Abstract
Bone tissue arises from mesenchymal cells induced into the osteoblast lineage by essential transcription factors and signaling cascades. MicroRNAs regulate biological processes by binding to mRNA 3′-untranslated region (UTR) sequences to attenuate protein synthesis. Here we performed microRNA profiling and identified miRs that are up-regulated through stages of osteoblast differentiation. Among these are the miR-29, miR-let-7, and miR-26 families that target many collagens and extracellular matrix proteins. We find that miR-29b supports osteoblast differentiation through several mechanisms. miR-29b decreased and anti-miR-29b increased activity of COL1A1, COL5A3, and COL4A2 3′-UTR sequences in reporter assays, as well as endogenous gene expression. These results support a mechanism for regulating collagen protein accumulation during the mineralization stage when miR-29b reaches peak levels. We propose that this mechanism prevents fibrosis and facilitates mineral deposition. Our studies further demonstrate that miR-29b promotes osteogenesis by directly down-regulating known inhibitors of osteoblast differentiation, HDAC4, TGFβ3, ACVR2A, CTNNBIP1, and DUSP2 proteins through binding to target 3′-UTR sequences in their mRNAs. Thus, miR-29b is a key regulator of development of the osteoblast phenotype by targeting anti-osteogenic factors and modulating bone extracellular matrix proteins.
Skeletal development and bone remodeling require stringent control of gene expression for osteoprogenitor lineage cells to progress through stages of differentiation. Osteoprogenitors proliferate then become mature osteoblasts to secrete an extracellular matrix (ECM),3 largely of collagen type I. Specialized noncollagenous proteins are also synthesized for promoting mineralization of the tissue that results in the final stage of cell differentiation to osteocytes. Osteoprogenitor cells are directed to this fate by a number of developmental signals and tissue-specific transcription factors for induction of the phenotype. Runx2 (Cbfa1/AML3) and Osterix transcription factors play crucial roles in osteoblast differentiation and bone formation at multiple levels (1–4). As an initial marker of the osteogenic cell lineage, Runx2 has epigenetic functions and contributes to activated expression of principal osteoblast-related genes (2). In addition, several signaling pathways including Wnt/β-catenin (5, 6), bone morphogenetic protein (7, 8), JAK/STAT (9, 10), and MAPK (10) have all been reported to promote osteogenesis in vivo and in vitro. In contrast to the number of osteogenic activating factors that are known, comparatively few negative regulators of bone formation have been identified.
MicroRNAs (miRNAs), an abundant class of noncoding small (∼22 nucleotides) RNAs, have emerged as key regulators of diverse biological and pathological processes, including developmental timing, organogenesis, apoptosis, cell proliferation, and differentiation (11–13). Many miRNAs have been characterized in relation to tumorigenesis (14, 15), but only a few studies have examined the role of miRNAs in the formation of skeletal tissues (16–19). Recently, we have shown that the bone morphogenetic protein 2, an inducer of osteogenesis, down-regulates a group of miRs to relieve the negative control on bone formation in nonosseous cells (20). This study established the importance of miRNA repression of alternative cell fate pathways for commitment to the osteoprogenitor cell phenotype (20). The specific miRNAs that contribute to regulation of the osteoblast differentiation process from the committed cell to the mature osteocyte in a mineralized bone matrix remain to be identified.
In this study, we performed microRNA profiling studies during stages of osteoblast differentiation. During maturation of murine calvaria-derived preosteoblasts (MC3T3 cells), we find the majority of miRs become up-regulated during the mineralization stage. Our studies focused on characterization of miR-29b, one of the most robustly expressed microRNAs throughout osteoblast differentiation. We find that miR-29b is a positive regulator of osteoblast differentiation by down-regulating inhibitory factors of osteogenic signaling pathways and controlling expression of collagen in differentiated osteoblasts.
Other studies have identified members of the miR-29 family as tumor suppressors that are down-regulated in association with various cancers (21–24) and activate p53 (25). One study showed miR-29 was related to metastasis in nasopharyngeal and lung cancers through targeting of extracellular matrix collagens (24). miR-29 was also reported to be down-regulated after a myocardial infarct and contributed to fibrosis of the tissue (26). However, functions of miR-29 in relation to normal cellular processes have not been identified.
EXPERIMENTAL PROCEDURES
Cell Culture
Primary fetal rat calvaria osteoblasts were isolated by sequential collagenase digestion as previously described (27). Two mouse osteoblastic cell lines, the parental MC3T3-E1 cells (28–30) and MC3T3-E1 clone 4 from American Type Culture Collection (Manassas, VA) (31), were maintained in α-minimum essential medium supplemented with 10% fetal bovine serum and 100 units/ml of penicillin and 100 μg/ml of streptomycin. For differentiation studies, rat calvaria osteoblasts and MC3T3 cells were seeded in 6-well or 100-mm dishes and were fed at confluence (0 time) with the above medium containing differentiation supplements 10 mm β-glycerophosphate (Sigma) and 50 μg/ml of ascorbic acid (Sigma) (30). The osteogenic medium was changed every 2 days, and the cells were harvested at the indicated times (0, 4, 7, 10, 14, 21, and 28 days) after differentiation was induced. Cell layers were either harvested for protein and mRNA or fixed in 2% paraformaldehyde for histochemical detection of alkaline phosphatase (AlkP) (using Sigma reagents) and mineral by the von Kossa stain (3% silver nitrate in H20) as previously described (6, 27).
Western Blot
Whole cell lysate preparation and Western blot analysis were performed as described previously (6). The primary antibodies used were as follows: mouse monoclonal anti-Runx2 (1:2000) (R & D Systems), goat polyclonal anti-actin (1:5000) (Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-Cdk2 (1:2000) (Santa Cruz Biotechnology, Inc.), goat polyclonal anti-collagen α1 type I (1:2000) (Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-HDAC4 (1:2000) (Santa Cruz Biotechnology, Inc.), and rabbit polyclonal anti-TGFβ3 (1:1000) (Santa Cruz Biotechnology, Inc.). Membranes were incubated for 1 h at room temperature with primary antibody in 5% milk and subsequently with horseradish peroxidase-conjugated secondary antibody. The signal was detected using a chemiluminescence detection kit (PerkinElmer Life Sciences).
miRNA Microarray and Data Analysis
RNA of MC3T3 cells at 0, 7, 14, 21, and 28 days following induced differentiation were used for miRNA array analysis. Microarray procedures and data analysis were performed at the Ohio State University as described previously (32). For microRNA precursor quantitation, the fold change of each time point was obtained by normalizing Log2 fluorescence with Log2 fluorescence of 0 day. The normalized fold changes were analyzed by using dChip software.
Real Time PCR
Total RNA was isolated from MC3T3 cells using TRIzol reagent (Invitrogen) per the manufacturer's instructions and was purified with the DNA-free RNA kit (Zymo Research Corp.). RNA (1 μg) of each time point was used to make cDNA through a reverse transcription reaction with oligo(dT) primers. The cDNA was then used for real time PCR with SYBR chemistry (Applied Biosystems, Inc., Foster City, CA). mRNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. The primers used for amplification are listed in Table 1.
TABLE 1.
Nucleotide sequences of primers used for quantitative RT-PCR detection
| Runx2 | 5′-CGCCCCTCCCTGAACTCT-3′ (Forward) |
| 5′-TGCCTGCCTGGGATCTGTA-3′ (Reverse) | |
| Alkaline phosphatase | 5′-TTGTGCGAGAGAAAGGAGA-3′ (Forward) |
| 5′-GTTTCAGGGCATTTTTCAAGGT-3′ (Reverse) | |
| Osteocalcin | 5′-CTGACAAAGCCTTCATGTCCAA-3′ (Forward) |
| 5′-GCGCCGGAGTCTGTTCACTA-3′ (Reverse) | |
| HDAC4 | 5′-ATCAGAGACCCAATGCCAAT-3′ (Forward) |
| 5′-AGCAGGTTTGACGCCTACAG-3′ (Reverse) | |
| TGFβ3 | 5′-AACCTGGAGGAGAACTGCTG-3′ (Forward) |
| 5′-CTCTGGGTTCAGGGTGTTGT-3′ (Reverse) | |
| Col1a1 | 5′-CCCAAGGAAAAGAAGCACGTC-3′ (Forward) |
| 5′-AGGTCAGCTGGATAGCGACATC-3′ (Reverse) | |
| Col5a3 | 5′-AGGGACCAACTGGGAAGAGT-3′ (Forward) |
| 5′-AAAGTCAGAGGCAGCCACAT-3′ (Reverse) | |
| Col4a2 | 5′-CGGAGTTTGTGGATCGGATA-3′ (Forward) |
| 5′-CCCCCGTTACACTCGATAAA-3′ (Reverse) |
Northern Blot
Total RNA isolated from osteoblasts at indicated days using TRIzol, (Invitrogen) was separated on a 12% acrylamide/urea gel and was transferred onto Hybond-XL membranes (Amersham Biosciences). The 5′-end-labeled miR-29b oligonucleotide probe (5′-AACACTGATTTCAAATGGTGCTA-3′) was used to hybridized the membranes in Rapid-Hyb Buffer (Amersham Biosciences) following the manufacturer's instructions. The blots were reprobed with labeled U6 oligonucleotide (ATATGGAACGCTTCACGAATT) as a control for equal loading. Band intensity of the hybridization signals on x-ray film were measured using AlphaImager software (Alpha Innotech Corp., San Leandro, CA). The relative expressions of the specific miRNA samples were normalized to U6 RNA expression and plotted as fold change setting 0 day to 1.
DNA Constructs
For functional analysis of miR-29b, partial segments (200–300 nucleotides) of the mRNA 3′-UTR containing the miR-29b-binding sequences for HDAC4, TGFβ3, ACVR2A, CTNNBIP1, DUSP2, COL1A1, COL5A3, and COL4A2 genes were PCR-amplified from mouse genomic DNA utilizing the forward and reverse primers (Table 2). The PCR product was then subcloned into the SpeI-MluI site downstream of the stop codon in the pMIR-REPORT Firefly Luciferase reporter vector (Ambion). The correct orientation of 3′-UTR fragments in the plasmid DNA constructs were further confirmed by sequencing.
TABLE 2.
Nucleotide sequences of primers for construct of plasmids
| Gene | Primer sequences |
|---|---|
| HDAC4 | 5′-GGACTAGTCCCGAAGCTGCTGTTCTCTCCT-3′ (Forward) |
| 5′-CGACGCGTCGCACAGCACCCCACTAAGGTT-3′ (Reverse) | |
| TGFB3 | 5′-GGACTAGTCCCATATCTTGCACTGCCTGGA-3′ (Forward) |
| 5′-CGACGCGTCGCATCCATGATTCCCCAAAAA-3′ (Reverse) | |
| ACVR2A | 5′-GGACTAGTCCTTTCTTTTGCAGGGCATTTT-3′ (Forward) |
| 5′-CGACGCGTCGGGCACAGTCTCTGTTCTGGA-3′ (Reverse) | |
| CTNNBIP1 | 5′-GGACTAGTCCGTGGTAGCAAACCACCGTCT-3′ (Forward) |
| 5′-CGACGCGTCGACCAAACCCGCATCTTACTG-3′ (Reverse) | |
| DUSP2 | 5′-GGACTAGTCCCACAGCTCTGGCTTTGACTG-3′ (Forward) |
| 5′-CGACGCGTCGGGAGGAAGTTGGGGAGAGAG-3′ (Reverse) | |
| COL1A1 | 5′-GGACTAGTCCCCCCAAGAACCTGACAACTT-3′ (Forward) |
| 5′-CGACGCGTCGTCCCCTCCCAAAGTCTTCTT-3′ (Reverse) | |
| COL5A3 | 5′-GGACTAGTCCACTCGGCTCCATCTGCTTTA-3′ (Forward) |
| 5′-CGACGCGTCGTGGAATCTCTCACCGTACCC-3′ (Reverse) | |
| COL4A2 | 5′-GGACTAGTCCCCGTGGTACCTGCCACTACT-3′ (Forward) |
| 5′-CGACGCGTCGGAATGAGTGCTGGTGCAGAA-3′ (Reverse) |
Luciferase Assay
For luciferase activity analysis, the double-stranded RNAs that mimic endogenous mmu-miR-29b (pre-miR), as well as the microRNA inhibitor Anti-miR-29b designed to inhibit endogenous miR-29b. Control negative miR (Control 1) and anti-miR control were purchased from Ambion and used in all transfections. Transient transfection of MC3T3 cells (8 × 105 cells/well) was carried out in 6-well plates with Lipofectamine 2000 (Invitrogen) following the manufacturer's instruction. The cells were co-transfected with 200 ng of the luciferase constructs, 5 ng of phRL-null (Promega) Renilla luciferase plasmid, and 100 nm microRNAs for 36 h, and luciferase assays were performed with Dual-luciferase reporter assay system (Promega) per the manufacturer's instructions. Luminescent signal was quantified by luminometer (Glomax, Promega), and each value from firefly luciferase construct was normalized by Renilla luciferase assay.
Transfection Assay
MC3T3-E1 cells at 30–50% confluence were transfected with mmu-miR-29b RNA and miRNA negative Control 1 at 50 or 100 nm concentration with oligofectamine (Invitrogen) following the manufacturer's instructions. The cells were harvested 48 h after transfection for protein and mRNA analysis. For functional studies examining the effects of miR-29b on differentiation, MC3T3 cells were transfected at 60–70% confluency with microRNAs for 72 h for cells to reach confluency and then cultured with differentiation-inducing α-minimum essential medium described as above for 4 and 7 days. Osteoblasts were fixed for histochemical detection of alkaline phosphatase or harvested for mRNA and protein analysis.
RESULTS
Up-regulation of microRNAs during MC3T3 Osteoblast Differentiation
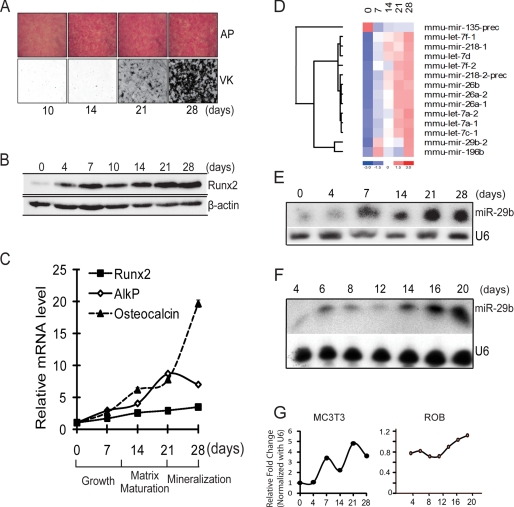
MC3T3 preosteoblastic cells derived from fetal mouse calvaria undergo maturation when cultured in osteogenic medium (Fig. 1). Robust activity of the early osteoblast marker AlkP begins in the matrix maturation period with cellular multi-layering from days 7–14 (Fig. 1A, top panel). The von Kossa silver stain (Fig. 1A, lower panel) shows the extent of mineralization throughout the cell layer from days 21 to 28. Runx2, the primary essential transcription factor for bone formation (33), is at low protein levels in the proliferating cells (days 0–4), up-regulates at confluency (day 7), and increases during differentiation (Fig. 1B). Quantitative RT-PCR results show the typical mRNA expression profile of early stage osteogenic markers Runx2 and AlkP and the mineralization stage marker osteocalcin at the five time points selected for microRNA profiling (Fig. 1C).
FIGURE 1.
miR-29b expression profile during MC3T3 osteoblast differentiation. A, MC3T3 preosteoblasts cells cultured in differentiation medium for 28 days. Histochemical staining of alkaline phosphatase (AP) activity and von Kossa (VK) for mineral deposition at 10, 14, 21, and 28 days is shown. B, Western blot for Runx2 protein which increases during osteoblast differentiation. β-Actin protein was used as control. C, quantitative mRNA normalized by GAPDH for osteoblastic markers Runx2, AlkP, and osteocalcin on selected days during MC3T3 osteoblast differentiation used in miR profiling studies. D, significantly changed microRNAs that putatively target extracellular cellular matrix genes. Total RNA of MC3T3 cells during differentiation time points (0, 7, 14, 21, and 28 days) was used for miRNA microarray analysis. Relative fold changes of the microRNAs were hierarchically clustered by using dChip software. E and F, representative Northern blot analysis of miR-29b using total RNA isolated from mouse (MC3T3) (E) and rat primary osteoblasts (F), which were induced to differentiate. U6 RNA was used as a loading control. G, densitometric quantitation of miR-29b in indicated osteoblasts normalized to U6. The average volumes from two different MC3T3 studies are shown, and one time course from primary osteoblasts is shown.
To understand the potential involvement of miRNAs in osteoblast differentiation, we analyzed the expression of miRs using an established microarray platform (32). Of the significantly expressed and statistically changed miRNAs, 58 were up-regulated from days 7 to 28 with maximal expression during the mineralization stage (supplemental Fig. S1A). Ten miRs were down-regulated during osteoblast differentiation (supplemental Fig. S1B). In the up-regulated group, miR-29b was among the most significantly changed. Interestingly, miR-29b and 14 others including the family of miR-let-7 and miR-26 (Fig. 1D) were all predicted to target many mRNAs encoding extracellular matrix proteins (supplemental Table S1). We validated the expression profile of miR-29b in three osteoblast cell models. Expression of mature miR-29b increased from low levels at day 0 to maximum levels on day 28 of MC3T3 cell differentiation, with a decline at 14 days (Fig. 1, E and G, left panel). This temporal pattern of miR29b expression was confirmed in primary rat calvaria osteoblast cells (Fig. 1, F and G, right panel) with a slight decline on days 8–12 when proliferation ceased in the multilayered cell nodules (34). In both osteoblast models, miR-29b increased during the period of mineral deposition. We also examined the MC3T3 clone 4, which was selected for its high capacity to mineralize and find that miR-29b expression increased from day 0 to day 28 but exhibited a more continuous profile (supplemental Fig. S1C). Together these expression profiles suggest a requirement for miR-29b to support osteoblast differentiation at multiple stages.
miR-29b Promotes Osteogenic Differentiation
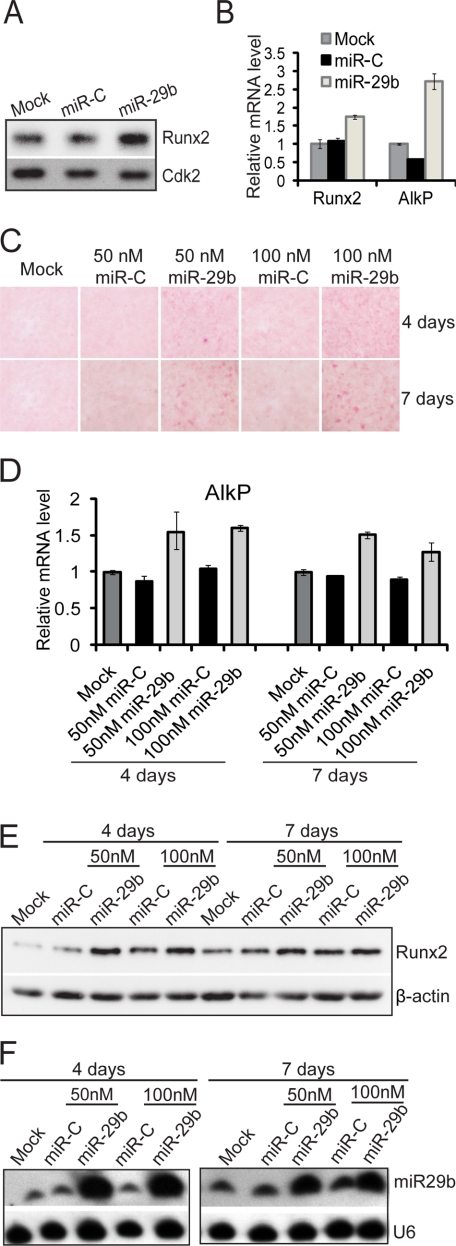
We addressed the functional activity of miR-29b in osteoblasts by overexpression studies (Fig. 2). miR-29b induced protein and mRNA expression of Runx2 and AlkP by 48 h (Fig. 2, A and B). We examined the long term effect of miR-29b in promoting osteoblast differentiation by transfection of miR-29b in preosteoblasts for 72 h until cells came to confluency. At this time, differentiation was induced for 4 and 7 days. Histological analysis showed that AlkP activity (Fig. 2C) and mRNA (Fig. 2D) were increased. Runx2 protein levels were also elevated by miR-29b, reflecting enhanced differentiation at both time points (Fig. 2E). Fig. 2F shows that exogenous miR-29b cellular levels were sustained throughout the 7 days of differentiation. These findings indicate that miR-29b increases osteogenic differentiation.
FIGURE 2.
miR-29b activates osteogenic differentiation in osteoblasts. A and B, MC3T3 osteoblast cells were transfected with miR-29b mature miRNA oligonucleotides (50 nm), miRNA negative Control 1 (miR-C) (50 nm), or transfection reagent only (Mock) for 48 h for protein and RNA analysis. Both expression of Runx2 protein (A) and Runx2 and AlkP mRNA were up-regulated in miR-29b overexpressing cells (B). C–E, miR-29b promotes osteoblast differentiation. After transfection with 50 or 100 nm miR-29b or miRNA Control (miR-C) for 72 h, confluent MC3T3 cells were treated with α-minimum essential medium containing 10 mm β-glycerophosphate and 50 μg/ml ascorbic acid for 4 or 7 days. The cells were fixed in 2% paraformaldehyde for histochemical detection of alkaline phosphatase (C). The mRNA level of AlkP was detected by quantitative PCR normalized by GAPDH (D). Runx2 and β-actin protein (as control) were detected by Western blot (E). F, Northern blot analysis shows endogenous (miR-C lanes) and overexpressed miR-29b levels in the samples shown in E. U6RNA is the loading control.
miR-29b Targets Negative Regulators of Osteoblast Differentiation
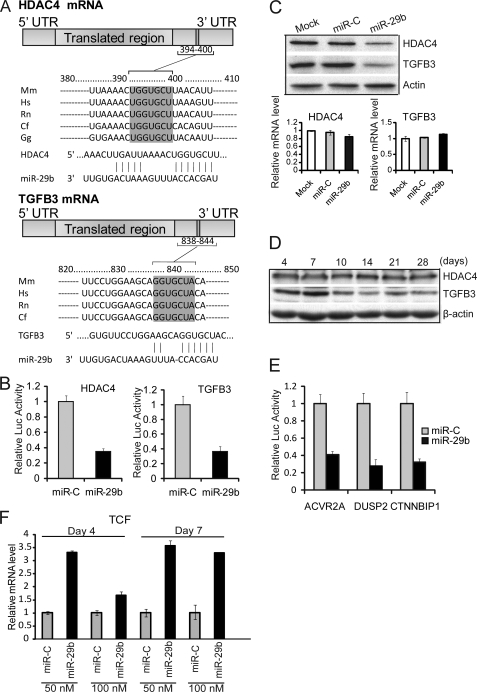
Our results suggest that miR-29b functions as a positive regulator of osteoblastogenesis. Therefore, we examined three data bases (TargetScan, PicTar, and miRanda) for high probability miR-29b targets that were proven in vivo inhibitors of osteoblast activities. Five putative miR-29 targets were selected for further study: HDAC4, TGFβ3, ACVR2A, CTNNBIP1, and DUSP2 (18, 35–42).
Histone deacetylase 4 (HDAC4) and TGFβ3 have been demonstrated to function as negative regulators in chondrocytes and osteoblasts (18, 35–38). One putative target site of miR-29b is predicted in the 3′-UTR of both HDAC4 and TGFβ3 mRNAs. The complementary pairing between the miR-29b “seed region” and the 3′-UTR is highly conserved in vertebrate species (Fig. 3A). The 3′-UTR reporter assays and HDAC4 and TGFβ3 endogenous protein levels were both repressed by miR-29b (Fig. 3B), indicating a block in protein translation (Fig. 3C, upper panel). As expected for miRNAs, no significant change in HDAC4 and TGFβ3 mRNA levels was found (Fig. 3C, lower panel). Together, these results provide evidence that both HDAC4 and TGFβ3 are direct targets of miR-29b mediated translational repression. Therefore, we examined protein levels of these factors during osteoblast differentiation. We observed that HDAC4 protein levels remained at a constitutive level from days 4 to 28 (Fig. 3D). However, TGFβ3 protein decreased after day 7 of osteoblast differentiation, consistent withmiR-29b increased levels and the potent inhibitory effects of TGFβ on osteoblast regulatory mechanisms (35).
FIGURE 3.
miR-29b targets negative regulators of osteoblast differentiation. A, one putative target site of miR-29b predicted by the TargetScan program was contained in the HDAC4 or TGFβ3 mRNA 3′-UTR, and both were highly conserved in vertebrate species. The numbers represent the position of the “seed region” match to miR-29b within the UTR sequences. Mm, mouse; Hs, human; Rn, rat; Cf, dog; Gg, chicken. B, MC3T3 cells were co-transfected with 100 nm RNA of miR-29b or miRNA control (miR-C), phRL-null (Renilla plasmid), and the luciferase constructs (firefly) carrying HDAC4 3′-UTR or TGFβ3 3′-UTR. Luciferase assays were performed 36 h after transfection. The ratio of reporter Firefly to control Renilla luciferase in relative luminescence units was plotted. The error bars represent the standard error for n = 3. C, 30–50% confluent osteoblast cell line MC3T3 was transfected with 100 nm miR-29b RNA, miRNA negative control, or transfection reagent only (Mock). Protein and RNA analysis was performed at 48 h after transfection. HDAC4, TGFβ3, and β-actin (as control) protein were monitored by Western blot. HDAC4 and TGFβ3 mRNA level were detected by quantitative RT-PCR normalized by GAPDH. D, HDAC4 and TGFβ3 expression during osteoblast differentiation. Differentiating MC3T3 time points were analyzed by Western for HDAC4, TGFβ3, and β-actin (as control) protein. E, MC3T3 cells were co-transfected with reagents in the panel, and the luciferase constructs carrying 3′-UTR of activin A receptor type IIA, DUSP2, or CTNNBIP1. Functional activity of the luciferase reporter plasmid was assessed as described above for B. The values represent the means ± S.E. for n = 3. F, shown is the effect of miR-29b overexpression on endogenous mRNA levels of TCF1, a marker of Wnt/β-catenin signaling that is inhibited by CTNNBIP1 (see supplemental Fig. S2, B). TCF1 is increased during osteoblast differentiation (days 4 and 7). The experimental design is described for Fig. 2C.
The 3′-UTR luciferase assay also indicated that miR-29b targeted activin A receptor type IIA, CTNNBIP1, and DUSP2 (Fig. 3E). All of these factors are known to be involved in the inhibition of osteoblast differentiation (35, 37–42); supplemental Fig. S2 further details the linkage of these genes to osteogenic pathways. We tested the consequence of miR-29b on Wnt signaling, which is known to promote bone formation (Fig. 3F). We expressed miR-29b at two concentrations, followed by differentiation conditions as described in Fig. 2D. This study showed that 4 and 7 days after initiation of osteoblast maturation, the TCF transcriptional mediator of Wnt-β-catenin signaling is up-regulated, consistent with miR-29b down-regulation of the Wnt pathway inhibitor CTNNBIP1. We conclude from these results that miR-29b inhibits multiple negative regulators of osteoblastogenesis and thereby facilitates progression of differentiation.
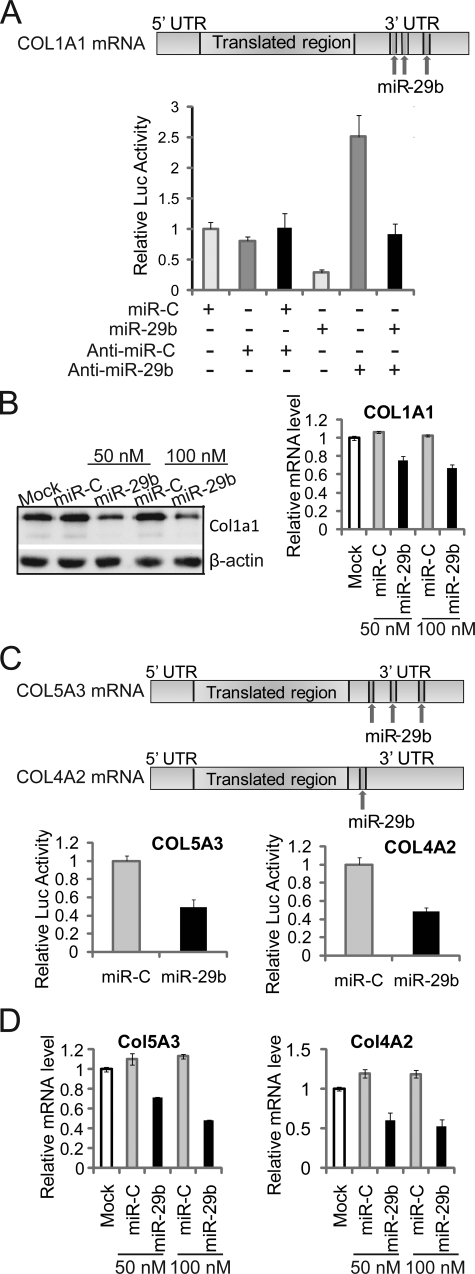
Collagen Genes Are Directly Targeted by miR-29b
miR-29b is predicted to target a large number of collagen genes (supplemental Table S1), which was counter-intuitive to the well established requirement for synthesis of a collagen type I ECM to promote osteoblast differentiation (31, 43). supplemental Tables S1 and S2) shows all collagens in the miR data bases that are putative targets of other miRs identified in our profile. This finding suggests that a cohort of microRNAs tightly regulate collagen protein accumulation in the bone ECM. Three putative binding sites for miR-29b are predicted in the Col1a1 mRNA 3′-UTR (Fig. 4A), and sites are evolutionarily conserved among vertebrate species. Our transfection assay results showed that ectopic miR-29b in osteoblasts significantly repressed the luciferase activity of reporter plasmids carrying the Col1a1 3′-UTR. Complementary to this effect, anti-miR-29b activated the luciferase activity of the reporter plasmid and antagonized the effect of miR-29b on the reporters (Fig. 4A). Col1a1 mRNA and protein were down-regulated in miR-29b overexpressing MC3T3 cells (Fig. 4B), which verifies that miR-29 directly targets Col1a1. Furthermore, the finding that Col1a1 mRNA was decreased indicates that mRNA stability was also affected by miR-29b. Luciferase assays (Fig. 4C) and decreased mRNA levels (Fig. 4D) also validated that miR-29b directly targets other collagen genes, including Col5a3, a minor component of bone tissue, and Col4a2, a basement membrane collagen.
FIGURE 4.
miR-29b functions as an attenuator for collagen in differentiated osteoblasts. A, Col1a1 is a direct target of miR-29b. Three putative target sites of miR-29b are predicted to be in Col1a1 mRNA 3′-UTR by TargetScan, PicTar, and miRanda program (upper panel). MC3T3 cells were co-transfected with the luciferase constructs, phRL-null (Renilla plasmid) and 100 nm RNA oligonucleotides of miR-29b, miRNA negative control (miR-C), Anti-miR-29b or Anti-miR negative control (Anti-miR-C), as shown in the lower panel. Luciferase assays were performed 36 h after transfection. The ratio of reporter (firefly) to control phRL-null plasmid (Renilla) in relative luminescence units was plotted. The error bars represent the standard error for n = 3. B, miR-29b overexpression inhibits Col1a1 expression in osteoblasts. 30–50% confluent MC3T3 osteoblast cells were transfected with 50 or 100 nm miR-29b RNA oligonucleotides or miRNA negative control (miR-C). The cells were harvested for protein and RNA analysis at 48 h after transfection. Col1A1 and β-actin (as control) protein were monitored by Western blot (left panel) and Col1a1 mRNA level was detected by quantitative RT-PCR normalized by GAPDH (right panel). C, miR-29b target Col5a3 and Col4a2 directly by mRNA 3′-UTR. Three and one putative target site for miR-29b is contained in the Col5a3 and Col4a2 mRNA 3′-UTR, respectively. MC3T3 cells were co-transfected with luciferase constructs carrying Col5a3 or Col4a2 3′-UTR, phRL-null, miR-29b, or miRNA negative control (miR-C). Luciferase activity assays and analysis were described as in A. D, mRNA of Col5a3 and Col4a2 were detected in MC3T3 overexpressed with RNA oligonucleotides as described for B.
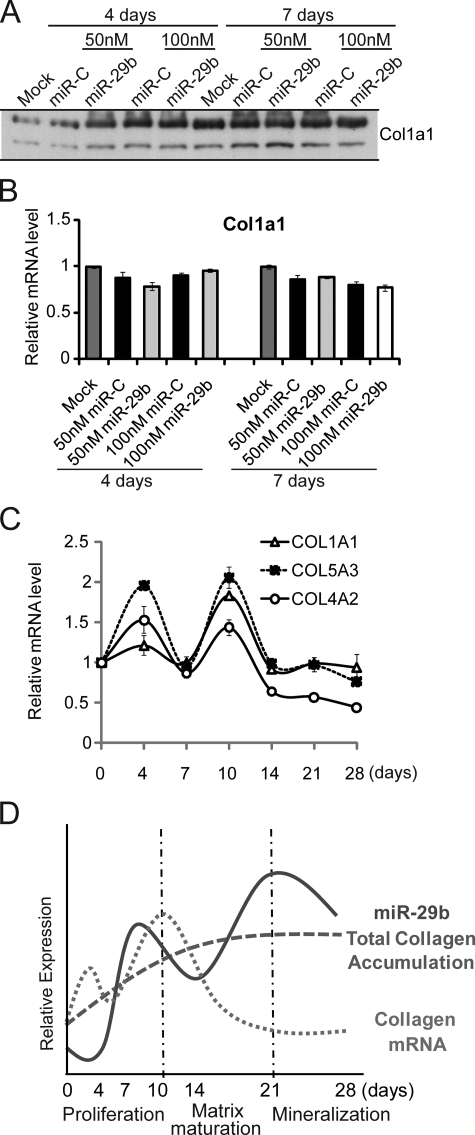
Collagen protein synthesis and secretion is necessary for osteogenesis and occurs in vitro during the proliferation stage of osteoblasts for formation of the ECM. Collagen then matures to “cross-linked” fibrils (29, 44). Osteoblasts on the bone surface in vivo produce the ECM. Because miR-29b is expressed throughout differentiation, we sought to resolve the question as to whether miR-29b might repress the negative regulators of bone formation in immature osteoblasts (early stages of differentiation), without affecting collagen. We therefore monitored collagen synthesis in post-confluent osteoblasts after induction of osteoblast differentiation (Fig. 5). Neither collagen protein nor mRNA expression was affected by miR-29b in the post-confluent early differentiated osteoblast, days 4 and 7 (Fig. 5, A and B). Indeed, from days 4 to 7, collagen increased to the same extent in miR-C- and miR-29b-treated cells, and mRNA expression was not altered. As shown in Fig. 5C, the three collagen gene targets of miR-29b show a similar mRNA profile during the complete course of osteoblast differentiation. A slight decline in collagen mRNAs is observed at monolayer confluency (day 7), followed by robust expression with cellular multilayering during the matrix maturation period (days 8–14). A rapid decline occurs at the onset of mineralization after day 14 (Fig. 1E), consistent with the increase in miR-29b. The reciprocal expression of collagen mRNA and miR-29b (Fig. 5D) is schematically represented for murine cell lines and rat primary osteoblasts. This profile suggests that collagen genes are preferentially negatively regulated by the higher endogenous levels of miR-29b expressed at the mineralization stage, when a steady state of protein accumulation is observed.
FIGURE 5.
miR-29b regulates collagen expression in differentiated osteoblasts. A and B, miR-29b has no effect on collagen gene expression in early osteoblast differentiation. MC3T3 cells were transfected with 50 or 100 nm miR-29b RNA mimics or miRNA Control (miR-C) and cultured with differentiation-inducing medium described as in Fig. 2C. Col1a1 protein was monitored by Western blot (A), and mRNA levels detected by quantitative PCR normalized by GAPDH (B). C, mRNA expression levels of Col1a1, Col5a3, and Col4a2 during MC3T3 osteogenic differentiation. Differentiation was initiated 3 days after plating cells (day 0). The cells were harvested at indicated time points for quantitative RT-PCR normalized to GAPDH. D, reciprocal expression of miR-29b and COL Type I (days 21–28); miR-29b quantitated from Northern blot analyses; total collagen measured from densitometry of Western blots; COL Type I mRNA determined by quantitative PCR.
DISCUSSION
In this study, we provide evidence for the concept that miR-29b mediates translational inhibition and alters the levels of critical regulators of biological pathways, thereby providing a mechanism for spatiotemporal control of developmental and homeostatic events during osteogenesis, consistent with other findings (45–47). We have identified a substantial number of microRNAs that are increased during progression of osteoblast differentiation, most significantly at the stage of mineral deposition characteristic of osteocytes. Among these, miR-29b was functionally characterized as a positive regulator of osteoblastogenesis. The key finding of our study is that miR-29b regulates osteoblast differentiation and function by suppressing inhibitors of cell signaling pathways and/or transcriptional programs required for osteogenesis, as well as by attenuating collagen gene expression during extracellular matrix maturation.
Our study indicates that miR-29b targets several negative regulators of osteogenic differentiation, including TGFβ3, HDAC4, ACTVR2A, CTNNBIP1, and DUSP2, which impinge on signal transduction pathways promoting osteogenesis in vivo (e.g. including Smad, ERK, p38 MAPK, and Wnt) (5, 35, 37–42) (see supplemental Fig. S2). For example, ACVR2A is a member of the TGFβ superfamily that inhibits differentiation of fetal rat calvarial osteoblasts and matrix mineralization in human osteoblast cultures (39, 40). CTNNBIP1 (also known as ICAT) inhibits β-catenin-mediated transcription by binding to β-catenin and preventing interactions with TCF/LEF factors (5, 42). DUSP2 inactivates and anchors ERK within the nucleus (41), as well as blocks an inhibitor of ERK signaling during osteoblast differentiation (48) (supplemental Fig. S2). Consistent with the importance of these miR-29b targets in controlling osteogenesis, we show that exogenous expression of miR-29b promotes osteoblastic differentiation, as reflected by stimulated Wnt signaling and increased levels of osteogenic marker genes, including Runx2 and alkaline phosphatase. Hence, the ability of miR-29b to promote osteoblast differentiation can be attributed to relieving the negative effects of anti-osteogenic cell signaling pathways.
Apart from effects on cell signaling, several miR-29b targets are known to inhibit osteoblast differentiation by directly affecting the functional activity of Runx2 (Cbfa1/AML3), which is essential for differentiation of osteoblast lineage cells and bone formation in vivo (1, 33, 49, 50). Endogenous TGFβ signaling suppresses maturation of osteoblastic mesenchymal cells by mechanisms that either reduce Runx2 cellular protein and activity or induce Runx2 to undergo Smurf-mediated degradation in vivo (35, 37, 38, 51). HDAC4 is a co-repressor of Runx2 and deacetylates Runx2 in hypertrophic chondrocytes during bone development (36). Thus, our data indicate that the osteogenic function of Runx2 is at least in part controlled by miR-29b.
The third principal result from our study is that miR-29b attenuates expression of collagen genes that encode essential proteins of the bone extracellular matrix. Both major and minor collagens are miR-29b targets (supplemental Table S2). We show that suppression of collagen gene expression is not just a secondary effect of miR-29b inhibiting osteoblast differentiation but the direct consequence of miR-29b interacting with the 3′-UTR of collagens to block translation. We observed a striking reciprocal relationship between miR-29b (up-regulated) and type I collagen synthesis (down-regulated) at the late stage of differentiation during the mineralization period. This miR-29b-dependent suppression of collagen may facilitate maturation of the collagen fibrillar matrix for mineral deposition (52, 53).
One biological implication of miR-29b-dependent suppression of collagen gene expression is that miR-29b may prevent fibrosis of bone tissue, similar to its proposed role in cardiac fibrosis following myocardial infarct (26).
Attenuation of collagen synthesis by miR-29b may allow ordered collagen fibril maturation to facilitate mineral deposition (52). In bone, collagens are the major ECM proteins with about 95% type I and 5% type V collagens assembled into heterofibrils (55), and genetic defects in the appropriate alignment of collagen fibrils results in specific bones disorders (56–58). Thus, the biological role of miR-29b as an “osteo-miR” may be to protect bone from fibrosis and/or restrict type I collagen protein accumulation to facilitate ordered deposition of mineral associated with collagen fibrils.
The finding of our studies that miR-29b does not repress collagen protein in immature osteoblasts but causes decreased collagen mRNA levels at later stages of differentiation may be explained by alternative mRNA splicing that results in different collagen 3′-UTRs. Although animal miRNAs mainly inhibit translation, they can also induce significant degradation of mRNA targets (11, 12, 59–63). Almost half of mammalian genes generate multiple mRNA isoforms differing in their 3′-UTRs by alternative cleavage and polyadenylation. Sandberg et al. (64) showed that in proliferating primary murine CD4+ T lymphocytes, 3′-UTR of some mRNAs were shortened, which led to fewer microRNA target sites (64). COL1A2 mRNA has been validated to be processed to produce different 3′-UTR by alterative polyadenylation signals (65). We identified mouse COL1A1 mRNA 3′-UTR within the expressed sequence tag data base (54) and found that two polyadenylation signals (AAUAAA) were contained in the 3′-UTR (supplemental Fig. S3). Furthermore, three putative target sites of miR-29b are located between these two polyadenylation signals. We therefore postulate that in the early proliferation period of MC3T3 differentiation, the COL1A1 mRNA 3′-UTR is processed with the proximal polyadenylation signal, resulting in the loss of miR-29b target sites. However, in the late maturation and mineralization periods, the COL1A1 mRNA 3′-UTR was processed with the distal polyadenylation signal and therefore includes the miR-29b targets that regulate collagen repression.
In summary, we conclude that during osteoblast differentiation, miR-29b has multiple functions. On the one hand, miR-29b promotes osteoblast differentiation by targeting negative regulators of osteogenic pathways and thereby contributes to establishment of osteoblast differentiation. On the other hand, miR-29b functions as an attenuator of collagen synthesis in mature osteoblasts to maintain the differentiated phenotype.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AR039588 (to G. S. S.) and R37 DE012528 (to J. B. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1, S2 and S3.
- ECM
- extracellular matrix
- UTR
- untranslated region
- STAT
- signal transducers and activators of transcription
- MAPK
- mitogen-activated protein kinase
- miRNA
- microRNA
- AlkP
- alkaline phosphatase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- RT
- reverse transcription
- HDAC
- histone deacetylase
- TGF
- transforming growth factor
- CTNNBIP
- catenin β-interacting protein
- DUSP
- dual specificity phosphatase
- ERK
- extracellular signal-regulated kinase.
REFERENCES
- 1.Lian J. B., Stein G. S., Javed A., van Wijnen A. J., Stein J. L., Montecino M., Hassan M. Q., Gaur T., Lengner C. J., Young D. W. ( 2006) Rev. Endocr. Metab. Disord. 7, 1– 16 [DOI] [PubMed] [Google Scholar]
- 2.Stein G. S., Zaidi S. K., Stein J. L., Lian J. B., van Wijnen A. J., Montecino M., Young D. W., Javed A., Pratap J., Choi J. Y., Ali S. A., Pande S., Hassan M. Q. ( 2008) J. Cell. Biochem. 104, 2016– 2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komori T. ( 2006) J. Cell. Biochem. 99, 1233– 1239 [DOI] [PubMed] [Google Scholar]
- 4.Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B. ( 2002) Cell 108, 17– 29 [DOI] [PubMed] [Google Scholar]
- 5.Bodine P. V., Komm B. S. ( 2006) Rev. Endocr. Metab. Disord. 7, 33– 39 [DOI] [PubMed] [Google Scholar]
- 6.Gaur T., Lengner C. J., Hovhannisyan H., Bhat R. A., Bodine P. V. N., Komm B. S., Javed A., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. ( 2005) J. Biol. Chem. 280, 33132– 33140 [DOI] [PubMed] [Google Scholar]
- 7.Deng Z. L., Sharff K. A., Tang N., Song W. X., Luo J., Luo X., Chen J., Bennett E., Reid R., Manning D., Xue A., Montag A. G., Luu H. H., Haydon R. C., He T. C. ( 2008) Front. Biosci. 13, 2001– 2021 [DOI] [PubMed] [Google Scholar]
- 8.Bandyopadhyay A., Tsuji K., Cox K., Harfe B. D., Rosen V., Tabin C. J. ( 2006) PLoS. Genet. 2, e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy J. B., Schindler C., Raz R., Levy D. E., Baron R., Horowitz M. C. ( 1996) Endocrinology 137, 1159– 1165 [DOI] [PubMed] [Google Scholar]
- 10.Ge C., Xiao G., Jiang D., Franceschi R. T. ( 2007) J. Cell Biol. 176, 709– 718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L., Fan J., Belasco J. G. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 4034– 4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filipowicz W., Bhattacharyya S. N., Sonenberg N. ( 2008) Nat. Rev. Genet. 9, 102– 114 [DOI] [PubMed] [Google Scholar]
- 13.Stefani G., Slack F. J. ( 2008) Nat. Rev. Mol. Cell Biol. 9, 219– 230 [DOI] [PubMed] [Google Scholar]
- 14.Ma L., Weinberg R. A. ( 2007) Cell Cycle 7, 570– 572 [DOI] [PubMed] [Google Scholar]
- 15.Croce C. M. ( 2008) N. Engl. J. Med. 358, 502– 511 [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T., Lu J., Cobb B. S., Rodda S. J., McMahon A. P., Schipani E., Merkenschlager M., Kronenberg H. M. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 1949– 1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harfe B. D., McManus M. T., Mansfield J. H., Hornstein E., Tabin C. J. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 10898– 10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuddenham L., Wheeler G., Ntounia-Fousara S., Waters J., Hajihosseini M. K., Clark I., Dalmay T. ( 2006) FEBS Lett. 580, 4214– 4217 [DOI] [PubMed] [Google Scholar]
- 19.Luzi E., Marini F., Sala S. C., Tognarini I., Galli G., Brandi M. L. ( 2008) J. Bone Miner. Res. 23, 287– 295 [DOI] [PubMed] [Google Scholar]
- 20.Li Z., Hassan M. Q., Volinia S., van Wijnen A. J., Stein J. L., Croce C. M., Lian J. B., Stein G. S. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 13906– 13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., Liu S., Alder H., Costinean S., Fernandez-Cymering C., Volinia S., Guler G., Morrison C. D., Chan K. K., Marcucci G., Calin G. A., Huebner K., Croce C. M. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 15805– 15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mott J. L., Kobayashi S., Bronk S. F., Gores G. J. ( 2007) Oncogene 26, 6133– 6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pekarsky Y., Santanam U., Cimmino A., Palamarchuk A., Efanov A., Maximov V., Volinia S., Alder H., Liu C. G., Rassenti L., Calin G. A., Hagan J. P., Kipps T., Croce C. M. ( 2006) Cancer Res. 66, 11590– 11593 [DOI] [PubMed] [Google Scholar]
- 24.Sengupta S., den Boon J. A., Chen I. H., Newton M. A., Stanhope S. A., Cheng Y. J., Chen C. J., Hildesheim A., Sugden B., Ahlquist P. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 5874– 5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S. Y., Lee J. H., Ha M., Nam J. W., Kim V. N. ( 2008) Nat. Struct. Mol. Biol. 16, 23– 29 [DOI] [PubMed] [Google Scholar]
- 26.van Rooij E., Sutherland L. B., Thatcher J. E., DiMaio J. M., Naseem R. H., Marshall W. S., Hill J. A., Olson E. N. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 13027– 13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassan M. Q., Tare R. S., Lee S., Mandeville M., Morasso M. I., Javed A., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. ( 2006) J. Biol. Chem. 281, 40515– 40526 [DOI] [PubMed] [Google Scholar]
- 28.Sudo H., Kodama H.-A., Amagai Y., Yamamoto S., Kasai S. ( 1983) J. Cell Biol. 96, 191– 198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quarles L. D., Yohay D. A., Lever L. W., Caton R., Wenstrup R. J. ( 1992) J. Bone Miner. Res. 7, 683– 692 [DOI] [PubMed] [Google Scholar]
- 30.Choi J.-Y., Lee B.-H., Song K.-B., Park R.-W., Kim I.-S., Sohn K.-Y., Jo J.-S., Ryoo H.-M. ( 1996) J. Cell. Biochem. 61, 609– 618 [DOI] [PubMed] [Google Scholar]
- 31.Franceschi R. T., Iyer B. S., Cui Y. ( 1994) J. Bone Miner. Res. 9, 843– 854 [DOI] [PubMed] [Google Scholar]
- 32.Liu C. G., Calin G. A., Meloon B., Gamliel N., Sevignani C., Ferracin M., Dumitru C. D., Shimizu M., Zupo S., Dono M., Alder H., Bullrich F., Negrini M., Croce C. M. ( 2004) Proc. Natl. Acad. Sci. U. S. A. 101, 9740– 9744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y.-H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. ( 1997) Cell 89, 755– 764 [DOI] [PubMed] [Google Scholar]
- 34.Owen T. A., Aronow M., Shalhoub V., Barone L. M., Wilming L., Tassinari M. S., Kennedy M. B., Pockwinse S., Lian J. B., Stein G. S. ( 1990) J. Cell. Physiol. 143, 420– 430 [DOI] [PubMed] [Google Scholar]
- 35.Kang J. S., Alliston T., Delston R., Derynck R. ( 2005) EMBO J. 24, 2543– 2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vega R. B., Matsuda K., Oh J., Barbosa A. C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J. M., Richardson J. A., Karsenty G., Olson E. N. ( 2004) Cell 119, 555– 566 [DOI] [PubMed] [Google Scholar]
- 37.Jeon E. J., Lee K. Y., Choi N. S., Lee M. H., Kim H. N., Jin Y. H., Ryoo H. M., Choi J. Y., Yoshida M., Nishino N., Oh B. C., Lee K. S., Lee Y. H., Bae S. C. ( 2006) J. Biol. Chem. 281, 16502– 16511 [DOI] [PubMed] [Google Scholar]
- 38.Maeda S., Hayashi M., Komiya S., Imamura T., Miyazono K. ( 2004) EMBO J. 23, 552– 563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikenoue T., Jingushi S., Urabe K., Okazaki K., Iwamoto Y. ( 1999) J. Cell. Biochem. 75, 206– 214 [DOI] [PubMed] [Google Scholar]
- 40.Eijken M., Swagemakers S., Koedam M., Steenbergen C., Derkx P., Uitterlinden A. G., van der Spek P. J., Visser J. A., de Jong F. H., Pols H. A., Van Leeuwen J. P. ( 2007) FASEB J. 21, 2949– 2960 [DOI] [PubMed] [Google Scholar]
- 41.Caunt C. J., Rivers C. A., Conway-Campbell B. L., Norman M. R., McArdle C. A. ( 2008) J. Biol. Chem. 283, 6241– 6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daniels D. L., Weis W. I. ( 2002) Mol. Cell 10, 573– 584 [DOI] [PubMed] [Google Scholar]
- 43.Lynch M. P., Stein J. L., Stein G. S., Lian J. B. ( 1995) Exp. Cell Res. 216, 35– 45 [DOI] [PubMed] [Google Scholar]
- 44.Gerstenfeld L. C., Riva A., Hodgens K., Eyre D. R., Landis W. J. ( 1993) J. Bone Miner. Res. 8, 1031– 1043 [DOI] [PubMed] [Google Scholar]
- 45.He L., Hannon G. J. ( 2004) Nat. Rev. Genet. 5, 522– 531 [DOI] [PubMed] [Google Scholar]
- 46.Ambros V. ( 2004) Nature 431, 350– 355 [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y., Srivastava D. ( 2007) Trends Biochem. Sci. 32, 189– 197 [DOI] [PubMed] [Google Scholar]
- 48.Raucci A., Bellosta P., Grassi R., Basilico C., Mansukhani A. ( 2008) J. Cell. Physiol. 215, 442– 451 [DOI] [PubMed] [Google Scholar]
- 49.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. ( 1997) Cell 89, 747– 754 [DOI] [PubMed] [Google Scholar]
- 50.Banerjee C., McCabe L. R., Choi J.-Y., Hiebert S. W., Stein J. L., Stein G. S., Lian J. B. ( 1997) J. Cell. Biochem. 66, 1– 8 [DOI] [PubMed] [Google Scholar]
- 51.Zhao M., Qiao M., Oyajobi B. O., Mundy G. R., Chen D. ( 2003) J. Biol. Chem. 278, 27939– 27944 [DOI] [PubMed] [Google Scholar]
- 52.Glimcher M. J. ( 1989) Anat. Rec. 224, 139– 153 [DOI] [PubMed] [Google Scholar]
- 53.Burger C., Zhou H. W., Wang H., Sics I., Hsiao B. S., Chu B., Graham L., Glimcher M. J. ( 2008) Biophys. J. 95, 1985– 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beaudoing E., Gautheret D. ( 2001) Genome Res. 11, 1520– 1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wenstrup R. J., Florer J. B., Davidson J. M., Phillips C. L., Pfeiffer B. J., Menezes D. W., Chervoneva I., Birk D. E. ( 2006) J. Biol. Chem. 281, 12888– 12895 [DOI] [PubMed] [Google Scholar]
- 56.Brenner R. E., Vetter U., Stoss H., Muller P. K., Teller W. M. ( 1993) Eur. J. Pediatr. 152, 505– 508 [DOI] [PubMed] [Google Scholar]
- 57.Sarathchandra P., Pope F. M., Kayser M. V., Ali S. Y. ( 2000) J. Pathol. 192, 385– 395 [DOI] [PubMed] [Google Scholar]
- 58.Stewart T. L., Roschger P., Misof B. M., Mann V., Fratzl P., Klaushofer K., Aspden R., Ralston S. H. ( 2005) Calcif. Tissue Int. 77, 113– 118 [DOI] [PubMed] [Google Scholar]
- 59.Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A. E. ( 2005) Cell 122, 553– 563 [DOI] [PubMed] [Google Scholar]
- 60.Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di P. F., Lin S. C., Gram H., Han J. ( 2005) Cell 120, 623– 634 [DOI] [PubMed] [Google Scholar]
- 61.Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. ( 2005) Nature 433, 769– 773 [DOI] [PubMed] [Google Scholar]
- 62.Giraldez A. J., Mishima Y., Rihel J., Grocock R. J., Van D. S., Inoue K., Enright A. J., Schier A. F. ( 2006) Science 312, 75– 79 [DOI] [PubMed] [Google Scholar]
- 63.Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. ( 2006) Genes Dev. 20, 1885– 1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B. ( 2008) Science 320, 1643– 1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Natalizio B. J., Muniz L. C., Arhin G. K., Wilusz J., Lutz C. S. ( 2002) J. Biol. Chem. 277, 42733– 42740 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.