δ(waaC lpxL lpxM lpxP) and lipid IVA precursor of ΔwaaA was found to be substituted by P-EtN, but not l-Ara4N, under LPS-modifying growth conditions. Deletion of late acyltransferases in ΔwaaC or deletion of the waaA gene resulted in constitutively elevated levels of periplasmic protease HtrA, due to additive induction of the envelope stress responsive CpxR/A two-component system and σE pathway.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Media
Bacterial strains and plasmids used in this study are described in Table 1. Luria-Bertani (LB) broth, M9, and 121 phosphate-limiting minimal media were prepared as described (16, 17). When necessary, media were supplemented with ampicillin (100 μg ml−1), tetracycline (10 μg ml−1), kanamycin (50 μg ml−1), spectinomycin (50 μg ml−1), or chloramphenicol (20 μg ml−1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains/plasmids | Relevant characteristic | Reference or source |
|---|---|---|
| Strains | ||
| W3110 | λ−,IN (rrnD-rrnE)1,rph-1 | E. coli Genetic Stock Center, Yale |
| BW25113 | lacIqrrnBT14ΔlacZWJ16hsdR514ΔaraBADAH33 | 18 |
| ΔrhaBADLD78 | ||
| GK1111 | W3110 Δlac | This study |
| WBB01 | JC7623waaCFtet | 45 |
| SR7330 | W3110lpxP<>aph | This study |
| SR7465 | W3110lpxP<>frt lpxM<>frt | This study |
| SR7751 | W3110 ΔwaaCFtet | This study |
| SR7770 | W3110lpxM<>cat | This study |
| SR7774 | W3110lpxM<>frt lpxL<>aph | This study |
| SR7781 | W3110lpxM<>frt lpxP<>frt lpxL<>aph | This study |
| SR7787 | SR7751lpxL<>aph | This study |
| SR7790 | SR7751lpxM<>aph | This study |
| SR7825 | W3110waaCFtetlpxM<>frt lpxL<>aph | This study |
| SR7870 | W3110waaCFtetlpxM<>frt lpxP<>frt lpxL<>aph | This study |
| SR7870 | SR7781 ΔwaaCFtet | This study |
| SR8035 | W3110waaC<>aph | This study |
| SR8101 | W3110lpxL<>cat | This study |
| SR8163 | W3110lpxP<>cat | This study |
| SR8233 | W3110waaC<>cat | This study |
| SR8258 | W3110lpxP<>ada | This study |
| SR8277 | SR7774lpxP<>ada | This study |
| SR8352 | W3110lpxM<>frt lpxP<>frt waaC<>aphlpxL<>cat | This study |
| SR8129 | GK1111 ϕ (rpoHP3-lacZ) | This study |
| SR8190 | SR8129waaC<>aph | This study |
| SR8165 | SR8129lpxL<>aph | This study |
| SR8159 | SR8129lpxL<>cat | This study |
| SR8168 | SR8129lpxM<>aph | This study |
| SR8160 | SR8129lpxM<>cat | This study |
| SR8162 | SR8129lpxP<>cat | This study |
| SR8225 | SR8129waaC<>aphlpxL<>cat | This study |
| SR8221 | SR8129waaC<>aphlpxM<>cat | This study |
| SR8481 | SR8129lpxM<>cat lpxL<>aph | This study |
| SR8510 | SR8129lpxM<>cat lpxL<>aph lpxP<>ada | This study |
| SR7718 | BW25113 ϕ (pcpxP-lacZ) | This study |
| SR8201 | SR7718waaCFtet | This study |
| SR8177 | SR7718lpxL<>cat | This study |
| SR8180 | SR7718lpxM<>cat | This study |
| SR8212 | SR7718waaCFtetlpxL<>cat | This study |
| SR8209 | SR7718waaCFtetlpxM<>cat | This study |
| SR8352 | SR8277waaC<>cat | This study |
| SR8356 | SR7781waaC (pmsbA+) | This study |
| SR8433 | SR8356 pmsbA (L279V) | This study |
| SR8437 | SR8356 pmsbA (A510P) | This study |
| SR8478 | SR7870msbA (D498V) | This study |
| SR8445 | SR7781waaC (psurA+) | This study |
| SR8469 | GK1162waaC<>cat | This study |
| SR8432 | SR7781 (pmsbA+) | This study |
| SR8614 | BW25113waaA<>aph | This study |
| SR8621 | SR8129waaA<>aph | This study |
| SR8647 | SR7718waaA<>cat | This study |
| GK1077 | W3110lpxM<>aph | This study |
| GK1162 | W3110eptB<>aph | This study |
| GK1275 | W3110lpxL<>aph | This study |
| GK1395 | W3110eptA<>aph | This study |
| GK1400 | W3110basR<>aph | This study |
| GK1735 | W3110eptB<>frt basR<>aph waaC<>cat | This study |
| GK1753 | GK1395waaC<>cat | This study |
| Plasmids | ||
| pCP20 | Contains temperature-sensitive replicon and a thermally inducible FLP recombinase | E. coli Genetic Stock Center, Yale |
| pKD3 | oriR6Kγ,bla (AmpR),kan,rgnB (Ter),cat | E. coli Genetic Stock Center, Yale (18) |
| pKD13 | oriR6Kγ,bla (AmpR),kan,rgnB (Ter) | E. coli Genetic Stock Center, Yale (18) |
| pKD46 | araBp-gam-bet-exo,bla (AmpR),repA101 (ts)oriR101 | E. coli Genetic Stock Center, Yale (18) |
| pRS415 | lacZYA transcriptional fusion vector AmpR | |
| pSE420 | AmpRexpression vectorlacIQ | Invitrogen |
| pCA24N | CmRexpression vectorlacIQ | (19) |
| pCL1921 | oripSC101 specR | (46) |
| pGK1663 | pRS415::(eptA-lacZ) | This study |
| pGK1665 | pRS415::(arnB-lacZ) | This study |
| pSR7409 | msbAcmRin pCA24N | This study |
| pSR8262 | msbAampRin pSE420 | This study |
| pSR2777 | surAampR | (13) |
Generation of Null Mutations and Construction of Their Combinations
Non-polar antibiotic-free deletion mutations of various genes were generated using the λ Red recombinase/FLP-mediated recombination system (18). The coding sequence of each gene was replaced with either the kanamycin (aph) or chloramphenicol (cat) resistance cassette flanked by FRT recognition sequences using plasmids pKD13 and pKD3 as templates (18), and recombined on the chromosome of BW25113 containing the λ Red recombinase-encoding plasmid pKD46. Gene replacements and their exact chromosomal locations were verified by PCR. All the initial single gene disruptions were made on minimal M9 medium at 30 °C, except for the waaA deletion, which was constructed at 21 °C. Deletion mutations were then transduced into W3110 selecting for antibiotic markers. Multiple null combinations were made through a series of transductions using bacteriophage T4-mediated transductions, followed by the removal of the aph or cat cassettes. The initial lpxM lpxP double null combinations were built using M9 minimal medium and limited growth at 37 °C to facilitate excision of the aph cassette and removal of Ts FLP recombinase plasmid pCP20 without causing accumulation of suppressors. The rationale for the first construction of the Δ(lpxP lpxM) strain was that such a genetic knock-out combination does not confer any known growth defects (8). SR7465 Δ(lpxM lpxP) was then used as a recipient to further transduce waaC::cm, waaC::kan, or waaCF tet::6, or lpxL (aph or cat) deletion mutations. As a control another set of strains was constructed by replacing lpxL- and lpxP-coding sequences with cat and spectinomycin (ada) cassettes and then transferred to the chromosome, following λ Red recombineering. Recombinants were selected on minimal M9 plates with appropriate antibiotics and used as donors to transduce deletion alleles in lpxM mutants. This resulted in the construction of lpxM lpxL::cm lpxP::spec (SR8277) and similar derivatives. All the transductions were performed in parallel at 30 or 23 °C on minimal M9 medium. Finally, strains lacking all of the late acyltransferase genes with the addition of deletion of waaC, SR7870 (lpxL lpxP lpxM waaCF::tet) and SR8352 (lpxL lpxP lpxM waaC::cm) constructed by transductions, were obtained only at 23 °C on the M9 minimal medium. Control transduction consisted of using lpxL lpxM lpxP null but carrying either waaC or msbA genes on the plasmid as a recipient SR8432 for bringing in the ΔwaaC::cm allele at 23 and 30 °C (Tables 1 and 3). Identical sets of single and multiple deletion strains were constructed in isogenic W3110 (GK1111) and BW25113 strains carrying single-copy promoter fusions to the rpoHP3 and cpxP promoter (Table 1). Disruptions and null allelic combinations involving eptA, basR, and eptB were constructed in the same manner as described above (Table 1). Finally, suppressor-free deletions of the waaA gene on chromosomes of W3110 and BW25113 were constructed by substitution of the entire coding sequence with either aph or cat cassettes. Such recombinants were constructed on the M9 minimal medium at 21 °C and subsequently transduced under the same growth conditions, resulting in SR8614, SR8621, and SR8647 (Table 1). Coding sequences of lpxL, waaC, and msbA genes were PCR amplified and cloned in tightly controlled expression vectors pSE420 (Invitrogen) and pCA24N (19).
TABLE 3.
Colony forming ability of ΔwaaC derivatives in transductional combinations with OMP/LPS-specific factors and suppression by MsbA variants
| Transductional efficiency |
||||
|---|---|---|---|---|
| M9 |
LB |
|||
| 30 °C | 37 °C | 30 °C | 37 °C | |
| Δ(lpxM lpxP) + pmsbA++ ΔlpxL | ++a | ++ | ++ | ++ |
| Δ(lpxL lpxM lpxP) + pmsbA++ ΔwaaC | ++ | +b | ++ | −g |
| Δ(lpxL lpxM lpxP) + pmsbAL279V + ΔwaaC | ++ | ++ | ++ | ++ |
| Δ(lpxL lpxM lpxP) + pmsbAA510P + ΔwaaC | ++ | ++ | ++ | ++ |
| Δ(lpxL lpxM lpxP)msbAcD498V + ΔwaaC | ++ | ++ | ++ | ++ |
| Δ(lpxL lpxM lpxP) + ΔsurA | ++ | NDd | ND | ND |
| Δskp+ ΔwaaC | ++ | ++ | ++ | ++ |
| ΔfkpA+ ΔwaaC | ++ | ++ | ++ | ++ |
| ΔppiD+ ΔwaaC | ++ | ++ | ++ | ++ |
| surA::kan +waaC::cm | FCe | FC | FC | FC |
| surA::cm+waaC::kan | FC | FC | FC | FC |
| surA::kan + psurA++waaC::cm | ++ | ++ | ++ | ++ |
| htrA::tet+waaC::cm | ++ | ++ | ++ | ++ |
| htrA::tet+waaC::kan | ++ | + | ++ | + |
| Δhfq+ ΔwaaC | ++ | ++ | ++ | + |
| ΔrybB+ ΔwaaC | ++ | ++ | ++ | ++ |
| ΔyfgL+ ΔwaaC | ++ | ++ | ++ | + |
| M9 |
LB |
|||
| 21 °C | 23 °C | 30 °C | 37 °C | |
| Δ(lpxL lpxM lpxP) + ΔwaaC | ++ | ++ | − | − |
| Wild type + ΔwaaA | ++ | ++ | +f | − |
a++, ≥500 colonies.
b+, 100–500 colonies, but small in size.
c Chromosomal MsbA D498V mutation.
d ND, not determined.
e FC, few colonies <10.
f Small colony size.
g −, inability to support colony forming ability.
LPS Extraction and Growth Analysis
Bacterial cultures were grown with shaking in M9 liquid medium with appropriate antibiotics at 23 °C to early log phase. To compare growth rates, cultures were washed in 10 mm MgSO4 and resuspended at an optical density A600 of 0.01 in LB and M9 minimal medium. Cultures were further incubated at 23, 30, 37, 39, and 43 °C with shaking. The optical density A600 was recorded at various times. For LPS extraction, bacterial cultures were grown under permissive growth conditions of 23 or 30 °C either in M9 minimal medium (non-modifying) or in 121 medium (LPS-modifying medium) until optical density A600 of 0.3 to 0.6 was reached. Cultures (400 ml) were harvested by centrifugation at 7000 × g for 30 min and dried. LPS was extracted by the phenol/chloroform/petroleum ether procedure (20) and lyophilized. For LPS analysis, lyophilized material was dispersed in water by sonication and resuspended at a concentration of 2 mg ml−1. Lipid IVA and its derivatives were extracted from 200-ml cultures of ΔwaaA mutants grown either in LB or different types of minimal media at either 30 or 21 °C, following the procedure described for isolation of LPS from deep-rough mutants (21). Glycerophospholipids and the lipid A mixture was resuspended in a chloroform/methanol mixture (4:1, v/v) at a concentration of 2 mg ml−1. For detection of chemotype 1 μg of purified LPS or a portion of whole cell lysate treated with proteinase K was applied to a 16.5% Tricine gel. Gels were silver stained for LPS analysis. TLC immunostaining for verification of the absence of Kdo but presence of tetraacylated lipid A in ΔwaaA, monoclonal antibodies A20 or A6 were used as described (22–24), using compounds 406, 506, and Re LPS as controls.
Mass Spectrometry
Electrospray ionization Fourier transform-ion cyclotron (ESI FT-ICR) mass spectrometry was performed in negative ion mode using an APEX II Bruker Daltonics, equipped with a 7-tesla actively shielded magnet and an Apollo ion source. Samples at a concentration of ∼10 ng μl−1 were sprayed at a flow rate of 2 μl min−1 as described (25). Capillary entrance voltage was set to 3.8 kV, and dry gas temperature to 200 °C. For unspecific fragmentation the DC offset (collision voltage) of the quadruple interface was set from 5 to 30 V. Under these conditions the labile linkage between lipid A and the core oligosaccharide is cleaved. If not otherwise stated the mass spectra were charge deconvoluted and mass numbers given refer to the monoisotopic masses of the neutral molecules. Mass calibration was done externally by well characterized similar compounds of known structure. Mass accuracy was better than 5 ppm.
Western Blot Analysis
Cultures were grown at either 21 or 23 °C for 24–48 h in 30 ml of M9 minimal medium, harvested by centrifugation at 3000 × g for 10 min, and resuspended in SDS lysis buffer. Proteins were resolved by 12% SDS-PAGE. After electrophoresis, proteins were blotted to nitrocellulose membrane. HtrA, PpiD, RseB, DnaK, and FtsZ proteins were detected with the respective antibody (26, 27).
β-Galactosidase Assays
The activity of CpxR/A and RpoE pathways in waaA, waaC, lpxL, and lpxM mutants, and in derivatives with their null allelic combinations were analyzed in strains carrying either rpoHP3-lacZ or cpxP-lacZ promoter fusions in a single copy on the chromosome. The construction of rpoHP3-lacZ or cpxP-lacZ and other RpoE-regulated promoter fusions has been previously described (28). Putative BasS/R-regulated promoter regions of the eptA-basS-basR operon, and arn operon were amplified by PCR, using primers listed in supplemental Table S1. The amplified PCR products were cloned in pRS415 vector and transferred to chromosome as described previously for other promoter fusions (13, 17). Isogenic bacterial strains, carrying promoter fusions, were grown in M9 medium, at 21, 23, or 30 °C, harvested by centrifugation, and diluted to an A600 of ≈0.03 in M9 or 121 medium. Cultures were allowed to grow for another 90 min and β-galactosidase activity was measured in Miller units at different growth intervals. At least four independent cultures were assayed for each mutant and the corresponding isogenic parent.
RESULTS
Construction of Chromosomal Deletions of lpxL, lpxM, lpxP, and waaC Genes and Their Combinations
To construct and characterize E. coli strains that synthesize LPS with only Kdo2-lipid IVA, deletions in genes encoding the late acyltransferases, namely lauroyl-, myristoyl-, and palmitoleoyl acyltransferases, and heptosyltransferase I were constructed as described under “Experimental Procedures.” In one case Δ(lpxL lpxM lpxP) strains (SR7781 and SR8277), constructed by a combination of individual deletions of the respective genes at 30 °C on M9 medium, were used to receive waaCF::tet or waaC::cm using T4-mediated transductions, resulting in SR7870 and SR8352, respectively. Alternatively, Δ(lpxM lpxP waaC) (SR7807), synthesizing Kdo2-lipid Apenta, was used as recipient to transduce lpxL::kan, resulting in SR7877. Transductions were performed at 23 and 30 °C in the presence or absence of plasmids expressing either the waaC or lpxL genes. In all cases viable transductants, resulting in Δ(waaC lpxL lpxM lpxP), were obtained at 23 °C on minimal medium but not on the LB medium, when waaC or lpxL genes were not expressed from the plasmid. The frequency of transductions at 23 °C was similar in the presence or absence of waaC/lpxL plasmids (Table 3). No viable transductants could be obtained at 30 °C even in slow-growth conditions of minimal medium in the absence of the waaC plasmid. However, in the presence of waaC plasmid, viable transductants were obtained at 23 and 30 °C at the same frequency. Taken together, these results suggest that a viable suppressor-free Δ(waaC lpxL lpxM lpxP) mutant synthesizing Kdo2-lipid IVA can be constructed under slow-growth conditions of minimal medium at 23 °C, but not at or above 30 °C even on minimal medium. However, as described in later sections, Δ(waaC lpxL lpxM lpxP) could be constructed even on rich medium at both 23 and 30 °C, when extra copies of MsbA were provided in trans.
Growth Properties of the Mutant Strains at Different Temperatures
Consistent with the reported Ts phenotype of rfaD mutants (12), ΔwaaC mutants exhibited a temperature-sensitive growth phenotype at or above 43 °C. Furthermore, Δ(waaC lpxM) strain exhibited an additive temperature-sensitive phenotype above 39 °C, which is permissive for both ΔwaaC and ΔlpxM bacteria. Significantly, the growth of Δ(waaC lpxL) on rich medium was severely compromised and supported the colony forming ability only at 23 or 30 °C, but not above such temperatures on minimal medium, with synthetic lethality on rich medium. Furthermore, the addition of ΔlpxM to such as strain Δ(waaC lpxL) caused even higher growth reduction in the temperature range of 23 to 30 °C (Table 2). The colony forming ability of Δ(waaC lpxL lpxM lpxP) and growth in liquid culture was restricted strictly to slow-growth conditions, within a narrow temperature range with slower growth rates (Table 2). The growth occurred on minimal medium at 23 °C with inability to form colonies at temperatures at or above 30 °C. These results demonstrate that E. coli strains, which synthesize the Kdo2-lipid IVA predicted LPS structure, can sustain growth in genetically suppressor-free backgrounds within a limited growth range unless Kdo2-lipid IVA is more efficiently transported.
TABLE 2.
Growth rate h−1(μ) of the wild-type and ΔwaaC derivatives with and without MsbA overexpression
| M9 |
LB |
|||
|---|---|---|---|---|
| 23 °C | 30 °C | 23 °C | 30 °C | |
| Wild type | 0.40 | 0.52 | 0.61 | 0.99 |
| ΔwaaC | 0.31 | 0.51 | 0.60 | 0.98 |
| Δ(waaClpxL) | 0.20 | 0.18 | 0.50 | 0.34 |
| Δ(waaClpxM) | 0.22 | 0.30 | 0.31 | 0.47 |
| Δ(waaClpxL lpxM) | 0.15 | 0.17 | 0.26 | 0.24 |
| Δ(waaClpxL lpxM lpxP) | 0.14 | NLa | NL | NC,bNL |
| Δ(lpxL lpxM lpxP) | 0.29 | 0.41 | 0.43 | NC, NL |
| LB |
||||
| 23 °C | 30 °C | 37 °C | ||
| Wild type | 0.60 | 1.01 | 1.34 | |
| Δ(lpxL lpxM lpxP) + pmsbA+ | NDc | 0.65 | 0.67 | |
| Δ(waaC lpxL lpxM lpxP) + pmsbA+ | 0.54 | 0.45 | ND | |
a NL, non-linear growth and hence data not used to calculate growth rate.
b NC, inability to support colony forming ability.
c ND, not determined.
The waaA Gene Is Dispensable at or below 30 °C
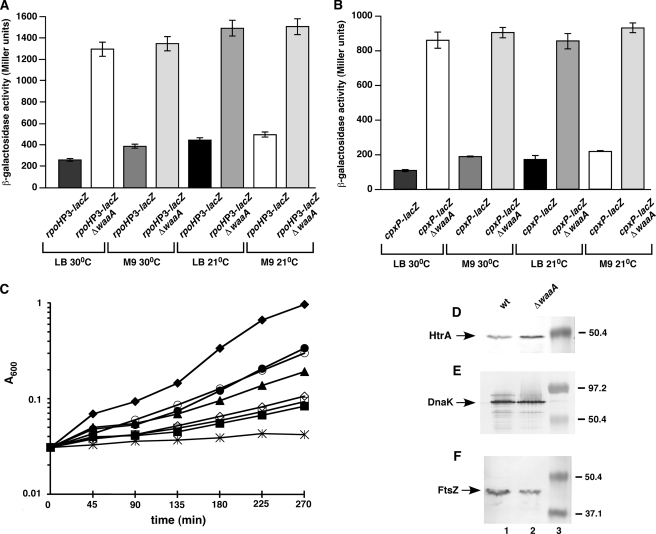
Because viable Δ(waaC lpxL lpxM lpxP) strains capable of synthesizing only Kdo2-lipid IVA could be constructed under slow-growth conditions, we wondered if under the same growth conditions, a suppressor-free chromosomal deletion in the otherwise essential waaA gene can be constructed. This allowed us to test if Kdo attachment to lipid IVA is absolutely required for viability of E. coli. Strains with viable non-polar deletions in the waaA gene were obtained on minimal medium at 21–23 °C in W3110 and BW25113 genetic backgrounds (SR8614, SR8621, and SR8647). Such strains grew even on LB medium up to 30 °C, although with a smaller colony size than the parental wild-type (Fig. 11C), unlike Δ(waaC lpxL lpxM lpxP) strains, which did not form colonies on rich LB medium (Table 3). Thus, under slow-growth conditions and at low temperatures the waaA gene is dispensable for E. coli viability, although such strains grew poorly. These should be to date the first viable suppressor-free deletions in the waaA gene in E. coli K-12. However, ΔwaaA mutants exhibited severe membrane defects, resulting in the inability to form colonies on MacConkey agar, extremely sensitive to antibiotics, detergents, and chelating agents like EDTA (data not shown). ΔwaaA were also found to have cell division defects and lower amounts of the cell division protein FtsZ (Fig. 11F). To rule out any accumulation of suppressors under slow-growth conditions (30 °C or below) besides transductions, genomic DNA of 3 independent ΔwaaA strains was prepared and found to have no mutations either in the msbA or yhjD genes, the two potential genes in which suppressors could arise. Furthermore, subsequent analysis of LPS of ΔwaaA mutants confirmed the presence of free lipid IVA precursors and its derivatives, lacking any Kdo and further glycosylation (see below). This was also verified by TLC immuno-overlay, using the Kdo-specific monoclonal antibody A20 (23) (data not shown).
FIGURE 11.
Induction of stress response pathways upon deletion of the waaA gene. Isogenic bacterial cultures of ΔwaaA and its parental wild-type carrying either single-copy chromosomal rpoHP3-lacZ fusion (A) or cpxP-lacZ fusion (B) were grown at 21 °C in M9 medium to early log phase. Cultures were washed and diluted to an A600 of ∼0.02 and grown at either 21 or 30 °C in LB and M9 medium and analyzed for β-galactosidase activity as described in the legend to Fig. 10. Average of four independent measurements is shown. Error bars represent S.E. of four independent measurements. C, the same cultures used in A were simultaneously monitored for bacterial growth by measuring optical density at A600. Symbols: ♦, wild-type 30 °C LB; ▴, ΔwaaA 30 °C LB; ●, wild-type 30 °C M9; ■, ΔwaaA 30 °C M9; ○, wild-type 21 °C LB; □, ΔwaaA 21 °C LB; ◇, wt 21 °C M9; ★, ΔwaaA 21 °C M9 indicate the respective strains and growth conditions. D–F, cultures of wild-type and ΔwaaA were grown at 21 °C in M9 medium to logarithmic phase, harvested by centrifugation, and equivalent amounts of cells were used to prepare whole cell lysates. Proteins were resolved by 12% SDS-PAGE and transferred to nitrocellulose membranes. Blots were probed with antiserum raised against HtrA (D), DnaK (E), and FtsZ (F). The genotype of each strain used is indicated on the top of panel D. The position of molecular weight standards from pre-stained molecular markers (Bio-Rad) is indicated in D–F (lane 3).
Δ(waaC lpxL lpxM lpxP) Mutants Exhibit Defects in Cell Division
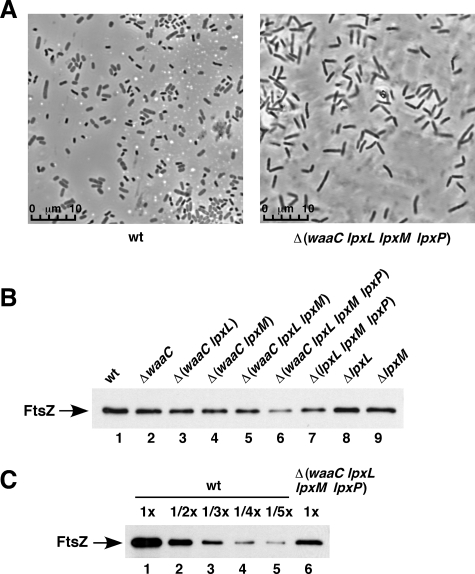
Because Δ(waaC lpxL lpxM lpxP) exhibited a slow-growth phenotype as compared with the parental strain even under permissive growth conditions, we examined cellular morphology. Δ(waaC lpxL lpxM lpxP) mutant bacteria formed short filaments with ∼2–3-fold longer dimension than the normal cell shape of the isogenic wild-type strain (Fig. 1A). This could be further attributed to impairment of cell division machinery, with an apparent 50% decrease in the levels of FtsZ in Δ(waaC lpxL lpxM lpxP) with only Kdo2-lipid IVA LPS (Fig. 1C). Thus, the cell division defects in Δ(waaC lpxL lpxM lpxP) can be ascribed to limiting amounts of the key cell division protein FtsZ.
FIGURE 1.
Cell division defects of ΔwaaC and its derivatives. A, cultures of wild-type (wt) and Δ(waaC lpxL lpxM lpxP) were grown in M9 minimal medium at 23 °C to early log phase and samples were fixed and visualized by confocal microscopy. B, aliquots of cultures grown as described above at 23 °C in M9 medium of waaC and its derivatives with mutations in lpxL or lpxM, or triple null Δ(lpxL lpxM lpxP) were harvested by centrifugation. Equivalent amounts of cells were lysed in SDS-sample buffer and proteins were resolved on 12% SDS-PAGE. Proteins were transferred to nitrocellulose membrane and probed with antisera raised against FtsZ. C, lysed samples of wild-type prepared as described above were diluted as indicated, and applied on 12% SDS-PAGE along with the undiluted sample from Δ(waaC lpxL lpxM lpxP) and analyzed for FtsZ levels.
msbA Overexpression Suppresses Δ(lpxL lpxM lpxP) Triple Mutants but Only a Partial Rescue for Δ(waaC lpxL lpxM lpxP)
Because the growth of lpxL mutants, which predominantly contain tetraacylated LPS, can be suppressed by overexpression of the msbA gene product (29), we tested if the MsbA in high dosage could suppress conditional lethality of Δ(lpxL lpxM lpxP) and Δ(waaC lpxL lpxM lpxP) strains. Introduction of plasmid, bearing the msbA gene with the inducible promoter, in Δ(lpxL lpxM lpxP) restored their growth and colony forming ability in the presence of inducer (isopropyl β-d-thiogalactopyranoside) even at 37 °C on rich medium (faster growing conditions), where Δ(lpxL lpxM lpxP) mutants are incapable of growth (Tables 2 and 3).
FIGURE 2.
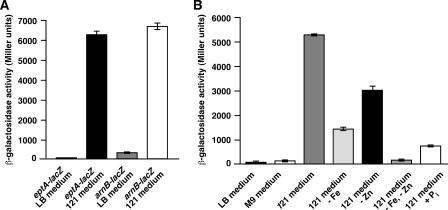
Induction of transcription of eptA and arnB promoters in 121 medium. Cultures of E. coli strain GK1111 carrying eptA-lacZ and arnB-lacZ promoter fusions were grown to early log phase in LB medium at 37 °C, washed, and adjusted to OD of 0.02 at 600 nm in LB, M9, or 121 medium. Aliquots of samples were drawn at different intervals and analyzed for β-galactosidase activity. Data of one representative set are presented (A). Cultures of GK1111 carrying eptA-lacZ promoter fusion were analyzed for β-galactosidase activity in LB, M9, 121 medium, or 121 with or without supplementation of various components as indicated for the various bars (B). Error bars represent S.E. of four independent measurements.
To verify if msbA overexpression would also restore growth in Δ(waaC lpxL lpxM lpxP) mutants, we first transduced at 23 °C the waaC null allele in strain SR7781 (lpxL lpxM lpxP) carrying the msbA gene on a plasmid. Such transductants were then tested for their ability to grow at different temperatures. Δ(waaC lpxL lpxM lpxP) with msbA overexpression grew nearly to wild-type levels at 23 and 30 °C both on rich and minimal medium (Tables 2 and 3). However, a similar complementation could not be observed at 37 °C, indicating partial suppression.
Mutations in the msbA Gene That Elevate Kdo2-Lipid IVA Mutants
Because mild overexpression of the msbA gene allowed growth of Δ(waaC lpxL lpxM lpxP) mutants at 30 °C, we looked for suppressor mutations under the same conditions that could rescue growth at 37 °C on LB medium. Temperature-resistant survivors at 37 °C of SR7870, carrying the msbA on plasmids, were obtained at a frequency of ∼10−4. Plasmid DNA was prepared from individual temperature-resistant clones and used to confirm if the suppressor mutation was in the coding region of the plasmid-encoded msbA gene and their ability to breed true upon retransformation. Three independent clones identified changes in the msbA-coding sequence at residues L279V, D498V, and A510P, respectively. Among these, the D498V clone was found to suppress SR7870 and SR7781, even without the presence of inducer, thus at the low level of MsbA expression. Thus, we constructed a chromosomal msbA-498Val mutation in SR7870 (Table 3), which supported the colony forming ability up to 37 °C in LB and minimal media, leading us to conclude that this mutant allele can suppress the Δ(waaC lpxL lpxP lpxM) mutant at the chromosomal copy.
Analysis of LPS Composition of ΔwaaC and Its Derivatives in Minimal Medium with and without Induction of LPS Modifications
The chemotype of the LPS of waaC mutant and its derivatives deleted for genes encoding the late acyltransferases such as lauroyl, myristoyl, and palmitoleoyl acyltransferase was determined by mass spectrometry. LPS was extracted from bacterial cultures grown at permissive temperatures, either in the lipid A non-modifying M9 medium or 121 medium, which we found induces lipid A and core modifications that are commonly observed in polymyxin-resistant mutants. In E. coli, it is known that lipid A modifications by P-EtN and l-Ara4N are under control of the inducible two-component system BasS/R (30, 31). We chose minimal medium, because it is permissive for growth of all these mutants, including ΔwaaA and Δ(waaC lpxL lpxM lpxP), and provides suppressor-free conditions.
To establish the molecular basis of lipid A modifications in 121 medium, basS/R-regulated eptA-lacZ and arnB-lacZ promoter fusions were constructed and assayed for transcriptional activity. Shift of cultures, carrying either arnB-lacZ or eptA-lacZ promoter fusions, from LB or M9 medium to 121 medium resulted in 20–90-fold induction of β-galactosidase activity (Fig. 2A). This induction in 121 medium was attributed to the cumulative effect of phosphate limitation and the simultaneous presence of non-toxic amounts (20 μm each) of Fe3+ and Zn2+ ions (Fig. 2B). When 121 medium was supplemented with 2 mm K2HPO4, the transcriptional activity of the eptA-lacZ was reduced nearly 6-fold (Fig. 2B). However, when Fe3+ and Zn2+ were omitted from 121 medium, the induction of eptA-lacZ fusion was drastically reduced, even in the phosphate-limiting conditions (Fig. 2B). Because the eptA gene is transcribed as an eptA-basS-basR operon, induction of the eptA promoter should activate all members of the BasS/R two-component system. Thus, using 121 medium growth conditions allowed us to monitor efficiency of LPS translocation, given some of the modifications are supposed to occur after translocation on the periplasmic side and serve as good markers for lipid A translocation (32, 33). This was important for analyses of the LPS of ΔwaaA and Δ(waaC lpxL lpxM lpxP) mutant strains, synthesizing free lipid IVA precursor and Kdo2-lipid IVA, respectively, given tetraacylated LPS as being a poor substrate for the LPS transporter MsbA (33).
The Lipid A Modifications of Wild-type and Isogenic ΔlpxL, ΔlpxM, and Δ(lpxL lpxM lpxP) Mutants in 121 Medium
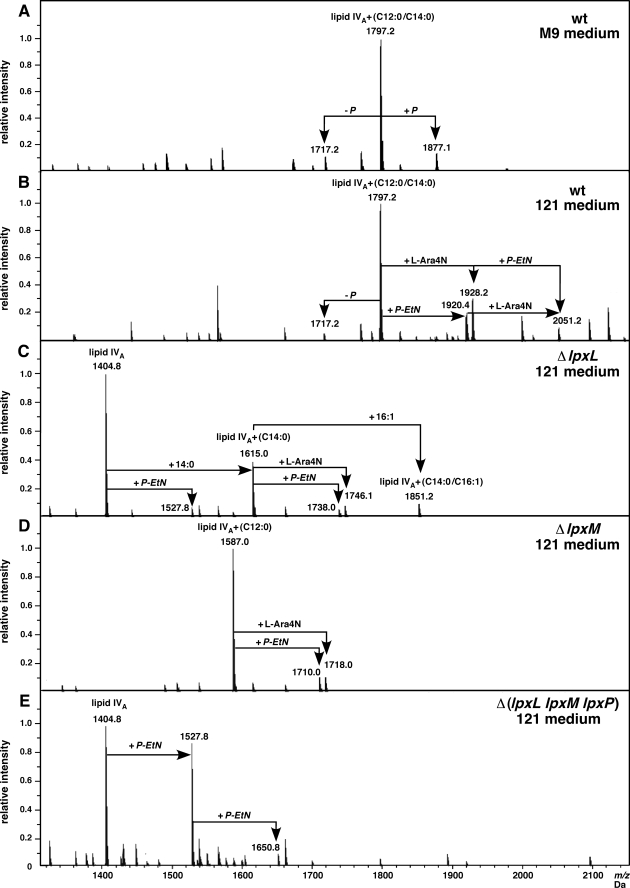
All LPS isolates were analyzed in their native, non-derivatized form by high resolution ESI FT-ICR MS under soft ionization conditions to obtain the intact molecular species and after unspecific fragmentation, generating Y-fragment ions comprising the heterogeneous lipid A part structures. The lipid A part of LPS of the wild-type E. coli K-12 grown in M9 revealed mass peaks of 1796.2 Da corresponding to hexaacylated lipid A and another peak with 1877.1 Da, which can be attributed to the substitution by 1-pyrophosphate (Figs. 3 and 4A). However, the lipid A part of LPS of wild-type bacteria grown in 121 medium, in addition to the presence of the hexaacylated 1,4′-bisphosphorylated species (1796.2 Da), also contained molecular species with P-EtN (1920.4 Da), l-Ara4N (1928.2 Da), and both substitutions (2051.2 Da) (Fig. 4B). Overall, these data confirm that the lipid A part of LPS from the wild-type bacteria grown in 121 medium contains P-EtN and l-Ara4N, due to the induction of basS/R-induced transcription of genes whose products mediate such additions.
FIGURE 4.
Charge deconvoluted ESI FT-ICR mass spectra in negative ion mode depicting lipid A modifications of LPS from wild-type (wt) or its derivatives with deletions of late acyltransferases genes grown at 30 °C. Part of the negative ion mass spectra of the native LPS after unspecific fragmentation leading to cleavage of the labile lipid A-Kdo linkage are presented. Spectra of lipid A section from LPS extracted from wild-type grown in M9 (A) or 121 medium (B) and its derivatives carrying deletions of ΔlpxL (C), ΔlpxM (D), and Δ(lpxL lpxM lpxP) (E) all grown in 121 minimal medium. Only the mass peaks corresponding to the lipid A part are marked and substitutions with P-EtN and/or l-Ara4N are indicated.
Deletion of the lpxM gene, encoding myristoyltransferase, resulted in the synthesis of the pentaacylated lipid A species, characterized by a peak at 1587.0 Da. However, two additional peaks with 1710.0 and 1718.0 Da (Fig. 4D) were observed corresponding to the addition of P-EtN or l-Ara4N to the pentaacylated lipid A, respectively. Deletion of the lpxL gene, encoding lauroyltransferase, revealed tetra-, penta-, and hexaacylated lipid A (peaks with 1404.8, 1615.0, and 1851.2 Da, respectively) (Fig. 4C). The mass peak corresponding to hexaacylated lipid A can arise due to the addition of one secondary myristate and one secondary pamitoleate. Tetraacylated lipid A was also found to be modified by P-EtN, giving rise to a peak at 1527.8 Da. Similarly, pentaacylated lipid A (with mass of 1615.0 Da) was also modified by P-EtN and l-Ara4N (Fig. 4D). Thus, the pentaacylated lipid A species in ΔlpxL, as well as in ΔlpxM, seem to be modified in 121 medium growth conditions. However, under the same growth conditions tetraacylated lipid A was not found to be modified by l-Ara4N (Fig. 4, C and E). The tetraacylated lipid A in Δ(lpxL lpxM lpxP) was found to be modified with 1 or 2 P-EtN residues as revealed by mass peaks at 1527.8 and 1650.8 Da, respectively (Fig. 4E). However, no species containing l-Ara4N could be detected. These results help explain the poor translocational ability of tetraacylated LPS export and hence limited access to modification systems, like l-Ara4N addition, which operate upon translocation across the inner membrane.
Kdo-independent Acylation of Lipid IVA Precursor in ΔwaaA by Late Acyltransferases
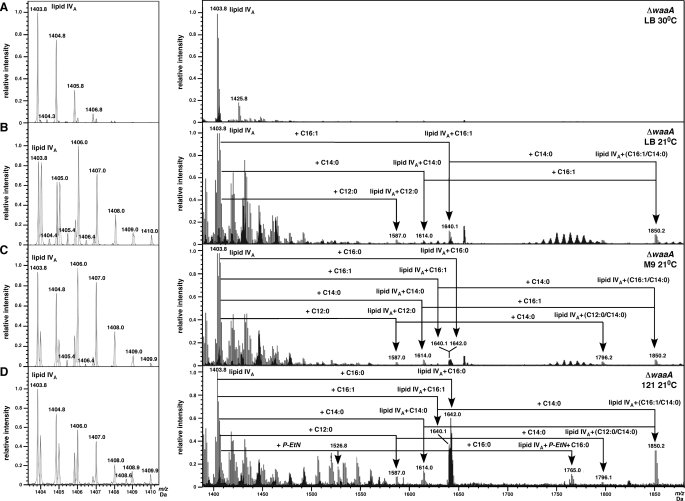
Lipid A composition of ΔwaaA mutants grown at either 21 or 30 °C in different growth medium was determined from the extracts containing a mixture of glycerophospholipids and lipid A. These growth conditions are permissive for suppressor-free ΔwaaA without requiring extra copies of the msbA gene. Due to the isolation procedure, phospholipids were not removed and are also present in the spectra. For example, the peak at m/z 1404.0 could be either a phospholipid dimer or a cardiolipin (Fig. 5). The non-deconvoluted mass spectrum of LPS from ΔwaaA grown at 30 °C in LB medium revealed an ion peak (M − H+)− at m/z 1403.8 Da, corresponding to the structure of tetraacylated 1,4′-bisphosphorylated lipid IVA precursor (Figs. 3 and 5A). Under these growth conditions no detectable penta- or hexaacylated derivatives of lipid IVA were observed. However, analysis of LPS/lipid A, extracted from ΔwaaA mutants grown at 21 °C, revealed a more complex composition of free lipid A. In addition to the ion peak, corresponding to tetraacylated lipid IVA, ion peaks corresponding to penta- and hexaacylated derivatives were also present, despite the lack of Kdo transferase. Thus, lipid A extracted from ΔwaaA, grown in LB medium at 21 °C, also revealed ion peak at m/z 1640.1 Da, corresponding to the predicted incorporation of the C16:1 secondary palmitoleate (Fig. 5B). Furthermore, ion peaks at m/z 1614.0 and 1850.2 Da indicated an addition of myristate to lipid IVA precursor and conversion of lipid IVA + C16:1, to the hexaacylated derivative of lipid IVA C16:1 + C14:0, respectively.
FIGURE 5.
ESI FT-ICR mass spectra in negative ion mode of LPS isolated from waaA deletion grown in LB medium at 30 °C (A) and 21 °C in LB medium (B), in M9 medium (C), and 121 medium (D). The left inset shows in detailed part of the spectra containing tetraacylated lipid IVA precursor. Mass numbers correspond to (M − H+)− ions. Addition of lauroyl, myristoyl, palmitoleate, palmitoyl, and P-EtN moieties to lipid IVA precursor and their derivatives are drawn schematically. These spectra also contain intensive signals for phospholipids. The ion peak at m/z 1404.0 in the inset could correspond to either phospholipid dimmer or a cardiolipin.
Analyses of lipid A extracted from ΔwaaA, grown at 21 °C in M9 or 121 medium, revealed the presence of four different ion peaks, corresponding to the synthesis of distinct pentaacylated lipid IVA derivatives, besides lipid IVA precursor (Fig. 5, C and D). Out of these, ion peaks at m/z 1640.1 and 1642.0 Da correspond to the addition of palmitoleate and the palmitoyl moiety to lipid IVA precursor. The other two ion peaks at m/z 1587.0 and 1615.0 Da represent the characteristic incorporation of secondary laurate and myristate groups to (R)-3-hydroxymyristate chains located at the 2′ and 3′ positions of the distal glucosoamine, respectively (Figs. 3 and 5).
The presence of palmitoyl-modified species in ΔwaaA, represented by ion peaks at m/z 1642.0 Da (Fig. 5, C and D), indicates that the lipid IVA precursor, synthesized at 21 °C in the minimal medium, is translocated to the outer membrane, because it requires the activity of the outer membrane PagP enzyme. Furthermore, the lipid IVA precursor was found to be modified by P-EtN, corresponding to ion peaks at m/z 1526.8 Da (lipid IVA + P-EtN) and 1765.0 Da (lipid IVA + P-EtN + C16:0) (Fig. 5D) in ΔwaaA grown in 121 medium. Like the hexaacylated lipid A represented by the ion peak at m/z 1850.2 Da, with the predicted composition of lipid IVA + C16:1 + C14:0, observed in LB grown ΔwaaA at 21 °C, also accumulated in 121 medium growth conditions (Fig. 5D). The Kdo-independent incorporation of some secondary laurate group in minimal medium at 21 °C also resulted in synthesis of the typical hexaacylated lipid A (ion peak at m/z 1796. 1 Da) with the predicted composition of lipid IVA + C16:1 + C14:0, due to further addition of the myristate acyl chain (Fig. 5, C and D). Thus, at low temperatures in slow-growth conditions in vivo lipid IVA can be used as the substrate for late acyltransferases independent of Kdo attachment. Of these, interesting is the incorporation of the LpxP-dependent C16:1 palmitoleate moiety, whose synthesis and incorporation is favored at low temperatures. This can be ascribed to a significant burst in the activity of RpoE in ΔwaaA, resulting in elevated RpoE-dependent lpxP transcription (Figs. 5 and 11).
Mass Spectrometry of Intact LPS from ΔwaaC and Its Isogenic Derivatives Lacking the Late Acyltransferases Grown in Non-modifying or Modifying Minimal Medium
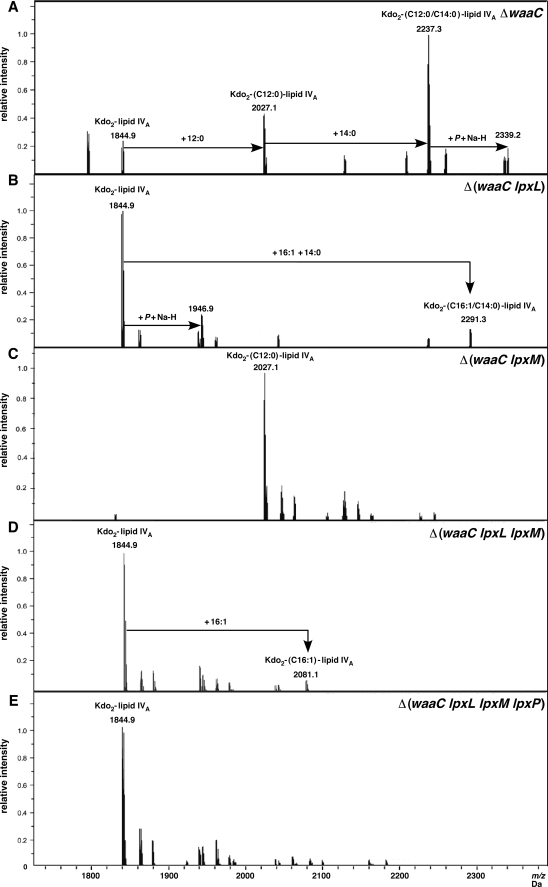
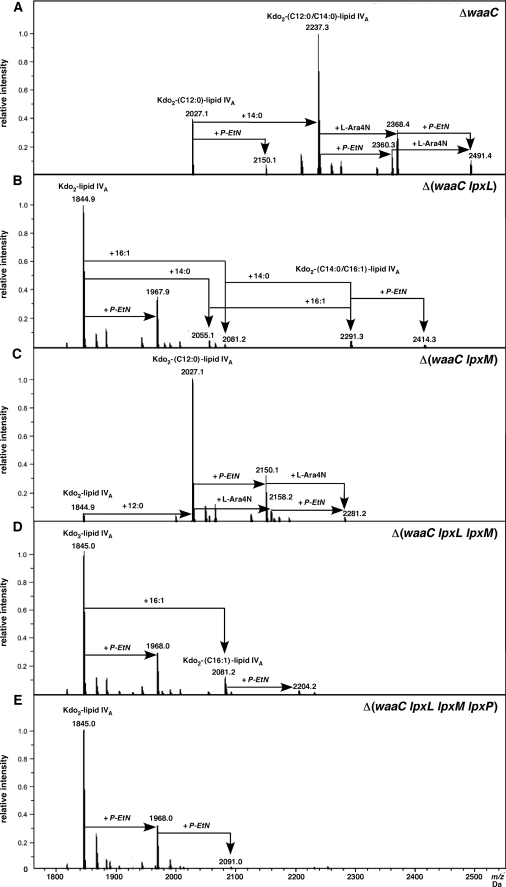
The composition of LPS of ΔwaaC in either phosphate-rich M9 medium (LPS non-modifying) or 121 medium (LPS modifying conditions) revealed the common mass peak of 2237.3 Da. This is in agreement with the composition of Kdo2-lipid Ahexa 1,4′-bisphosphate (Figs. 6 and 7A). The other common peak at 2027.1 Da could be interpreted as Kdo2-lauroyl-lipid IVA. However, LPS of ΔwaaC from 121 medium also revealed mass peaks of 2368.4 (addition of l-Ara4N) and 2491.4 Da (addition of l-Ara4N and P-EtN) (Fig. 7A). Modification with only P-EtN on Kdo2-lipid Ahexa or Kdo2-lipid Apenta was also revealed with peaks at 2360.3 and 2150.1 Da, respectively (Fig. 7A). Besides the differences in the presence of the P-EtN and l-Ara4N species on Kdo2-lipid Ahexa and Kdo2-lipid Apenta, the species corresponding to Kdo2-lipid IVA was conspicuously missing in the LPS of ΔwaaC mutants from 121 medium as compared with its presence in the M9 medium.
FIGURE 6.
Charge deconvoluted ESI FT-ICR mass spectra in negative ion mode of LPS isolated from waaC deletion (A) and its derivatives with deletions in genes encoding late acyltransferases (B–E) grown in permissive conditions at 23 °C in M9 minimal medium. The origin of the lipid A species by the addition of lauroyl, myristoyl, and palmitoleate are drawn schematically. Mass numbers are the monoisotopic masses of major peaks. Minor peaks, mostly corresponding to the substitution by phosphate or sodium adducts, are not labeled.
FIGURE 7.
Charge deconvoluted ES FT-ICR mass spectra in negative ion mode of LPS from waaC and its derivatives grown under LPS-modifying conditions in 121 medium. A, ΔwaaC, proposed structure of Kdo2-lipid A with assignment of P-EtN and l-Ara4N addition. B, Δ(waaC lpxL), the predicted composition of Kdo2-lipid IVA and Kdo2-lipid IVA with the addition of myristoyl and palmitoleate residues with P-EtN additions are shown. C, Δ(waaC lpxM), predicted structure of P-EtN and l-Ara4N modified Kdo2-myristoyl-lipid IVA. D, Δ(waaC lpxL lpxM) assignments of Kdo2-lipid IVA-P-EtN and Kdo2-palmitoleate-lipid IVA with P-EtN. E, Δ(waaC lpxL lpxM lpxP) with two P-EtN residues. Note the attachment of P-EtN in B–D can also occur on the outer Kdo.
LPS extracted from Δ(waaC lpxL), grown in either M9 or 121, revealed a common mass peak of 1844.9 Da, characteristic of Kdo2-lipid IVA (Figs. 6B and 7B). Consistent with the known substitution of palmitoleate (cisΔ9C16:1) to the same position of the missing laurate (C12:0) in lpxL mutants at or below 30 °C, a compound of 2291.3 Da corresponded to hexaacylated LPS, representing Kdo2-lipid IVA C16:1, C14:0 was observed. Such a hexaacylated compound was totally absent in Δ(waaC lpxL lpxM lpxP) (Figs. 6E and 7E). The LPS of Δ(waaC lpxL) grown in 121 medium also revealed peaks at 2055.1 and 2081.2 Da, corresponding to Kdo2-lipid IVA containing an additional myristate or palmitoleate, respectively. Interestingly, species representing modification by P-EtN on Kdo2-lipid IVA or Kdo2-lipid IVA C16:1, C14:0 forms in Δ(waaC lpxL) LPS were also observed with peaks at 1967.9 and 2414.3 Da, respectively (Fig. 7B). Thus, such structures seemed to be accessible and act as substrates for P-EtN modification.
LPS of Δ(waaC lpxM) revealed a common peak at 2027.1 Da, both under M9 and 121 growth conditions (Figs. 6C and 7C), corresponding to the expected structure of the Kdo2-lauroyl-lipid IVA. However, two additional peaks at 2150.1 and 2281.2 Da correspond to Kdo2-lauroyl-lipid IVA with the addition of P-EtN and P-EtN+l-Ara4N, respectively, were detected in LPS from Δ(waaC lpxM) grown in 121 medium. The substitution of Kdo2-myristoyl-lipid IVA by P-EtN and l-Ara4N indicates the normal transport of such pentaacylated LPS. These results support the observed lack of severe growth defects in Δ(waaC lpxM) as compared with Δ(waaC lpxL) derivatives (Table 2).
LPS extracted from Δ(waaC lpxL lpxM) confirmed the induction and incorporation of a secondary palmitoleate residue due to LpxP activity, resulting in the presence of both Kdo2-lipid IVA and Kdo2-palmitoleate-lipid IVA with peaks at 1845.0 and 2081.2 Da, respectively (Figs. 6D and 7D). Furthermore, both of these LPS species were also found to be modified by single P-EtN residues with peaks at 1968.0 and 2204.2 Da, respectively (Fig. 7D).
The authenticity of the suppressor-free strain Δ(waaC lpxL lpxM lpxP) was established by the presence of only the tetraacylated form with Kdo2-lipid IVA, corresponding to the main peak at 1845.0 Da, without any hexa- or pentaacylated derivatives. Remarkably, Δ(waaC lpxL lpxM lpxP) LPS also contains a single (1968.0 Da) as well as two P-EtN additions (2091.0 Da) (Fig. 7E). The presence of 2 P-EtN residues could be due to stronger induction of the RpoE-regulated eptB gene, as shown by our transcriptional induction of the rpoE regulon in such mutants (Fig. 10). Because 121 medium contains 2 mm CaCl2, the addition of P-EtN is more likely to occur on the outer Kdo residue, which was not observed in M9 medium, which contains only 0.1 mm CaCl2. However, a fraction of lipid A Δ(waaC lpxL lpxM lpxP) might contain two P-EtN on lipid A as seen in fragmentation spectra (data not shown). Overall, these analyses confirm the absence of l-Ara4N in the LPS of Δ(waaC lpxL), Δ(waaC lpxL lpxM lpxP), although P-EtN-modified derivatives could be observed (Fig. 7, B, D, and E). Thus, modification with l-Ara4N serves as a more stringent marker than by the P-EtN for poor translocation efficiency of tetraacylated lipid A.
FIGURE 10.
Induction of RpoE and CpxR/A pathways. Exponentially grown cultures of wild-type K-12 strain W3110 or its derivatives lacking waaC, lpxL, and lpxM alone and their deletion combinations with lpxP, carrying either rpoHP3-lacZ (A) or cpxP-lacZ fusion (B) on the chromosome were grown in M9 minimal medium under permissive growth conditions. Cultures were diluted to an A600 of ∼0.02 and grown at 30 °C in the same medium. Aliquots of samples were drawn and analyzed for β-galactosidase activity. Error bars represent S.E. of four independent measurements. C–E, cultures of wild-type, ΔwaaC, and its derivatives were grown at 23 °C in M9 medium to logarithmic phase, harvested by centrifugation, and equivalent amounts of cells were used to prepare whole cell lysates. Proteins were resolved by 12% SDS-PAGE and transferred to nitrocellulose membranes. Blots were probed with antiserum raised against HtrA (C), RseB (D), and PpiD (E). The genotype of each strain used is indicated.
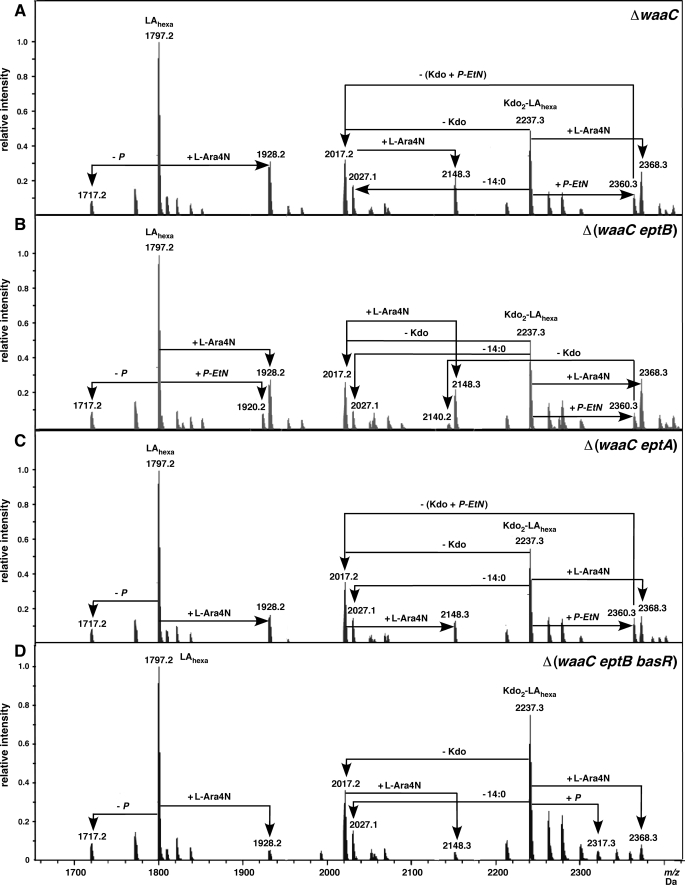
The Origin of P-EtN and l-Ara4N in ΔwaaC and Its Derivatives
We expected in ΔwaaC lipid A, as observed in the wild-type lipid A, both P-EtN and l-Ara4N substitution in 121 medium growth conditions. However, LPS of ΔwaaC contained mostly l-Ara4N substitution on the lipid A (Figs. 7 and 8). The P-EtN substitution in ΔwaaC, giving rise to peaks at 2360.3 and 2491.4 Da, seemed to arise mostly from the P-EtN addition to the Kdo region. We addressed the presence of P-EtN either on Kdo or lipid A, and l-Ara4N in waaC derivatives. Thus, LPS from strains Δ(waaC eptA), Δ(waaC eptB), and Δ(waaC basR eptB) were analyzed. LPS of ΔwaaC and its derivatives revealed a common occurrence of species with a peak at 1928.2 Da, corresponding to l-Ara4N substitution on lipid Ahexa (Fig. 8). In the LPS of Δ(waaC eptB) peaks at 1920.2 Da could arise due to P-EtN substitution on lipid A, consistent with the presumed function of EptA phosphoethanolamine transferase. This mass peak is missing in the LPS of Δ(waaC eptA), consistent with lack of EptA phosphoethanolamine transferase in this strain. Hence, these data suggest, depending on the presence of Ca2+ in growth medium (34) and genetic composition, P-EtN can be present on both lipid A and Kdo. However, P-EtN substitution on Kdo in ΔwaaC is favored due to increased σE-dependent transcription of the eptB gene in ΔwaaC. Quite surprising is the presence of a peak at 1928.2 Da in Δ(waaC eptB basR) LPS, which is ascribed to the l-Ara4N substitution. These results argue that genes, whose products are involved in l-Ara4N synthesis and transfer, can also be induced independent of basS/R regulon in 121 medium.
FIGURE 8.
Origin of P-EtN in waaC LPS. Charge deconvoluted ES FT-ICR mass spectra in negative ion mode of Re LPS derivatives with moderate unspecific fragmentation, showing the complete structure as well as lipid A fragments. Spectra of LPS from isogenic ΔwaaC (A), Δ(waaC eptB) (B), Δ(waaC eptA) (C), and Δ(waaC basR eptB) (D). All cultures were grown at 30 °C in 121 minimal medium. The mass numbers given are those of monoisotopic peaks. In all spectra the major peak with a mass of 1797.2 Da corresponds to the hexaacylated 1,4′-bisphosphate lipid A. In panel B the substitution of lipid A with both P-EtN and l-Ara4N is indicated. Fragmentation schemes along with the observed masses are indicated. Unlabeled peaks mostly correspond to Na+ adducts.
Analysis of unspecific fragmentation, leading to cleavage of labile lipid A Kdo linkage of these LPS, revealed the peak at 2140.2 Da in Δ(waaC eptB) (Fig. 8). This can arise from the parent molecule with a mass of 2360.3 Da (Kdo2-lipid Ahexa-P-EtN) by loss of 1 Kdo residue. These results support the presence of P-EtN on lipid A in Δ(waaC eptB), because phosphoethanolamine transferase EptB, which mediates P-EtN addition to the outer Kdo, is absent. This analyses revealed peaks at 2237.3 and 2368.9 Da, corresponding to Kdo2-lipid Ahexa and Kdo2-lipid Ahexa-l-Ara4N, respectively, in all 4 samples. Another peak at 2360.3 Da was present in all, except Δ(waaC basR eptB). This could arise from substitution with P-EtN to Kdo2-lipid Ahexa either on lipid A or Kdo in ΔwaaC. However, in Δ(waaC eptB) this species can only be attributed to the addition of P-EtN to lipid A. In contrast, this species in Δ(waaC eptA) represents the presence of P-EtN on the Kdo. Thus, depending upon the relative induction and activation of EptB in waaC derivatives, P-EtN can be found either on Kdo or the lipid A, or both.
LPS with Tetraacylated Lipid A Leads to Alteration of the Core Glycoform Composition in Modifying Medium
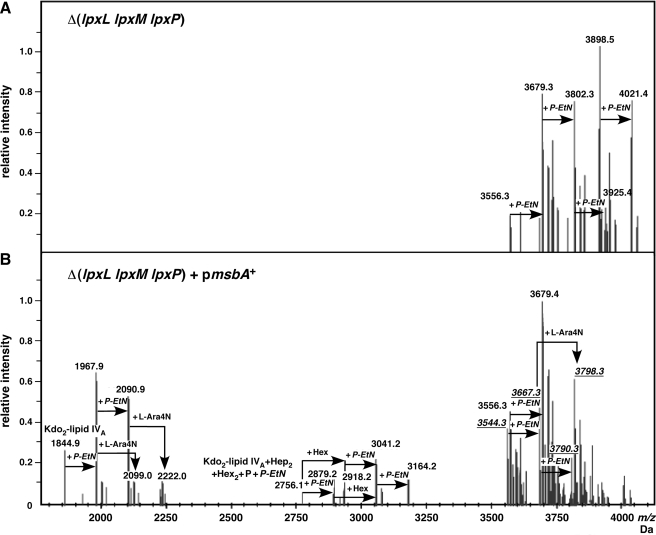
Wild-type E. coli K-12 is known to contain multiple glycoforms of the core oligosaccharide (35). One minor form, which contains 3 Kdo residues, has been described that is associated with truncation of the outer core after the Gal residue (35, 36). Here, we found that LPS of the Δ(lpxL lpxM lpxP) strain, deleted for all three late acyltransferases, hence synthesizing tetraacylated lipid A, also exhibited changes in the LPS core structure in 121 medium, but not in M9 medium (Fig. 9A). In 121 medium, LPS of Δ(lpxL lpxM lpxP) mutants possessed mostly derivatives of the glycoform containing 3 Kdo residues with rhamnose and up to 4 P-EtN residues, revealed by peaks at 3556.3, 3679.4, 3802.3, and 3925.4 Da (Fig. 9A). However, as expected from the lack of l-Ara4N in the lipid A part, no glycoform derivatives with substitution by l-Ara4N were found. The 4 P-EtN residues could arise from 2 substitutions in lipid A, one on Kdo and one on Hep I. Further additional compositional changes, which are common to 121 medium for the wild-type, were also observed in Δ(lpxL lpxM lpxP).
FIGURE 9.
Multicopy suppression leading to addition of l-Ara4N in the LPS of mutant synthesizing tetraacylated lipid A. Charge deconvoluted ES FT-ICR mass spectra in negative ion mode of native LPS of strain SR7781 Δ(lpxL lpxM lpxP) (A) and SR7781/pmsbA+ (B). Cultures were grown in 121 medium at 30 °C and LPS was extracted. Mass numbers refer to monoisotopic peaks with predicted composition, correlating with varying numbers of the substitution of P-EtN and/or l-Ara4N on the basic structure lipid IVA-3Kdo-Rha with 3Hep-3Hex are presented in bold. The mass numbers in italics and underlined in panel B refer to species corresponding to tetraacylated derivatives of glycoform II with 2 Kdo residues (35, 36), but with varying P-EtN and/or l-Ara4N substitutions. Both the spectra for panels A and B were recoded under the same conditions. In panel B accumulation of various intermediates with various substitutions following MsbA overexpression is depicted.
Because msbA overexpression suppressed the conditional lethality of Δ(lpxL lpxM lpxP), we also examined LPS composition of such derivatives. Under MsbA overproducing conditions, lipid A could be substituted by l-Ara4N as is evident from the peak at 2222.0 Da, corresponding to species with the presumed structure of [Kdo2-P-EtN-lipid IVA] with 1-residue P-EtN and l-Ara4N molecules (Fig. 9B). These results are in accordance with more efficient transport of tetraacylated lipid A under the conditions of MsbA overproduction. In the presence of msbA overexpression, LPS extracted from the Δ(lpxL lpxM lpxP) triple mutant contained glycoforms with 2 as well as 3 Kdo residues. Tetraacylated LPS glycoforms containing 2 Kdo residues corresponded to peaks at 3544.3, 3667.3, and 3790.4 Da, each with 1 P-EtN residue. The peak at 3798.3 Da corresponded to the incorporation of l-Ara4N, consistent with the data from the lipid A part. These glycoforms with 2 Kdo residues contained a complete core. However, the derivatives with 3 Kdo had characteristic truncation of the outer core (36). Some new glycoforms were also found with the predicted structure of lipid Atetra-Kdo2-Hep2-Hex2-P1 and 1 or 2 P-EtN with peaks at 2756.1 and 2879.2 Da, respectively. This seemed to act as precursor for the addition of 1 Hex (3041.2 Da) and 1 P-EtN (3164.2 Da) as well as its modification to the glycoform containing 3 Kdo residues and rhamnose. The restoration of synthesis of glycoforms with 2 Kdo and a complete core by msbA overexpression in the Δ(lpxL lpxM lpxP) mutant confirms the suppression of other growth defects.
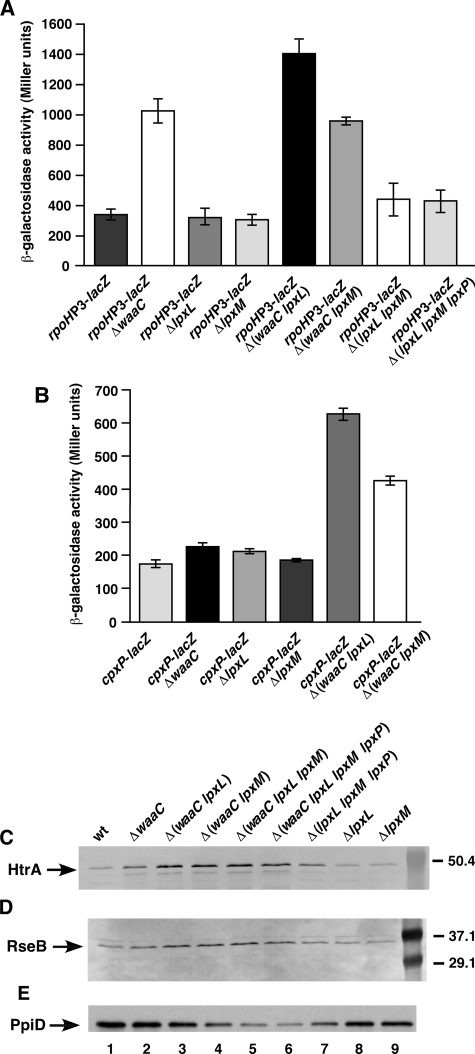
Synergistic Induction of the CpxR/A and σE Pathways of Extracytoplasmic Signal Transduction in waaC and Its Derivatives
The physiological response to the synthesis of Kdo2-lipid Ahexa, Kdo2-lipid Apenta, or Kdo2-lipid IVA was determined by following the effects on envelope stress responsive pathways under the control of RpoE and CpxR/A two-component system. The responses of these two principal pathways were assayed by measuring transcriptional activities of rpoHP3-lacZ and cpxP-lacZ promoter fusions. The rpoHP3 promoter activity reflects the impact on RpoE activity being solely transcribed by EσE RNA polymerase (26), although the cpxP promoter is under the transcriptional control of the CpxR/A two-component system of signal transduction (28). Significantly, ΔwaaC mutants showed constitutive 3-fold elevated levels of the rpoHP3 promoter even at the permissive temperature of 30 °C and in growth conditions that do not result in any known stress (M9 medium), indicating constitutive induction of the RpoE pathway (Fig. 10A). However, deletion of neither lpxL nor lpxM genes alone resulted in any significant increase in the activity of the rpoHP3 promoter, which is different from an earlier report (37) (Fig. 10A). Only a small increase in rpoHP3-lacZ activity (up to 30%) was observed in Δ(lpxL lpxM) or Δ(lpxL lpxM lpxP) mutants (Fig. 10A). Thus, underacylation of the lipid A in E. coli, synthesizing the complete LPS core, does not induce σE-dependent transcriptional response significantly, but LPS core truncation due to waaC deletion causes strong σE induction.
However, strains synthesizing tetraacylated lipid IVA with Re LPS Δ(waaC lpxL) exhibited a synergistic additive induction of RpoE activity. This was reflected by more than 4-fold elevated transcriptional activity of the rpoHP3 promoter (Fig. 10A). Curiously, the activity of the rpoHP3 promoter in Δ(waaC lpxM), synthesizing Kdo2-lipid Apenta, remained at the same level as that of the waaC mutant alone with a 3-fold increase as compared with the isogenic wild-type or the ΔlpxM mutant alone. Taken together, these findings explain the severe growth defects and constitutive elevated levels of RpoE activity in strains carrying Δ(waaC lpxL) deletion combinations synthesizing primarily Kdo2-lipid IVA derivatives.
Regarding Cpx two-component system activation, deletion of the waaC gene or lpxL, or lpxM, did not significantly induce cpxP promoter activity (Fig. 10B). Only a modest increase of about 30% in the basal level activity of the cpxP promoter was observed in ΔwaaC. However, a significant 3- to 4-fold induction was observed when either ΔlpxL or ΔlpxM were combined with the waaC deletion (Fig. 10B). The maximal increase again with cpxP promoter activation, like in case with the σE-dependent promoter, was observed in Δ(waaC lpxL) mutational combination. Overall these results revealed a strong induction of the CpxR/A pathway of signal transduction in mutants that synthesize Kdo2-lipid IVA or Kdo2-lipid Apenta, due to a combination of waaC deletion with either lpxL or lpxM deletions. However, individual lack of either lpxL or lpxM genes did not significantly induce either Cpx or RpoE pathways.
These results were further corroborated by Western blot analysis of two downstream indicator proteins (HtrA and RseB) of these two pathways of signal transduction. Transcription of the htrA gene is induced upon induction of both RpoE pathways as well as the CpxR/A two-component system (26). RseB serves as an indicator for the RpoE pathway alone, because its encoding gene is cotranscribed as the rpoE-rseArseBrseC operon, which is positively autoregulated by RpoE (38, 39). Even under permissive growth conditions of 23 °C in M9 medium, Δ(waaC lpxL), Δ(waaC lpxM), and Δ(waaC lpxL lpxM lpxP) showed a significant increase in the levels of HtrA and RseB proteins (Fig. 10, C and D), further strengthening our conclusions based on transcriptional assays with reporter promoter fusions. The higher levels of HtrA, as compared with that of RseB, are consistent with the synergistic control of htrA transcription by σE and CpxR/A pathways, leading to its additive induction. Increase in RseB only reflects activation of the RpoE operon. It should be noted that levels of RpoE operon are tightly regulated not only positively at the transcriptional level, but also negatively by RseA and RseB (38, 39).
Induction of Signal Transduction in ΔwaaA
Because the LPS of the ΔwaaA mutant comprised either mostly free lipid IVA precursor or lipid IVA derivatives with late secondary acyl chains incorporated, depending upon the growth medium and temperature, we analyzed its impact on envelope responsive RpoE and Cpx pathways. The ΔwaaA suppressor-free deletions were constructed in strains containing σE- and CpxR/A-regulated promoter fusions. Under the permissive growth conditions, both in M9 and LB medium, at either 21 or 30 °C, pronounced 3–6-fold induction of the σE-dependent rpoHP3 promoter was observed (Fig. 11A). Of significance is the more enhanced magnitude of RpoE induction (6-fold) in ΔwaaA grown in LB medium under the conditions when mostly lipid IVA precursor accumulated without any further acylation by late acyltransferases. However, at lower temperature (21 °C), when not only tetra- but also penta- and hexaacylated lipid IVA derivatives were observed, induction of the σE-dependent promoter rpoHP3 was somewhat dampened, although still more than 3-fold higher than the wild-type. Given the activity of the rpoHP3 promoter in the Δ(lpxL lpxM lpxP) mutant strain was hardly induced by 30–40% (Fig. 10) as compared with a 3–6-fold induction in ΔwaaA, further reinforcing the requirement of Kdo and glycosylation of lipid A derivatives, which is Kdo-dependent, for proper outer membrane function. This induction of RpoE in ΔwaaA mutants was further supported by the increased accumulation of products of RpoE regulon members such as HtrA (Fig. 11D).
Despite the increased complexity of lipid A composition in M9 medium-grown waaA mutants, as compared with that grown in LB medium at 21 °C due to the incorporation of C16:0 palmitoyl acyl chain, no further increase in σE activation was observed (Fig. 11A). Because no such lipid IVA modification by palmitoylation was observed in LB medium either at 21 or 30 °C (Fig. 5) indicates that σE activity is not induced by PagP-dependent lipid A palmitoylation. Thus, in waaA mutants RpoE activation results from accumulation of free lipid IVA derivatives lacking any glycosylation.
The response of CpxR/A-dependent cpxP promoter was even more pronounced in ΔwaaA mutants. The cpxP promoter was activated in LB medium, ∼8-fold at 30 °C and around 5-fold at 21 °C. In M9 medium ΔwaaA also exhibited ∼4.5–5-fold induction of the cpxP promoter at either 21 or 30 °C (Fig. 11B). This massive induction of the cpxP promoter in ΔwaaA mutants under permissive growth conditions is unprecedented in E. coli upon a single gene deletion. As a control, the amounts of the RpoH-regulated heat shock protein DnaK were not significantly altered (Fig. 11E), indicating that the cytoplasmic misfolding responsive RpoH pathway is not significantly affected in ΔwaaA. Overall these results show that ΔwaaA mutation, resulting in the synthesis of free lipid IVA precursor and its derivatives without any glycosylation, alters the primarily envelope responsive stress pathways and not the cytoplasmic protein misfolding pathway represented here by the lack of induction of the RpoH-dependent DnaK heat shock protein.
Requirement of SurA for the Viability of waaC Mutants
Deep-rough mutants are known to have impaired OM, including reduced amounts of OMP content (11, 13). To identify the limiting factors that confer this phenotype, we constructed a series of mutational combinations with null mutations in genes whose products are involved in maintaining the correct OMP content. These include mutations in the genes encoding periplasmic folding factors with a presumed role in OMP maturation (skp, surA, ppiD, and fkpA), one of the key components of the OMP biogenesis complex (yfgL), periplasmic protease (htrA), and OMP synthesis regulators like hfq and small non-coding RNA gene rybB. Among these, the combination Δ(surA waaC) turned out to be a uniquely lethal combination both at 30 and 37 °C in medium supporting slow (minimal medium) or rapid growth (LB medium). A double null chromosomal Δ(surA waaC) mutant could be constructed only if the surA gene was present in trans on a plasmid. To authenticate these results, two different null alleles of waaC and surA each were tested in reciprocal transductions (Table 3). These results provide an explanation for our previous isolation of the surA gene as a multicopy suppressor in restoring the proper amounts of OMP in a deep-rough rfaD mutant (13). In contrast, a surA null mutation could be transduced at a normal frequency in Δ(lpxL lpxM lpxP) (Table 3), arguing a more critical requirement of SurA in waaC mutants rather than in strains synthesizing tetraacylated LPS. Interestingly, Δ(htrA waaC) and Δ(yfgL waaC) could be constructed at both temperatures, although the colony size at 37 °C was significantly reduced. Deletion of yfgL results in compromised OMP, but does not seem to be as crucial as the SurA requirement under the conditions that effect LPS biogenesis. These results indicate a primary role for SurA in maturation of OMP and being a limiting factor in deep-rough ΔwaaC.
Because the transcription of surA, htrA, and skp genes, encoding various envelope-folding factors, is positively regulated by σE and/or CpxR/A systems, the amounts of their products also increased in ΔwaaC and its combinations with ΔlpxL (Fig. 10). To gain further insights in limiting factors, we looked at the amounts of other periplasmic folding factors that might become limiting in ΔwaaC and its derivatives. Western blot analyses of total cell extracts from cultures grown under permissive conditions was performed and probed for the amounts of various proteins. This analyses revealed that PpiD levels were reduced in deletion derivatives of the waaC gene, particularly in Δ(waaC lpxL lpxM) and Δ(waaC lpxL lpxM lpxP) (Fig. 10E, lanes 5 and 6). As controls ΔlpxL or ΔlpxM mutants exhibited PpiD levels comparable with the wild-type (Fig. 10E). These results are in agreement with in vitro binding of PpiD with peptide libraries derived from OMP.5 This reduction in PpiD amounts can partly explain the reduction in OMP amounts in Re mutants coupled with synthetic lethality of waaC surA. PpiD has also been recently shown to bind early nascent secretory precursors as they emerge from the Sec translocon (40).
DISCUSSION
Up to now, suppressor-free E. coli strains that have minimal LPS structure are either mutants that synthesize Re LPS, i.e. Kdo2 hexaacylated lipid A, or strains with tetraacylated lipid A having a complete LPS core. In this work, we constructed several Re mutants (ΔwaaC derivatives) that lack one or more of the late acyltransferases under slow-growth conditions (minimal medium at 23 °C) to avoid accumulation of extragenic suppressors. This led to the construction of viable suppressor-free Δ(waaC lpxL lpxM lpxP), which synthesize only Kdo2-lipid IVA LPS. To further identify the minimal LPS structure required for viability, we constructed strains deleted for the waaA gene, which encodes Kdo transferase, under the same slow-growth conditions. Such suppressor-free ΔwaaA deletion strains were viable up to 30 °C and did not require under such growth conditions additional copies of the lipid A transporter MsbA and were shown to have no suppressor mutations. The structure of LPS of such strains was confirmed by mass spectrometric analyses and shown to lack Kdo and at 30 °C primarily synthesized lipid IVA. Thus, acyl-oxyacyl residues within the lipid A, Kdo attachment to lipid A, as well as further core sugar substituents that are on Kdo, are not absolutely essential for growth under slow-growth conditions of minimal medium at low temperatures. This would be up to now the minimal LPS structure that does not require either extra copies of the msbA gene or any suppressors for sustaining E. coli viability. However, Δ(waaC lpxL lpxM lpxP) and ΔwaaA exhibited a very narrow growth range, with defects in cell division. Such suppressor-free strains could only be constructed in slow-growth conditions of minimal medium at low temperatures. Δ(waaC lpxL lpxM lpxP), even at the low temperature of 23 °C, did not grow at wild-type rates unless extra copies of the msbA gene were provided.
ΔwaaA mutants were predicted to synthesize only the free lipid IVA precursor species, without the incorporation of late secondary acyl chains. Indeed at 30 °C, ΔwaaA accumulated only lipid IVA precursors without any noticeable amounts of late secondary acyl chains. However, mass spectrometric analyses of LPS from ΔwaaA grown at 21 °C revealed incorporation of σE-dependent cold shock-inducible (12 °C), acyl carrier protein-dependent palmitoleate (C16:1) in the tetraacylated lipid IVA precursor. This modified lipid IVA derivative with palmitoleate served in vivo as a precursor to generate hexaacylated species with lipid IVA + palmitoleate and myristate derivatives. Presence and incorporation of the unsaturated C16:1 palmitoleate acyl chain indicates that Kdo attachment to lipid IVA may not be absolutely required for its incorporation. However, such a Kdo-independent incorporation of the C16:1 acyl chain seems to be restricted to conditions that lead to its substantial induction, due to transcriptional activation of σE regulon including the lpxP gene in ΔwaaA and LpxP activity at temperatures below 30 °C. These results support the concept of unsaturated fatty acid incorporation in homeoviscous adaptation (41). Because such a pentaacylated derivative of free lipid IVA precursor with palmitoleate in place of laurate also seemed to be a in vivo substrate for addition of myristate, argues that this late acylation step also does not require the absolute presence of Kdo at low temperatures. Furthermore, mass peaks corresponding to laurate C12:0 acyl chain incorporation were also observed in the free lipid IVA precursor at 21 °C, particularly in slow-growth conditions of minimal medium, but not at 30 °C. These results indicate that under slow-growth conditions at temperatures around 21 °C, late acylation can occur without attachment of Kdo to the lipid IVA precursor. Because we observed the lipid IVA precursor to be modified by P-EtN when LPS-modifying growth conditions of 121 medium were used, implies that lipid IVA can be translocated at basal levels of MsbA under slow-growth conditions to support OM biogenesis without accumulation of any suppressors. Thus, our suppressor-free waaA mutants should be ideal for further detailed studies on the in vivo and in vitro lipid A trafficking and kinetics of late acyltransferase-dependent incorporation of secondary acyl chains.
Growth of suppressor-free ΔwaaA in minimal medium at low temperature also caused additional lipid IVA modification. This is reflected by acylation with the secondary palmitate chain, which occurs at position 2 on the proximal glucosoamine. This lipid A modification occurs in the outer membrane by the PagP enzyme, which uses glycerophospholipids as its acyl donor (42). Because the PagP enzyme in E. coli is usually latent and is mostly activated upon breach of the OM permeability barrier indicates the increased presence of phospholipid patches in the OM of ΔwaaA when grown at low temperatures in minimal medium. However, ΔwaaA revealed palmitoyl modification only in minimal medium at low temperatures, but not in LB medium either at 21 or 30 °C indicating influence of growth conditions.
LPS of Δ(waaC lpxM) was found to be substituted by both P-EtN and l-Ara4N in 121 medium. However, under the same conditions, LPS of Δ(waaC lpxL), Δ(waaC lpxL lpxM), Δ(waaC lpxL lpxM lpxP), Δ(lpxL lpxM lpxP), and free lipid IVA derivatives from ΔwaaA did not show l-Ara4N addition, although P-EtN substitution occurred. This lack of l-Ara4N substitution in tetraacylated lipid A or lipid IVA precursors reflects the main growth defect due to their presumed retarded translocation to the outer membrane, because l-Ara4N addition occurs only in the periplasm, supports our results, and is consistent with the requirement of MsbA for translocation. These results provide evidence that our Kdo2-lipid IVA and waaA mutants do not contain any extragenic suppressors, which allowed rapid transport of the lipid A. However, l-Ara4N could be incorporated in the tetraacylated lipid A of Δ(lpxL lpxM lpxP) mutants when extra copies of the msbA gene were provided in trans. Taken together our results suggest that the addition of l-Ara4N define the more stringent requirement for rapid translocation of lipid IVA to the periplasm than P-EtN, because both modifications are presumed to occur in the periplasm.
Consistent with a requirement for lipid IVA translocation to be necessary for viability of E. coli, we showed that near wild-type growth could be restored when msbA is overexpressed in Δ(lpxL lpxM lpxP) mutants. Furthermore, even the growth of Δ(waaC lpxL lpxM lpxP) mutants could be partially restored in fast-growing conditions, when expression of the msbA gene was enhanced from an exogenous-controlled promoter. Interestingly, we could isolate plasmid-born suppressor mutations in the msbA gene, which showed a better suppression of Δ(waaC lpxL lpxM lpxP) mutants. Three such alterations, Asp-498 to Val-498, Ala-510 to Pro-510, and Leu-279 to Val-279 changes were characterized. Among these, Val-498 mutation in a single chromosomal copy was found to effectively restore growth of Δ(waaC lpxL lpxM lpxP) mutants even at 37 °C in both minimal and rich medium. The Asp-498 residue is part of the linker region to Walker box II and is highly conserved in a majority of MsbA homologues. The other two residues are also relatively well conserved. Leu-279 is located in EL loop 3, which connects TM5 and TM6 and is in close vicinity of residue Ala-270. Mutation of residue Ala-270 to Thr-270 in MsbA has been shown to cause a block in the lipid A transport (43). Ala-510 is located within the Walker box II (44). Further in vitro work will be required to understand their exact mechanism of suppression and properties of such MsbA variants.
Analyses of LPS of ΔwaaC, Δ(waaC eptB), Δ(waaC eptA), and Δ(waaC basR eptB) mutants revealed that P-EtN substituents can occur both on lipid A or Kdo in ΔwaaC from the structural point of view. However, ΔwaaC have elevated RpoE activity, hence the synthesis of EptB is favored, thus explaining the preferred Kdo-P-EtN in ΔwaaC. Curiously, lipid A of Δ(waaC basR eptB) mutants revealed the presence of l-Ara4N from LPS extracted from bacteria grown in 121 medium. Thus, in E. coli l-Ara4N modification of lipid A can occur in part without induction of the BasS/R two-component system and may involve an additional mechanism of regulation.
Interestingly, ΔwaaA mutants synthesizing free lipid IVA precursor and its unglycosylated derivatives and ΔwaaC derivatives with Kdo2-lipid IVA or Kdo2-lipid Apenta LPS, exhibited a dramatic basal level induction of envelope responsive signal transduction pathways, even under permissive growth conditions. The most striking was the induction of the CpxR/A two-component system, because its activation had not been linked to either truncations in LPS or underacylation of LPS. Absence of either LpxL or LpxM, or WaaC alone did not induce significantly the CpxR/A two-component system. However, either combination of Δ(waaC lpxL) or Δ(waaC lpxM) caused strong induction of this pathway of signal transduction. Among these, the most significant induction was observed in Δ(waaC lpxL), consistent with their more severe growth defects. Furthermore, ΔwaaC mutant alone also exhibited constitutively elevated levels of the RpoE pathway. However, a further additive effect in Δ(waaC lpxL) combinations, but not in Δ(waaC lpxM), was observed. This induction mirrors the growth defects of these corresponding mutants, known ability of pentaacylated lipid A translocation, and underlies the severe envelope defects in Kdo2-lipid IVA mutants. Consistent with the principal mechanism of OMP sensing by RpoE (27) and lack of any known major OMP content/folding defects, individual lpxL or lpxM mutants did not show elevated RpoE activity. However, strains lacking all the late acyltransferases, Δ(lpxL lpxM lpxP) mutants, revealed a mild 30 to 40% increase in RpoE activity, however, to a much lower extent than the waaC mutant, which have known major defects in OMP maturation.
ΔwaaA mutants grown at 30 °C in LB medium exhibited up to 8-fold strong induction of both RpoE and CpxR/A pathways, conditions under which only free lipid IVA precursor without any penta- and hexaacylated derivatives accumulated. Although at 21 °C, overall RpoE- and CpxR/A-regulated promoter activities increased, but the fold-induction was reduced from 8- to 5-fold and 6- to 3-fold for cpxP and rpoHP3 promoters, respectively, in comparison to their activity at 30 °C. This suppression can be due to incorporation of palmitoleate secondary acyl chain and further acylation by myristate, generating penta- and hexaacylated lipid IVA derivatives, which could confer some adaptive advantage. Still the 3- to 5-fold increase in these stress regulons, even at 21 °C growth conditions, is significantly more than that observed in Δ(lpxL lpxM lpxP) under similar growth conditions, indicating that accumulation of free lipid A derivatives and lack of glycosylation induces this hyperinduction of envelope stress response. Furthermore, conditions leading to PagP-dependent palmitoyl modification of lipid IVA derivatives did not induce σE activity more than that observed, when no such lipid A modification was observed in ΔwaaA. Overall, these results demonstrate that both RpoE and Cpx signal transduction pathways are highly induced upon accumulation of free lipid IVA precursors lacking any glycosylation due to waaA deletion.
An additional aspect of signal transduction induction observed in this study revealed that Δ(lpxL lpxM lpxP), synthesizing tetraacylated lipid A but with the intact waaC gene, exclusively synthesized the glycoform with 3 Kdo with a characteristic truncation in the outer core (35, 36). However, synthesis and accumulation of this glycoform was observed only when Δ(lpxL lpxM lpxP) were grown in 121 medium, but not in the phosphate-rich M9 medium. This indicates that modifications of tetraacylated lipid A also influence core structure alterations that promote incorporation of a third Kdo and biosynthesis. Furthermore, overexpression of MsbA was found to restore the accumulation of the usual glycoform with 2 Kdo residues as well as incorporation of l-Ara4N.
We also investigated the limiting factors in waaC mutants and potential reasons for defects in OMP maturation in deep-rough mutants. Construction of a series of waaC null combinations with null mutations in known periplasmic folding factors or those that regulate them, revealed that the waaC surA null combination is synthetically lethal, whereas Δ(waaC htrA), Δ(waaC skp), Δ(waaC ppiD), and Δ(waaC yfgL) were viable. These results support our earlier isolation of the surA gene as a multicopy suppressor of the rfaD mutant, which like waaC mutants synthesize Re LPS (13). Thus, one of the factors that is limiting and causing OMP defects could be due to the requirement of higher amounts of SurA, presumably to accelerate folding of porin monomers. Another reason for waaC surA synthetic lethality could result from constitutively alleviated levels of RpoE and the consequent increase of HtrA amounts in waaC mutants. Increased HtrA protein levels could cause higher turnover of intermediates of OMP folding, causing a decrease in overall amounts of OMP. Our results also suggest that known decreased OMP amounts could partly be due to a decrease in the levels of PpiD, a periplasmic OMP folding factor. Taken together, these results suggest both, SurA, PpiD for accelerating OMP folding/maturation, and MsbA for LPS export, are limiting in bacteria that synthesize Re LPS with tetraacylated lipid A. Such mutants should allow further insight into the mechanisms and factors involved in lipid A export and OMP maturation. waaA and waaC mutants could be further exploited to search for additional components that are required for OMP maturation. ΔwaaA and Δ(waaC lpxL lpxM lpxP) mutants should also be of potential use in identifying novel antibiotics. Given our results, Kdo-independent late acylation of the lipid IVA precursor in ΔwaaA mutants will be useful for further investigation of in vivo and in vitro studies to examine specificity of these acyltransferases. It will be of interest to obtain extragenic suppressors that restore wild-type growth and in converse to identify lethal combinations with other mutation(s) in ΔwaaA background.
Supplementary Material
Acknowledgments
The skillful technical assistance of I. von Cube, B. Kunz, and V. Susott is gratefully acknowledged. We also thank Thomas Scholzen for expert help with microscopy.
This work was supported in part by Deutsche Forsch ungs ge mein schaft Grant DFG LI 448/4 (to B. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
M. Liebscher, C. Schiene-Fischer, G. Fischer, and S. Raina, unpublished results.
- LPS
- lipopolysaccharide
- Kdo
- 3-deoxy-α-d-manno-oct-2-ulosonic acid
- OMP
- outer membrane proteins
- P-EtN
- phosphoethanolamine
- l-Ara4N
- 4-amino-4-deoxy-l-arabinose
- ESI FT-ICR MS
- electrospray ionization Fourier transform-ion cyclotron mass spectrometry
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Raetz C. R. H., Whitfield C. ( 2002) Annu. Rev. Biochem. 71, 635– 700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gronow S., Brade H. ( 2001) J. Endotoxin Res. 7, 3– 23 [PubMed] [Google Scholar]
- 3.Belunis C. J., Raetz C. R. H. ( 1992) J. Biol. Chem. 267, 9988– 9997 [PubMed] [Google Scholar]
- 4.Gronow S., Brabetz W., Brade H. ( 2000) Eur. J. Biochem. 267, 6602– 6611 [DOI] [PubMed] [Google Scholar]
- 5.Clementz T., Zhou Z., Raetz C. R. H. ( 1997) J. Biol. Chem. 272, 10353– 10360 [DOI] [PubMed] [Google Scholar]
- 6.Clementz T., Bednarski J. J., Raetz C. R. H. ( 1996) J. Biol. Chem. 271, 12095– 12102 [DOI] [PubMed] [Google Scholar]
- 7.Carty S. M., Sreekumar K. R., Raetz C. R. H. ( 1999) J. Biol. Chem. 274, 9677– 9685 [DOI] [PubMed] [Google Scholar]
- 8.Vorachek-Warren M. K., Ramirez S., Cotter R. J., Raetz C. R. H. ( 2002) J. Biol. Chem. 277, 14194– 14205 [DOI] [PubMed] [Google Scholar]
- 9.Karow M., Raina S., Georgopoulos C., Fayet O. ( 1991) Res. Microbiol. 142, 289– 294 [DOI] [PubMed] [Google Scholar]
- 10.Karow M., Fayet O., Cegielska A., Ziegelhoffer T., Georgopoulos C. ( 1991) J. Bacteriol. 173, 741– 750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnaitman C. A., Klena J. D. ( 1993) Microbiol. Rev. 57, 655– 682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raina S., Georgopoulos C. ( 1991) Nucleic Acids Res. 19, 3811– 3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Missiakas D., Betton J.-M., Raina S. ( 1996) Mol. Microbiol. 21, 871– 884 [DOI] [PubMed] [Google Scholar]
- 14.Meredith T. C., Aggarwal P., Mamat U., Lindner B., Woodard R. W. ( 2006) ACS Chem. Biol. 1, 33– 42 [DOI] [PubMed] [Google Scholar]
- 15.Mamat U., Meredith T. C., Aggarwal P., Kühl A., Kirchhoff P., Lindner B., Hanuszkiewicz A., Sun J., Holst O., Woodard R. W. ( 2007) Mol. Microbiol. 67, 633– 648 [DOI] [PubMed] [Google Scholar]
- 16.Torriani A. ( 1960) Biochim. Biophys. Acta 38, 460– 469 [DOI] [PubMed] [Google Scholar]
- 17.Missiakas D., Raina S. ( 1997) EMBO J. 16, 1670– 1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datsenko K. A., Wanner B. L. ( 2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640– 6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. ( 2006) Mol. Syst. Biol. 2, 0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galanos C., Lüderitz O., Westphal O. ( 1969) Eur. J. Biochem. 9, 245– 249 [DOI] [PubMed] [Google Scholar]
- 21.Kanipes M. I., Lin S., Cotter R. J., Raetz C. R. H. ( 2001) J. Biol. Chem. 276, 1156– 1163 [DOI] [PubMed] [Google Scholar]
- 22.Kuhn H.-M., Brade L., Appelmelk B. J., Kusumoto S., Rietschel E. T., Brade H. ( 1992) Infect. Immun. 60, 2201– 2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozalski A., Brade L., Kosma P., Appelmelk B. J., Krogmann C., Brade H. ( 1989) Infect. Immun. 57, 2645– 2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller-Loennies S., Gronow S., Brade L., MacKenzie R., Kosma P., Brade H. ( 2006) Glycobiology 16, 184– 196 [DOI] [PubMed] [Google Scholar]
- 25.Kondakova A., Lindner B. ( 2005) Eur. J. Mass. Spectrom. 11, 535– 546 [DOI] [PubMed] [Google Scholar]
- 26.Raina S., Missiakas D., Georgopoulos C. ( 1995) EMBO J. 14, 1043– 1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dartigalongue C., Raina S. ( 1998) EMBO J. 17, 3968– 3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dartigalongue C., Missiakas D., Raina S. ( 2001) J. Biol. Chem. 276, 20866– 20875 [DOI] [PubMed] [Google Scholar]
- 29.Zhou Z., White K. A., Polissi A., Georgopoulos C., Raetz C. R. H. ( 1998) J. Biol. Chem. 273, 12466– 12475 [DOI] [PubMed] [Google Scholar]
- 30.Hagiwara D., Yamashino T., Mizuno T. ( 2004) Biosci. Biotechnol. Biochem. 68, 1758– 1767 [DOI] [PubMed] [Google Scholar]
- 31.Tran A. X., Lester M. E., Stead C. M., Raetz C. R. H., Maskell D. J., McGrath S. C., Cotter R. J., Trent M. S. ( 2005) J. Biol. Chem. 280, 28186– 28194 [DOI] [PubMed] [Google Scholar]
- 32.Raetz C. R. H., Reynolds C. M., Trent M. S., Bishop R. E. ( 2007) Annu. Rev. Biochem. 76, 295– 329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doerrler W. T., Gibbons H. S., Raetz C. R. H. ( 2004) J. Biol. Chem. 279, 45102– 45109 [DOI] [PubMed] [Google Scholar]
- 34.Reynolds C. M., Kalb S. R., Cotter R. J., Raetz C. R. H. ( 2005) J. Biol. Chem. 280, 21202– 21211 [DOI] [PubMed] [Google Scholar]
- 35.Müller-Loennies S., Lindner B., Brade H. ( 2003) J. Biol. Chem. 278, 34090– 34101 [DOI] [PubMed] [Google Scholar]
- 36.Frirdich E., Lindner B., Holst O., Whitfield C. ( 2003) J. Bacteriol. 185, 1659– 1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam C., Missiakas D. ( 2005) Mol. Microbiol. 55, 1403– 1412 [DOI] [PubMed] [Google Scholar]
- 38.Missiakas D., Mayer M. P., Lemaire M., Georgopoulos C., Raina S. ( 1997) Mol. Microbiol. 24, 355– 371 [DOI] [PubMed] [Google Scholar]
- 39.De Las Peñas A., Connolly L., Gross C. A. ( 1997) Mol. Microbiol. 24, 373– 385 [DOI] [PubMed] [Google Scholar]
- 40.Antonoaea R., Fürst M., Nishiyama K.-i., Müller M. ( 2008) Biochemistry 47, 5649– 5656 [DOI] [PubMed] [Google Scholar]
- 41.Vorachek-Warren M. K., Carty S. M., Lin S., Cotter R. J., Raetz C. R. H. ( 2002) J. Biol. Chem. 277, 14186– 14193 [DOI] [PubMed] [Google Scholar]
- 42.Bishop R. E., Gibbons H. S., Guina T., Trent M. S., Miller S. I., Raetz C. R. H. ( 2000) EMBO J. 19, 5071– 5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doerrler W. T., Raetz C. R. H. ( 2002) J. Biol. Chem. 277, 36697– 36705 [DOI] [PubMed] [Google Scholar]
- 44.Ward A., Reyes C. L., Yu J., Roth C. B., Chang G. ( 2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19005– 19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brabetz W., Müller-Loennies S., Holst O., Brade H. ( 1997) Eur. J. Biochem. 247, 716– 724 [DOI] [PubMed] [Google Scholar]
- 46.Lerner C. G., Inouye M. ( 1990) Nucleic Acids Res. 18, 4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.