Abstract
A previous bioinformatics study identified a putative PY-NLS in the yeast transcription factor Tfg2p (Suel, K. E., Gu, H., and Chook, Y. M. (2008) PLoS Biol. 6, e137). In this study, we validate Tfg2p as a Kap104p substrate and examine the energetic organization of its PY-NLS. The Tfg2p PY-NLS can target a heterologous protein into the cell nucleus through interactions with Kap104p. Surprisingly, full-length Tfg2p is still localized to the nucleus of Kap104p temperature-sensitive cells and, similarly, Tfg2p with a mutated PY-NLS is nuclear in wild-type cells. Other Karyopherinβs (Kapβs) such as Kap108p and Kap120p also bind Tfg2p and may import it into the nucleus. More importantly, we demonstrate that Tfg2p is retained in the nucleus through DNA binding. Mutations of DNA binding residues relieve nuclear retention and unmask the role of Kap104p in Tfg2p nuclear import. More generally, steady-state localization of a nuclear protein is dictated by its nuclear import and export activities as well as its interactions in the nucleus and the cytoplasm.
The majority of nucleocytoplasmic transport is mediated by Karyopherinβ proteins (Kapβ,2 importins/exportins). There are 19 Kapβs in humans and 14 in yeast. Ten of the yeast Kapβs import substrates from the cytoplasm to the nucleus (2, 3). Import Kapβs recognize and bind substrates via nuclear localization signals (NLSs) for transport through the nuclear pore complex. Once inside the nucleus, import Kapβs bind the small GTPase RanGTP and release their substrates (4–7). Only a few substrates have been identified for each Kapβ, and many yeast proteins are imported by more than one karyopherin.
Each import Kapβ recognizes a different set of substrates with distinct NLSs. The best characterized NLS is the short, basic classic NLS (cNLS), which is recognized by the Kapα/Kapβ1 heterodimer (yeast Kap60p/Kap95p) (5). Monopartite cNLSs consist of a single cluster of basic residues with a consensus sequence of K(K/R)X(K/R), whereas bipartite NLSs have two clusters of basic residues separated by 10–12 amino acids (8–15). The cNLS is a relatively small well defined NLS with concentrated binding energy.
In contrast to the small monopartite cNLS, the PY-NLS recognized by Kapβ2 (Kap104p in yeast) is a larger linear signal that is quite diverse in sequence (1, 16). Structural and biochemical studies of Kapβ2 bound to the NLS of heterogeneous nuclear ribonucleoprotein A1 revealed that PY-NLSs should be 1) structurally disordered in the native substrate, 2) have overall basic charge, and 3) contain an N-terminal hydrophobic or basic motif and a C-terminal (R/K/H)X2–5PY motif (16). These rules are general among eukaryotes as the Saccharomyces cerevisiae homolog Kap104p recognizes PY-NLSs in its two substrates, Hrp1p and Nab2p (1, 17–21). However, unlike human Kapβ2, Kap104p recognizes only the basic but not the hydrophobic PY-NLS subclass (1).
Recent thermodynamic analyses of Kap104p-PY-NLS interactions revealed biophysical properties that govern binding affinity (1). The PY-NLS is a modular signal that contains at least three energetically significant binding epitopes. Epitope 1 is the N-terminal basic/hydrophobic motif, epitope 2 is the arginine, lysine, or histidine residue of the (R/K/H)X2–5PY motif, and epitope 3 is the PY motif. Hrp1p and Nab2p have an aromatic residue two residues N-terminal of the PY motif that also contributes to binding energy and is potentially an extension of epitope 3 (1). This residue has not been included in the consensus sequence due to the small numbers of validated substrates. Each linear epitope can also accommodate large sequence diversity, can contribute differently to overall binding energy in different NLSs, and are energetically quasi-independent.
A bioinformatics search identified several putative Kap104p substrates that contain PY-NLSs (1). Among these potential Kap104p substrates is Tfg2p, which is one of three subunits of the yeast general transcription factor TFIIF. TFIIF is involved in the initiation and elongation of RNA polymerase II-mediated gene transcription, and mutations in TFIIF can cause upstream shifts in start site utilization (22, 23). Additionally, TFIIF stimulates early phosphodiester bond formation and stabilizes a short DNA-RNA hybrid in the active center of RNA polymerase II (24). In cells, Tfg2p exists in a large assembly with RNA polymerase II proteins and two other TFIIF subunits, Tfg1p and Tfg3p (25). Tfg1p and Tfg2p are essential for cell viability and are homologous to human RAP74 and RAP30, respectively, whereas Tfg3p is not essential and has no known human homolog (26). Tfg2p contains an N-terminal Tfg1p interaction domain (residues 1–230) and a C-terminal winged helix DNA binding domain (residues 280–400) (see Fig. 1A) (26–29).
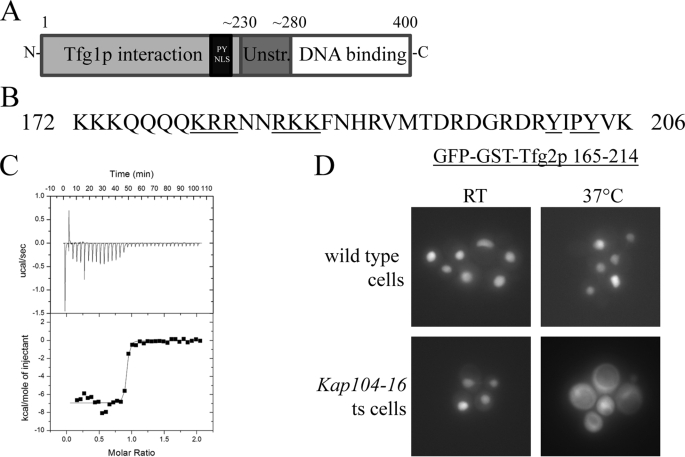
FIGURE 1.
Tfg2p has a PY-NLS that is recognized by Kap104p. A, domain organization of Tfg2p. B, sequence of the PY-NLS. Residues mutated in Fig. 2 are underlined. C, ITC profile of MBP-Tfg2p PY-NLS injected into Kap104p. D, wild-type and kap104–16 temperature-sensitive cells expressing GFP-GST-Tfg2p PY-NLS were analyzed by fluorescence microscopy at permissive (room temperature) and non-permissive (37 °C) temperatures.
The scarcity of Kap104p substrates hinders additional evaluation and refinement of the rules and physical properties that govern PY-NLS recognition. Here we validate Tfg2p as a substrate of Kap104p and examine the physical properties of its PY-NLS. We show that the PY-NLS is a functional targeting signal in cells, and its import is primarily mediated by Kap104p. However, full-length Tfg2p accumulates in the nucleus even in the absence of Kap104p-mediated nuclear import. When Kap104p import is compromised, other Kapβs such as Kap108p and Kap120p may import Tfg2p into the nucleus where it is retained through its DNA binding domain. Mutations of DNA binding residues alleviate nuclear retention and reveal Kap104p as the primary nuclear import pathway for Tfg2p.
EXPERIMENTAL PROCEDURES
Plasmids
Full-length Tfg2 was cloned by PCR from a S. cerevisiae genomic library (Novagen) and subcloned into the SpeI and SmaI sites of a modified pRS415 (CEN6, ARS, LEU2, and APR) shuttle vector containing an ADH1 promoter and a C-terminal GFP gene (30). The PY-NLS and other Tfg2 fragments were cloned into the pRS415 plasmid containing N-terminal GFP and GST genes using inserted BamHI and NotI restriction sites. GGSGG linkers were placed between the genes. The PY-NLS was also subcloned into the BamHI and NotI sites of the pGEX-Tev vector for recombinant protein expression in Escherichia coli. All point mutations were cloned using the QuikChange method (Stratagene) and confirmed by nucleotide sequencing. All Kapβ genes (except Kap104, which was a gift from J. Aitchison) were obtained by PCR from the S. cerevisiae genomic library and subcloned into pGEX-Tev vectors.
Cell Culture and Microscopy
BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) cells (31) harboring pRS415 plasmids were grown at 30 °C overnight in SC-leu media. Cells were then transferred to a 1.5% low melting agarose pad made with SC-leu media in a coverslip bottom dish (Willco from Ted Pella). Cells were visualized on an Olympus IX-81 inverted microscope at room temperature with a 60× phase objective with fluorescence capability (numerical aperture = 1.35). Images were acquired with a Hamamatsu ORCA-ER camera using Image-Pro Plus software (Media Cybernetics) and were also analyzed using Image Pro. All the cell images in individual figure panels were taken on the same day, and GFP signals in the panels were displayed using the same fluorescence scale. To obtain a nucleus:cytoplasm ratio, the mean fluorescence intensity was measured in the nucleus and cytoplasm using a 36-pixel box for at least 50 cells of each mutant.
Kap104–16 temperature-sensitive cells (a gift from J. Aitchison) were grown at room temperature in SC-leu-trp media overnight (20). Cells at the permissive temperature were analyzed as above. The remaining cells were shifted to 37 °C for 1 h and then transferred to a pre-warmed agarose pad. Cells were observed in a 37 °C temperature chamber and analyzed as above.
Protein Expression, Purification, and Binding Assays
GST-Kapβ proteins were expressed in E. coli BL21(DE3) cells and lysed using an EmulsiFlex-C5 homogenizer (Avestin). After centrifugation, the supernatant was applied to glutathione-Sepharose (Amersham Biosciences) and washed extensively, protein was eluted with 20 mm glutathione, and the GST tag was cleaved using Tev protease. The protein was further purified by ion-exchange chromatography (Amersham Biosciences). The final product was subjected to nickel affinity chromatography to remove excess Tev. GST fusions of Tfg2p PY-NLSs and Tfg2p full-length protein were expressed in E. coli BL21(DE3) cells and lysed by sonication. The protein was immobilized on glutathione-Sepharose for binding assays, which were preformed as in a previous study (1). For each assay 40 μg of recombinant Kapβ was added to ∼10 μg of GST-Tfg2p immobilized on 20 μl of glutathione-Sepharose.
Isothermal Titration Calorimetry
Affinity of MBP-Tfg2p PY-NLS binding to Kap104p was determined by ITC using a MicroCal Omega VP-ITC calorimeter. Proteins were dialyzed in 20 mm Tris, pH 7.5, 100 mm NaCl, 2 mm β-mercaptoethanol, and 10% glycerol. Thirty-five 8-μl injections of 210 μm MBP-Tfg2p PY-NLS were titrated into 23 μm Kap104p at 20 °C. Data were plotted and analyzed with a single site binding model using MicroCal Origin software (7.0).
RESULTS
Tfg2p Contains a PY-NLS Recognized by Kap104p
A bioinformatics search identified a putative PY-NLS in Tfg2p (1). In addition, a previous high throughput mass spectrometric protein complex identification study identified Tfg2p along with Nab2p, Hrp1p, and Ran as a binding partner of Kap104p (32). Residues 165–214 of Tfg2p match the PY-NLS consensus sequence and are located in a long loop in the Tfg1p interaction domain. Sequence alignment with RAP30, the human homolog of Tfg2p, suggests that this loop is an insertion in the yeast protein that is absent in RAP30 (supplemental Fig. S1, A and B). Epitope 1 of the PY-NLS would correspond to two stretches of basic residues, 179KRR181 and 184RKK186, whereas epitope 2 is Arg200 (Fig. 1B). Epitope 3 is located at 203PY204, with a predicted additional binding hotspot at Y201A. Epitope 1 of the Tfg2p NLS differs from the epitope 1 in Hrp1p and Nab2p, but epitope 3 in all three proteins ((Y/F)XPY) are almost identical.
We had shown recently that the predicted PY-NLS and full-length Tfg2p bound Kap104p and were dissociated by RanGTP in vitro (1). Tfg2p bound Kap104p with higher affinity than either Nab2p or Hrp1p. Tfg2p bound Kap104p with a dissociation constant or KD of <10 nm as measured by ITC, at least four times stronger than Nab2p or Hrp1p (Fig. 1C). The Tfg2p PY-NLS is able to dissociate Kap104p from immobilized GST-Nab2p NLS to the similar degree as RanGTP (supplemental Fig. S2). Although these data show that Tfg2p PY-NLS and Kap104p interact in vitro, it is not known if the PY-NLS-like sequence in Tfg2p is a functional NLS that is capable of targeting Tfg2p to the cell nucleus.
To determine if the Tfg2p PY-NLS is a functional Kap104p-dependent NLS in cells, a GFP-GST reporter was fused to the N terminus of Tfg2p PY-NLS. GST was added to make the reporter larger to prevent free diffusion into the nucleus. The fusion protein was transformed into wild-type and kap104–16 temperature-sensitive cells (20). At the permissive temperature, GFP-GST-Tfg2p PY-NLS is localized to the nucleus in both cell types (Fig. 1D). However, at the non-permissive temperature GFP-GST-Tfg2p PY-NLS mislocalized to the cytoplasm in the kap104–16 temperature-sensitive cells. Thus, the Tfg2p PY-NLS is a functional targeting signal that is transported by Kap104p into the nucleus.
Tfg2p PY-NLS Has a Strong Epitope 3
The three linear epitopes of PY-NLSs can contribute differently to total binding energy in different substrates (1). We performed site-directed mutagenesis of the Tfg2p PY-NLS to examine energetic contributions of the three linear epitopes to Kap104p binding. Recombinant Kap104p bound immobilized wild-type GST-Tfg2p PY-NLS stoichiometrically (Fig. 2A), and ITC data showed a 1:1 Kap104p:Tfg2p PY-NLS binding ratio (Fig. 1C). Alanine mutations within either stretches of basic residues, 179KRR181 or 184RKK186, had no visible effect on Kap104p binding. Mutagenesis of Tyr201 to alanine resulted in a modest decrease in binding, whereas the 203PY204/AA mutant showed a large decrease in Kap104p binding (Fig. 2A).
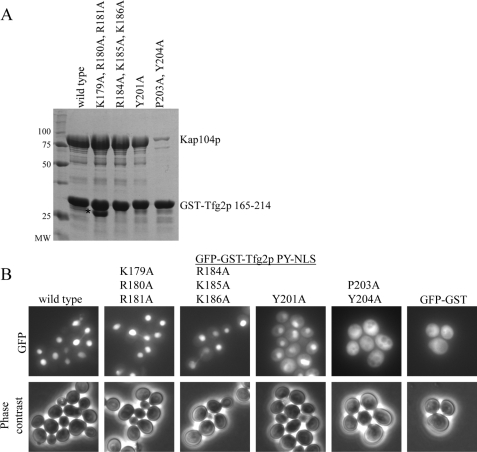
FIGURE 2.
Tfg2p has a strong epitope 3. A, binding assays of recombinant Kap104p with immobilized wild-type or mutant Tfg2p PY-NLS. Proteins were visualized by Coomassie staining. The asterisk labels a degradation product. B, cellular localization of GFP-GST-Tfg2 PY-NLS mutants in wild-type yeast cells analyzed by fluorescence microscopy and phase contrast. GFP-GST is included as a control.
GFP-GST-Tfg2 PY-NLS fusion proteins with the above point mutations were visualized in wild-type cells to examine the effects of the mutations on cellular localization. Wild-type Tfg2p PY-NLS is localized to the nucleus with a nucleus to cytoplasm ratio (N:C) of 4:1 (Fig. 2B). K179A,R180A,R181A and R184A,K185A,K186A mutants are predominantly nuclear with N:C ratios of 3.7:1 and 3.2:1, respectively (S.E. < 0.14 for each mutant). These data are in agreement with the in vitro data that showed no decrease in Kap104p binding. These mutants also confirm that these basic residues do not form a bipartite cNLS within the PY-NLS. Mutations in epitope 3 that decreased Kap104p binding also caused protein mislocalization in cells. GFP-GST-Tfg2p Y201A showed significant cytoplasmic localization, with an N:C ratio of 1.9:1, and the fusion protein harboring 203PY204/AA mutations is predominantly cytoplasmic (Fig. 2B). Therefore, the Tfg2p NLS has an energetically strong epitope 3 with a PY motif that is necessary for steady-state nuclear localization.
Only Kap104p Binding Is Affected by Mutations in the PY Motif
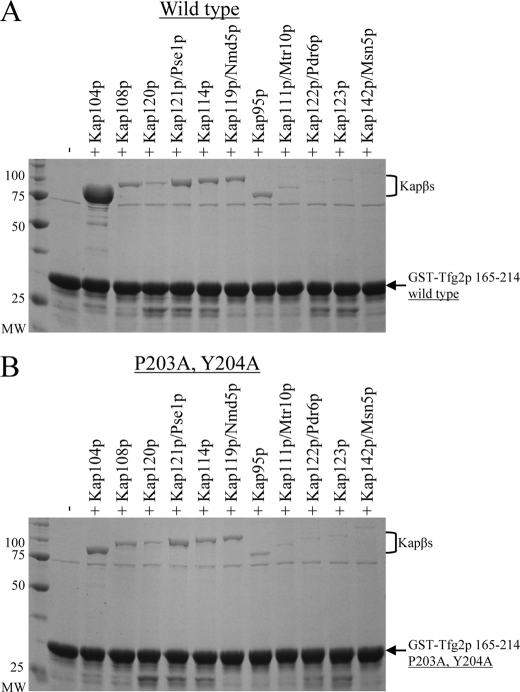
The 203PY204/AA mutations mislocalized Tfg2p PY- NLS to the cytoplasm. We wanted to see if this effect was solely due to a decrease in Kap104p binding or if the PY motif may be utilized by another import Kapβ. We cloned and purified all 10 import Kapβs and the bidirectional transporter Msn5 for in vitro binding assays (33). Kap104p is the only Kapβ that bound stoichiometrically to wild-type PY-NLS (Fig. 3A). Kap108p, Kap121p, Kap114p, Kap119p, and Kap95p bound the PY-NLS more weakly. Mutagenesis of the PY motif only caused a decrease in Kap104p binding and did not affect other Kapβ binding (Fig. 3B). Other karyopherins weakly recognize the PY-NLS, but the PY motif is only necessary for recognition by Kap104p. Therefore, mislocalization of Tfg2p PY-NLS caused by the 203PY204/AA mutations can be attributed solely to a decrease in Kap104p binding and nuclear import.
FIGURE 3.
Only Kap104p binding is decreased by mutations in the PY motif. A, binding assay of recombinant Kapβs with immobilized wild-type Tfg2p PY-NLS. B, same assay as in A, except with 203PY204/AA mutation in the PY-NLS. Proteins were visualized by Coomassie staining.
Nuclear Import of Full-length Tfg2 Appears Unaffected by Loss of Kap104p Recognition
Although the PY-NLS of Tfg2p is localized to the nucleus, the localization of full-length Tfg2p is unknown. Full-length Tfg2p was tagged at the C terminus with GFP, and its localization was examined in wild-type yeast cells. Wild-type Tfg2p is nuclear at steady state (Fig. 4A). Surprisingly, the 203PY204/AA mutations did not affect the localization of Tfg2p-GFP (Fig. 4A), suggesting that nuclear import of Tfg2p may not be entirely dependent on Kap104p.
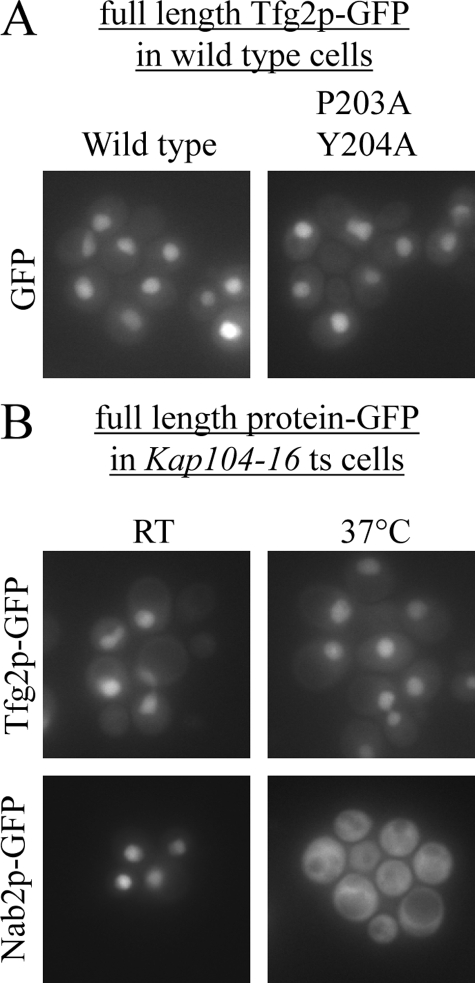
FIGURE 4.
Nuclear import of full-length Tfg2p is not dependent on Kap104p. A, wild-type yeast cells expressing either wild-type full-length Tfg2p-GFP fusion protein or 203PY204/AA mutant fusion protein were analyzed by fluorescence microscopy. B, Kap104–16 temperature-sensitive cells expressing either Tfg2p-GFP or Nab2p-GFP were analyzed by fluorescence microscopy at permissive and non-permissive temperatures.
To examine if nuclear accumulation of full-length Tfg2p is dependent on Kap104p, Tfg2p-GFP was transformed into kap104–16 temperature-sensitive cells. At both permissive and non-permissive temperatures, Tfg2p-GFP is nuclear (Fig. 4B). Nab2p-GFP was used as a positive control to ensure that the kap104–16 temperature-sensitive cells had not reverted. In the same experiment, Nab2p-GFP is mislocalized to the cytoplasm at non-permissive temperature. Nuclear import and accumulation of full-length Tfg2p is therefore not solely dependent on Kap104p.
Other Karyopherins Bind Tfg2p
We examined if other Kapβs can also bind and import Tfg2p into the nucleus. The best characterized import pathway is the Kap95p/Kap60p (Kapβ1/Kapα) pathway utilizing cNLS recognition by Kap60p. Examination of the Tfg2p protein sequence revealed a putative monopartite cNLS (356KKLK359) at the C terminus of Tfg2p (supplemental Fig. S3A). Immobilized GST-Tfg2p-(341–374)-bound Kap60p and alanine mutagenesis of the three lysine residues abolished Kap60p binding (supplemental Fig. S3B). We made these mutations in the background of the 203PY204/AA mutations in full-length Tfg2p-GFP and examined its localization in wild-type cells. The mutant Tfg2p (356KK357/AA, K359A, 203PY204/AA)-GFP fusion protein accumulated in the nucleus (supplemental Fig. S3C). Additionally, GFP-GST-Tfg2p 341- 374 was cytoplasmic (data not shown) suggesting that these residues (356KKLK359) are not a functional cNLS in cells.
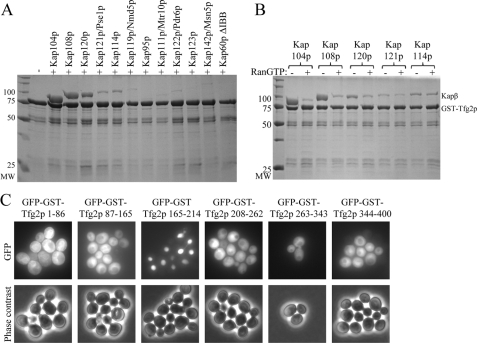
All ten import Karyopherin βs and the bidirectional transporter Msn5p were used in binding assays with immobilized full-length Tfg2p. Kap108p and Kap120p bound Tfg2p, whereas Kap121p and Kap114p bound Tfg2p weakly (Fig. 5A). None of the karyopherins bound GST (supplemental Fig. S4). We used RanGTP to test specificity of the interactions. RanGTP effectively dissociated Kap104p and Kap108p from Tfg2p, and it also dissociated a small fraction of Kap120p from Tfg2p. In contrast, RanGTP did not affect interactions of Tfg2p with Kap121p or Kap114p, indicating that these interactions are not specific (Fig. 5B). Therefore, among the additional ten Karyopherin βs tested, it appears that only Kap108p and Kap120p can function as alternative nuclear import factors for Tfg2p.
FIGURE 5.
Other karyopherins bind Tfg2p, but they do not import it into the nucleus. A, binding assay of recombinant karyopherins with immobilized full-length Tfg2p. Proteins were visualized by Coomassie stain. B, binding assay of recombinant karyopherins with immobilized full-length Tfg2p in the presence and absence of RanGTP. Proteins were visualized by Coomassie stain. C, GFP-GST-Tfg2p fragments were expressed in wild-type yeast cells and analyzed by fluorescence microscopy and phase contrast. Only the PY-NLS (165–214) is nuclear.
The weak interactions observed between Kap108p, Kap120p, and the Tfg2p PY-NLS (Fig. 3A) suggest that the two Kapβs may recognize different NLSs in Tfg2p. We cloned GFP-GST fusions of several Tfg2p fragments and visualized their localization in wild-type yeast cells (Fig. 5C). These fragments, included large N- and C-terminal fragments of Tfg2p, which do not contain the PY-NLS (1–165 and 208–400, data not shown) and smaller fragments that span entire secondary structure elements of the protein (based on alignment with RAP30 (supplemental Fig. S1A)). All of the fragments were cytoplasmic at steady state, suggesting that, although Kap108p and Kap120p bind Tfg2p, they do not import it into the nucleus at discernable rates. However, it remains a possibility that the Tfg2p fragments are misfolded or that NLSs recognized by other Kapβs partially overlap with the PY-NLS.
To examine the roles of other Kapβs in the nuclear import of Tfg2p we examined the localization of wild-type Tfg2p-GFP and the 203PY204/AA mutant in Δkap108, Δkap114, Δkap120, and kap121–34 temperature-sensitive strains (data not shown). Both wild-type and mutant Tfg2p-GFP were nuclear in every strain tested implying that these individual Kapβs are not necessary for import of Tfg2p in the absence of Kap104p-mediated import. These data suggest that more than two Kapβs are involved in importing Tfg2p into the nucleus or perhaps Tfg2p is imported via an indirect mechanism.
The DNA Binding Domain of Tfg2p Is Required for Its Nuclear Accumulation
Tfg2p binds Tfg1p and DNA through its Tfg1p interaction domain (residues 1–230) and DNA binding domain (residues 280–400), respectively (Fig. 1A). Tfg2p also interacts with at least seven polymerase subunits (25), but details of these interactions are still unclear. It is possible that nuclear accumulation of Tfg2p is mediated through its interaction with Tfg1p, DNA, or polymerase subunits.
Deletion of RAP30 residues 15–30 abolished its interaction with the Tfg1p human homolog (34). We deleted the first 86 residues of Tfg2p (corresponding to the first 36 residues of RAP30) and visualized its localization in cells. Both the wild-type and PY mutant of Tfg2p-(87–400)-GFP were nuclear (Fig. 6A). A larger N-terminal truncation of Tfg2p also had no effect on localization (Fig. 6A) implying that interaction with Tfg1p is not required for Tfg2p import.
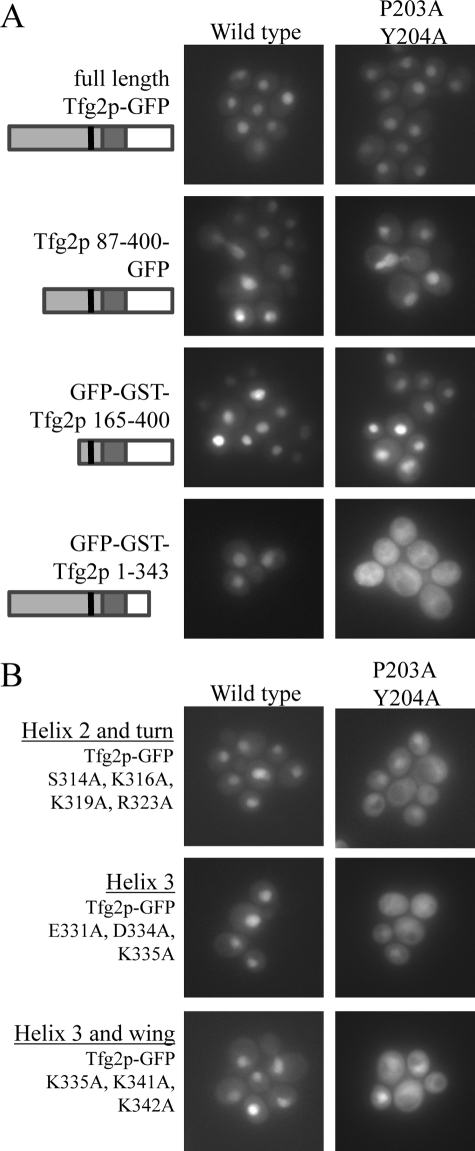
FIGURE 6.
The DNA binding domain of Tfg2p is required for nuclear accumulation. A, wild-type cells expressing N-terminal and C-terminal truncations of Tfg2p tagged with either GFP or GFP-GST as indicated in the figure. Localization of proteins with 203PY204/AA mutations are in the right panels and localization of wild-type proteins are in the left panels. Cells were analyzed by fluorescence microscopy. A small domain structure is under each label in corresponding shades of gray as to Fig. 1A. B, wild-type cells expressing GFP-tagged full-length Tfg2p harboring point mutations in the DNA binding domain in either a wild-type background or 203PY204/AA background. Cells were analyzed by fluorescence microscopy.
To examine the role of DNA binding in Tfg2p nuclear accumulation, we made C-terminal truncations of Tfg2p-(1–214, 1–262, and 1–343) to disrupt its DNA binding domain and test localization of the mutants in the absence and presence of the 203PY204/AA mutations. Although DNA binding domain mutants and the PY mutant of full-length Tfg2p were nuclear, combined DNA binding domain and PY mutations mislocalized the proteins to the cytoplasm (Fig. 6A). Even a truncation of only 56 residues in Tfg2p-(1–343) had a dramatic effect on localization. These data show that the DNA binding domain of Tfg2p is necessary for nuclear accumulation.
Tfg2 Is Sequestered in the Nucleus via DNA Binding
The DNA binding domains of Tfg2p and RAP30 share 80% sequence similarity and the NMR structure of the RAP30 DBD places it in the winged helix turn helix family (29). The domain has three N-terminal α-helices (helix 1–3) followed by a 2-strand β-sheet (supplemental Fig. S5). A 5-residue loop between two strands is usually referred to as the “wing” (29). The DNA-binding surface of RAP30 was previously mapped by DNA titration and 15N-1H correlation NMR spectroscopy (29). We used the RAP30 data to mutate DNA binding residues in the highly homologous Tfg2p (supplemental Fig. S1A). We made four site-specific DBD mutants of Tfg2p: 1) the helix 1-Tfg2p mutant with residues Arg293, Lys296, and Lys297 on one side of helix 1 mutated to alanines, 2) the helix 2-Tfg2p mutant with residues Ser314, Lys316, Lys319, and Arg323 of helix 2 mutated, 3) the helix 3-Tfg2p mutant with residues Glu331, Asp334, and Lys335 in helix 3 mutated, and 4) the helix 3-wing-Tfg2p mutant with residues Lys335 of helix 3, Lys341 and Lys342 of the wing mutated. The Tfg2p DBD mutations had varying effects on DNA binding as analyzed by size exclusion chromatography (supplemental Fig. S6). Mutations in helix 1 caused a modest decrease in DNA·DBD complex formation, whereas mutations in helix 3 and wing and helix 2 and turn significantly decreased DNA binding.
We analyzed cellular localization of these mutants in a wild-type NLS or 203PY204/AA background (Fig. 6B). Mutations within helices 2 and 3 and the wing did not affect nuclear localization in the wild-type NLS background. However, when these DNA-binding mutations were coupled with the 203PY204/AA mutations, the Tfg2p mutants were mislocalized to the cytoplasm (Fig. 6B). Mutations within helix α1 had no effect on localization (data not shown). Cellular localization of DBD mutants correlate well with the degree of DNA binding disruption (supplemental Fig. S6). These mutations had no effect on other Kapβ interactions (supplemental Fig. S7). These data suggest that sequestration of Tfg2p in the nucleus through DNA binding accounts for its observed nuclear accumulation even when its primary nuclear import pathway Kap104p is compromised.
DISCUSSION
We show that Kap104p imports Tfg2p into the nucleus. Tfg2p is the first PY-NLS-bearing substrate identified through a bioinformatics search to be validated in vivo. Many human PY-NLSs were validated as NLSs in vivo prior to their identification as PY-NLSs (1). We show that the PY-NLS of Tfg2p is sufficient to target GFP-GST to the nucleus of wild type but not Kap104p temperature-sensitive cells, and mutation of the PY motif mislocalized the NLS to the cytoplasm. Binding assays confirmed that mislocalization of mutant PY-NLS was solely due to loss of recognition by Kap104p and not other Kapβs. However, disruption of Kap104p-mediated import, either in Kap104p temperature-sensitive cells or by mutagenesis of the PY motif, did not mislocalize the full-length protein. Instead, Tfg2p was mislocalized to the cytoplasm only when both DNA binding residues and the PY motif were mutated. It appears that Tfg2p is still imported into the nucleus probably via a lower affinity and slower backup method when Kap104p-mediated transport is disrupted. Tfg2p can bind other Kapβs. Its PY-NLS interacts weakly with Kap108p, Kap121p, Kap114p, Kap119p, and Kap95p, and it also binds Kap108p and Kap120p through unidentified NLSs. Tfg2p may be imported into the nucleus by any of these karyopherins, albeit inefficiently. However, once full-length Tfg2p gets in the nucleus, it binds DNA and other members of the PIC and is retained there. Importance of the Kap104p import pathway in targeting Tfg2p to the nucleus is revealed only when DNA binding and thus nuclear retention is disrupted.
Identification of Tfg2p as the third Kap104p substrate reinforces the physical properties we have previously found to govern PY-NLS recognition by Kap104p. The PY-NLS is a modular signal with three epitopes, each of which contributes differently to total binding energy in different NLSs. Like another Kap104p substrate, Hrp1p, Tfg2p has a strong epitope 3 and a weak epitope 1. Similarly, its PY dipeptide is necessary for Kap104p binding and nuclear import by Kap104p (1, 18). In contrast, another Kap104p substrate, Nab2p, has a strong epitope 1 and a medium strength epitope 3. Mutagenesis of Nab2p's epitope 3 only slightly mislocalized the protein to the cytoplasm (1). Despite different distribution of epitope strengths, Tfg2p, Hrp1p, and Nab2p all bind Kap104p with similar affinity (10–37 nm), consistent with the concept that combinatorial mixing of different epitope strengths can result in sequentially diverse but still functional NLSs.
We had previously predicted an additional binding hotspot at the aromatic residue two amino acids N-terminal of the PY motif. Alanine mutation of this residue in both Hrp1p and Nab2p decreased Kap104p binding affinity (1). Similarly, the Y201A mutant of Tfg2p PY-NLS showed decreased Kap104p binding and nuclear accumulation, suggesting that this residue is a binding hotspot. Because the positions two residues N-terminal of the PY motif in all three Kap104p substrates and several Kapβ2 substrates are occupied by aromatic residues, this sequence pattern could be used as an additional restraint for future substrate searches (1, 16).
Damelin et al. (35) suggested four criteria to verify a putative NLS as a functional targeting signal recognized by a specific Kapβ pathway (35). First, the NLS and full-length substrate must bind the Kapβ directly and be dissociated by RanGTP. Second, the sequence must be necessary for import such that deletion or mutagenesis of the sequence should mislocalize the substrate. Third, the NLS must be sufficient to target another protein into the nucleus. Finally, mutations in the transport pathway should inhibit nuclear import of the substrate. These criteria work well under simple circumstances but validation according to the criteria can often be thwarted by complexities in the cell. In the case of Tfg2p, it was straightforward to invalidate the cNLS as a functional NLS, but it was significantly more difficult to validate the PY-NLS as a functional targeting sequence. Cellular functions of Tfg2p as a transcription factor and as a nuclear resident rather than a shuttling protein dictated additional experiments to verify it as a Kap104p substrate.
Verification of NLSs in other proteins has also been complicated by cellular conditions and exemplifies the many caveats of the above validation criteria. Import substrates may be transported into the nucleus in a piggyback manner through interactions with another substrate, be imported by more than one karyopherin, be retained in the nucleus, or imported only when modified appropriately. For example, although Kap121p imports Nop1p into the nucleus, Leslie et al. (36) did not see mislocalization in a Kap121p temperature-sensitive strain, because Nop1p is retained in the nucleolus. They had to first inactivate Kap121p and then turn on Nop1p expression using a galactose-inducible promoter to see mislocalization of Nop1p (36). Many yeast proteins are imported into the nucleus by more than one mechanism. Leslie et al. (36) also showed that Sof1p can be imported directly by Kap121p or through a piggyback mechanism where it binds Nop1p. Additionally, many proteins such as histones and ribosomal proteins are imported into the nucleus by more than one Kapβ (37–39). Post-translational modifications can also affect protein localization, making it more difficult to determine how they are imported into the nucleus. The criteria outlined by Damelin et al. are useful guidelines and can be modified according to cellular functions of individual substrates.
The steady-state localization of a nuclear protein is not solely dictated by the presence or absence of functional nuclear import or export signals. Strengths of the respective targeting signals and strengths of interactions with other binding partners in either the nucleus or the cytoplasm (nuclear or cytoplasmic retention) all contribute to protein localization. Retention in cellular compartments may result from signal masking, anchorage to cellular structures, or promotion of degradation (40). Additionally, localization is often altered in response to changes in the extra- or intracellular environment such as resulting from stimuli, stress, and variations in the cell cycle or developmental period (41, 42). Here, we show the example of Tfg2p whose nuclear steady-state localization is a result of a combination of nuclear import through its PY-NLS and nuclear retention through its DNA binding domain.
Many of the Kapβs responsible for nuclear import of general transcription factors in yeast have been identified. Kap114p imports TFIIB and the TATA-binding protein (43–45); Kap119p imports TFIIS and Kap122p imports TFIIA (46, 47). These Kapβs also import proteins other than transcription factors. For example, Kap114p also imports proteins involved in chromatin assembly such as histones H2A, H2B, and chromatin assembly factor Nap1p (39, 48). The two previously identified Kap104p substrates, Hrp1p and Nab2p, are both mRNA-binding proteins. In vivo validation of Tfg2p as a PY-NLS bearing substrate of Kap104p implies that Kap104p substrates are not limited to mRNA-binding proteins. Similarly, 16 of the 17 validated human Kapβ2 substrates bind RNA, and of these 3 are also transcription factors (49). Both Kap104p and Kapβ2 appear to transport a range of functionally related substrates such as transcription factors and RNA-binding or -processing proteins. However many more Kap104p and Kapβ2 substrates will need to be identified and validated before we can truly define a functional network that relates the groups. Cellular and energetic dissections of the Tfg2p PY-NLS corroborates recently reported physical properties that govern Kap104p-PY-NLS interactions and suggests additional sequence restraints, which will be useful for large-scale substrate searches in the future (1).
Supplementary Material
Acknowledgments
We thank T. Cagatay and G. Süel for microscopy assistance, J. Kang for yeast strains and assistance, J. Aitchison for the kap104–16 and kap121–34 strains and kap104 gene, and L. Pemberton for the Δkap108, Δkap120, and Δkap114 strains. We thank G. Süel for discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM069909 and 5-T32-GM008297. This work was also supported by Welch Foundation Grant I-1532 and the University of Texas Southwestern Endowed Scholars Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- Kapβ
- Karyopherinβ protein
- DBD
- DNA-binding domain
- NLS
- nuclear localization signal
- cNLS
- classic NLS
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1.Suel K. E., Gu H., Chook Y. M. ( 2008) PLoS Biol. 6, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mosammaparast N., Pemberton L. F. ( 2004) Trends Cell Biol. 14, 547– 556 [DOI] [PubMed] [Google Scholar]
- 3.Fried H., Kutay U. ( 2003) Cell Mol. Life Sci. 60, 1659– 1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chook Y. M., Blobel G. ( 2001) Curr. Opin. Struct. Biol. 11, 703– 715 [DOI] [PubMed] [Google Scholar]
- 5.Conti E., Izaurralde E. ( 2001) Curr. Opin. Cell Biol. 13, 310– 319 [DOI] [PubMed] [Google Scholar]
- 6.Gorlich D., Kutay U. ( 1999) Annu. Rev. Cell Dev. Biol. 15, 607– 660 [DOI] [PubMed] [Google Scholar]
- 7.Weis K. ( 2003) Cell 112, 441– 451 [DOI] [PubMed] [Google Scholar]
- 8.Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. ( 1998) Cell 94, 193– 204 [DOI] [PubMed] [Google Scholar]
- 9.Hodel M. R., Corbett A. H., Hodel A. E. ( 2001) J. Biol. Chem. 276, 1317– 1325 [DOI] [PubMed] [Google Scholar]
- 10.Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. ( 2007) J. Biol. Chem. 282, 5101– 5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catimel B., Teh T., Fontes M. R., Jennings I. G., Jans D. A., Howlett G. J., Nice E. C., Kobe B. ( 2001) J. Biol. Chem. 276, 34189– 34198 [DOI] [PubMed] [Google Scholar]
- 12.Kalderon D., Richardson W. D., Markham A. F., Smith A. E. ( 1984) Nature 311, 33– 38 [DOI] [PubMed] [Google Scholar]
- 13.Fontes M. R., Teh T., Jans D., Brinkworth R. I., Kobe B. ( 2003) J. Biol. Chem. 278, 27981– 27987 [DOI] [PubMed] [Google Scholar]
- 14.Fontes M. R., Teh T., Kobe B. ( 2000) J. Mol. Biol. 297, 1183– 1194 [DOI] [PubMed] [Google Scholar]
- 15.Conti E., Kuriyan J. ( 2000) Structure 8, 329– 338 [DOI] [PubMed] [Google Scholar]
- 16.Lee B. J., Cansizoglu A. E., Suel K. E., Louis T. H., Zhang Z., Chook Y. M. ( 2006) Cell 126, 543– 558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siomi M. C., Fromont M., Rain J. C., Wan L., Wang F., Legrain P., Dreyfuss G. ( 1998) Mol. Cell. Biol. 18, 4141– 4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange A., Mills R. E., Devine S. E., Corbett A. H. ( 2008) J. Biol. Chem. 283, 12926– 12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Truant R., Fridell R. A., Benson R. E., Bogerd H., Cullen B. R. ( 1998) Mol. Cell. Biol. 18, 1449– 1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aitchison J. D., Blobel G., Rout M. P. ( 1996) Science 274, 624– 627 [DOI] [PubMed] [Google Scholar]
- 21.Lee D. C., Aitchison J. D. ( 1999) J. Biol. Chem. 274, 29031– 29037 [DOI] [PubMed] [Google Scholar]
- 22.Ghazy M. A., Brodie S. A., Ammerman M. L., Ziegler L. M., Ponticelli A. S. ( 2004) Mol. Cell. Biol. 24, 10975– 10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freire-Picos M. A., Krishnamurthy S., Sun Z. W., Hampsey M. ( 2005) Nucleic Acids Res. 33, 5045– 5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khaperskyy D. A., Ammerman M. L., Majovski R. C., Ponticelli A. S. ( 2008) Mol. Cell. Biol. 28, 3757– 3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung W. H., Craighead J. L., Chang W. H., Ezeokonkwo C., Bareket-Samish A., Kornberg R. D., Asturias F. J. ( 2003) Mol. Cell 12, 1003– 1013 [DOI] [PubMed] [Google Scholar]
- 26.Henry N. L., Campbell A. M., Feaver W. J., Poon D., Weil P. A., Kornberg R. D. ( 1994) Genes Dev. 8, 2868– 2878 [DOI] [PubMed] [Google Scholar]
- 27.Gaiser F., Tan S., Richmond T. J. ( 2000) J. Mol. Biol. 302, 1119– 1127 [DOI] [PubMed] [Google Scholar]
- 28.Chen H. T., Warfield L., Hahn S. ( 2007) Nat. Struct. Mol. Biol. 14, 696– 703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groft C. M., Uljon S. N., Wang R., Werner M. H. ( 1998) Proc. Natl. Acad. Sci. U. S. A. 95, 9117– 9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sikorski R. S., Hieter P. ( 1989) Genetics 122, 19– 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. ( 1998) Yeast 14, 115– 132 [DOI] [PubMed] [Google Scholar]
- 32.Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sorensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. ( 2002) Nature 415, 180– 183 [DOI] [PubMed] [Google Scholar]
- 33.Yoshida K., Blobel G. ( 2001) J. Cell Biol. 152, 729– 740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan S., Conaway R. C., Conaway J. W. ( 1995) Proc. Natl. Acad. Sci. U. S. A. 92, 6042– 6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Damelin M., Silver P. A., Corbett A. H. ( 2002) Methods Enzymol. 351, 587– 607 [DOI] [PubMed] [Google Scholar]
- 36.Leslie D. M., Zhang W., Timney B. L., Chait B. T., Rout M. P., Wozniak R. W., Aitchison J. D. ( 2004) Mol. Cell. Biol. 24, 8487– 8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rout M. P., Blobel G., Aitchison J. D. ( 1997) Cell 89, 715– 725 [DOI] [PubMed] [Google Scholar]
- 38.Mosammaparast N., Guo Y., Shabanowitz J., Hunt D. F., Pemberton L. F. ( 2002) J. Biol. Chem. 277, 862– 868 [DOI] [PubMed] [Google Scholar]
- 39.Mosammaparast N., Jackson K. R., Guo Y., Brame C. J., Shabanowitz J., Hunt D. F., Pemberton L. F. ( 2001) J. Cell Biol. 153, 251– 262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ziegler E. C., Ghosh S. ( 2005) Sci. STKE 2005, RE6. [DOI] [PubMed] [Google Scholar]
- 41.Makhnevych T., Lusk C. P., Anderson A. M., Aitchison J. D., Wozniak R. W. ( 2003) Cell 115, 813– 823 [DOI] [PubMed] [Google Scholar]
- 42.Cardoso M. C., Leonhardt H. ( 1999) J. Cell Biol. 147, 25– 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hodges J. L., Leslie J. H., Mosammaparast N., Guo Y., Shabanowitz J., Hunt D. F., Pemberton L. F. ( 2005) Mol. Biol. Cell 16, 3200– 3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morehouse H., Buratowski R. M., Silver P. A., Buratowski S. ( 1999) Proc. Natl. Acad. Sci. U. S. A. 96, 12542– 12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pemberton L. F., Rosenblum J. S., Blobel G. ( 1999) J. Cell Biol. 145, 1407– 1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albertini M., Pemberton L. F., Rosenblum J. S., Blobel G. ( 1998) J. Cell Biol. 143, 1447– 1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Titov A. A., Blobel G. ( 1999) J. Cell Biol. 147, 235– 246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mosammaparast N., Ewart C. S., Pemberton L. F. ( 2002) EMBO J. 21, 6527– 6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertolotti A., Melot T., Acker J., Vigneron M., Delattre O., Tora L. ( 1998) Mol. Cell. Biol. 18, 1489– 1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.