Abstract
Processing of the 3′ terminus of tRNA in many organisms is carried out by an endoribonuclease termed RNase Z or 3′-tRNase, which cleaves after the discriminator nucleotide to allow addition of the universal -CCA sequence. In some eubacteria, such as Escherichia coli, the -CCA sequence is encoded in all known tRNA genes. Nevertheless, an RNase Z homologue (RNase BN) is still present, even though its action is not needed for tRNA maturation. To help identify which RNA molecules might be potential substrates for RNase BN, we carried out a detailed examination of its specificity and catalytic potential using a variety of synthetic substrates. We show here that RNase BN is active on both double- and single-stranded RNA but that duplex RNA is preferred. The enzyme displays a profound base specificity, showing no activity on runs of C residues. RNase BN is strongly inhibited by the presence of a 3′-CCA sequence or a 3′-phosphoryl group. Digestion by RNase BN leads to 3-mers as the limit products, but the rate slows on molecules shorter than 10 nucleotides in length. Most interestingly, RNase BN acts as a distributive exoribonuclease on some substrates, releasing mononucleotides and a ladder of digestion products. However, RNase BN also cleaves endonucleolytically, releasing 3′ fragments as short as 4 nucleotides. Although the presence of a 3′-phosphoryl group abolishes exoribonuclease action, it has no effect on the endoribonucleolytic cleavages. These data suggest that RNase BN may differ from other members of the RNase Z family, and they provide important information to be considered in identifying a physiological role for this enzyme.
Maturation of tRNA precursors requires the removal of 5′ and 3′ precursor-specific sequences to generate the mature, functional tRNA (1). In eukaryotes, archaea, and certain eubacteria, the 3′-processing step is carried out by an endoribonuclease termed RNase Z or 3′-tRNase (2–6). However, in some bacteria, such as Escherichia coli, removal of 3′ extra residues is catalyzed by any of a number of exoribonucleases (7, 8). The major determinant for which mode of 3′-processing is utilized appears to be whether or not the universal 3′-terminal CCA sequence is encoded (2, 9). Thus, for those tRNA precursors in which the CCA sequence is absent, endonucleolytic cleavage by RNase Z right after the discriminator nucleotide generates a substrate for subsequent CCA addition by tRNA nucleotidyltransferase (1–3, 10). In view of this role for RNase Z in 3′-tRNA maturation, it is surprising that E. coli, an organism in which the CCA sequence is encoded in all tRNA genes (2), nevertheless contains an RNase Z homologue (11), because its action would appear not to be necessary. In fact, the physiological function of this enzyme in E. coli remains unclear, because mutants lacking this protein have no obvious growth phenotype (12). Hence, there is considerable interest in understanding the enzymatic capabilities of this enzyme.
The E. coli RNase Z homologue initially was identified as a zinc phosphodiesterase (11) encoded by the elaC gene (now called rbn) (13). Subsequent work showed that the protein also displayed endoribonuclease activity on certain tRNA precursors in vitro (6, 14). However, more recent studies revealed that this protein actually is RNase BN, an enzyme originally discovered in 1983 and shown to be essential for maturation of those bacteriophage T4 tRNA precursors that lack a CCA sequence (15, 16). Using synthetic mimics of these T4 tRNA precursors, RNase BN was found to remove their 3′-terminal residue as a mononucleotide to generate a substrate for tRNA nucleotidyltransferase. Based on these reactions RNase BN was originally thought to be an exoribonuclease (13, 15, 17). However, subsequent work by us and others showed that it can act as an endoribonuclease on tRNA precursors (13, 18). RNase BN is required for maturation of tRNA precursors in E. coli mutant strains devoid of all other 3′-tRNA maturation exoribonucleases, although it is the least efficient RNase in this regard (7, 19). Thus, under normal circumstances, it is unlikely that RNase BN functions in maturation of tRNA in vivo except in phage T4-infected cells (15, 16).
To obtain additional information on what types of RNA molecules might be substrates for RNase BN and to clarify whether it is an exo- or endoribonuclease, we have carried out a detailed examination of its catalytic properties and substrate specificity. We show here that RNase BN has both exo- and endoribonuclease activity and that it can act on a wide variety of RNA substrates. These findings suggest that E. coli RNase BN may differ from other members of the RNase Z family of enzymes.
EXPERIMENTAL PROCEDURES
Materials
RNA oligonucleotides were synthesized by Dharmacon, Inc. T4 polynucleotide kinase and T4 RNA ligase were purchased from New England Biolabs, Inc. Calf intestine alkaline phosphatase was purchased from Fermentas. [γ-32P]ATP and 5′-[32P]pCp2 were obtained from PerkinElmer Life Sciences. SequaGel for denaturing urea-polyacrylamide gels was from National Diagnostics. DE81 chromatography paper was from Whatman. The His-Trap HP column was obtained from Amersham Biosciences. Bis(p-nitrophenyl) phosphate was purchased from Sigma. All of the other chemicals were reagent grade.
Overexpression and Purification of RNase BN
Strain BL21I−II−(DE3)/pLys carrying the pET-ElaC(His) plasmid (13) was grown at 37 °C in 1 liter of LB medium supplemented with 100 μg/ml ampicillin plus 34 μg/ml chloramphenicol to maintain the pLys plasmid. At an A600 of 0.6, 1 mm isopropyl β-d-thiogalactopyranoside was added, and growth was continued for another 2 h. The cells were harvested by centrifugation and washed with 0.9% NaCl. The cell pellet was frozen at −80 °C until use.
All of the steps of the purification procedure were carried out at 4 °C. The frozen pellet was resuspended in 8 ml of buffer M (20 mm Tris-Cl, pH 7.5, 100 mm KCl, 10% (v/v) glycerol). The cells were ruptured by two passes through an Aminco French press at 12,000 p.s.i. The resulting crude extract was centrifuged at 30,000 × g for 20 min at 4 °C. The supernatant fluid was loaded on a 5-ml His-Trap HP column charged with nickel and equilibrated with buffer M. The proteins were eluted by a gradient from 0 to 600 mm imidazole, and the fractions were analyzed by SDS-PAGE. The RNase BN-containing fractions were pooled and concentrated by ultrafiltration using Ultrafree Biomax-10K.
To determine the purity of the RNase BN preparation, 2.5 μg of purified protein was run on SDS-PAGE, followed by silver staining. Only a single band at ∼35 kDa was observed; no minor contaminating proteins were detected.
Substrate Preparation
Oligoribonucleotide substrates were deprotected according to the manufacturer's instructions. Unless stated otherwise, single-stranded oligoribonucleotide substrates were 5′-labeled with 32P using T4 polynucleotide kinase and [γ-32P]ATP. Duplex RNA substrates were prepared by mixing a 5′-32P-labeled oligoribonucleotide with a nonradioactive complementary oligoribonucleotide in a 1:1.2 molar ratio in the presence of 10 mm Tris-HCl (pH 8.0) and 20 mm KCl. The mixture was heated in a boiling water bath for 5 min and then allowed to cool slowly to room temperature to promote annealing.
The 34-mer (G5A29) or the 24-mer (usRNA1- 5′-GAG UGA CUA CCU CCA AGG CCC UUU-3′) (20) were labeled at their 3′ ends with 5′-[32P]pCp using T4 RNA ligase. Unincorporated [32P]pCp was removed using a Sephadex G25 column. The 3′-terminal phosphate was removed using calf intestine alkaline phosphatase. The reaction mixture was incubated at 37 °C for 30 min, after which calf intestine alkaline phosphatase was inactivated by heating the reaction mixture at 85 °C for 15 min. The G5A29[32P]pC substrate was annealed with a complementary strand in a 1:1.2 ratio to generate a 17-nt duplex with an 18-nt 3′ overhang.
Preparation of a Substrate with a 3′-Phosphoryl Terminus
An oligonucleotide substrate containing a 3′-phosphate terminus (A16P) was prepared by periodate oxidation of A17 in lysine HCl buffer (21).
Phosphodiesterase Assay
Phosphodiesterase activity of RNase BN was determined using bis(p-nitrophenyl) phosphate as substrate. Standard reaction conditions were 20 mm Tris-HCl (pH 7.4), 2 mm substrate, 2.0 μg of His-tagged RNase BN, and 0.2 mm of the indicated metal ion. EDTA, when present, was at 2 mm concentration. Release of p-nitrophenol (ϵ = 11500 m−1 cm−1 at pH 7.4) was continuously monitored for 3 min at 405 nm. One unit of activity corresponds to 1 μmol of p-nitrophenol liberated per minute at 37 °C.
Acid Soluble Assays
The assays were carried out in 50-μl reaction mixtures containing 20 mm HEPES (pH 6.5), 200 mm KCl, 0.2 mm CoCl2, 10 μm synthetic 34-mer RNA substrate (labeled at its 3′ end with [32P]C) and 1.5 μg of purified RNase BN. The reaction mixtures were incubated at 37 °C for 30 min. The reaction was stopped by the addition of 150 μl of 0.5% (w/v) yeast RNA and 200 μl of 10% trichloroacetic acid. The sample was placed in ice for 30 min and then centrifuged at 16,000 × g for 15 min at 4 °C. The radioactivity in 200 μl of the supernatant fraction was determined by liquid scintillation counting using a LS 6500 multipurpose scintillation counter (Beckman Coulter, Inc.).
Electrophoretic Activity Assays
The assays were typically carried out in 30-μl reaction mixtures containing 20 mm HEPES (pH 6.5), 200 mm KCl, 0.2 mm CoCl2, 10 μm oligoribonucleotide substrate, and 1.5 μg of purified enzyme, except as otherwise stated in the figure legends. The reaction mixtures were incubated at 37 °C. The portions were taken at the indicated times, and the reaction was terminated by addition of 2 volumes of gel loading buffer (95% formamide, 20 mm EDTA, 0.05% SDS, 0.025% bromphenol blue, and 0.025% xylene cyanol). The reaction products were resolved on denaturing 7.5 m urea, 20% polyacrylamide gels and visualized using a STORM 840 phosphorimaging device (GE Healthcare). Quantification was carried out using Image J (National Institutes of Health).
Paper Chromatography
Substrates labeled at the 3′ end with [32P]pCp with or without the 3′-terminal phosphate were used for this assay. Following incubation, the longer oligoribonucleotides were precipitated with 10% trichloroacetic acid. After centrifugation, the supernatant fraction was treated with ether to remove the trichloroacetic acid and then applied to DE81 ion exchange chromatography paper. Labeled products were resolved using 0.3 m ammonium formate (pH 3.1). The paper was then allowed to air dry. Each lane was cut into strips of 1 cm, and the radioactivity associated with each strip was determined in a scintillation counter. Standards were run in parallel with the test samples to identify the products.
RESULTS
Phosphodiesterase Activity of RNase BN
E. coli RNase BN is unusual among the RNase Z family of enzymes in that it displays efficient phosphodiesterase activity against bis(p-nitrophenyl) phosphate and thymidine-5′-p-nitrophenyl phosphate (11). RNase BN is also unusual in that its activity against tRNA-type substrates is highest in the presence of Co2+ (13, 15, 17). In as much as the phosphodiesterase activity is relevant to whether RNase BN is an exoribonuclease, it was of interest to determine whether the enhanced activity in the presence of Co2+ extended to a phosphodiesterase substrate.
The data for Experiment 1 in Table 1 show that at 0.2 mm metal ion, Co2+ and Mn2+ were the most efficient in stimulating activity against bis(p-nitrophenyl) phosphate, whereas a variety of other ions had little or no effect; Cu2+ was actually inhibitory. The addition of EDTA reduced the basal activity, suggesting that a stimulatory metal ion already was associated with the purified enzyme, possibly the catalytic site zinc ions (11). Measurement of activity with an artificial double-stranded RNA substrate (G5A29:C5U17) indicated that Co2+ and Mn2+ were the most effective stimulatory ions, with Co2+ even surpassing Mn2+ (data not shown). Based on these data, Co2+ was used for all the subsequent experiments reported here.
TABLE 1.
Effect of metal ions on the phosphodiesterase activity of RNase BN
Phosphodiesterase activity was determined as described under “Experimental Procedures.” The ions, when added to the reaction mixture, were present at 0.2 mm. In Experiment 1, untreated enzyme was used; in Experiment 2, apo-enzyme was prepared by removal of the existing metal ion with EDTA, followed by extensive dialysis and incubation of the apo-enzyme with the indicated ion for 15, 30, and 60 min. The values shown are for the 60-min time point.
| Addition | Relative activitya |
|---|---|
| Experiment 1 | |
| None | 35 ± 3 |
| EDTA | 11 ± 2 |
| Co2+ | 100 |
| Mn2+ | 95 ± 4 |
| Mg2+ | 54 ± 3 |
| Fe2+ | 34 ± 2 |
| Zn2+ | 28 ± 1 |
| Cd2+ | 28 ± 2 |
| Cu2+ | 8 ± 1 |
| Experiment 2 | |
| None | 2 ± 0.5 |
| Mg2+ | 15 ± 1 |
| Mn2+ | 23 ± 2 |
| Zn2+ | 26 ± 2 |
| Co2+ | 100 |
a Activity with Co2+ was set at 100 in each experiment.
It should be noted that earlier results of Vogel et al. (11) indicated that Zn2+ was the most effective cation for stimulating phosphodiesterase activity, although Co2+ was not tested. In those experiments the authors first prepared apo-enzyme by removing the existing metal ion by chelation with EDTA and adding back various test cations. We repeated that procedure, testing Co2+ as well. As shown in Table 1, Experiment 2, the use of that protocol led to the same conclusion that Co2+ was the most effective cation, stimulating phosphodiesterase activity 4-fold more than Zn2+.
RNA Substrate Specificity
In early work, RNase BN was shown to act on tRNA-type substrates (15, 17). However, its specificity was quite unusual. RNase BN removed the 3′-terminal nucleotide from tRNA-CU and tRNA-CA, whereas it was inactive with tRNA-CC, and tRNA-CCA was a very poor substrate. Interestingly, RNase BN could act on the E. coli tRNA precursor analogue, tRNA-CCA-Cn, but relatively poorly compared with other RNases. These data were confirmed by experiments showing that RNase BN processed tRNA precursors lacking (15, 16) or containing (8, 19) the 3′-terminal CCA sequence in vivo. Other recent experiments showed that the E. coli enzyme could also act on some unstructured, synthetic RNAs, although most of these studies were carried out at 52 °C (20). To examine in more detail the catalytic potential of RNase BN, we have carried out a systematic analysis of substrate specificity using a variety of defined, synthetic RNAs that enabled us to study separately various structural parameters.
For these experiments, the substrates were labeled at either their 5′- or 3′-termini, and the action of RNase BN was followed by use of either a gel electrophoretic assay or an acid-soluble assay as described under “Experimental Procedures.” With these procedures the products of RNase BN action could be easily visualized and quantitated.
Effect of RNA Secondary Structure
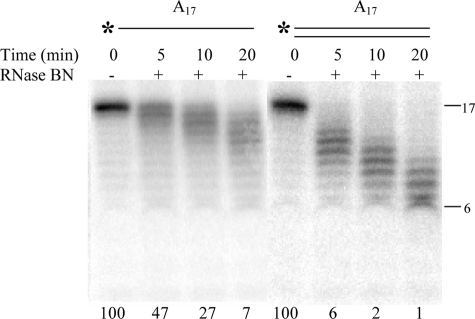
We first turned our attention to the influence of substrate secondary structure. We compared the ability of RNase BN to digest 5′-[32P]A17 with the same substrate complexed to complementary U17 under identical assay conditions. As shown in Fig. 1, single-stranded A17 was a substrate for the enzyme, confirming that RNase BN can act on simple, unstructured RNA molecules. Interestingly, the double-stranded RNA was an even better substrate, disappearing at least twice as rapidly. Note that for both substrates, a ladder of products was generated, consistent with a distributive, exonucleolytic mode of action for RNase BN. This point will be examined in detail below.
FIGURE 1.
Comparison of single- versus double-stranded RNA cleavage by RNase BN. The reactions were carried out as described under “Experimental Procedures” with the addition of 1.5 μg of RNase BN and 10 μm 5′-32P-labeled single- or completely double-stranded A17 substrate. Portions of 5 μl were taken at 5, 10, and 20 min, as indicated. The value at the bottom of each lane represents the percentage of initial substrate remaining.
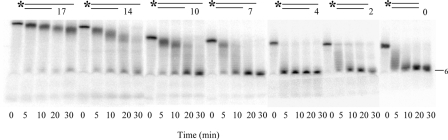
The importance of secondary structure was further emphasized by the data shown in Fig. 2. In this experiment a series of 5′-[32P]G5An oligomers were hybridized with C5U12, resulting in duplex molecules with 3′-A overhangs ranging in length from 0 to 17 nt. Comparison of the rate of reaction with these substrates showed that a duplex RNA with a single-stranded 3′-overhang of 4 nt was the most efficient substrate and that increasing the length of the overhang reduced RNase BN activity. The fact that molecules with only a 2-nt overhang or without any overhang also could be digested is most likely due to thermal breathing at the 3′-termini of these substrates.
FIGURE 2.
Effect of different length of 3′-overhang on RNase BN activity. The reactions were carried out as described for Fig. 1 except that the amount of enzyme was 1.6 μg. Portions of 5 μl were withdrawn after 5, 10, 20, and 30 min as indicated at the bottom of each lane.
Effect of Base Composition
RNase BN displays a dramatic base specificity in its action. Under conditions in which A17 is an effective substrate, no activity was observed with either U17 or C17 (Fig. 3A). Upon elevation of the enzyme level 3-fold, some degradation of U17 could be observed (Fig. 3B). Based on these assays, we estimate that RNase BN action on U17 is at least 6-fold slower than that on A17. A more pronounced effect was observed with C17 as substrate. Even with five times as much enzyme as was used with A17, no digestion was observed over a period of 30 min (Fig. 3B); under these conditions, 90% of A17 is removed. These data indicate that E. coli RNase BN is strongly inhibited by a run of C residues.
FIGURE 3.
RNase BN action on RNA homopolymers. The assay conditions were the same as described in Fig. 1. A, RNase BN was present at 1.5 μg. B, RNase BN was present at 4.5 μg for U17 and 7.5 μg for C17. The value at the bottom of each lane represents the percentage of initial substrate remaining.
Effect of CCA Sequence at 3′ Terminus
RNase Z enzymes from a variety of organisms are known to be inhibited by the 3′-CCA sequence present in tRNAs and certain tRNA precursors (22, 23). Although RNase BN can act on such molecules both in vitro and in vivo, the influence of the presence of CCA residues has not been carefully examined. To determine what the effect of a -CCA sequence might be on RNase BN catalysis, two substrates were constructed from the 17-mer, G5A12, one terminating with a 3′-CCA and the second containing five additional A residues following the -CCA. These RNAs were treated with RNase BN in the presence or absence of the oligomer C5U12, which is complementary to the first 17 residues of the substrate.
As can be seen in Fig. 4, the presence of the -CCA sequence dramatically inhibits the action of RNase BN on both single-stranded and duplex RNA. The five A residues following the -CCA sequence could be removed rapidly, but digestion then ceased almost completely. Likewise, RNAs terminating with -CCA were poor substrates. In all cases, a product 1 nt shorter also could be generated, indicating that the A residue of the -CCA sequence was removed but at a slow rate. In the absence of the -CCA sequence (Fig. 4C), RNase BN could easily digest the 17-nt double-stranded RNA. These data demonstrate directly that a -CCA sequence acts as a barrier to RNase BN action, consistent with its inability to digest through a series of C residues. Based on these data, it appears that just two C residues may be sufficient to inhibit the enzyme. Moreover, these C residues strongly affect action on the adjacent nucleotide because that A residue is removed much more slowly than the five additional A residues that follow it.
FIGURE 4.
Effect of CCA sequence at the 3′ end of RNA on RNase BN activity. Two different RNA substrates were constructed, one containing the CCA sequence at the 3′ terminus of [32P]G5A12 and one containing five A residues following the CCA. Duplex RNAs were annealed with the unlabeled complementary strand, U12C5, as described under “Experimental Procedures.” The reactions were carried out as described in Fig. 1. A, cleavage of single-stranded G5A12CCA and single-stranded G5A12CCA-A5. B, cleavage of duplex G5A12CCA and duplex G5A12CCA-A5 with 3′-overhang. C, cleavage of duplex G5A12.
Length Requirement and Limit Product of RNase BN Digestion
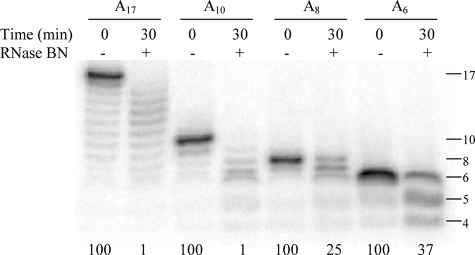
Inspection of the data presented in Figs. 1–4 suggested that the shortest product observed is 6 nt in length. This may indicate that RNase BN is not able to act on short oligonucleotides. To examine this point in more detail, oligonucleotides of different lengths, A17, A10, A8, and A6 were treated with RNase BN, and the products were analyzed (Fig. 5). As already shown above, A17 was an effective substrate, disappearing almost completely in 30 min. Likewise A10 was efficiently degraded. In contrast, A8 and A6 were degraded more slowly, and degradation of A10 also slowed markedly in this size range. Nevertheless, products as short as A3 could be generated (data not shown). These data clearly show that RNase BN can function as a distributive exoribonuclease even on very short oligonucleotides. However, the rate of digestion decreases as the substrate length is reduced below ∼10 nt.
FIGURE 5.
Length requirement for RNase BN digestion. Four substrates of different lengths, A17, A10, A8, and A6 were assayed for 30 min as described in the legend to Fig. 1. The value at the bottom of each lane represents the percentage of initial substrate remaining.
Effect of 3′-Phosphoryl Group
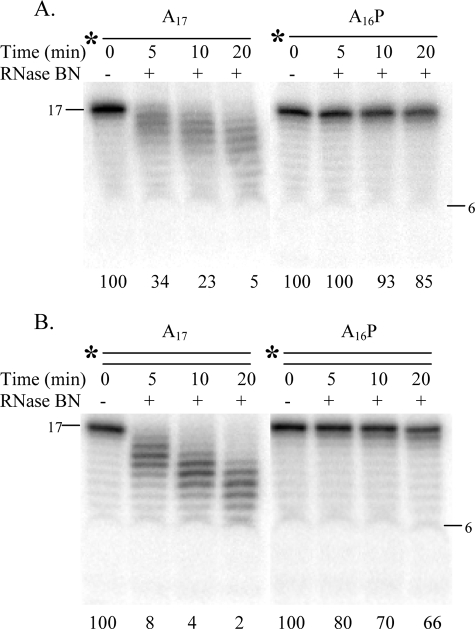
The high level of exoribonuclease activity exhibited by RNase BN prompted us to examine whether vicinal hydroxyl groups at the 3′ terminus of the RNA substrate were important for RNase action. To answer this question, the terminal residue of A17 was removed by periodate oxidation in the presence of lysine to generate A16P. The data in Fig. 6 indicate that the presence of the 3′-phosphoryl group severely reduces the rate of hydrolysis, both of single-stranded (Fig. 6A) and double-stranded substrates, although slow removal of a single residue can be seen (Fig. 6B). Interestingly, with these homopolymeric substrates, there is no indication of any endoribonucleolytic action.
FIGURE 6.
Effect of 3′-phosphoryl group on RNase BN activity. Single-stranded (A) and double-stranded (B) A17 and A16P were assayed as described in the legend to Fig. 1. The value at the bottom of each lane represents the percentage of initial substrate remaining.
To better quantitate the difference in activity between a 3′-phosphorylated and a dephosphorylated molecule, [32P]pCp was added to the 3′ terminus of G5A29 with RNA ligase. The 3′-terminal phosphate was then removed from a portion of these molecules, and activity with the two 32P-labeled RNAs was compared by determination of the release of acid soluble radioactivity. Thus, this assay measures the rate of removal of the 3′-terminal C residue from either a 3′-phosphorylated or dephosphorylated molecule. Based on this measurement, over a 30-min time period, release of radioactivity from the 3′-phosphorylated substrate was only 13% of that with its unphosphorylated counterpart (data not shown). These data confirm that the presence of a 3′-phosphoryl group dramatically inhibits RNase BN action.
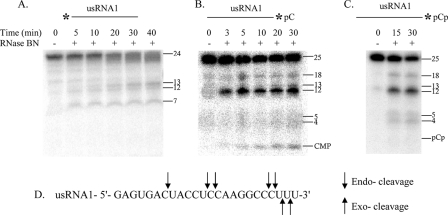
Mode of Action of RNase BN
The data presented here using synthetic RNA molecules support the conclusion that RNase BN can act as a distributive exoribonuclease. However, because it is generally assumed that members of the RNase Z family are endoribonucleases (5), we wished to examine this point in more detail. For this purpose, we made use of a 24-nt synthetic RNA (usRNA1) previously studied by Shibata et al. (20) with which it was concluded that the E. coli enzyme is an endoribonuclease. This RNA was labeled either at its 5′ end with 32P or at its 3′ end with [32P]pCp or [32P]pC (following phosphatase treatment of the pCp-labeled molecule). Each of these RNAs was treated with RNase BN, and the products were analyzed either by gel electrophoresis of the total sample or by paper chromatography of the acid-soluble fraction.
Treatment of the 5′-32P-labeled substrate with RNase BN (Fig. 7A) revealed a series of major products 1–4 nt shorter than the starting material and also products 13, 12, and 7 nt in length. If these 5′-products were generated by endonucleolytic digestion, one would expect the corresponding 3′-fragments from the substrates labeled at the 3′ terminus, and these products were observed (Fig. 7, B and C). Thus, taking into account the one additional 3′-pC(p) residue, fragments of the expected size, 12, 13, and 18 nt were generated. Also, fragments 4 and 5 nt in length were observed, corresponding to the expected 3′ fragments of the products in Fig. 7A that had 3 or 4 residues removed. These data confirm the conclusions of Shibata et al. (20) that the enzyme can act as an endoribonuclease.
FIGURE 7.
Mode of action of RNase BN. usRNA1 was assayed as described under “Experimental Procedures.” A, 5′-32P-labeled usRNA1. B, usRNA1-[32P]pC. C, usRNA1-[32P]pCp. D, diagram of endo- and exo-cleavages by RNase BN on usRNA1. Length markers are noted on the right side of each panel.
On the other hand, the data indicate that RNase BN also functions as an exoribonuclease. From Fig. 7B, it can be seen that the 3′-[32P]pC residue is released as CMP, and this was confirmed by paper chromatography of the acid soluble fraction (data not shown). Moreover, there are no bands in Fig. 7B corresponding to the di- and trinucleotides. Thus, the 22- and 23-mers observed in Fig. 7A must have been generated by exonucleolytic trimming, rather than by endonucleolytic cleavage. In contrast, when the 3′-phosphoryl group from the added [32P]pCp was retained (Fig. 7C), the CMP band disappeared, and there was no band corresponding to pCp. Likewise, no CMP or pCp was detected by paper chromatography (data not shown). Production of the endoribonucleolytic cleavage products was unaffected. Thus, although the presence of a 3′-phosphoryl group inhibits the exoribonuclease activity of RNase BN, its endoribonucleolytic function continues. From these data it is clear that RNase BN can function both as an exo- and an endoribonuclease and that it can do so on a single substrate.
It should be noted that the exoribonuclease activity of RNase BN differs from that of any known E. coli exoribonuclease. It is most active in the presence of Co2+ at a slightly acidic pH, and its substrate specificity does not correspond to any other RNase. This, coupled with the high degree of purity of the RNase BN preparation, makes it extremely unlikely that the exoribonuclease activity could be due to a contaminating enzyme.
DISCUSSION
The data presented here provide important insights into the catalytic potential of the E. coli member of the RNase Z family, RNase BN. Using a variety of synthetic RNA or chromogenic substrates, we have been able to systematically analyze factors and structural determinants that affect RNase BN activity. Based on this analysis, the E. coli enzyme was found to differ in certain respects from RNase Z homologues in other organisms. Thus, Co2+ is the preferred cation, both for phosphodiesterase activity and for action on RNA. RNase BN also displays a marked base specificity, is more active on double-stranded than single-stranded RNA, and is relatively inactive on short RNAs, and its activity is severely reduced by a 3′-phosphoryl group or a 3′-CCA sequence. Most interestingly, RNase BN is both a distributive exoribonuclease and an endoribonuclease, even on the same substrate. These data expand upon and clarify observations made by us and others over the years regarding the E. coli enzyme.
In early work, our laboratory showed that RNase BN efficiently removed the 3′-terminal residue from tRNA-CA and tRNA-CU but was essentially inactive on tRNA-CC (15, 17). This high degree of specificity on the tRNA substrates can now be understood in the context of the inability of the enzyme to remove even a single C residue from a C17 substrate. Based on the tRNA data, just two C residues at the 3′ end apparently are sufficient to block RNase BN action. Interestingly, the removal of the terminal AMP residue from tRNA-CCA also was inefficient (13, 15). Likewise, as shown here, removal of the A residue from CCA-terminated, single- and double-stranded 17-mers was inefficient compared with trimming of the A residues following the -CCA sequence. This led to a pronounced pause at the CCA triplet and suggests that the presence of two C residues also impairs removal of the adjacent nucleotide.
Exactly how two C residues or the CCA sequence act to prevent RNase Z action is not well understood. Based on the structure of Bacillus subtilis RNase Z, it was suggested that the amino group of C-74 in tRNA clashes with the loop between strands β1 and β2 (24). In contrast, the Thermotoga maritima enzyme, which is not affected by the CCA sequence (6), has a much shorter loop suggesting that it may be involved in the CCA effect. Studies with the Drosophila RNase Z showed that the presence of the CCA sequence in mature tRNA reduced kcat ∼80-fold with little effect on Km compared with a tRNA precursor lacking the CCA residues (25). However, other examples have been reported in which the CCA sequence can be removed by RNase Z enzymes (28, 29). Clearly, further studies are required to unravel the anti-determinant effect of the CCA sequence observed with certain RNase Zs.
The finding that RNase BN can act on A17, A10, and U17, as well as usRNA1 of Shibata et al. (20), confirms and extends those authors' conclusion that the E. coli enzyme can act on unstructured RNA. Our data show that a molecule as short as A4 is a substrate for RNase BN but that the rate of digestion of molecules shorter than 10 nt slows markedly. This suggests either that short oligomers bind weakly and dissociate before catalysis can occur or that their size precludes proper placement or orientation at the catalytic site. RNase BN is a dimer, and its crystal structure (26), and that of the B. subtilis enzyme (24), have indicated that an RNA substrate binds primarily to one subunit, whereas catalysis is carried out by the active site of the other subunit (24, 27). The proteins also contain an exosite or flexible arm, which is thought to bind and clamp the RNA substrate across the top surface of the protein dimer (14). We speculate that shorter oligonucleotides may not be of sufficient length to be stabilized on the enzyme by the RNA binding site and the flexible arm. On the other hand, because other small molecules such as bis(p-nitrophenyl) phosphate and thymidine-5′-nitrophenyl phosphate are substrates for RNase BN, they must be bound with sufficient stability at the catalytic site to be acted upon. Most likely, this occurs because these substrates typically are present in the millimolar range, whereas RNA substrates are used in the micromolar range for in vitro assays. Thus, even weak binding within the catalytic site may be sufficient to allow action on the chromogenic substrates.
An interesting feature of RNase BN action is its increased activity on a double-stranded substrate compared with its single-stranded counterpart. Of the model substrates examined, the most active was one with a 4-nt single-stranded 3′-overhang, although a completely double-stranded molecule also served as a substrate. These observations are consistent with the conclusion that binding to the basic patch/flexible arm favors duplex RNA but that access to the catalytic site requires a stretch of single-stranded nucleotides. This latter point is also suggested by the crystal structure of the enzyme (24). Consequently, the fact that a completely duplex RNA is a substrate argues that it must partially open to allow as many as 4 nt to become single-stranded. Further work will be needed to understand why duplex RNA is a preferred substrate and how RNAs interact with the flexible arm.
The most important finding of our studies is that RNase BN is both a distributive exoribonuclease and an endoribonuclease. Early work from our laboratory had shown that RNase BN could remove a single 3′-nucleotide from a variety of tRNA substrates (15), and on that basis the enzyme was deemed to be an exoribonuclease. Subsequent work by us and others (13, 20) indicated that RNase BN has endoribonuclease activity, in keeping with other members of the RNase Z family. In fact, it has been suggested that the apparent exoribonuclease activity on tRNA substrates is simply a manifestation of its endoribonuclease activity cleaving after a C nucleotide (5). The data presented here clearly show that this cannot be the case and that RNase BN is also an exoribonuclease. Given its phosphodiesterase activity, its action as an exoribonuclease is not surprising. It is not yet known whether RNase BN is an atypical member of the RNase Z family or whether additional studies of other RNase Zs may unearth exoribonuclease activity among them as well. Nevertheless, it is extremely interesting to ask how an RNase can act as both an exo- and endoribonuclease.
RNase BN, as all RNase Z members, belongs to the β-lactamase family of metallo-hydrolases (5). Two other RNases of this family have also been shown to act as both exo- and endoribonucleases. These are RNase J from B. subtilis (31) and CPSF-73 from mammalian cells (32), an RNase that functions in histone mRNA processing. Interestingly, each of these enzymes is a 5′ to 3′ exoribonuclease, in contrast to RNase BN, which acts 3′ to 5′.
Although RNase BN can act on tRNA precursors in vivo under very special conditions (absence of other tRNA processing enzymes or phage infection), the true physiological role of this enzyme in E. coli remains unclear. It has been shown that the absence of RNase BN can slightly lengthen mRNA half-life and that this effect is much more pronounced when RNase E is also missing (30). This suggests a role for RNase BN in mRNA decay. On the other hand, the E. coli enzyme has also been suggested to remove amino acids and the CCA terminus from aminoacyl-tRNAs (29), a very unusual reaction. Clearly, much more work will be needed to determine the role(s) of RNase BN in E. coli. The studies presented here will aid in this analysis because they provide insights into the catalytic potential of RNase BN to be considered in evaluating its physiological role. These include preference for duplex RNA, inability to digest through adjacent C residues, and exoribonuclease activity. Knowledge of these catalytic properties should prove useful for identifying its in vivo substrates.
Acknowledgments
We thank Dr. Helen A. Vincent and Dr. Mattias Lovgren for helpful discussions and comments. We also thank Dr. Arun Malhotra, Dr. Chaitanya Jain, Dr. Kenneth Rudd, Dr. Georgeta Basturea, and Christie Taylor for reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM16317.
- pCp
- cytidine 3′,5′-bis(phosphate)
- nt
- nucleotide(s).
REFERENCES
- 1.Deutscher M. P. ( 1995) in tRNA: Structure, Biosynthesis, and Function ( Soll D., Rajbhandary U. L. eds) pp. 51– 65, American Society for Microbiology, Washington, D.C [Google Scholar]
- 2.Deutscher M. P. ( 1990) Prog. Nucleic Acid Res. Mol. Biol. 39, 209– 240 [DOI] [PubMed] [Google Scholar]
- 3.Vogel A., Schilling O., Späth B., Marchfelder A. ( 2005) Biol. Chem. 386, 1253– 1264 [DOI] [PubMed] [Google Scholar]
- 4.Schierling K., Rösch S., Rupprecht R., Schiffer S., Marchfelder A. ( 2002) J. Mol. Biol. 316, 895– 902 [DOI] [PubMed] [Google Scholar]
- 5.Redko Y., de la Sierra-Gallay I. L., Condon C. ( 2007) Nat. Rev. Microbiol. 5, 278– 286 [DOI] [PubMed] [Google Scholar]
- 6.Minagawa A., Takaku H., Takagi M., Nashimoto M. ( 2004) J. Biol. Chem. 279, 15688– 15697 [DOI] [PubMed] [Google Scholar]
- 7.Li Z., Deutscher M. P. ( 1996) Cell 86, 503– 512 [DOI] [PubMed] [Google Scholar]
- 8.Reuven N. B., Deutscher M. P. ( 1993) FASEB J. 7, 143– 148 [DOI] [PubMed] [Google Scholar]
- 9.Schiffer S., Rösch S., Marchfelder A. ( 2002) EMBO J. 21, 2769– 2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiner A. M. ( 2004) Curr. Biol. 14, 883– 885 [Google Scholar]
- 11.Vogel A., Schilling O., Niecke M., Bettmer J., Meyer-Klaucke W. ( 2002) J. Biol. Chem. 277, 29078– 29085 [DOI] [PubMed] [Google Scholar]
- 12.Schilling O., Rüggeberg S., Vogel A., Rittner N., Weichert S., Schmidt S., Doig S., Franz T., Benes V., Andrews S. C., Baum M., Meyer-Klaucke W. ( 2004) Biochem. Biophys. Res. Commun. 320, 1365– 1373 [DOI] [PubMed] [Google Scholar]
- 13.Ezraty B., Dahlgren B., Deutscher M. P. ( 2005) J. Biol. Chem. 280, 16542– 16545 [DOI] [PubMed] [Google Scholar]
- 14.Schilling O., Späth B., Kostelecky B., Marchfelder A., Meyer-Klaucke W., Vogel A. ( 2005) J. Biol. Chem. 280, 17857– 17862 [DOI] [PubMed] [Google Scholar]
- 15.Asha P. K., Blouin R. T., Zaniewski R., Deutscher M. P. ( 1983) Proc. Natl. Acad. Sci. U. S. A. 80, 3301– 3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidman J. G., Schmidt F. J., Foss K., McClain W. H. ( 1975) Cell 5, 389– 400 [DOI] [PubMed] [Google Scholar]
- 17.Deutscher M. P. ( 1993) J. Bacteriol. 175, 4577– 4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbarbary R. A., Takaku H., Nashimoto M. ( 2008) Biochim. Biophys. Acta 1784, 2079– 2085 [DOI] [PubMed] [Google Scholar]
- 19.Kelly K. O., Deutscher M. P. ( 1992) J. Bacteriol. 174, 6682– 6684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata H. S., Minagawa A., Takaku H., Takagi M., Nashimoto M. ( 2006) Biochemistry 45, 5486– 5492 [DOI] [PubMed] [Google Scholar]
- 21.Uziel M. ( 1973) Biochemistry 12, 938– 942 [DOI] [PubMed] [Google Scholar]
- 22.Pellegrini O., Nezzar J., Marchfelder A., Putzer H., Condon C. ( 2003) EMBO J. 22, 4534– 4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohan A., Whyte S., Wang X., Nashimoto M., Levinger L. ( 1999) RNA 5, 245– 256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Sierra-Gallay I. L., Pellegrini O., Condon C. ( 2005) Nature 433, 657– 661 [DOI] [PubMed] [Google Scholar]
- 25.Zareen N., Hopkinson A., Levinger L. ( 2006) RNA 12, 1104– 1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostelecky B., Pohl E., Vogel A., Schilling O., Meyer-Klaucke W. ( 2006) J. Bacteriol. 188, 1607– 1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Sierra-Gallay I. L., Mathy N., Pellegrini O., Condon C. ( 2006) Nat. Struct. Mol. Biol. 13, 376– 377 [DOI] [PubMed] [Google Scholar]
- 28.Schiffer S., Rösch S., Marchfelder A. ( 2003) Biol. Chem. 384, 333– 342 [DOI] [PubMed] [Google Scholar]
- 29.Takaku H., Nashimoto M. ( 2008) Genes Cells 13, 1087– 1097 [DOI] [PubMed] [Google Scholar]
- 30.Perwez T., Kushner S. R. ( 2006) Mol. Microbiol. 60, 723– 737 [DOI] [PubMed] [Google Scholar]
- 31.de la Sierra-Gallay I. L., Zig. L., Jamalli A., Putzer H. ( 2008) Nat. Struct. Mol. Biol. 15, 206– 212 [DOI] [PubMed] [Google Scholar]
- 32.Yang X. C., Sullivan K. D., Marzluff W. F., Dominski Z. ( 2009) Mol. Cell. Biol. 29, 31– 42 [DOI] [PMC free article] [PubMed] [Google Scholar]