Abstract
Parathyroid hormone-related protein (PTHrP) plays a vital role in the embryonic development of the skeleton and other tissues. When it is produced in excess by cancers it can cause hypercalcemia, and its local production by breast cancer cells has been implicated in the pathogenesis of bone metastasis formation in that disease. Antibodies have been developed that neutralize the action of PTHrP through its receptor, parathyroid hormone receptor 1, without influencing parathyroid hormone action through the same receptor. Such neutralizing antibodies against PTHrP are therapeutically effective in animal models of the humoral hypercalcemia of malignancy and of bone metastasis formation. We have determined the crystal structure of the complex between PTHrP (residues 1–108) and a neutralizing monoclonal anti-PTHrP antibody that reveals the only point of contact is an α-helical structure extending from residues 14–29. Another striking feature is that the same residues that interact with the antibody also interact with parathyroid hormone receptor 1, showing that the antibody and the receptor binding site on the hormone closely overlap. The structure explains how the antibody discriminates between the two hormones and provides information that could be used in the development of novel agonists and antagonists of their common receptor.
The discovery of parathyroid hormone (PTH)6 -related protein (PTHrP) as the cause of hypercalcemia in many patients with cancer provided new insights into the pathogenesis of the skeletal complications of malignancy (1). It revealed PTHrP as a previously unrecognized hormone, related in evolution to the calcium-regulating PTH, but important in the pathogenesis of the humoral hypercalcemia of malignancy, a syndrome in which hypercalcemia occurs without evident bone metastases. Whereas PTH consists of 84 amino acids, human PTHrP has three alternative splice products of 139, 141, and 173 residues. Apart from 8 of the first 13 residues of PTH and PTHrP being identical, there is no significant identity between these peptides (2). PTHrP actively promotes bone resorption, doing so in a manner identical to that of PTH by acting upon the receptor (PTH1R) it shares with PTH. The PTH1R is located on cells of the osteoblast lineage, which program the formation and activation of osteoclasts, and on cells of the kidney tubule, through which both PTHrP and PTH promote cyclic AMP and phosphorus excretion but reduce calcium excretion. Other actions of PTHrP that reflect those of PTH include the ability to relax vascular and other smooth muscle. This response may reflect a physiological function of PTHrP rather than of PTH and is consistent with PTHrP production and local action on smooth muscles at various sites (3).
The first 34 amino acids of each hormone contain the full biological activities of both PTH and of PTHrP to activate the PTH1R (4). The sequences of PTHrP and PTH between residues 14 and 34 are interesting in that, although they are not homologous, nevertheless they appear to be critical for binding of each to the seven transmembrane G protein-coupled receptor, PTH1R (4). Within the first 34 amino acids of PTH and PTHrP two functional regions have been revealed based on structural and cross-linking studies (5–8). These studies have indicated that the C-terminal half of the first 34 residues of each hormone comprises the high affinity binding domain, interacting with the N-terminal portion of the extracellular domain of the receptor. The N-terminal half of each hormone activates the receptor through contact points on the extracellular loops and juxtamembrane regions (9).
Despite their equal ability to activate through the PTH1R, it was clear from the earliest work, even with antibodies against peptides within the first 14 residues of PTHrP, that highly specific antibodies could be generated that discriminate between PTH and PTHrP (10). Likewise, polyclonal antibodies against PTHrP-(1–34) that neutralized its effects completely in vitro in promotion of cyclic AMP production in response to PTHrP without any detectable neutralizing effect on PTH were used to prevent and to treat hypercalcemia in nude mice bearing xenografts of PTHrP-secreting human cancers (11, 12). Similar results were obtained with a neutralizing mouse monoclonal antibody against PTHrP (13). Subsequently, after the finding that breast cancer metastases to bone were enriched in PTHrP production (14), Guise and Mundy (15) used an experimental model in nude mice in which human breast cancer cells grow as lytic deposits in bone after intracardiac injection and showed that PTHrP production by the cancers contributed to the process of tumor establishment and growth in bone by promoting osteoclast formation and bone resorption. Furthermore, the tumor establishment and growth in bone could be prevented by treating the mice with a monoclonal antibody against PTHrP (16) or with a bisphosphonate (17) to inhibit bone resorption.
The efficacy of anti-PTHrP antibodies in treating both humoral-mediated hypercalcemia in cancer and bone metastasis formation and growth in mouse models raises the prospect of humanized forms of these antibodies being used as therapeutic agents in these diseases in human subjects, and preclinical data have been obtained in support of that (18, 19). With that in mind, the present project was undertaken in which we have made use of a monoclonal antibody prepared against human PTHrP (residues 1–34), which neutralizes the actions of PTHrP through PTH1R without any action against PTH. The antibody has been complexed with recombinant human PTHrP (residues 1–108) to generate crystals that have been used to analyze the three-dimensional structure with the aim of discovering the structural basis of neutralization of PTHrP action by the antibody.
EXPERIMENTAL PROCEDURES
Cell Line
The rat osteosarcoma cell line, UMR106, was used for the bioassay of PTHrP using the cyclic AMP response.
Establishment of a Mouse Anti-human PTHrP-(1–34) Antibody
The antibody was developed against synthetic PTHrP-(1–34) and shown to be effective in treating hypercalcemia in a nude mouse model of the humoral hypercalcemia of malignancy (13). The cells were re-cloned and the hybridoma, #23-57-137-1, was established as a monoclonal cell line by Mitsubishi Kagaku BCL. The monoclonal antibody derived in this way was shown to be effective in lowering blood calcium in nude mice bearing a hypercalcemic human lung cancer-producing PTHrP (20). The hybridoma cells were transplanted into the abdominal cavity of Balb/c mice to produce ascites, and the antibody in the ascites was purified by using protein A-Sepharose column chromatography.
Cloning and Sequencing of Mouse Anti-PTHrP Antibody
mRNA from hybridoma #23-57-137-1 was prepared using the Quick Prep mRNA purification kit (GE Healthcare). cDNA synthesis was then carried out using avian myeloblastosis virus reverse transcriptase, with the extracted mRNA as the template and appropriate primers for the H and L chains using the 5′-AmpliFINDER RACE kit (Clontech, Palo Alto, CA). The specific genes for the antibody variable regions were amplified by PCR. The DNA fragments were digested with the restriction enzymes XmaI and EcoRI. Cloning at the EcoRI and XmaI sites on pUC19 was then carried out using DNA Ligation kit Version 2 (Takara Shuzo). The DNA sequence was determined by the dye terminator cycle-sequencing method using the 373 DNA Sequencer (PE Biosystems).
Specificity of the Mouse Anti-PTHrP Antibody
The binding specificity of the antibody was investigated using an enzyme-linked immunosorbent assay. In brief, 96-well plates were coated with 1 μg·ml−1 of human PTHrP-(1–34) (Peptide Institute Inc., Osaka, Japan). After washing, the samples were serially diluted and added to each well, and then, after washing, alkaline phosphatase-conjugated goat anti-mouse IgG (#62-6622; Zymed Laboratories Inc., San Francisco, CA) was added. After incubation and washing, phosphatase substrate (Sigma 104 Phosphatase Substrate) was added, and the optical density at 405/620 nm was then measured.
In Vitro Neutralizing Activity of the Anti-PTHrP Antibody
The activity of the mouse anti-PTHrP monoclonal antibody was assessed in vitro by determining its ability to neutralize the effect of PTHrP in promoting cyclic adenosine mono-phosphate (cAMP) formation in the PTH/PTHrP-responsive osteosarcoma cell line, UMR106. The cells in 12-well plates were preincubated for 20 min in α-modification of Eagle's medium (α-MEM) containing 0.1% (w/v) bovine serum albumin and 1 mm isobutylmethylxanthine. Cells were subsequently incubated for 12 min in the presence or absence of PTHrP preparations with or without monoclonal antibody at 0.630 μg/ml, then washed with phosphate-buffered saline, and intracellular cAMP was extracted with ethanol. Samples were evaporated to dryness, and cAMP levels were determined by radioimmunoassay as described (21).
Protein Preparation and Crystallization
Fab fragments of #23-57-137-1 were prepared by briefly digesting the antibody with papain and separating the Fab fragments from the undigested antibody and Fc fragments using protein A-Sepharose. Recombinant human PTHrP1–108 was expressed and purified from Escherichia coli (22). An excess of PTHrP1–108 was incubated with the Fab fragment at 20 °C for 2 h, and the resultant PTHrP1–108·Fab complex was purified by size exclusion chromatography using a Superdex 75 10/30 column (GE Biosciences) previously equilibrated in 20 mm MES buffer, pH 6.0, containing 150 mm sodium chloride, 0.02% (v/v) Tween 20, and 0.02% (w/v) sodium azide. The complex was buffer-exchanged into 10 mm MES, pH 6.0, containing 0.02% sodium azide and concentrated to 5 mg·ml−1 using a Centricon-10 concentrator (Millipore, Beverley, MA). Crystals of the complex were grown in hanging drops at 20 °C with 38% (v/v) polyethylene glycol 400 as the precipitant and buffered with 100 mm Tris at pH 8.7.
Data Collection
An x-ray diffraction data set to 2.0 Å resolution was collected at beamline 14-BM-C (Advanced Photon Source, Chicago, IL) from a single frozen crystal at 100 K with no need of an additional cryoprotectant. The data were processed using the HKL program package (23). The data collection statistics are given in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics
The values in parentheses are for the highest resolution bin. r.m.s.d., root mean square deviation.
| Data collection | |
| Temperature (K) | 100 |
| Space group | P21212 |
| Cell dimensions | |
| a (Å) | 72.6 |
| b (Å) | 96.3 |
| c (Å) | 88.5 |
| Maximum resolution (Å) | 2.0 (2.07−2.0) |
| No. of crystals | 1 |
| No. of observations | 331,990 |
| No. of unique reflections | 42,014 |
| Data completeness (%) | 100 (100) |
| Rsym (%)a | 8.3 (69.6) |
| I/σI | 27.8 (4.1) |
| Multiplicity | 7.8 (7.5) |
| Refinement | |
| Non-hydrogen atoms | |
| Protein | 3392 |
| Solvent | 204 |
| Resolution (Å) | 2.0 |
| Rcryst.b (%) | 22.4 (33.2) |
| Rfreec (%) | 27.0 (33.9) |
| r.m.s.d. from ideal geometry: | |
| Bonds (Å) | 0.017 |
| Angles (°) | 1.9 |
| Dihedrals (°) | 27.2 |
| Impropers (°) | 1.14 |
| Bonded Bs (Å2) | |
| Main chain | 2.18 |
| Side chain | 3.07 |
| Mean B (protein) (Å2) | |
| Main chain | 39.3 (heavy chain), 42.4 (light chain), 37.4 (PTHrP) |
| Side chain | 40.7 (heavy chain), 44.5 (light chain), 40.8 (PTHrP) |
| Mean B (solvent) (Å2) | 42.0 |
| Residues in regions of Ramachandran plotd (%) | |
| Most favored regions | 89.7 |
| Additionally allowed | 9.5 |
| Generously allowed | 0.5 (2 residues) |
| Disallowed | 0.3 (1 residue) |
a Rsym = Σh,k,lΣi|Ii− 〈I〉|/|〈I〉|, whereIi is the intensity for the ith measurement of a symmetry related reflection with indicesh,k,l.
b Rcryst. = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where Fobs and Fcalcare the observed and calculated structure factor amplitudes, respectively.
c Rfree was calculated with 10% of the diffraction data that were selected randomly and not used throughout refinement.
d Regions defined in Laskowski et al. (27).
Structure Determination
Molecular replacement calculations were performed with the program AMoRe (24) using data in the 12 to 4 Å resolution bin. The best model for the molecular replacement calculations was chosen by matching the sequences for the variable domains of the heavy and light chains of the Fab against Protein Data Bank entries using a BLAST search (www.ncbi.nlm.nih.gov). The best hit for the variable domain of the heavy chain was the PDB entry 1IGT with 77% pairwise sequence identity (83% including similar residues), and the best hit for the variable domain of the light chain was the PDB entry 1IGC with 43% pairwise sequence identity (60% including similar residues). The molecular replacement solution was subjected to rigid body refinement in the resolution shell from 30 to 4 Å, yielding an R-factor of 49.5% (Rfree of 50.5%). The elbow angle of the heavy chain of the final model of the Fab was found to be 15° more acute than that in entry 1IGT, causing a deviation of up to 16 Å between the equivalent atoms of each model. The change in the elbow angle of the light chain was about 3°, resulting in deviations up to 2.7 Å between each model. The Fab model was then refined using CNS (25). After four cycles of model building and refinement, clear density for the helical region of the antigen could be seen. Residues 14–31 of PTHrP were built into the model (with the interpretation being confirmed by omit maps), and further refinement cycles were performed until convergence. Structural coordinates have been deposited in the Protein Data Bank under accession code 3FFD.
RESULTS
In Vitro and in Vivo Neutralizing Capacity of the Anti-PTHrP Antibody
In previous work the mouse monoclonal antibody (2.5 μg/ml) has been shown to inhibit by 92.5% the action of PTHrP-(1–34) in promoting cAMP production by osteosarcoma cells without any effect on the response to PTH-(1–34) (26). In the present work this efficacy was confirmed, and blockade of responses to longer PTHrP sequences is shown. Thus, cAMP levels in response to 1 nm PTHrP-(1–34), -(1–84), and -(1–108) were 282 ± 69, 168 ± 44, and 227 ± 4 pmol per well, respectively. In the presence of antibody these levels were 4 ± 2, 3 ± 1, and 4 ± 0.2 pmol/ml, respectively. In separate experiments the antibody showed no neutralizing effect against human PTH-(1–34), with the latter at 1000-fold higher concentration than PTHrP-(1–34) (data not shown). When the anti-PTHrP monoclonal was used to treat in vivo nude mice that had been rendered hypercalcemic by PAN-7-JCK cells (derived from a human pancreatic cancer associated with hypercalcemia and high production of PTHrP (26)), the hypercalcemia was corrected, and the calcium control was maintained within the normal range throughout the experiment. The humanized form of the antibody was similarly effective in the same series of experiments. It also suppressed growth of lytic bone deposits in mice after intracardiac injection of MDA-MB-231 cells (18) as well as preventing cachexia in mice bearing the LC6-JCK human lung cancer xenograft, independent of the calcium-lowering effect of the antibody (19).
Structure Determination
The structure of PTHrP-(1–108) complexed to its neutralizing antibody was solved by molecular replacement and refined to a resolution of 2.0 Å. The final model includes residues from 14–31 of PTHrP, residues 1–131 and 140–215 of the Fab heavy chain, residues 1–215 of the Fab light chain, and 204 water molecules. The model has been refined to a crystallographic R-factor of 22.4% (R-free of 27.0%) and is of good quality with 89.7% of residues in the most favorable regions of the Ramachandran plot. The stereochemical quality of the final model is good (Table 1), and other stereochemical parameters such as side chain χ angle values, peptide bond planarity, α carbon tetrahedral distortions, and non-bonded interactions are all better than or within the allowed ranges according to PROCHECK (27).
Overall Structure
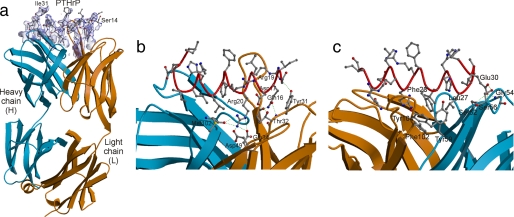
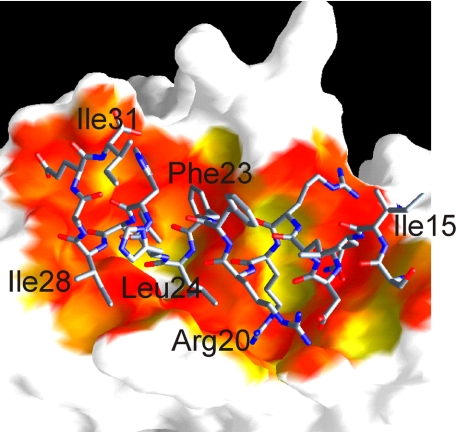
Of 108 residues, only residues 14–31 of PTHrP were visible in the final electron density map, with residues 14–29 adopting a helical conformation (Fig. 1a). The crystal lattice is formed solely by interactions between the Fab molecules, and there are large solvent channels in the crystal that would accommodate the disordered parts of the hormone. The N-terminal end of the helix forms interactions via a number of hydrogen bonds and salt bridges with residues of the antibody light chain (Fig. 1b), whereas the C-terminal end of the helix makes a number of hydrophobic interactions with residues of the antibody heavy chain (Fig. 1c). The antibody makes use of all complementarity-determining regions for antigen binding. In total, there are 18 hydrogen bonds or salt bridges and 22 hydrophobic interactions observed between antibody and antigen, indicating a rather tight association. There are only four water-mediated contacts identified between antibody and antigen; that is, between the carbonyl group of Thr-30 of the light chain and the amide of Asp-17 of PTHrP, between the hydroxyl of Tyr-104 of the heavy chain and the side chain amino of Gln-16 of PTHrP, between the hydroxyl of Thr-97 of the light chain and the main chain carbonyl of Arg-19 of PTHrP, and between the amides of Ile-15 and Gln-16 of PTHrP and the hydroxyl group of the side chain of Thr-30 of the light chain. The interaction of antibody and antigen causes a solvent-accessible area of 904 Å2 to be buried, a typical value for antibody-antigen interactions. The fit between hormone and antibody is highly complementary (Fig. 2).
FIGURE 1.
Structure of the PTHrP-antibody complex. a, view of the final difference (Fobs − Fcalc) electron density map (blue transparent surface) in the vicinity of the hormone, at 2.0 Å resolution, calculated using protein phases derived from the final model after omitting the hormone (and surrounding atoms within 3 Å of the hormone) and performing a round of simulated annealing to remove bias. The map is contoured at 3σ with the final model overlaid upon it. b and c, view of the complex with the hormone shown in red worm (main chain) and ball-and-stick (side chain) fashion and the antibody drawn in ribbon style (the heavy chain is in blue, and light chain is in orange). b, polar interactions between the N-terminal end of the hormone with antibody. c, polar and hydrophobic interactions between the C-terminal end of the hormone with antibody. All figures were drawn using MOLSCRIPT (42) and RASTER3D (43). The electron density figure was displayed with the help of CONSCRIPT (44).
FIGURE 2.
Surface representation of the PTHrP binding site colored according to surface complementarity (40). The most complimentary fits are shown in red, yellow is average, and white denotes very poor or no fit. The image was generated with GRASP (45) and RASTER3D (43).
Comparison to Uncomplexed PTHrP and PTH Structures
The observation of a helical region between PTHrP residues 14–29 is consistent with published structural studies of PTHrP and PTH (5–7). Previous NMR studies reported that the region in either protein of residues 1–34 is folded into two short helices consisting of approximately residues 3–11 and 16–30, connected by a flexible linker (5–7). Of particular note, the linker residue Gly-12 is strictly conserved throughout PTH/PTHrP sequences from different species. Whether there are any interactions between these two helices has been a matter of controversy (28–30). The crystal structure reported here does not support the tightly folded hairpin structure suggested by some workers (5, 7, 28). It is also markedly different from the crystal structure of PTH where residues 1–34 adopt a slightly bent helical structure and where there is no evidence of any flexibility in the region between residues 11 and 14 (30).
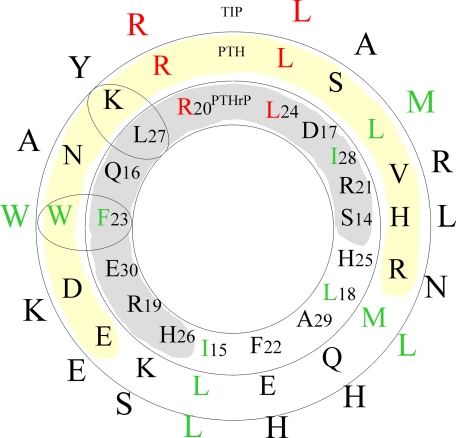
The C-terminal regions (residues 15–34) of PTHrP and PTH share only moderate sequence similarity with only 2 identical residues (Fig. 3). Nevertheless this region in both hormones is required for high affinity (in the low nanomolar range) binding to the extracellular N-terminal domain of PTH1R. The shortest native N-terminal peptide that retains full binding consists of residues 1–31, and residues between 15 and 34 can inhibit binding of the full-length peptide. The structure described herein and those reported elsewhere (5–7) consistently show that residues 14–29 form a single helix.
FIGURE 3.
Helical wheel diagram of the N-terminal region (residues 14–29) of human PTHrP, PTH, and the homologue, Tip39, a hypothalamic ligand for PTH2R (29, 33). Strictly conserved residues are highlighted in red, and conservative substitutions are in green. The shaded region denotes the helical face that interacts with both antibody and receptor.
DISCUSSION
The biological assay data confirm the selectivity of the monoclonal antibody for PTHrP and not PTH and the capacity of this antibody to neutralize the effects of PTHrP both in vitro and in vivo. The mouse antibody used in this work was used to develop humanized forms, with no significant differences among these molecules in inhibiting PTHrP action in vitro or cancer-induced hypercalcemia in vivo (26). The KD for binding of antibody to PTHrP-(1–34) was calculated at 1.02 × 10−10 m, and the epitope for binding was shown to be between residues 20 and 30, with some contribution from residues 15–20 (26). The region between residues 14 and 34 of PTH has been shown to be responsible for binding to the PTH1R (31), although all residues between 1 and 34 of the native (1–34) sequences of PTH and PTHrP are required for full signaling. Several substitutions in shorter analogs have yielded peptides with substantial activity, some with virtually full agonist activity (32). Interestingly, the neutralizing antibody described here specifically recognizes and complexes with residues 14–31 even though a much larger form of the molecule, PTHrP-(1–108), has been presented to it in the crystallization studies. We saw no electron density for the hormone beyond residue 31, indicating the rest of the molecule is highly mobile. NMR studies of longer length PTHrP show that the hormone adopts little, if any, regular structure beyond this region in solution7 in accord with our results.
A number of studies have identified specific residues of PTHrP involved in receptor binding. Arg-20 and Leu-24 are the only two residues that are strictly conserved among PTH/PTHrP sequences in the C-terminal region (between residues 14 and 31) (Fig. 3). Bearing in mind that PTH and PTHrP bind to the same receptor, these positions are then most likely important for receptor binding. In the crystal structure Arg-20 and Leu-24 make extensive interactions with the antibody. Residues Phe-23, Leu-24, and Ile-28 are intolerant to substitution by polar residues (31, 33). Photo-affinity cross-linking experiments of all residues between 22 and 35 suggest that Phe-23, Leu-27, and Ile-28 are positioned close to the receptor, and photolabeling of Leu-24 caused a 10-fold reduction in receptor binding (8). All three residues form extensive van der Waals interactions with the antibody. On the other hand residues Lys-26, Gln-29, and Asp-30 of PTH can be mutated without effect on receptor binding (34).
Studies of PTH/PTHrP chimeras have led to the identification of sites of receptor binding at position 19 of PTH that are incompatible (34). Position 19 in PTHrP is an arginine, whereas in PTH it is a glutamate. In the crystal structure Arg-19 is in hydrogen bonding distance of the hydroxyl of Tyr-31 of the light chain and forms a salt bridge with Asp-96 and is immediately adjacent to Arg-20 that appears to be playing a critical role in mediating 4 hydrogen bonds with the antibody. In PTHrP Glu-30 forms hydrogen bonds to the side chain hydroxyl of Ser-52 and the backbone of Gly-54, both of the heavy chain. It is easy to envisage that replacement of Glu-30 by aspartic acid in PTH (Fig. 3) could readily retain these interactions through use of a bound water molecule. All these observations suggest that the interaction surface of PTHrP used by the neutralizing antibody is the same surface used by the hormone in interacting with its receptor.
The region between residues 15 and 34 of either hormone contains the principal determinants of binding to the receptor, and indeed such a fragment can inhibit binding of either natural ligand (34). These observations suggest that either hormone binds to the same location on the receptor despite bearing only two identical residues. A helical wheel analysis of the PTHrP structure (Fig. 3) provides a molecular explanation for this puzzle; one face of the helix is more conserved (four strictly conserved or conservative substitutions), and it is residues from this face that interact with antibody or receptor. Thus, we propose that the conformation of this region when bound to the receptor is likely to remain α-helical as seen in the structure here.
In support of this hypothesis is the recently published crystal structure of PTH (residues 15–34) bound to the extracellular domain of the human PTH1 receptor (35). The structure reveals that PTH binds as a single continuous helix into a hydrophobic groove formed by a three-layer α-β-βα-fold. The “hot dog in a bun” analogy used by the authors is reminiscent of the way that PTHrP is recognized by the neutralizing antibody described here. Although the secondary structural elements used to recognize hormone are very different between receptor and antibody, there are a number of similarities. In both cases ∼900 Å2 of solvent-accessible surface area is buried on hormone binding, and the surface complementarity fits (36) are very similar (data not shown). The receptor structure confirms that the antibody does recognize the same surface of the hormone that the receptor uses. A number of PTH residues (Val-21, Trp-23, Leu-24, Leu-28, Val-31) form a hydrophobic face on the helix that interacts with the hydrophobic groove of PTH1R, anchored by polar interactions at the N and C termini of the PTH fragment. In a similar manner, PTHrP is recognized by its monoclonal antibody. The equivalent core residues in PTHrP (Phe-23, Leu-24, Ile-28) interact with a hydrophobic pocket in the antibody (Fig. 1c). PTH residue Arg-20, completely buried in the complex structure, forms salt bridges and hydrogen bonds to the receptor as the same residue in PTHrP does with the antibody (Fig. 1b). It should be noted that the side chains of conserved residues Arg-20 and Leu-24 adopt different conformations in the different hormone complex structures.
PTH and PTHrP are equally well recognized by the same PTH1 receptor. The only data of which we are aware concerning the relative affinities of PTH and PTHrP for the PTH1R were based on receptor binding and fluorescence resonance energy transfer-based kinetic analysis (37, 38). These indicated that PTH-(1–34) bound more strongly than PTHrP-(1–36) to a high affinity conformational state of the PTH1R and, furthermore, that mutation of His-5 to Ile-5 in PTHrP-(1–36) resulted in receptor interaction indistinguishable from that of PTH-(1–34). Given the bulkiness of antibodies, the binding site of a neutralizing antibody toward PTHrP need not overlap with the receptor binding site to achieve the selectivity toward PTHrP and still interfere with the receptor binding. Nevertheless, the binding site in our structure of PTHrP-(1–108) in complex with the selective neutralizing antibody closely overlaps the binding site to the receptor. We can now begin to consider the molecular basis by which certain antibodies discriminate between PTHrP and PTH. The consistent finding that antibodies can readily be prepared against PTHrP that do not recognize PTH, even in great excess, has been very helpful in the development of discriminatory plasma assays and in the study of tissue distribution of PTHrP using immunohistology (2). The same specificity has been applied to a number of polyclonal antisera that have been used in vivo (12, 39) and to the monoclonal antibodies used by Sato et al. (13) and Guise et al. (16). Such discrimination would be essential for an antibody to be used therapeutically because as the excess PTHrP is neutralized and serum calcium returns to normal, the suppressed PTH level rises, and normal physiological controls return. It is worth noting in this regard that a neutralizing antibody is aimed at preventing the deleterious effect of excess PTHrP, acting on the skeleton generally and on the kidney to cause the humoral hypercalcemia of malignancy. On the other hand PTH plays an important role in normal calcium homeostasis and, additionally, acts as an extremely effective anabolic therapy on the skeleton when administered by daily injection to subjects with osteoporosis (40). Thus, structural information concerning molecular mechanisms of inhibition of PTHrP-receptor interaction can usefully complement other studies of the detailed interaction of both ligands with PTH1R.
There are a number of factors that explain how the antibody discriminates between PTHrP and PTH even though they are both recognized by the same receptor. First, we superimposed the PTHrP structure onto the PTH1-receptor complex structure (data not shown). We find that PTHrP docks well into the ligand binding groove of the receptor as might be expected, with PTH possessing an extra turn of helix at its C-terminal end. We then superimposed the two complex structures, which reveals that the extra turn of helix in PTH sterically clashes with a complementarity-determining region of the antibody, providing a compelling explanation for the antibody specificity. Another factor could be related to residue 23 that is a phenylalanine in PTHrP but a tryptophan in PTH. The phenylalanine residue is located in a tight hydrophobic pocket of the antibody made up of residue Tyr-59 of the heavy chain and Thr-97 and Phe-102 of the light chain (Fig. 1c). A tryptophan residue would be too bulky to fit in this pocket without local readjustments. Modeling based on the PTH-PTH1R structures suggests the side chain would clash with Tyr-104 of the antibody heavy chain. In addition, the substitution of Leu-27 in PTHrP by a lysine residue in PTH would contribute to the disruption of the hydrophobic pocket as its side chain is immediately adjacent to the side chain of Phe-23 (Fig. 1c). Conversely, in the PTH-receptor structure Leu-27 superimposes with the aliphatic portion of the lysine residue, thus maintaining key van der Waals interactions with the receptor.
A means of antagonizing the deleterious effects of PTHrP in contributing to the skeletal complications of cancer could be an attractive therapeutic approach even bearing in mind the possibility of actions of PTHrP at earlier stages of cancer that could impair invasion (39, 41). The first step in evaluating this possibility is the use of humanized monoclonal anti-PTHrP. What could be even more attractive is the prospect of using the structural information obtained from analysis of the PTHrP-antibody complex together with that obtained from other approaches to ligand structure and interaction with receptor to design small molecule mimetics of the antibody or mimetics of PTHrP that would compete for receptor binding without causing signaling.
Acknowledgments
We thank Dr. Harry Tong and other staff at BioCARS for help with data collection during our visit to the Advanced Photon Source.
This work was supported in part by grants from the Cancer Council of Victoria and National Health and Medical Research Council of Australia with infrastructural support from the Australian Cancer Research Foundation Rational Drug Discovery Facility. This work, including use of the BioCARS sector, was also supported by the Australian Synchrotron Research Program, which is funded by the Commonwealth of Australia. Use of the Advanced Photon Source was supported by the United States Department of Energy, Basic Energy Sciences, Office of Energy Research.
The atomic coordinates and structure factors (code 3FFD) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
J. A. Barden, unpublished results.
- PTH
- parathyroid hormone
- PTHrP
- parathyroid hormone related protein
- PTH1R
- parathyroid hormone receptor 1
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Suva L. J., Winslow G. A., Wettenhall R. E., Hammonds R. G., Moseley J. M., Difenbach-Jagger H., Rodda C. P., Kemp B. E., Rodriquez H., Chen E. Y., Hudson P. J., Martin T. J., Wood W. I. ( 1987) Science 237, 893– 896 [DOI] [PubMed] [Google Scholar]
- 2.Moseley J. M., Gillespie M. T. ( 1995) CRC Crit. Rev. Clin. Lab. Sci 32, 299– 343 [DOI] [PubMed] [Google Scholar]
- 3.Clemens T. L., Cormier S., Eichinger A., Endlich K., Fiaschi-Taesch N., Fischer E., Friedman P. A., Karaplis A. C., Massfelder T., Rossert J., Schlüter K. D., Silve C., Stewart A. F., Takane K., Helwig J. J. ( 2001) Br. J. Pharmacol 134, 1113– 1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mannstadt M., Jüppner H., Gardella T. J. ( 1999) Am. J. Physiol. Renal Physiol 277, 665– 675 [DOI] [PubMed] [Google Scholar]
- 5.Barden J. A., Kemp B. E. ( 1989) Eur. J. Biochem 184, 379– 394 [DOI] [PubMed] [Google Scholar]
- 6.Barden J. A., Cuthbertson R. M., Jia-Zhen W., Moseley J. M., Kemp B. E. ( 1997) J. Biol. Chem 272, 29572– 29578 [DOI] [PubMed] [Google Scholar]
- 7.Chen Z., Xu P., Barbier J. R., Willick G., Ni F. ( 2000) Biochemistry 39, 12766– 12777 [DOI] [PubMed] [Google Scholar]
- 8.Gensure R. C., Gardella T. J., Jüppner H. ( 2001) J. Biol. Chem 276, 28650– 28658 [DOI] [PubMed] [Google Scholar]
- 9.Bergwitz C., Gardella T. J., Flannery M. R., Potts J. T., Jr., Kronenberg H. M., Goldring S. R., Jüppner H. ( 1996) J. Biol. Chem 271, 26469– 26472 [DOI] [PubMed] [Google Scholar]
- 10.Moseley J. M., Kubota M., Diefenbach-Jagger H., Wettenhall R. E., Kemp B. E., Suva L. J., Rodda C. P., Ebeling P. R., Hudson P. J., Zajac J. D., Martin T. J. ( 1987) Proc. Natl. Acad. Sci. U. S. A 84, 5048– 5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kukreja S. C., Rosol. T. J., Wimbiscus S. A., Shevrin D. H., Grill V., Barengolts E. I., Martin T. J. ( 1990) Endocrinology 127, 305– 310 [DOI] [PubMed] [Google Scholar]
- 12.Kukreja S. C., Shevrin D. H., Wimbiscus S. A., Ebeling P. R., Danks J. A., Rodda C. P., Wood W. I., Martin T. J. ( 1988) J. Clin. Investig 82, 1798– 1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato K., Yamakawa Y., Shizume K., Satoh T., Nohtomi K., Demura H., Akatsu T., Nagata N., Kasahara T., Ohkawa H., Ohsumi K. ( 1993) J. Bone Miner. Res 8, 849– 860 [DOI] [PubMed] [Google Scholar]
- 14.Powell G. J., Southby J., Danks J. A., Stillwell R. G., Hayman J. A., Henderson M. A., Bennett R. C., Martin T. J. ( 1991) Cancer Res 51, 3059– 3061 [PubMed] [Google Scholar]
- 15.Guise T. A., Mundy G. R. ( 1996) Curr. Opin. Nephrol. Hypertens 5, 307– 315 [DOI] [PubMed] [Google Scholar]
- 16.Guise T. A., Yin J. J., Taylor S. D., Kumagai Y., Dallas M., Boyce B. F., Yoneda T., Mundy G. R. ( 1996) J. Clin. Investig 98, 1544– 1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoneda T., Michigami T., Yi B., Williams P. J., Niewolna M., Hiraga T. ( 2000) Cancer 88, 2979– 2988 [DOI] [PubMed] [Google Scholar]
- 18.Saito H., Tsunenari T., Onuma E., Sato K., Ogata E., Yamada-Okabe H. ( 2005) Anticancer Res 25, 3817– 3823 [PubMed] [Google Scholar]
- 19.Onuma E., Tsunenari T., Saito H., Sato K., Yamada-Okabe H., Ogata E. ( 2005) Int. J. Cancer 116, 471– 478 [DOI] [PubMed] [Google Scholar]
- 20.Iguchi H., Onuma E., Sato K., Sato K., Ogata E. ( 2001) Int. J. Cancer 94, 24– 27 [DOI] [PubMed] [Google Scholar]
- 21.Suda N., Gillespie M. T., Traianedes K., Zhou H., Ho P. W. M., Hards D. K., Allan E. H., Martin T. J., Moseley J. M. ( 1996) J. Cell. Physiol 166, 94– 104 [DOI] [PubMed] [Google Scholar]
- 22.Hammonds R. G., Jr., McKay P., Winslow G. A., Diefenbach-Jagger H., Grill V., Glatz J., Rodda C. P., Moseley J. M., Wood W. I., Martin T. J. ( 1989) J. Biol. Chem 264, 14806– 14811 [PubMed] [Google Scholar]
- 23.Otwinowski Z., Minor W. ( 1997) Methods Enzymol 276, 307– 326 [DOI] [PubMed] [Google Scholar]
- 24.Navaza J. ( 1994) Acta Crystallogr. Sect. A 50, 157– 163 [Google Scholar]
- 25.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. ( 1998) Acta Crystallogr. Sect. D 54, 905– 921 [DOI] [PubMed] [Google Scholar]
- 26.Onuma E., Sato K., Saito H., Tsunenari T., Ishii K., Esaki K., Yabuta N., Wakahara Y., Yamada-Okabe H., Ogata E. ( 2004) Anticancer Res 24, 2665– 2674 [PubMed] [Google Scholar]
- 27.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. ( 1993) J. Appl. Crystallogr 26, 283– 291 [Google Scholar]
- 28.Pellegrini M., Royo M., Rosenblatt M., Chorev M., Mierke D. F. ( 1998) J. Biol. Chem 273, 10420– 10427 [DOI] [PubMed] [Google Scholar]
- 29.Gardella T. J., Jüppner H. ( 2001) Trends Endocrinol. Metab 12, 210– 217 [DOI] [PubMed] [Google Scholar]
- 30.Jin L., Briggs S. L., Chandrasekhar S., Chirgadze N. Y., Clawson D. K., Schevitz R. W., Smiley D. L., Tashjian A. H., Zhang F. ( 2000) J. Biol. Chem 275, 27238– 27244 [DOI] [PubMed] [Google Scholar]
- 31.Gardella T. J., Wilson A. K., Keutmann H. T., Oberstein R., Potts J. T., Jr., Kronenberg M., Nussbaum S. R. ( 1993) Endocrinology 132, 2024– 2030 [DOI] [PubMed] [Google Scholar]
- 32.Shimizu M., Carter P. H., Khatri A., Potts J. T., Jr., Gardella T. J. ( 2001) Endocrinology 142, 3068– 3074 [DOI] [PubMed] [Google Scholar]
- 33.Gardella T. J., Luck M. D., Jensen G. S., Usdin T. B., Jüppner H. ( 1996) J. Biol. Chem 271, 19888– 19893 [DOI] [PubMed] [Google Scholar]
- 34.Gardella T. J., Luck M. D., Wilson A. K., Keutmann H. T., Nussbaum S. R., Potts J. T., Jr., Kronenberg H. M. ( 1995) J. Biol. Chem 270, 6584– 6588 [DOI] [PubMed] [Google Scholar]
- 35.Pioszak A. A., Xu H. E. ( 2008) Proc. Natl. Acad. Sci. U. S. A 105, 5034– 5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence M. C., Colman P. M. ( 1993) J. Mol. Biol 234, 946– 950 [DOI] [PubMed] [Google Scholar]
- 37.Dean T., Vilardaga J.-P., Potts J. T., Jr., Gardella T. J. ( 2008) Mol. Endocrinol 22, 156– 166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dean T., Linglart A., Mahon M. J., Bastepe M., Jüppner H., Potts J. T., Jr., Gardella T. J. ( 2006) Mol. Endocrinol 20, 931– 942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henderson M. A., Danks J. A., Slavin J., Byrnes G. B., Choong P. M. F., Spillane J. B., Hopper J. L., Martin T. ( 2006) Cancer Res 66, 2250– 2256 [DOI] [PubMed] [Google Scholar]
- 40.Neer R. M., Arnaud C. D., Zanchetta J. R., Prince R., Gaich G. A., Reginster J. Y., Hodsman A. B., Eriksen E. F., Ish-Shalom S., Genant H. K., Wang O., Mitlak B. H. ( 2001) N. Engl. J. Med 344, 1434– 1441 [DOI] [PubMed] [Google Scholar]
- 41.Bakre M. M., Zhu Y., Yin H., Burton D. W., Terkeltaub R., Deftos L. J., Varner J. A. ( 2002) Nat. Med 8, 995– 1003 [DOI] [PubMed] [Google Scholar]
- 42.Kraulis P. J. ( 1991) J. Appl. Crystallogr 24, 946– 950 [Google Scholar]
- 43.Merritt E. A., Bacon D. J. ( 1997) Methods Enzymol 277, 505– 524 [DOI] [PubMed] [Google Scholar]
- 44.Lawrence M. C., Bourke P. ( 2000) J. Appl. Crystallogr 33, 990– 991 [Google Scholar]
- 45.Nicholls A., Sharp K. A., Honig B. ( 1991) Proteins 11, 281– 296 [DOI] [PubMed] [Google Scholar]