Abstract
Cleavage of phosphatidylinositol (PI) to inositol 1,2-(cyclic)-phosphate (cIP) and cIP hydrolysis to inositol 1-phosphate by Bacillus thuringiensis phosphatidylinositol-specific phospholipase C are activated by the enzyme binding to phosphatidylcholine (PC) surfaces. Part of this reflects improved binding of the protein to interfaces. However, crystallographic analysis of an interfacially impaired phosphatidylinositol-specific phospholipase (W47A/W242A) suggested protein dimerization might occur on the membrane. In the W47A/W242A dimer, four tyrosine residues from one monomer interact with the same tyrosine cluster of the other, forming a tight dimer interface close to the membrane binding regions. We have constructed mutant proteins in which two or more of these tyrosine residues have been replaced with serine. Phospholipid binding and enzymatic activity of these mutants have been examined to assess the importance of these residues to enzyme function. Replacing two tyrosines had small effects on enzyme activity. However, removal of three or four tyrosine residues weakened PC binding and reduced PI cleavage by the enzyme as well as PC activation of cIP hydrolysis. Crystal structures of Y247S/Y251S in the absence and presence of myo-inositol as well as Y246S/Y247S/Y248S/Y251S indicate that both mutant proteins crystallized as monomers, were very similar to one another, and had no change in the active site region. Kinetic assays, lipid binding, and structural results indicate that either (i) a specific PC binding site, critical for vesicle activities and cIP activation, has been impaired, or (ii) the reduced dimerization potential for Y246S/Y247S/Y248S and Y246S/Y247S/Y248S/Y251S is responsible for their reduced catalytic activity in all assay systems.
A detailed description of peripheral protein attachment to membranes is often difficult to achieve yet crucial to understanding the functions of these proteins. Few examples exist where a well defined docked structure has been determined. The phosphatidylinositol-specific phospholipase C (PI-PLC)2 enzymes from several Bacillus species are good structural models for the catalytic domain of the larger and more complex mammalian enzymes that play key roles in PI signal transduction (1–3). The enzyme from Bacillus thuringiensis closely resembles the catalytic barrel of PLCδ1 (1). It can be activated toward both PI cleavage to cIP and cIP hydrolysis to I-1-P by non-substrate phosphatidylcholine (PC) (4–6). Studies have shown that tryptophan residues in two discrete surface structural features, short helix B and a longer loop, are needed for the enzyme to bind to activating PC surfaces and for optimal catalytic activity (7, 8). A crystal structure (9) of an interfacially impaired mutant, W47A/W242A, showed that the enzyme could form dimers with a string of tyrosine residues (Tyr246, Tyr247, Tyr248, and Tyr251), forming the hydrophobic core of this dimer interface. The W47A/W242A dimer structure could not be formed by replacing the alanine at positions 47 and 242 with the original tryptophan residues without significant conformational rearrangements of the loop and helix B residues. Fluorescence quenching experiments were consistent with dimer formation of pyrene-labeled Cys mutants placed in the vicinity of the dimer interface (9). However, the hydrophobic probes introduced could drive formation of a dimer that does not occur with wild type protein and is not directly relevant to membrane binding and catalysis.
To further explore the relevance of this surface region of the protein to PI-PLC activity, we have constructed a series of mutants converting the tyrosine residues in the W47A/W242A dimer interface to serines (Y247S/Y251S, Y246S/Y247S/Y248S, and Y246S/Y247S/Y248S/Y251S). As more tyrosines are replaced by serine, there is a decrease in catalytic activity, both phosphotransferase cleavage of PI to cIP and diacylglycerol (in a variety of assay systems) and cyclic phosphodiesterase hydrolysis of cIP to I-1-P. There is an impairment of protein binding to PC vesicles when two or more tyrosine residues are removed that correlates with reduced specific activity in assays with PI/PC vesicles. Binding of the mutant proteins to diC7PC micelles has also been examined (intrinsic fluorescence and 31P NMR methods) and provides evidence that this interaction is very different when three or four tyrosines have been replaced by serine. Crystal structures of two of the mutant enzymes, Y247S/Y251S and Y246S/Y247S/Y248S/Y251S, have been solved, and these show that the proteins are very similar to the very closely related Bacillus cereus PI-PLC (10) in that they fail to crystallize as dimers (presumably because helix B and Trp242 prevent close approach of two monomers (9)). Their active sites also superimpose well on that of the B. cereus PI-PLC. The only difference is in the loss of Tyr side chains in the area that forms the crystallographic dimer interface in W47A/W242A. These results can be interpreted as evidence for either (i) surface-induced dimerization of the PI-PLC or (ii) identification of a very specific PC binding site on the protein.
EXPERIMENTAL PROCEDURES
Chemicals
1-Palmitoyl-2-oleoylphosphatidylcholine (POPC), l-α-phosphatidylinositol (PI) from bovine liver, and the short chain lipids dihexanoyl-PC (diC6PC) and diheptanoyl-PC (diC7PC) were purchased from Avanti Polar Lipids, Inc. and used without further purification. Dihexanoyl phosphatidic acid (diC6PA) was prepared from diC6PC by phospholipase D cleavage. D2O was purchased from Sigma. cIP was generated enzymatically from crude PI using PI-PLC as described previously (4). All other chemicals were reagent grade.
PI-PLC Mutations, Overexpression, and Purification
Because all four Tyr residues contribute to stability of the W47A/W242A dimer, single mutations converting each one to Ser were not prepared. Instead, a minimum of two Tyr were replaced. The primers were designed for use with the Exsite mutagenesis kit (Stratagene) to produce the double mutants Y246S/Y248S and Y247S/Y251S. The plasmid containing the double mutant Y247S/Y251S went through another round of Exsite mutagenesis to introduce mutations replacing Tyr246 and Tyr248 (to form Y246S/Y247S/Y248S/Y251S); the gene for Y246S/Y248S was further mutated to introduce Y247S (generating Y246S/Y247S/Y248S). All mutant PI-PLC genes were sequenced to make sure the correct mutations were incorporated and that no other mutation had arisen. Recombinant wild type B. thuringiensis PI-PLC (rPI-PLC) and the tyrosine mutants overexpressed well in Escherichia coli and were purified with two chromatographic steps (elution on a Q-Sepharose fast flow column followed by a phenyl-Sepharose column) (7).
Circular Dichroism
Wild type and mutant PI-PLC secondary structure and thermal stability as monitored by the thermal denaturation transition (Tm) were measured using an AVIV 202 CD spectrophotometer equipped with a thermoelectric sample temperature controller as described previously (8, 11). Comparison of secondary structure for wild type and mutant PI-PLC in this solution used wavelength scans from 265 down to 195 nm with protein (0.2–0.3 mg/ml) in a 0.1-cm cell at 25 °C. Secondary structure content was estimated with CDNN (12, 13).
PI-PLC Kinetic Assays
The activity of these PI-PLC mutants toward PI (typically 4–8 mm) dispersed in either Triton X-100 (1:2 PI/Triton X-100 unless otherwise noted) or in diC7PC (1:4 PI/diC7PC unless otherwise noted) micelles, PI co-sonicated with PC (3:1 PI/PC), and toward cIP in the absence or presence of 5 mm diC7PC was measured by 31P NMR spectroscopy as described previously (4, 7). Assays (in 25 mm HEPES, pH 7.5) were carried out at either fixed time points (picked so that <10% product was generated) or as a function of time (monitoring cIP intensity over 0.5–1 h) to estimate specific activity for the different mutants. The amount of PI-PLC used depended on whether the phosphotransferase or cyclic phosphodiesterase step was to be followed (4, 5, 7).
Fluorescence Spectroscopy
Intrinsic fluorescence measurements of PI-PLC as a function of added diC7PC were initially carried out on a Shimadzu RF5000U spectrofluorometer, then more recently using a Fluorolog R-3 spectrofluorometer. Measurements were carried out at 25 °C with ∼2 μm protein in 50 mm HEPES, pH 7.5, with 1 mm EDTA using an excitation wavelength of 290 nm and 5-nm excitation and emission slit widths. At this concentration of protein there was no detectable light scattering for any of the protein samples with PC (or diC6PA) micelles added. Changes in the fluorescence intensity were usually expressed as (I − I0)/I0, where I0 is the intensity of protein alone, and I is the intensity in the presence of an additive.
PI-PLC Vesicle Binding Assays
A vesicle binding assay based on centrifugation to separate free and vesicle-bound protein was used to characterize the partitioning of PI-PLC to PC interfaces (11). This assay used 10–15 μg/ml PI-PLC (ET) in 10 mm Tris HCl, pH 7.5. The POPC SUV concentrations ranged from 0 to 2 mm. Filtrates, containing free enzyme, were lyophilized and analyzed by SDS-PAGE. Band intensities were used to calculate the concentration of EF by comparing intensities from vesicle-containing samples to the ET value of the control. The concentration, EB, was then evaluated as ET − EF. Binding was analyzed using a simple binding isotherm and assuming the binding was cooperative: EB/ET = C(L0n/(L0n + Kdn)). The apparent Kd derived from this treatment, which is really the concentration of PC needed for half the maximum binding, allows a direct comparison of the extent to which the different mutant PLC proteins partition to PC vesicles.
31P NMR Line Width of DiC7PC in the Presence of PLC Enzymes
The line width of the diC7PC 31P resonance in the presence of PI-PLC was measured at 28 °C using a broadband probe in a Varian INOVA 500 spectrometer. Samples contained 3 mg/ml (0.085 mm) enzyme in 0.05 m HEPES with 1 mm EDTA, pH 7. The initial concentration of diC7PC was 0.5 mm; this was gradually increased to 20 mm. At each concentration, the width at half peak height was measured (5, 14).
High Resolution 31P Field Cycling
Field-cycling 31P T1 experiments were run at 22 °C on an unmodified Varian Unityplus 500 spectrometer at Brandeis University using a standard 10-mm Varian probe in a custom-built device which can move the sample, sealed in a standard 10-mm tube, from the sample probe up to or from a higher position within or just above the magnet, where the magnetic field is between 0.06 and 11.7 tesla (T) or can be varied with the aid of a small Helmholtz coil atop the magnet down to 0.002 T. The method and shuttling device are essentially as described previously, although improved by use of a stepper motor-based shuttle and many other new features (15, 16). Relaxation rates at each field strength were measured using 6–8 programmed delay times. Data were analyzed by curve-fitting to equations as described earlier (15–17). The results are parameterized quite well with only six constants. These are based on a standard “model-free” theory (18), which is not fully appropriate to membranes. The parameters are nevertheless useful as indicating general time scales of motion and structural preferences. Four of the parameters are derived from the high-field part of the measurement, above 0.08 T. These are: τc, which is the time scale (typically ∼5 ns) associated with a restricted, rotational diffusion process motion of an individual phospholipid; CH, which reports the contribution of chemical shift anisotropy (CSA) relaxation produced by sub-nanosecond motions within the lipid; CL, which is proportional to CSA relaxation produced by the part of the CSA relaxation remaining un-averaged by the above sub-nanosecond motions and resulting from the slower internal motions fluctuating with time scale τc; Rc(0), which is the dipolar-relaxation contribution resulting from 31P interacting with its nearest protons, mediated by the internal time-scale τc. Theory and other knowledge (15, 19) then allow us to estimate three other parameters from these, namely τhf, which is the time scale of the highest frequency motions mentioned above, Sc2, the square of the chemical shift anisotropy order parameter, and reff, which indicates the general size of the distance between the phosphorus and its nearest proton neighbors that relax it.
There is also a distinct new dispersion observed by extending measurements to very low fields (<0.03 T) that is easily defined on top of a base line of magnitude Rc(0) that is expected to be flat at these low fields. This base line can, thus, be subtracted to produce a pure dispersion. This dispersion is parameterized using Solomon theory with a correlation time τv, and an extrapolated rate in the zero field limit (17). We treat τv as equal to the rotational correlation time Dr/6, where Dr is the rotational diffusion constant of the individual lipid axes due to both Einstein-Debye diffusion of the entire phospholipid aggregate in the absence or presence of PI-PLC and translational diffusion of individual lipids in the aggregate. The area under this dispersion can be related to the spatial probability-density distributions of the closest protons to the phosphorus, averaged over a time long compared with the time scale of internal rotation τc. Because the detailed spatial distribution is unknown, we have used a rigid rotator model due to Woessner (20), with the axis assumed perpendicular to the aggregate surface (17). The R1 rate is then predicted to be proportional to (1–3 cos2θPH), where θPH is the angle between the director and the P-to-H vector. Only one of these four angles is reported here for PC micelles, as a comparison of data for dipalmitoylphosphatidylcholine vesicles with molecular dynamics simulations suggest the θPH ∼ 45° is likely to be the correct value for PC species (21).
X-ray Crystallography
Crystals of the PI-PLC Y247S/Y251S mutant were grown at 17 °C by vapor diffusion with hanging drop against a reservoir solution containing 9% (w/v) polyethylene glycol 8000, 0.2 m zinc acetate, and 0.1 m sodium cacodylate, pH 6.5. Later some crystals were soaked for 2 h in the mother liquor with 100 mm myo-inositol added to introduce a soluble substrate analog into the active site. Crystals of the Y246S/Y247S/Y248S/Y251S mutant were grown in 10% (w/v) polyethylene glycol 8000, 0.1 m MnCl2, and 50 mm MOPS, pH 7.0.
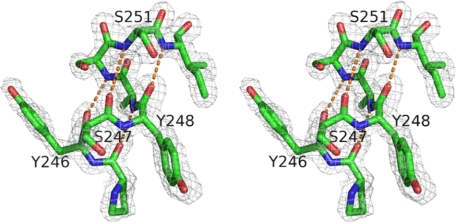
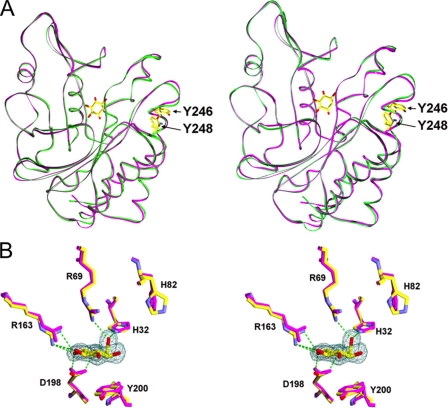
Data on crystals cooled to 80–100 K were collected at the Brookhaven National Laboratory Synchrotron X8C beamline. The diffraction data were indexed, integrated, and scaled using DENZO and SCALEPACK software (22). Phases for the Y247S/Y251S mutant structure were provided by molecular replacement using EPMR (23) and the coordinates from wild type B. cereus PI-PLC (PDB entry 1PTD) and the B. thuringiensis mutant W47A/W242A (PDB entry 2OR2). Phases of the Y246S/Y247S/Y248S/Y251S mutant and the myo-inositol-bound Y247S/Y251S structures were generated by molecular replacement using the solved Y247S/Y251S structure. The programs O (24) and CNS (25) were used for model building and refinement steps. Data collection and refinement statistics for the three structures are presented in Table 4. All structure figures were made with Pymol (26). An example of the electron density in the region of the mutation sites for Y247S/Y251S is shown in Fig. 1.
TABLE 4.
Crystallographic data and refinement statistics
| Y247S/Y251S | Y247S/Y251Smyo-inositol | Y246S/Y247S/Y248S/Y251S | |
|---|---|---|---|
| PDB ID code | 3EA1 | 3EA2 | 3EA3 |
| Diffraction data | |||
| Resolution (Å) | 1.75 | 1.95 | 1.78 |
| Space group | P212121 | P212121 | P1 |
| Unit cell | |||
| a (Å) | 59.643 | 59.518 | 47.666 |
| b (Å) | 97.064 | 97.226 | 56.352 |
| c (Å) | 112.799 | 112.727 | 59.852 |
| α (°) | 90 | 90 | 92.358 |
| β (°) | 90 | 90 | 99.381 |
| γ (°) | 90 | 90 | 113.214 |
| No. measured reflections | 62,818 | 45,672 | 51,149 |
| Completenessa (%) | 93.7 (97.5) | 93.9 (100) | 95.4 (92.8) |
| Redundancy | 5.9 | 6.9 | 2.0 |
| Rmergeb (%) | 6.9 (34.9) | 8.7 (34.8) | 5.4 (35.5) |
| Refinement | |||
| Resolution range (Å) | 50-1.75 | 50-1.95 | 50-1.78 |
| Rcrystc | 0.1972 | 0.1894 | 0.1850 |
| Rfreed | 0.2299 | 0.2264 | 0.2212 |
| No. of monomers in asymmetric units | 2 | 2 | 2 |
| No. of protein atoms | 4818 | 4818 | 4794 |
| No. of water molecules | 622 | 628 | 525 |
| No. of ions | 6 (Zn2+) | 6 (Zn2+) | 5 (Mn2+) |
| No. of ligand molecules | 0 | 2 (myo-inositol) | 0 |
| No. of atoms total | 5446 | 5465 | 5382 |
| r.m.s.d. angles (°) | 1.2989 | 1.2846 | 1.2932 |
| r.m.s.d. bonds (Å) | 0.0049 | 0.0053 | 0.0054 |
| Mean 〈B〉 overall (Å2) | 20.465 | 21.651 | 23.988 |
| Mean 〈B〉 protein (Å2) | 19.074 | 20.323 | 22.979 |
| Ramachandran plot (%) | |||
| Most favored | 86.6 | 86.9 | 86.9 |
| Additional allowed | 12.3 | 11.9 | 12.1 |
| Generously allowed | 0.8 | 0.8 | 0.6 |
| Disallowed | 0.4 | 0.4 | 0.4 |
a Completeness, redundancy, and Rmergereported for all reflections and for the highest resolution shell (values in parenthesis).
bRmergereported for all reflections and for the highest resolution shell (values in parenthesis). Rmerge = |Ii− 〈I〉|/ΣIi, whereIi is the intensity of an individual reflection, and 〈I〉 is the mean intensity of that reflection.
c Rcryst = Σ‖Fp| − |Fcalc‖/Σ|Fp|, where |Fcalc| and |Fp| are the calculated and observed structure factors, respectively.
d Rfreewas as defined in Brüngeret al. (25).
FIGURE 1.
Stereo structure of the mutation sites in B. thuringiensis Y247S/Y251S PI-PLC. Shown in the gray network is the electron density (from the simulated-annealing composite omit map and contoured at 1.2σ) for the residues in mutation sites. The helix hydrogen-bond network is shown as orange dotted lines.
RESULTS
Overexpression of Tyr Ridge Mutants; Secondary Structure and Thermostability
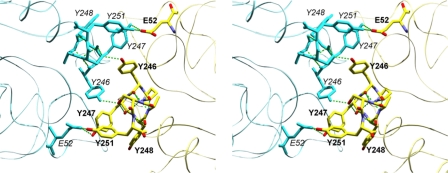
Four tyrosines in W47A/W242A, Tyr246, Tyr247, Tyr248, and Tyr251, form the hydrophobic core of the dimer interface (Fig. 2). In rPI-PLC protein, their replacement with serine residues should dramatically weaken any membrane-induced dimerization, assuming the dimer is similar to W47A/W242A, but maintain any hydrogen-bonding interactions with solvent or phospholipid head groups. If, instead of direct dimer formation, these residues are involved in specific binding of PC molecules (possibly via a Tyr π-cation interaction with the choline moiety as has been observed in peptide systems (27)), both activity and PC binding might also be affected.
FIGURE 2.
Stereoview showing tyrosine mutation sites at the crystallographic dimer interface of B. thuringiensis PI-PLC mutant, W47A/W242A (PDB code 2OR2). Subunit A is shown with Corey-Pauling-Koltun coloration, labels are in bold. subunit B is shown in cyan, and labels are in italics. Hydrogen bonds shown as green dashed lines.
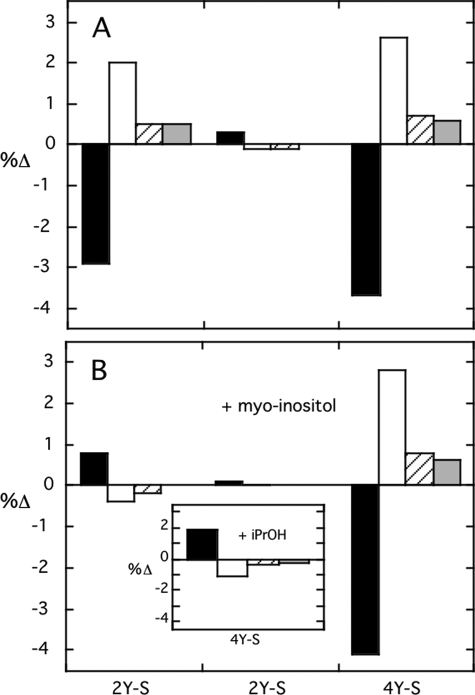
The mutated PI-PLC proteins Y247S/Y251S, Y246S/Y247S/Y248S, and Y246S/Y247S/Y248S/Y251S were examined by CD for changes in secondary structure. Y246S/Y247S/Y248S exhibited a far-UV spectrum equivalent to rPI-PLC. However, both Y247S/Y251S and Y246S/Y247S/Y248S/Y251S appeared to have less α-helix and more β-sheet (Fig. 3A) at low concentrations in 10 mm borate buffer. Upon the addition of myo-inositol (a soluble portion of the substrate molecule that binds, although not with terribly high affinity, to the active site) a CD spectrum identical to rPI-PLC was observed for Y247S/Y251S (Fig. 3B). The α-helix content for the quadruple Tyr mutant, Y246S/Y247S/Y248S/Y251S, dispersed in this solution was not recovered with added myo-inositol. For comparison, myo-inositol had no effect on the secondary structure of rPI-PLC or the triple tyrosine mutant. This suggested that occupation of the active site with the polar part of the substrate can affect the structure of helical regions in the protein and that the double tyrosine mutant likely resembles rPI-PLC under those conditions. The presence of 30% isopropyl alcohol has also been shown to activate rPI-PLC (28). This co-solvent has little effect on rPI-PLC secondary structure (if measured at 4 °C) (29); however, it did lead to recovery of α-helix for the quadruple mutant (Fig. 3B, inset). To the extent that a co-solvent system may mimic an interfacial environment of a membrane, these results indicate that conditions that lead to kinetic activation (e.g. isopropanol) tend to lead to changes in Y246S/Y247S/Y248S/Y251S secondary structure that approximate rPI-PLC.
FIGURE 3.
Change in secondary structure of tyrosine mutants compared with wild type recombinant PI-PLC in the (A) absence or (B) presence of 60 mmmyo-inositol. Secondary structure motifs are indicated as follows: (filled squares) α-helix, (open squares) β-sheet, (diagonal lines) β-turn, and (gray squares) random coil. Mutant proteins are abbreviated as follows: 2Y-S, Y247S/Y251S; 3Y-S, Y246S/Y247S/Y248S; 4Y-S, Y246S/Y247S/Y248S/Y251S. The inset in B shows the change in secondary structure for Y246S/Y247S/Y248S/Y251S in the presence of 30% isopropanol (iPrOH).
Specific Activities of Tyr Mutants
PI-PLC enzymes catalyze the hydrolysis of PI to I-1-P in two steps (cleavage of PI to water-soluble cIP, then hydrolysis of cIP to I-1-P) that can be analyzed separately. Y247S/Y251S exhibited specific activity toward PI solubilized in diC7PC (1:4 PI/diC7PC) (Table 1), comparable with wild type enzyme. Changing the detergent matrix to Triton X-100 instead of an activating PC molecule leads to reduced specific activity for rPI-PLC, typically 30–40% (4). However, Y247S/Y251S phosphotransferase activity with PI/Triton X-100 micelles was noticeably lower than that of rPI-PLC. In comparison to Y247S/Y251S, the other tyrosine mutants had even lower specific activity toward PI whether solubilized in diC7PC or Triton X-100 (Table 1). A similar trend was observed for cleavage of PI/PC (4:1) vesicles; Y247S/Y251S was closest to rPI-PLC in cleavage ability, whereas the mutants with three or four of the tyrosines removed were much less active.
TABLE 1.
Specific activities of surface tyrosine mutants towards PI and cIP in diverse assay systems
| PI-PLC | Specific activitya |
||||
|---|---|---|---|---|---|
| PI/diC7PC (1:4) | PI/Triton X-100 (1:2) | PI/PC (4:1) SUVs | cIP | cIP/diC7PC | |
| μmol min−1mg−1 | |||||
| Wild type | 560 ± 130 | 375 ± 56 | 9.5 ± 1.1 | 2.3 ± 0.5 | 73 ± 10 |
| Y247S/Y251S | 670 ± 100 | 239 ± 48 | 6.7 ± 0.9 | 0.6 ± 0.2 | 41 ± 8 |
| Y246S/Y247S/Y248S | 301 ± 21 | 98 ± 15 | 2.1 ± 0.4 | 0.2 ± 0.1 | 2.9 ± 0.7 |
| Y246S/Y247S/Y248S/Y251S | 112 ± 5 | 62 ± 12 | 1.6 ± 0.4 | 0.4 ± 0.1 | 0.8 ± 0.2 |
a In the phosphotransferase assays, the PI concentration was 8 mm with either 32 mmdiC7PC, 16 mmTriton X-100, or 2 mmPOPC (for the SUVs). The cyclic phosphodiesterase assays used 8 mmcIP in the absence and presence of 5 mmdiC7PC.
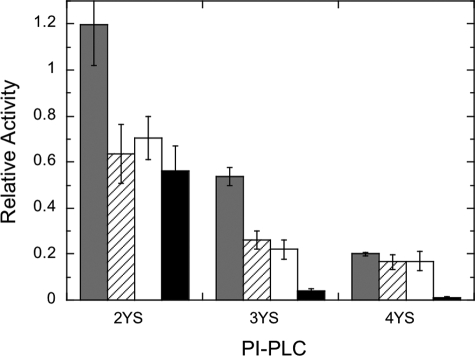
In the absence of an activating interface, the second step of the PI-PLC reaction, hydrolysis of cIP to I-1-P, was greatly reduced for all the tyrosine mutants compared with rPI-PLC. In the presence of diC7PC micelles, the specific activity of Y247S/Y251S increased dramatically, within a factor of two of the wild type enzyme-specific activity (Table 1). However, as more tyrosine residues were replaced by serine, the mutant PI-PLC enzymes exhibited a much smaller activation by diC7PC to the point where PC activation was virtually abolished for Y246S/Y247S/Y248S/Y251S. The larger effect on the phosphodiesterase activity compared with the phosphotransferase activity is better seen when the specific activity for each mutant enzyme is normalized to that of rPI-PLC for that assay system (Fig. 4). Hydrolysis of cIP in the presence of diC7PC is reduced to a much greater extent for Y246S/Y247S/Y248S and Y246S/Y247S/Y248S/Y251S compared with decreases in any of the PI cleavage assays.
FIGURE 4.
Relative activities of the three Tyr-to-Ser mutant enzymes compared with rPI-PLC. Data for Y247S/Y251S (2YS), Y246S/Y247S/Y248S (3YS), and Y246S/Y247S/Y248S/Y251S (4YS) are shown for PI (8 mm) cleavage in diC7PC micelles (gray bars), in Triton X-100 (16 mm) micelles (diagonal lines), or mixed with 2 mm PC in SUVs (open bars), and cIP (8 mm) hydrolysis with 5 mm diC7PC (black bars).
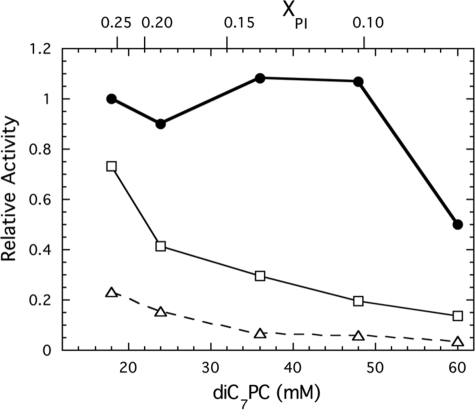
A striking kinetic feature of this bacterial PI-PLC is the lack of surface dilution inhibition until the mole fraction PI is reduced below 0.1 at fixed PI (4). Because removal of surface Tyr residues might alter this, we monitored the specific activity of the Tyr to Ser mutant enzymes in experiments where the bulk concentration of PI was held constant at 6 mm and the amount of diC7PC increased (Fig. 5). Y247S/Y251S, like rPI-PLC, has a moderate affinity for PI as 50% inhibition was not observed until a diC7PC/PI ratio of 10 (mole fraction of 0.091 PI). In contrast, the other two tyrosine mutants started at lower specific activities and showed a continuously decreasing activity (referenced to the value for 6 mm PI/18 mm diC7PC), as the diC7PC concentration was increased. Clearly, removal of three or four of the surface tyrosines that make up the core of the dimer interface in W47A/W242A reduces the activities of both steps of the reaction. The enhanced surface dilution suggests that removal of these Tyr reduces the affinity of the protein for interfacial substrate.
FIGURE 5.
Surface dilution kinetics for PI-PLC tyrosine mutant enzymes. Data for Y247S/Y251S (●), Y246S/Y247S/Y248S (□), and Y246S/Y247S/Y248S/Y251S (▵) are shown. The PI bulk concentration was fixed at 6 mm with the diC7PC increasing from 18 to 60 mm; the mole fraction of PI, XPI, in these mixed micelle assays is shown on the top of the graph.
Binding of Tyr Mutant Proteins to Phospholipid SUVs and diC7PC Micelles
The lower activities as tyrosine residues were replaced with serine could reflect a progressively weaker binding of PI-PLC to phospholipid interfaces. Previous work has shown that B. thuringiensis PI-PLC binds to PC vesicles with a large hydrophobic component, whereas it binds to pure anionic phospholipid vesicles through electrostatic interactions (11). Y247S/Y251S exhibited a 2-fold weaker affinity for PC vesicles than rPI-PLC (Table 2). Y246S/Y247S/Y248S presented an interesting case in that partitioning of the protein onto the vesicles very much depended on the protein concentration. At moderate protein concentrations (0.01–0.1 mg/ml, 0.29–2.9 μm), the protein bound to PC vesicles with an apparent KD of 0.5 ± 0.2 mm as measured by the centrifugation/filtration assay. When all four tyrosine residues were replaced by serine, partitioning of the enzyme on PC vesicles was considerably weaker (apparent KD ∼ 1 mm with the centrifugation/filtration assay). Although this binding assay is not very accurate for proteins with weak affinities for PC SUVs, it does stress that removal of these Tyr residues on the surface in a region that is below the barrel rim and not near the active site is clearly correlated with the lower activities in assay systems with PI/PC. However, specific activities progressively decreased for assay systems with diC7PC micelles as well so that interaction of these micelles with the proteins must be evaluated.
TABLE 2.
Apparent dissociation constants for PI-PLC tyrosine mutants binding to vesicles and short-chain phospholipid micelles
| PI-PLC | ApparentKDa |
||
|---|---|---|---|
| POPC SUV | diC7PCb | diC6PAb | |
| mm | |||
| Wild type | 0.075 ± 0.013 | 1.97 ± 0.02 | 17.0 ± 0.8 |
| Y247/251S | 0.16 ± 0.03 | 2.64 ± 0.09 | |
| Y246/247/248S | 0.5 ± 0.2c | 0.83 ± 0.04 | |
| Y246/247/248/251S | 1.0 ± 0.3c | 1.60 ± 0.16 | 14.9 ± 1.0 |
a Binding fit with a cooperative model (7).
b The apparent dissociation constant is the concentration of detergent needed for half the total change in fluorescence intensity. The CMC of diC7PC at is 1.5 mm(32); the CMC for diC6PA at pH 8.5 is 14 mm.
c The errors in measuring the partitioning of these two proteins onto the PC SUVs were particularly large and varied with small variations in protein concentration (0.05–0.2 mg/ml).
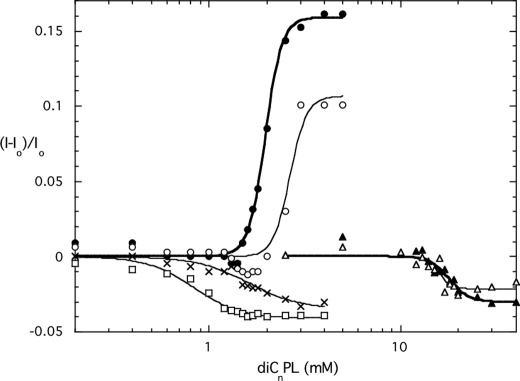
The intrinsic fluorescence of PI-PLC is sensitive to surface binding with the intensity increasing when PLC binds to activating PC interfaces (diC7PC or POPC SUVs) and decreasing when molecules bind at the active site (30, 31). This is a particularly useful method for characterizing PI-PLC binding to short-chain phospholipid micelles (8, 31). For rPI-PLC, the fluorescence intensity increases above the critical micelle concentration of the short-chain PC, and the steepness of the increase reflects the apparent binding constant of the enzyme to micellar phospholipids. Comparison of fluorescence changes for Y247S/Y215S binding to rPI-PLC protein showed that the double Tyr mutant protein had a weaker interaction with diC7PC micelles than rPI-PLC, consistent with vesicle partitioning assays (Fig. 6). Assuming the critical micelle concentration of diC7PC in the presence of the protein is comparable with the 1.5 mm value for pure diC7PC (32), we would estimate an apparent KD of 1.1 mm “micellar” PC for Y247S/Y251S compared with rPI-PLC, for which the KD is 0.5 mm micellar diC7PC. With Y246S/Y247S/Y248S and Y246S/Y247S/Y248S/Y251S, the fluorescence intensity did not increase with increasing micellar diC7PC; rather, it decreased (Fig. 6). In rPI-PLC, Trp242 has been identified as the major contributor to the increased intrinsic fluorescence upon PC binding (7). Y246S/Y247S/Y248S clearly binds to diC7PC, as there is a change in protein intrinsic fluorescence. The decrease in intensity as this mutant protein binds to diC7PC micelles must reflect a different conformational change upon diC7PC binding. Interestingly, the decreased fluorescence of Y246S/Y247S/Y248S and Y246S/Y247S/Y248S/Y251S was comparable in magnitude to that for diC6PA binding to rPI-PLC (Fig. 6). That anionic phospholipid binds competitively with PI at the active site (30, 31). Indeed, if the quadruple Tyr mutant protein is titrated with diC6PA, a change comparable with rPI-PLC was obtained. These intrinsic fluorescence changes indicate the Y246S/Y247S/Y248S and Y246S/Y247S/Y248S/Y251S proteins bind diC7PC but not in the same way as rPI-PLC and Y247S/Y251S. The differences correlate with the lower activities of the three and four Tyr-to-Ser enzymes.
FIGURE 6.
Change in intrinsic fluorescence of recombinant PI-PLC and mutants as a function of added short chain phospholipids. DiC7PC was added to rPI-PLC (●), Y247S/Y251S (○), Y246S/Y247S/Y248S (□), and Y246S/Y247S/Y248S/Y251S (×); diC6PA was added to rPI-PLC (▴) and Y246S/Y247S/Y248S/Y251S (▵). The fluorescence change is plotted as (I − Io)/Io where Io is the initial fluorescence of the sample in the absence of the short-chain phospholipid.
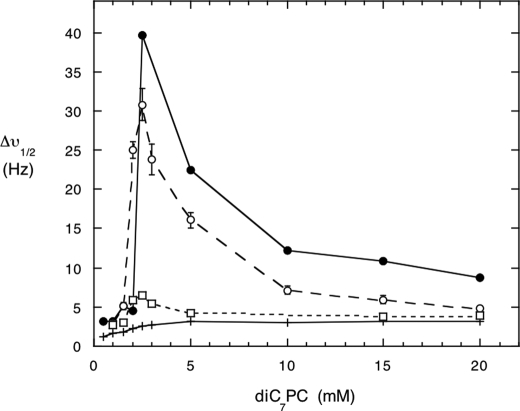
Another way of assessing PI-PLC binding to phospholipid micelles is to examine the 31P line width of the short-chain phospholipids, in this case diC7PC, in the presence of the enzyme (5, 14). The addition of enzyme to diC7PC causes a large increase in the 31P line width above the critical micelle concentration of the lipid. This increase is due in part to formation of large particles in this system (5, 33), although an intermediate exchange rate could also contribute to the large line width. The very active Y247S/Y251S enzyme also broadened the diC7PC 31P resonance, almost to the extent of rPI-PLC (Fig. 7). However, the addition of the triple mutant, Y246S/Y247S/Y248S, which binds to diC7PC, as monitored by the intrinsic fluorescence experiments, did not lead to large increases in PC line width; only small increases were seen. This reinforces that there is a pronounced difference in the way these two mutant proteins interact with diC7PC interfaces.
FIGURE 7.
31P line width for varying concentrations of diC7PC in the presence of PI-PLC enzymes. The diC7PC was titrated into a solution of 3 mg/ml rPI-PLC (●), Y247S/Y251S (○), or Y246S/Y247S/Y248S (□). For comparison, the line width of pure diC7PC is shown as (+) at each concentration; the critical micelle concentration for the pure lipid is 1.5 mm.
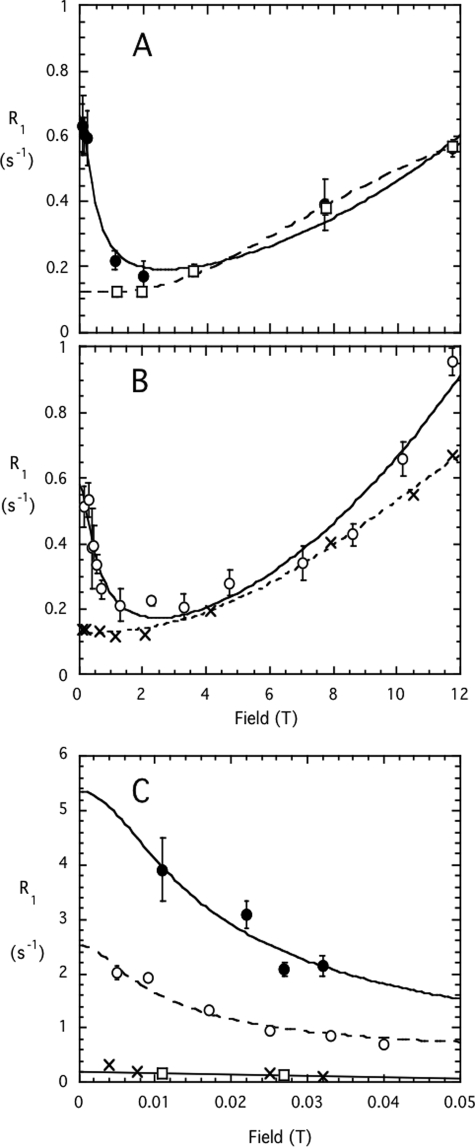
This difference in how rPI-PLC and Y246S/Y247S/Y248S interact with diC7PC was further explored using high resolution 31P NMR field cycling (Fig. 8). This is a unique technique that can probe the dynamics of phospholipids in aggregates and assess the effect of additives (protein, peptide, or other lipids) (16, 17). At a concentration of 3 mm, diC7PC forms rod-shaped polydisperse micelles with an average size of ∼60 monomers per micelle (34). Such a rod-shaped micelle would have a transverse radius of 18 Å and a length ∼80 Å. The 31P R1, the spin-lattice relaxation rate (1/T1), for 3 mm diC7PC in the absence of protein (open circles in Fig. 8B) was ∼ 0.9 s−1 at 11.7 T. R1 then decreased with a decreasing field to a minimum around 2 T and increased again as the field decreased below 2 T. The high field behavior reflects relaxation by chemical shift anisotropy (here R1 is proportional to the square of the field) with two characteristic correlation times; that is, a fast motion at 150–200 ps and a slower one (τc) of 5.8 ± 1.7 ns. An order parameter for the chemical shift anisotropy contribution can also be estimated, and the value for 3 mm diC7PC (0.26 for Sc2) was considerably smaller than obtained for PC in SUVs (16), consistent with the more disordered structure of micelles. As the field was decreased below 2 T, the increasing R1 found in all phospholipid aggregates indicates that dipolar relaxation of the 5–10-ns slower motion has now become the dominant relaxation mechanism. Extrapolation of R1 to zero field in this field region provides the parameter Rc(0), which reflects both τc, a correlation time for effective rotation/wobbling of the head group, and rPH, the effective P-H distance related to an 〈r−6〉6 average of distances r from P to H (16). Decreasing the field below 0.06 T (Fig. 8C) led to a substantial increase in the dipolar relaxation rate for diC7PC. From the shape of this very low field curve and knowing Rc(0) from the analysis of the R1 field dependence in the 0.08 to 11.74 T region, we can extract a correlation time, τv, that has components from both particle rotation and exchange (17). For this concentration of diC7PC, the τv was 0.36 ± 0.08 μs. We can obtain a rough estimate of the average particle size by relating τv to an isotropic rotational diffusion constant (τr = 1/(6Do) and Do = kT/(8 πηr3)) (35). The extracted r for the diC7PC micelles would then be 70 Å. Integration of the very low field dipolar contribution compared with the integration for dipolar relaxation over the entire field range examined reflects an averaged angle θPH for the P to glycerol CH2 with respect to the field (17). For the pure diC7PC micelles at this concentration, the average θPH was 45.6°. DiC7PC was previous examined at 10 mm, and the averaged angle was the same (45.4°) (17).
FIGURE 8.
Field dependence of the spin-lattice relaxation rate, R1, for short-chain phosphatidylcholines in the absence and presence of PI-PLC enzymes. Data covering the field range 0.06 to 11.7 T is shown in (A) for rPI-PLC (●) and Y246S/Y247S/Y248S (□) in the presence of 3 mm diC7PC and in B for 3 mm diC7PC (○) and 25 mm diC6PC (X). C shows at fields below 0.05 T. The lines drawn through the each data set represent best fits to equations discussed in Refs. 16 and 17).
In the presence of 3 mg/ml or 0.086 mm rPI-PLC (filled circles in Fig. 8A) there were small but significant changes in the field dependence profile from 0.08 up to 11.7 T. These corresponded to a slightly longer τc and a significantly increased Sc2 (Table 3). In the very low field region the τv correlation time for diC7PC in the presence of rPI-PLC was 0.22 ± 0.08 μs. The decrease in τv corresponds to a slightly smaller average radius for the particle (60 Å) compared with pure diC7PC micelles. However, the Rv(0) value was more than 2-fold higher (Fig. 8C). The increased Rv(0) with a slightly lower τv is consistent with a different average orientation of the POCH2 in the rPI-PLC·diC7PC complex (Table 3). The averaged θPH now corresponded to 37.8°. This is a very significant change in the average orientation of the P-to-glycerol C(3)-H vector in the particle formed with diC7PC and rPI-PLC.
TABLE 3.
Dynamic parameters extracted from the field dependence of the31P spin-lattice relaxation time of diC7PC and diC6PC and the effect of PI-PLC enzymes
| Samplea | Rc (0) | τc | rPH | Sc2 | τhf, psb | Rv (0) | τv | θPH |
|---|---|---|---|---|---|---|---|---|
| s−1 | ns | Å | s−1 | μs | ||||
| rPI-PLC + diC7PC | 0.66 ± 0.05 | 6.9 ± 2.6 | 3.15 | 0.26 | 100 | 4.69 ± 1.23 | 0.22 ± 0.08 | 37.8° |
| 3Y-S + diC7PC | 0.12 ± 0.03 | 1.2 ± 0.2 | 3.12 | 0.17 | 40 | —c | —c | —c |
| diC7PC | 0.58 ± 0.08 | 5.8 ± 1.7 | 3.12 | 0.14 | 150 | 1.94 ± 0.28 | 0.36 ± 0.08 | 45.6° |
| diC6PC | 0.13 ± 0.01 | 1.0 ± 0.5 | 2.99 | 0.13 | 60 | —c | —c | —c |
a The diC7PC concentration was 3 mm; the diC6PC concentration was 25 mm. The PI-PLC enzyme 3Y-S is the interfacial challenged triple mutant Y246S/Y247S/Y248S.
b Errors in estimating τhfdepend on errors in the Sc2term and are likely to be 30–40%; hence, the value for each short-chain PC in the absence and presence of protein are essentially the same.
c The very low field rise in R1is only to about 0.4 s−1; therefore, that we cannot accurately determine Rv(0) and τv(both are needed to derive θPH).
In contrast to rPI-PLC, Y246S/Y247S/Y248S had a very different effect on diC7PC parameters (open squares in Fig. 8A) at the same ratio of diC7PC/protein (35:1). The τc decreased significantly to 1.2 ns, Rc(0) was also much smaller than with rPI-PLC, and the extrapolated fast correlation time was also significantly shorter (Fig. 8A). The order parameter Sc2 was smaller than that observed with rPI-PLC but greater than the value for pure diC7PC at this concentration. Most interestingly, at very low fields there was only a small increase in R1, indicating that the diC7PC now forms small aggregates with the tyrosine mutant enzyme (Fig. 8C). These may resemble the detergent-solubilized integral membrane proteins now used for NMR structural studies in that the micelle contribution to particle size is quite small. The relaxation profile for diC7PC with the mutant Y246S/Y247S/Y248S PI-PLC enzyme is much more like that for diC6PC micelles (36), which form small nearly spherical aggregates of 19 ± 1 molecules. This cannot be the result of slow exchange between protein-bound diC7PC and free diC7PC, as going to lower and lower fields would shift such an equilibrium to the fast exchange regime. All these different binding studies indicate that as these surface Tyr are replaced by Ser, the interaction with PC molecules is significantly altered.
Structures of Y247S/Y251S and Y246S/Y247S/Y248S/ Y251S
To explore if removal of these surface tyrosine residues alters the PI-PLC active site, we determined the crystal structure of two of the tyrosine mutants, Y247S/Y251S (PDB entry 3EA1) and Y246S/Y247S/Y248S/Y251S (PDB entry 3EA3). Crystallographic data and refinement statistics are presented in Table 4. The Y247S/Y251S mutant crystallized in the space group of P212121 with two monomers in the asymmetric unit. The PI-PLC Y246S/Y247S/Y248S/Y251S mutant protein crystallized in space group P1; its asymmetric unit also contained two monomers. Both tyrosine-to-serine mutant structures are of high quality at 1.75 and 1.78 Å resolution (Table 4), and the individual monomers in both mutant structures closely resemble the PI-PLC structure from B. cereus (PDB entry 1PTD) (10). The proteins fold as an incomplete (βα)8 barrel (TIM-barrel) with an inner layer composed of eight β-strands surrounded by six major α-helices of the outer layer. As was the case with the structure of W47A/W242A (9), the last two residues of PI-PLC, Lys297 and Glu298, did not have observable electron density. The main-chain r.m.s.d. is 0.610 Å between the structures of the B. cereus PI-PLC and the Y247S/Y251S mutant and 0.654 Å between the B. cereus PI-PLC and the Y246S/Y247S/Y248S/Y251S (Table 5). The wild type-like double tyrosine mutant (Y247S/Y251S) and the PC interface-impaired quadruple mutant structures are even more similar to each other, with an r.m.s.d. of only 0.310 Å (Table 5). Structural alignments and comparisons indicate the mutations introduced little conformational change in the crystallized proteins (Fig. 9A).
TABLE 5.
Root mean square deviations (Å) of superimposed PI-PLC structures
2Y-S, Y247S/Y251S; 4Y-S, Y246S/Y247S/Y248S/Y251S; 2OR2-HB/RL is a monomer from the W47A/W242A dimer structure omitting helix B and the rim loop region containing Trp242in the comparison. Ins, inositol.
| PDB entrya | 2Y-S | 2Y-S (+Ins) | 4Y-S | 2OR2 | 2OR2-HB/RL |
|---|---|---|---|---|---|
| 1PTD | 0.610 | 0.654 | |||
| 1PTG (+Ins) | 0.597 | 0.592 | 0.629 | ||
| 2Y-S | 0.135 | 0.310 | 1.379 | 0.471 | |
| 4Y-S | 1.410 | 0.507 |
FIGURE 9.
Myo-inositol complex with B. thuringiensis PI-PLC Y247S/Y251S compared with corresponding complex from wild type B. cereus PI-PLC (PDB code 1PTG). The latter structure is shown in magenta in both A and B. A, backbone superposition with the Y247S/Y251S structure shown in green, myo-inositol (unlabeled) displayed in the active site, and non-mutated tyrosine side chains (labeled in bold) shown as markers to indicate tyrosine strip region. B, superposition of B. cereus PI-PLC active site residues with the B. thuringiensis PI-PLC Y247S/Y251S-myo-inositol complex shown in Corey-Pauling-Koltun coloring. Hydrogen bonds are shown as green dotted lines.
The helix B region and the rim loop in both mutant protein crystal structures are similar to the corresponding regions in the B. cereus monomer structure (Table 5), except that the position of Trp242 in the flexible rim loop was slightly shifted. The active sites of the B. cereus PI-PLC and both tyrosine mutants, Y247S/Y251S, with activity similar to wild type protein, and Y246S/Y247S/Y248S/Y251S, with greatly reduced interfacial activity, are superimposable except for His82, which is rotated by about 90° in both mutant protein crystal structures. A crystal of Y247S/Y251S PI-PLC was later soaked with 100 mm myo-inositol (PDB entry 3EA2) for comparison to the B. cereus PI-PLC structure with myo-inositol in the active site (PDB entry 1PTG) (10). The r.m.s.d. between those two structures is 0.597 Å, and the structural alignment between them reveals no significant conformational changes for active site residues; bound myo-inositol is also at the same position for the two protein structures (Fig. 9B). Therefore, in the crystal and in the absence of an interface, the tyrosine-to-serine mutations of PI-PLC appear to have little influence on the active site geometry either with or without an inositol moiety bound.
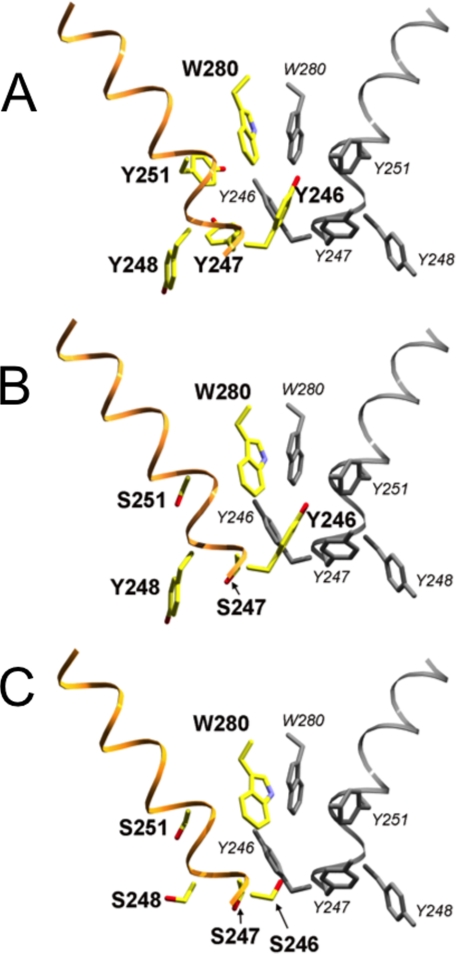
The PI-PLC W47A/W242A mutant was crystallized as a dimer, prompting the conjecture that the membrane state of the enzyme might be a dimer. As shown in Figs. 2 and 10A, Tyr246, Tyr247, Tyr248, and Tyr251 from each subunit of the dimer form a quasi-herringbone pattern that stabilizes the dimer interface (9). Although both Y247S/Y251S and Y246S/Y247S/Y248S/Y251S mutant monomers are structurally similar to a crystallographic monomer of the W47A/W242A PI-PLC dimer, neither tyrosine mutant crystallized as a dimer. For rPI-PLC, the lack of dimer formation in solution is presumably because helix B is still intact, and the hydrophobic tryptophan “plug” this feature forms in solution prevents close approach of another PI-PLC and subsequent dimerization via the surface tyrosine residues (9). A closer examination of the putative dimer interface region for the tyrosine mutants shows that the essential interactions cannot be made when four of the crucial tyrosine residues have been replaced by serine (Fig. 10C). However, Y247S/Y251S still has Tyr246 and Tyr248 as well as Trp280 to stabilize the hydrophobic core of a membrane-induced dimer interface (Fig. 10B). The remaining interactions may be adequate to allow dimers to form on the membrane surface. Tyr246 in particular may be a key interaction for a surface-induced dimer that resembles that of W47A/W242A.
FIGURE 10.
Location of tyrosine → serine substitutions in B. thuringiensis PI-PLC mutants relative to the crystallographic dimer interface observed in the W47A/W242A mutant (PDB code 2OR2). Backbone ribbon representation with key side chains is shown in stick representation. A, crystallographic dimer (2OR2), with the second subunit shown in gray. Y247S/Y251S (B) and Y246S/Y247S/Y248S/Y251S (C) mutants, shown in Corey-Pauling-Koltun coloration, have been superimposed onto the first subunit of the A dimer structure; the second subunit from A is included to complete the hypothetical dimer interface. This comparison supports the hypothesis that the more active Y247S/Y251S mutant, but not the catalytically impaired Y246S/Y247S/Y248S/Y251S mutant, retains the key tyrosine residues needed for dimer formation.
The high similarity between the structures of the double/quadruple mutant and the wild type PI-PLC indicate that the reduction in enzymatic activities of the triple and quadruple tyrosine mutant proteins and altered binding affinity toward PC vesicles is not likely to be caused by large conformational changes within the single monomer domain. Two possible explanations may explain how replacement of these surface Tyr residues with Ser reduces enzymatic activity; (i) removal of these tyrosine residues disrupts the transient dimer that forms when protein binds to PC-containing membranes, or (ii) these same tyrosine residues are critical parts of a defined PC binding site that aids in anchoring the protein as a monomer to interfaces.
DISCUSSION
Although the binding of peripheral proteins to membranes often has a strong electrostatic component, tryptophan insertion into the membrane is also frequently observed, particularly for interfacial binding to PC surfaces (37–39). Tryptophan residues are particularly prominent in the region of the bilayer interfaces in membrane proteins as well (40–42). Previous work has suggested that Trp47 and Trp242 of the B. thuringiensis PI-PLC insert into the membrane surface (7, 8) and that this interaction is facilitated by PC molecules present in the interface (11). Trp insertion in turn helps anchor the PI-PLC to vesicles and is ultimately connected to enzymatic activation in that assay system. A variety of biophysical techniques suggested that binding of the protein to the interface changed the enzyme conformation and shifted the enzyme to an “activated” form (4–6, 14). However, no specifics were available on the structure of that activated form.
Interaction of a phospholipid with a specific binding site on the protein has been detected with a wide variety of peripheral membrane proteins. It usually is identified by enhanced membrane association (e.g. C2 domains and Ca2+-mediated interaction with anionic phospholipids, PH domains, and phosphoinositides) or altered kinetics (if the protein is an enzyme). Unless the interaction is of fairly high affinity, localizing the binding site can be difficult and is usually explored with mutagenesis. Along with specific phospholipid binding, protein oligomerization may also provide a way of regulating function. There are integral membrane proteins that are purified as monomers but are believed to function as dimers or oligomers. For example, a tetracycline-divalent cation efflux protein TetL was suggested to exist as an oligomer rather than a monomer in the membrane based on SDS-PAGE and Western blot analysis of membrane samples (43). However, exploring membrane-induced oligomerization of peripheral membrane proteins, particularly where such aggregation might not be necessary for enzymatic activity but rather enhance catalysis, is very difficult. Chemical cross-linking only works if suitable chemical handles are available (14). Association and dissociation of the proteins from the membrane can also make trapping aggregates difficult.
Initial studies (4–6) with the B. thuringiensis PI-PLC suggested that kinetic activation of cIP (and monomeric diC4PI) hydrolysis by PC involved the protein binding to a PC interface at a site distinct from the active site. That interaction then induced a conformational change in the active site that enhanced catalysis of water-soluble substrates. Localization of a distinct binding region on PI-PLC was restricted to the observation that the Trp47 and Trp242 seemed to aid in binding of the protein to interfaces. More recent work, that of a crystal structure of a dimeric protein when the two key surface tryptophan residues were changed to alanine, suggested that the specific kinetic activation of PI-PLC by PC interfaces was the result of interface-induced dimerization of the protein (9). The extrapolation of that dimer to rPI-PLC posited that when the Trp side chains were present, a hydrophobic Trp plug centered on helix B prevents dimer formation in solution. Thus, the initial approach of the protein to the lipid interface would be as a monomer. Changes in this helix B feature as well as the rim of the βα-barrel as a result of partitioning of Trp47 into the membrane would displace the Trp plug and allow transient dimers to form on the surface. In the case of PI cleavage, the dimer could enhance catalytic activity by allowing each monomer to alternate substrate binding and catalysis. Although the product diacylglycerol could easily be released back into the bilayer, water-soluble cIP must be released into solution. This would require the rim of the barrel to be at least partially solvent-exposed. For a monomeric protein, release of cIP might destabilize membrane binding of the protein and cause the protein to dissociate from the membrane. Dimerization could effectively increase the affinity of the protein for substrate by increasing the residence time of the protein on the membrane, essentially promoting “scooting” rather than “hopping” of the enzyme (44, 45). However, anchoring of a PI-PLC monomer via a discrete PC binding site could also keep the protein on the surface long enough for processive catalysis. Can these two possibilities be distinguished or at least constrained with the mutagenesis experiments reported in this work?
A dimer interface similar to what we see with W47A/W242A would be held together by four key tyrosine residues, Tyr246, Tyr247, Tyr248, and Tyr251. Because these tyrosine residues are involved in both hydrophobic and hydrophilic binding between the two subunits, replacing some or all of these Tyr residues should impair dimer formation, reduce the affinity of the protein for interfaces, and reduce catalytic activity. The three different tyrosine mutants constructed to test the importance of PI-PLC dimer formation for interfacial activation of PI-PLC by PC surfaces definitely show that this region of the protein is linked to kinetic activation. At least in the crystal, removal of tyrosines does not alter the active site structure. In solution, there may be a small loss of helix in the area of the tyrosine strip, but at least for Y247S/Y251S this can be retrieved by occupation of the active site. The difference in crystal and solution conformations does highlight one important point; occupation of the active site can affect the secondary structure of the protein, at least around helix G. So if the dimer is formed, it could influence detailed arrangements of groups in the active site, at least enough to increase kcat.
Clearly, when the four key tyrosines are replaced by serine, the herringbone intersubunit interactions can no longer occur. Consistent with this, the quadruple mutant has reduced activity (and reduced PC affinity) in all assay systems. The other two tyrosine mutant enzymes would be expected to have different dimerization potentials. The double mutant Y247S/Y251S behaved kinetically almost the same as rPI-PLC. The crystal structure of this mutant suggests that whereas the putative dimer interface might be somewhat destabilized, many intersubunit interactions can still be formed to stabilize the dimer on a PC-containing membrane. Because occupation of the active by a ligand (myo-inositol) was shown to enhance structure in this protein, it is likely that a phospholipid at the active could work in concert with the Tyr residues in stabilizing, at least transiently, a surface-bound dimer.
The alternate explanation is that these Tyr form part of a well defined PC binding site on a monomer that is distinct from the interaction of Trp47 and Trp242 with interfaces. Tyr aromatic rings could form a binding site for the positively charged choline moiety. This region of PI-PLC is rich in surface Tyr, and several of them could stabilize PC binding via π-cation interactions. In that case one would also expect removal of the Tyr would dramatically reduce the enzyme activity, particularly for PI/PC cleavage and diC7PC activation of cIP.
The observation that both triple and quadruple mutant enzymes have dramatically altered interactions with diC7PC micelles provides clues into what happens to the protein when it binds activator lipids. Both these less active enzymes still bind diC7PC molecules with fairly good affinity, as assessed by the intrinsic fluorescence experiments. However, the change in intrinsic fluorescence of the protein upon binding to diC7PC is more in keeping with a ligand binding to the active site. In the field cycling experiments, Y246S/Y247S/Y248S, compared with rPI-PLC, converts the polydisperse rod-shaped diC7PC micelles to much smaller structures at a ratio of diC7PC to protein where the maximum line broadening is observed. At this total concentration of diC7PC to PI-PLC, 35:1, there are no larger micelles detected that coexist. A PI-PLC monomer could bind a relatively small and finite number of these diC7PC molecules to form a nearly spherical small mixed micelle. However, the very elongated dimer, like that observed for W47A/W242A, would certainly form larger aggregates. It would present an extended rim with a number of hydrophobic contacts.
If by replacing the Tyr with Ser we have removed a specific PC binding site, then there should be little difference in specific activity for PI cleavage in diC7PC versus Triton X-100 for the triple and quadruple Tyr mutant proteins. Yet there is a 2–3-fold decrease in the Triton X-100 assay system (even more pronounced than for rPI-PLC). If transient surface-bound dimers are the optimally active species, then they must form in PI/Triton X-100 as well as in PC-containing systems. The high relative activities for the Tyr mutants in both micelle systems compared with PI/PC SUV or cIP/diC7PC assay systems would then suggest that an amphiphilic molecule occupying the active site is also very important for biasing the distribution to dimers.
On balance, then, these experiments seem more consistent with a surface-activated dimer analogous to W47A/W242A. Could dimers form in the absence of interfaces? Some indication of this may be provided by examining the second step of the reaction, cIP hydrolysis. In solution in the absence of interfaces, monomer and a very small fraction of dimers of rPI-PLC could exist in equilibrium (indeed, in previous cross-linking experiments we could observe a very small amount of dimer for protein alone in solution (14), suggesting aggregates might occur). Removing any of the tyrosine residues that contribute to the dimer interface of W47A/W4242A would be expected to bias the equilibrium even more toward monomer. The reduced cIP hydrolysis rates for all the tyrosine-to-serine mutant enzymes in the absence of diC7PC are consistent with this possibility. If a diC7PC interface can still enhance dimerization, then the cIP hydrolysis rate should increase dramatically as observed for Y247S/Y251S. These kinetics are harder to explain with the postulate of a specific PC binding site. It is interesting that all the Tyr mutant cIP hydrolysis activities are similar (0.2 to 0.6 μmol min−1 mg−1) and considerably smaller than for rPI-PLC (2.3 μmol min−1 mg−1). If disruption of a PC binding site were the result of the Tyr replacements with Ser, then one would not expect much of difference between rPI-PLC and these mutant enzymes in the absence of diC7PC (and this is what is observed with examining the kinetics of W47A, W242A, and W47A/W242A (7), mutants that reduce binding to PC interfaces but would not alter a monomer-dimer equilibrium involving Tyr246, Tyr247, Tyr248, and Tyr251).
In summary, we have probed the importance of the strip of tyrosine residues that form the core of the dimer interface observed previously in the mutant protein W47A/W242A. The reduction in specific activity in all assay systems with the removal of three or four of these tyrosines can arise from either two effects. (i) The lack of the tyrosine strip disfavors the surface-induced dimer formation that is needed for optimal activity. (ii) Removal of these tyrosines abolishes a specific and critical PC binding site. Because both phosphotransferase and cyclic phosphodiesterase activities are reduced by these mutations (and the physical interaction of the mutant protein with PC interfaces is altered), we cannot absolutely distinguish between these two alternatives (and the definition of the activated form of the enzyme they imply). However, the evidence, particularly kinetic changes in PI/Triton X-100 and cIP in the absence of interfaces, appears more consistent with dimerization of this PI-PLC as the activated form of the enzyme.
This work was supported, in whole or in part, by National Institutes of Health Grant GM60418 (to M. F. R.).
The atomic coordinates and structure factors (codes 3EA1, 3EA2 and 3EA3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PI-PLC
- phosphatidylinositol (PI)-specific phospholipase C
- cIP
- 1,2-cyclic inositol phosphate
- rPI-PLC
- recombinant PI-PLC
- I-1-P
- d-inositol-1-phosphate
- PC
- phosphatidylcholine
- POPC
- 1-palmitoyl-2-oleoyl-PC
- diC7PC
- diheptanoylphosphatidylcholine
- r.m.s.d.
- root mean standard deviation
- SUV
- small unilamellar vesicle
- diC6PC
- dihexanoyl-PC
- diC6PA
- dihexanoylphosphatidic acid
- T
- tesla.
REFERENCES
- 1.Heinz D. W., Essen L. O., Williams R. L. ( 1998) J. Mol. Biol 275, 635– 650 [DOI] [PubMed] [Google Scholar]
- 2.Rebecchi M. J., Pentyala S. N. ( 2000) Physiol. Rev 80, 1291– 1335 [DOI] [PubMed] [Google Scholar]
- 3.Toker A. ( 2002) Cell. Mol. Life Sci 59, 761– 779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou C., Wu Y., Roberts M. F. ( 1997) Biochemistry 36, 347– 355 [DOI] [PubMed] [Google Scholar]
- 5.Zhou C., Qian X., Roberts M. F. ( 1997) Biochemistry 36, 10089– 10097 [DOI] [PubMed] [Google Scholar]
- 6.Qian X., Zhou C., Roberts M. F. ( 1998) Biochemistry 37, 6513– 6522 [DOI] [PubMed] [Google Scholar]
- 7.Feng J., Wehbi H., Roberts M. F. ( 2002) J. Biol. Chem 277, 19867– 19875 [DOI] [PubMed] [Google Scholar]
- 8.Feng J., Bradley W. D., Roberts M. F. ( 2003) J. Biol. Chem 278, 24651– 24657 [DOI] [PubMed] [Google Scholar]
- 9.Shao C., Shi X., Wehbi H., Zambonelli C., Head J. F., Seaton B. A., Roberts M. F. ( 2007) J. Biol. Chem 282, 9228– 9235 [DOI] [PubMed] [Google Scholar]
- 10.Heinz D. W., Ryan M., Bullock T. L., Griffith O. H. ( 1995) EMBO J 14, 3855– 3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wehbi H., Feng J., Kolbeck J., Ananthanarayanan B., Cho W., Roberts M. F. ( 2003) Biochemistry 42, 9374– 9382 [DOI] [PubMed] [Google Scholar]
- 12.Böhm G., Muhr R., Jaenicke R. ( 1992) Protein Eng 5, 191– 195 [DOI] [PubMed] [Google Scholar]
- 13.Andrade M. A., Chacón P., Merelo J. J., Morán F. ( 1993) Protein Eng 6, 383– 390 [DOI] [PubMed] [Google Scholar]
- 14.Zhang X., Wehbi H., Roberts M. F. ( 2004) J. Biol. Chem 279, 20490– 20500 [DOI] [PubMed] [Google Scholar]
- 15.Roberts M. F., Cui Q., Turner C. J., Case D. A., Redfield A. G. ( 2004) Biochemistry 43, 3637– 3650 [DOI] [PubMed] [Google Scholar]
- 16.Roberts M. F., Redfield A. G. ( 2004) J. Am. Chem. Soc 126, 13765– 13777 [DOI] [PubMed] [Google Scholar]
- 17.Roberts M. F., Redfield A. G. ( 2004) Proc. Natl. Acad. Sci. U. S. A 101, 17066– 17071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipari G., Szabo A. ( 1982) J. Am. Chem. Soc 104, 4546– 4559 [Google Scholar]
- 19.Herzfeld J., Griffin R. G., Haberkorn R. A. ( 1978) Biochemistry 17, 2711– 2718 [DOI] [PubMed] [Google Scholar]
- 20.Woessner D. E. ( 1995) in Encyclopedia of NMR ( Grant D. M. ed.) pp. 1068– 1083, John Wiley & Sons, Inc., New York [Google Scholar]
- 21.Klauda J. B., Roberts M. F., Redfield A. G., Brooks B. R., Pastor R. W. ( 2008) Biophys. J 94, 3074– 3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z., Minor W. ( 1997) Methods Enzymol 276, 306– 326 [DOI] [PubMed] [Google Scholar]
- 23.Kissinger C. R., Gehlhaar D. K., Fogel D. B. ( 1999) Acta Crystallogr. D Biol. Crystallogr 55, 484– 491 [DOI] [PubMed] [Google Scholar]
- 24.Jones T. A., Kjeldgaard M. ( 1992) O: The Manual, Uppsala Software Factory, Uppsala, Sweden [Google Scholar]
- 25.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. ( 1998) Acta Crystallogr. D Biol. Crystallogr 54, 905– 921 [DOI] [PubMed] [Google Scholar]
- 26.DeLano W. L. ( 2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 27.Tsou L. K., Tatko C. D., Waters M. L. ( 2002) J. Am. Chem. Soc 124, 14917– 14921 [DOI] [PubMed] [Google Scholar]
- 28.Wu Y., Roberts M. F. ( 1997) Biochemistry 36, 8514– 8521 [DOI] [PubMed] [Google Scholar]
- 29.Wehbi H., Feng J., Roberts M. F. ( 2003) Biochim. Biophys. Acta 1613, 15– 27 [DOI] [PubMed] [Google Scholar]
- 30.Volwerk J. J., Shashidhar M. S., Kuppe A., Griffith O. H. ( 1990) Biochemistry 29, 8056– 8062 [DOI] [PubMed] [Google Scholar]
- 31.Zhou C., Roberts M. F. ( 1998) Biochemistry 37, 16430– 16439 [DOI] [PubMed] [Google Scholar]
- 32.Bian J., Roberts M. F. ( 1992) J. Colloid Interface Sci 153, 420– 428 [Google Scholar]
- 33.Berg O. G., Yu B. Z., Apitz-Castro R. J., Jain M. K. ( 2004) Biochemistry 43, 2080– 2090 [DOI] [PubMed] [Google Scholar]
- 34.Lin T. L., Chen S. H., Gabriel N. E., Roberts M. F. ( 1987) J. Phys. Chem 91, 406– 413 [Google Scholar]
- 35.Barrall G. A., Schmidt-Rohr K., Lee Y. K., Landfester K., Zimmermann H., Chingas G. C., Pines A. ( 1995) J. Chem. Phys 104, 509– 520 [Google Scholar]
- 36.Lin T. L., Chen S. H., Gabriel N. E., Roberts M. F. ( 1986) J. Am. Chem. Soc 108, 3499– 3507 [Google Scholar]
- 37.Sopkova-De Oliveira Santos J., Fischer S., Guilbert C., Lewit-Bentley A., Smith J. C. ( 2000) Biochemistry 39, 14065– 14074 [DOI] [PubMed] [Google Scholar]
- 38.Ghosh A. K., Rukmini R., Chattopadhyay A. ( 1997) Biochemistry 36, 14291– 14305 [DOI] [PubMed] [Google Scholar]
- 39.Han S. K., Kim K. P., Koduri R., Bittova L., Munoz N. M., Leff A. R., Wilton D. C., Gelb M. H., Cho W. ( 1999) J. Biol. Chem 274, 11881– 11888 [DOI] [PubMed] [Google Scholar]
- 40.Wallace B. A., Janes R. W. ( 1999) Adv. Exp. Med. Biol 467, 789– 799 [DOI] [PubMed] [Google Scholar]
- 41.Hong H., Park S., Jiménez R. H., Rinehart D., Tamm L. K. ( 2007) J. Am. Chem. Soc 129, 8320– 8327 [DOI] [PubMed] [Google Scholar]
- 42.Schiffer M., Chang C. H., Stevens F. J. ( 1992) Protein Eng 5, 213– 214 [DOI] [PubMed] [Google Scholar]
- 43.Safferling M., Griffith H., Jin J., Sharp J., De Jesus M., Ng C., Krulwich T. A., Wang D. N. ( 2003) Biochemistry 42, 13969– 13976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain M. K., Berg O. G. ( 1989) Biochim. Biophys. Acta 1002, 127– 156 [DOI] [PubMed] [Google Scholar]
- 45.Gelb M. H., Min J. H., Jain M. K. ( 2000) Biochim. Biophys. Acta 1488, 20– 27 [DOI] [PubMed] [Google Scholar]