Abstract
tRNAs are transcribed as precursors and processed in a series of reactions culminating in aminoacylation and translation. Central to tRNA maturation, the 3′ end trailer can be endonucleolytically removed by tRNase Z. A flexible arm (FA) extruded from the body of tRNase Z consists of a structured ααββ hand that binds the elbow of pre-tRNA. Deleting the FA hand causes an almost 100-fold increase in Km with little change in kcat, establishing its contribution to substrate recognition/binding. Remarkably, a 40-residue Ala scan through the FA hand reveals a conserved leucine at the ascending stalk/hand boundary that causes practically the same increase in Km as the hand deletion, thus nearly eliminating its ability to bind substrate. Km also increases with substitutions in the GP (α4–α5) loop and at other conserved residues in the FA hand predicted to contact substrate based on the co-crystal structure. Substitutions that reduce kcat are clustered in the β10–β11 loop.
tRNAs are transcribed as precursors with a 5′ end leader and 3′ end trailer. The 5′ end leader is removed by RNase P. The 3′ end trailer can be endonucleolytically removed by tRNase Z, which cleaves following the unpaired nucleotide just beyond the 3′ side of the acceptor stem (the discriminator) leaving a 3′-OH ready for CCA addition. In some bacteria and in all archaea and eukaryotes (including their organelles), CCA at the 3′ end of mature tRNAs is not transcriptionally encoded, and a CCA-adding enzyme is required (1); endonucleolytic processing by tRNase Z is thus a precise and probably essential reaction in the pathway to a mature 3′ end (2, 3).
Interestingly, the 3′ end CCA is an anti-determinant for tRNase Z that discourages the recycling of mature tRNAs (4–7), although not in every case (8). Additional functions have been suggested for tRNase Z, including a possible role in human prostate cancer susceptibility (2, 9–12). In some instances, tRNase Z can recognize and cleave RNAs that are structurally related to pre-tRNAs with 3′ end extensions (10, 12).
tRNase Z is an ancient member of the β-lactamase superfamily of metal-dependent hydrolases (2, 13). The signature sequence of this family, the His domain (HXHXDH, Motif II), in conjunction with histidines in Motifs III and V and aspartate in Motif IV, contributes side chains that coordinate two divalent metal ions (14, 15). Additionally, the Glu side chain in HEAT and His in the HST loop (located between Motifs IV and V) apparently function as a pair to facilitate proton transfer at the final stage of reaction (16, 17). A Glu-His pair in CPSF-73, the long sought endonuclease responsible for pre-mRNA cleavage and a member of the tRNase Z class of RNA endonucleases (13, 16, 18), displays the same structure relative to the active site and presumably functions identically in catalysis.
Substitutions in Motifs II–V, HEAT, and HST loop residues did not show increases in Km (17, 19); thus, these residues apparently contribute to metal ion binding and catalysis without being involved with substrate recognition/binding. This interpretation is supported by the co-crystal structure (20), in which tRNase Z with a Motif II His→Ala substitution (introduced to reduce catalytic activity) bound only one metal ion/subunit but displayed structurally reasonable tRNA binding. Increases in Km observed with several of the substitutions in the PXKXRN loop and Motif I region (7) were suggested to be involved with CCA anti-determination and with substrate recognition and binding around the acceptor stem (7, 19). Results of previous studies of processing kinetics (7, 17, 19) left open for further detailed analysis the initiating substrate recognition/binding events leading to catalysis by tRNase Z, centered on the flexible arm (15, 20, 21).
Flexible Arm Is Only Present in Amino Portion of tRNase ZL
The flexible arm (FA)3 consists of a co-globular (ααββ) hand extruded and held apart from the body of tRNase Z by extended polypeptide stalks (15). This unusual structure confers the ability to recognize pre-tRNA substrate (20, 21). Thus, whereas CPSF-73 has a β-CASP domain that covers the active site like a flap that has to be unfolded by accessory proteins (13, 16), the FA of tRNase Z allows it to recognize tRNA and catalyze 3′ end cleavage without additional participants.
Long (tRNase ZL) and short (tRNase ZS) forms of tRNase Z are encoded by different genes (2, 22, 23). Both forms are present in human and other vertebrate genomes and in Arabidopsis and other higher plants. Only tRNase ZS is found in bacteria and archaea; only tRNase ZL is found in Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster. Sequence alignments suggest conserved FA structure and function between tRNase ZL and tRNase ZS (see Fig. 1), although they have different positions in the primary structure.
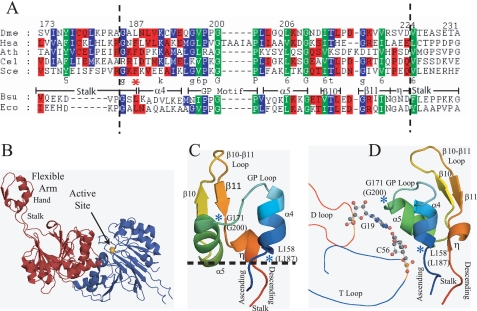
FIGURE 1.
Multiple sequence alignments of the flexible arm of tRNase Z and structure views of B. subtilis tRNase ZS and the flexible arm. A, the top five sequences are eukaryotic tRNases ZL (the one tRNase Z from D. melanogaster (Dme), H. sapiens (Hsa) tRNase ZL, one of several tRNases ZL from A. thaliana (Ath), and the one tRNase ZL from C. elegans (Cel) and from S. cerevisiae (Sce)) (accession numbers Q8MKW7, NP_060597, AAM51378, O4476, and NP013005.1, respectively). The lower two sequences are the bacterial tRNases ZS from B. subtilis (Bsu) and E. coli (Eco) (accession numbers P54548 and ZP_00726790, respectively). The numbering across the top of the eukaryotic tRNases ZL is based on presumed internal translation initiation at residue +24 (MAAT-) of the full-length D. melanogaster tRNase Z cDNA sequence (32). Structural designations (α4, α5, β10, and β11) are based on the B. subtilis (tRNase ZS) structure (15). η (residues 222–224 in D. melanogaster tRNase Z) refers to a short conserved 3/10 helix following β11. Dashed vertical lines indicate boundaries of the 40-residue Ala scan through the hand of the FA and the approximate boundaries of FA hand deletions. B, the two-subunit structure in which the blue subunit (A, lower right) has an intact active site containing the two metal ions (orange) and the red (B) subunit (upper left) has a structured flexible arm. C, an enlarged view of the FA is shown. Designations (α4, α5, β10, and β11) are from Ref. 15. η, a short conserved 3/10 helix following β11. The dashed line indicates the boundaries of the hand of the FA (ascending stalk-α4 and H-descending stalk). Blue asterisks mark the conserved leucine at the ascending stalk-α4 boundary (Leu158 in B. subtilis and Leu187 in D. melanogaster) and the conserved glycine at the GP loop-α5 boundary (Gly170 in B. subtilis and Gly200 in D. melanogaster). The GP loop is the α4–α5 loop and is rich in Gly and Pro residues. D, the contacts suggested by the co-crystal structure (Protein Data Bank code 2FK6) (20) are shown. The polypeptide is shown in the schematic. Selected region of tRNA in ribbon. Ball and stick view of Gly19/Cys56 tertiary base pair. Blue asterisks represent the contact surface including key residues (Leu187 and Gly200). Residue numbers are for B. subtilis tRNase Z; D. melanogaster residue numbers are in parentheses, e.g. Leu158(Leu187); Gly171(Gly200).
tRNase ZL may have arisen from a tandem gene duplication of tRNase ZS (2) with subsequent evolutionary adaptation. Human tRNase ZL displays between 1,500–2,000-fold higher catalytic efficiency than tRNase ZS (24). The short form has been more thoroughly studied (25), including the solution of three high resolution crystal structures (15, 26, 27) and a co-crystal structure with tRNA (20).
The active site is found entirely within the carboxyl-terminal two-fifths of tRNase ZL, which is a close homolog of tRNase ZS. The amino-terminal three-fifths of tRNase ZL displays weak homology with the carboxyl-terminal two-fifths and with tRNase ZS. Architectural relics from tRNase ZS were apparently retained in the amino-terminal portion of tRNase ZL (2), whereas residues involved with metal ion binding and catalysis were specifically replaced. Exceptionally, the FA of tRNase ZS occurs between the Motif III His and Motif IV Asp (twice in the homodimer), but only in the amino-terminal half of tRNase ZL between residues 173–232 (see Fig. 1). This adaptation, in which the FA occurs only in the amino portion of tRNase ZL and the active site is present only in the carboxyl-terminal part, could contribute to its expanded substrate range (10) and higher catalytic efficiency (24).
Sequence alignments were sufficient to identify the tRNase ZL FA (see Fig. 1) (21) despite the absence of additional structural information. Remarkably, deletion of the FA from S. cerevisiae tRNase Z abolished tRNA binding and catalysis without affecting its activity with the small molecule substrate bis(p-nitrophenyl)phosphate (21). The co-crystal structure shows contacts principally between α5 residues in the hand of the FA and the D/T loops (elbow) of tRNA (20). These results demonstrate that the FA of tRNase Z is involved with substrate recognition and binding remote from its active site and from the scissile bond of the substrate. Some of the substrate D/T loop substitutions in human tRNAArg also cause the Km for tRNase ZL to increase, suggesting effects on binding (28). The FA of Thermotoga maritima tRNase Z (a short form with an atypical cleavage site and FA) (29, 30) is dispensable, but some of the mutations affected its cleavage site (31).
D. melanogaster tRNase Z (a long form) with a deleted hand of the FA has a Km almost 100-fold higher than that of the wild type enzyme. A 40-residue Ala scan through the hand of the FA shows a cluster of residues centered on the β10–β11 loop with reduced kcat, increases in Km clustered in the GP (α4–α5) loop and at the boundary with α5, and additional Km increases scattered at other conserved positions suggested by the co-crystal structure (20) to contact substrate. Interestingly, substitution of a single residue (Leu187) at the ascending stalk-α4 boundary (a conserved residue and predicted contact) (20) causes almost as great an increase in Km as deletion of the entire FA hand, with little decrease in kcat.
EXPERIMENTAL PROCEDURES
tRNase Z Mutagenesis and Expression
D. melanogaster tRNase Z cDNA (accession number AY119279) was baculovirus-expressed from methionine 24 (suggested to be the translation start for the nuclear form of the enzyme) (32). Residues are thus numbered +1 from this methionine (19), hence Gly200 for a central conserved residue in the GP (α4–α5) loop of the FA (see Fig. 1). Overlap extension amplification for the Ala scan through the hand of the FA was achieved using complementary mismatched oligonucleotides (Sigma Genosys) with a GCC Ala codon substituted at each of 40 positions (Ala186 in wild type tRNase Z was substituted with Thr). ΔFA hand deletions were similarly constructed; ΔFA designations refer to the last residue retained on the amino-terminal side of the breakpoint and the first residue retained on the carboxyl-terminal side (e.g. ΔFA183/226 retains Arg183 and Thr226 with 42 residues deleted from Ala184 to Val225) (see Fig. 1). Versions of tRNase Z with FA deletions closer to the boundary between the stalk and the body of the enzyme (e.g. from 173/232 to 177/228) did not express (data not shown), suggesting that a finishing loop is required to avoid interfering with structure of the body of the protein.
Oligonucleotides were 33-nucleotides (nt) long (15 wild type nt on each side of the substitution). Forward and reverse primers for the unique internal BstEII (nt 470) and PflMI (nt 1149) subcloning sites were used to obtain A and B segments incorporating the mismatches. VENT DNA polymerase (New England Biolabs) was used for amplifications with an annealing temperature of 65° C. Longer oligonucleotides were used whenever an A or B segment failed to amplify. A/B segments were joined by overlap extension-amplification using the BstEII and PflMI primers and subcloned into the tRNase Z cDNA pFastBac-HTA (Invitrogen) transfer plasmid. Accuracy of construction was confirmed by DNA sequencing (Genewiz).
Bacmid DNA transfer, virus amplification, expression in insect Sf9 cells, and nickel chelate affinity purification of soluble tRNase Z were performed as described previously (19). Enzyme concentrations were determined using Bio-Rad dye reagent and confirmed by gel electrophoresis and fluorescent staining (Figs. 3A and 4A).
Substrate Preparation
D. melanogaster tRNAArg(UUA) (Fig. 3B), a canonical tRNA with a 17-nt 3′ end trailer, was prepared by T7 transcription of a template DraI digest (to obtain -UUU at the 3′ end of the 3′ end trailer), cleaved with a cis-acting hammerhead to obtain a mature 5′ end (as in Ref. 33), and gel-purified as described previously (17). Pre-tRNAArg was 5′ end-labeled with polynucleotide kinase and [γ-32P]ATP and repurified.
tRNase Z Processing Kinetics
Variant enzyme concentrations for processing kinetics were based on results of processing efficiencies (data not shown) determined using ∼10−10 m labeled pre-tRNAArg, between one and two orders of magnitude below the lowest Km observed (supplemental Table 1). At a low [S], V is pseudo-first order in [S], and % product/min (V/[S]) is a measure of reaction efficiency (kcat/Km). More than 50% of the substrate could be processed with sufficient wild type enzyme (data not shown).
For kinetic experiments, a constant trace amount of labeled substrate was supplemented with a concentration series of unlabeled pre-tRNAArg. The substrate concentration range (usually 2–100 nm for wild type (WT) tRNase Z) was adjusted so that the Km fell within the [S] range for each tRNase Z variant. RNA concentrations were independently determined in each experiment by running analytical lanes of the unlabeled RNA samples and comparing with standards. Percent product/min (V/[S]) decreases with increasing [S] (supplemental Fig. 1, C–E); V (obtained by multiplying V/[S] by [S]) increases with [S] as expected.
Enzyme concentrations were determined from original wild type and variant enzyme stocks (Bio-Rad assay). At the time of use, enzymes were diluted to 2 μg/μl (equivalent to 2.5 × 10−5 m) in the first tube of a dilution series. To refine the estimation of [E] in kinetic experiments, 2.5 μl of the subsequent 1:10 dilution (to 200 ng/μl, 25 μm) was electrophoresed on a protein gel (supplemental Fig. 1A) and compared with protein standards. Vmax (obtained from Eadie-Hofstee plots or by nonlinear regression analysis of Michaelis-Menten plots) (supplemental Fig. 1) was converted to kcat by dividing by [E].
Variant enzymes were used in two or more kinetic experiments (typically n = 3–4); a parallel experiment with wild type tRNase Z was included each time variant processing kinetics was performed. Calculations of values relative to WT were made between experiments with variant and wild type tRNases Z performed on the same day and, therefore, do not coincide with values calculated using the wild type tRNase Z data presented on row 1 of supplemental Table 1 (the means of all eighty kinetic experiments). Reproducible kinetic parameters were obtained with the lowest possible concentration of each variant enzyme (supplemental Fig. 1). Wild type tRNase Z was used at ∼25 pm, at least 1,000 times lower concentration than has been used by other laboratories (e.g. Refs. 31 and 34).
Multiple Sequence Alignments
Multiple sequence alignments (Fig. 1A) were prepared using ClustalW and displayed using GeneDoc. Structural designations for the hand of the FA (e.g. α4, α5, β10, and β11) are taken from the Bacillus subtilis tRNase ZS structure (Protein Data Bank code 1Y44) (15). Corresponding residue numbers are higher by 29 or 30 in the FA hand of D. melanogaster tRNase Z than in B. subtilis (e.g. D. melanogaster Leu187 corresponds to B. subtilis Leu158, and Gly200 corresponds to Gly171) (see alignments in Fig. 1) (15, 21). Residue colors signify identity or similarity, with green being the most conserved.
Structure Model Display of the Flexible Arm
The tRNase Z structure model (Fig. 1, B and C, and supplemental Fig. 3, A–C) (Protein Data Bank code 1Y44) (15) was visualized with PyMOL (35) using a schematic with ball and stick or space filling side chains. The tRNA in the co-crystal structure (Fig. 1D and supplemental Fig. 3D) (Protein Data Bank code 2FK6) (20) was displayed using ribbon with ball and stick or space filling side chains. Residue numbers are for B. subtilis tRNase Z; corresponding residue numbers for D. melanogaster tRNase Z are given in parentheses.
RESULTS
Multiple Sequence Alignments of the Flexible Arm
The flexible arm of tRNase Z consists of 35–40 residues in a globular, compact ααββη structure (the hand) (15, 27) extruded from the body of the enzyme and held apart from it by an extended two-stranded polypeptide stalk (Fig. 1). The sequence of the hand is more conserved than that of the stalks (Fig. 1A).
To further probe for residues and regions important for substrate recognition and binding, a FA hand deletion and an Ala scan through the hand of D. melanogaster tRNase Z (a long form) were analyzed using processing reaction kinetics. In the Ala scan, each of the 40 wild type residues (between the dashed lines in Fig. 1) was singly substituted with alanine, expressed using baculovirus, tested for processing efficiency (data not shown), and compared with the wild type enzyme in repeated kinetic experiments (Fig. 2) (cf. supplemental Fig. 1, supplemental Table 1, and supplemental Fig. 2). The Ala scan is bounded on the amino side by two residues of the ascending stalk and on the carboxyl side by three residues of a short conserved 3/10 helix (η in Fig. 1) following β11 and just preceding the descending stalk.
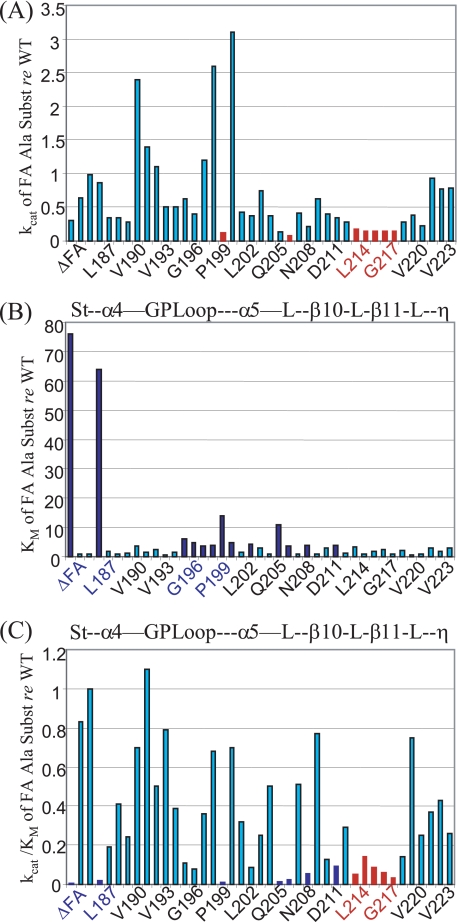
FIGURE 2.
Graphical representation of kinetic effects of the FA deletion and Ala substitutions relative to the wild type (from right three columns of supplemental Table 1). A–C, kcat, Km, and kcat/Km relative to wild type, respectively. S.E. are presented in supplemental Table 1. Abscissa, ΔFA and the 40 residues (Gly185–Asp224) (supplemental Table 1) with Ala substitutions. The designations between A and B and between B and C are structural elements based on alignment with B. subtilis tRNase ZS and the crystal structure (as in Figs. 1 and 2) (de la Sierra-Gallay et al. (15)). Colored residues and values on the bar graph indicate significant reductions in kcat (Gly200, Lys207, Lys214–Lys218) in red (A) and increases in Km (ΔFA, Leu187, Gly196–Pro201, Leu206, Lys207, Gly209, and Ile212) in dark blue (B). Substitutions that contribute to reduced catalytic efficiency (kcat/Km) are shown using the same colors as in C. See supplemental Fig. , which emphasizes the internal high Km relative to WT variants by suppressing the two highest values (ΔFA and L187A).
From the amino-terminal side, the sequence at the ascending stalk-α4 boundary (Gly185-X-Leu187) is conserved; Leu187 may be replaced by another bulky hydrophobic residue (Phe or Ile). α4 is an amphipathic helix with polar residues present at both ends and in the middle (n = 1, 4, and 7) with their side chains pointing toward the solvent and the tRNA, when present. Length, sequence, and position of the GP (α4–α5) loop are conserved with the consensus GXPXGP (sometimes GXPPGP), producing its characteristic shape (Fig. 1 and supplemental Fig. 3D), in which the polypeptide backbone in the vicinity of Gly200 runs parallel to the bases in the elbow of the tRNA (the tertiary Gly19-Cys56 base pair), producing favorable stacking interactions. α5 is moderately conserved, starting with two bulky hydrophobics and ending with a Gly, with a bulky hydrophobic residue and a Lys in between. The β10 consensus is two bulky hydrophobics flanking a Thr. The β10–β11 loop has a consensus Asp. β11 (G(K/R)x(I/V)) has a consensus Gly at its amino boundary followed by a consensus Lys or Arg, a nonconserved residue and a consensus hydrophobic (Ile or Val). The short 3/10 helix following β11 (η) ends with a consensus Asp, and the η-descending stalk boundary is marked by a bulky hydrophobic.
The original tRNase ZL sequence alignments (2) included a description of a conserved 10-residue Walker A motif that begins at the start of α5 (15). Several α5 residues were suggested to contact substrate based on the co-crystal structure (20). Additionally, a five-residue insertion sequence (TAAIA) close to the carboxyl-terminal border of the GP loop in Homo sapiens and other vertebrate tRNases ZL has not been further characterized.
Structure of the FA, Free and Bound to tRNA
The available structures of bacterial tRNases Z (short forms) (15, 26, 27) are similar; structure models presented here are taken from the first published structure (Fig. 1, B and C) (supplemental Fig. 3, A–C) (15) and the co-crystal structure (Fig. 1D and supplemental Fig. 3D) (20). The B subunit FA structure in free tRNase ZS is practically the same as when bound to tRNA (15, 20). These models provide the basis for secondary structure designations (from the amino to carboxyl terminus: ascending stalk-α4-GP loop-α5-β10-loop-β11-η-descending stalk). The structure of the hand (Fig. 1, B and C) (15) and the co-crystal structure with tRNA (Fig. 1D) (20) suggest that the hand directly binds the elbow of the tRNA (at a conserved distance from the scissile bond) through specific contacts and that the stalk establishes the distances from the hand to the body and active site. One side of the hand (the ascending stalk-α4 boundary, the GP loop-α5 boundary, and α5) faces the elbow of the tRNA. In Fig. 1C, the tRNA would be above the page. In Fig. 1D, the tRNA is on the left, giving the best view of all secondary structure elements. β10, the β10–β11 loop, and β11 are behind α4 and α5 and would not be expected to contact the tRNA. Conserved residues most important for substrate binding (Leu187 and Gly200) (Fig. 1, supplemental Fig. 1, and supplemental Table 1) are marked with blue asterisks.
Deletion of FA Hand Produces an ∼100-fold Increase in Km
A FA hand deletion was prepared to determine the contribution the FA hand makes to processing kinetics. Deletion of the FA hand close to its boundaries with the stalk (dashed lines in Fig. 1) does not interfere with stability and expression of tRNase ZL. FA hand deletions displayed processing efficiencies >100-fold lower than the wild type tRNase Z (data not shown). Processing kinetics was analyzed using ΔFA183/226 (supplemental Fig. 1).
The ΔFA hand variant has a Km ∼ 100× higher and a kcat ∼ 2× lower than wild type tRNase Z (Fig. 2). Because Km is roughly equivalent to an enzyme-substrate dissociation constant (KD), the hand of the FA thus contributes up to two orders of magnitude to the recognition/binding of substrate by wild type tRNase Z, which displays a Km of ∼3.7× 10−8 m (supplemental Table 1).
L187A Substitution at Ascending Stalk/Hand Boundary Reduces Substrate Binding Almost as Much as Deletion of FA Hand
A 40-residue Ala scan through the hand of the FA in which processing kinetics for each variant was compared with wild type enzyme (supplemental Table 1) revealed one residue (Leu187, equivalent to Leu158 in B. subtilis tRNase Z) (Fig. 1) in which substitution of alanine causes Km to increase almost as much as deletion of the entire FA hand, with barely any decrease in kcat (supplemental Fig. 1, compare E with D). As with the FA hand deletion, a higher concentration of L187A enzyme than that of WT tRNase Z had to be used to accommodate the lower catalytic efficiency of the variant; the [S] range for L187A experiments was raised to compensate for a Km between one and two orders higher than the wild type.
Other Changes in Km and kcat Arise from Clustered Ala Substitutions in the FA Hand
Although less striking, clusters of kinetic effects are observed with Ala substitutions in the FA hand (Fig. 2, cf. supplemental Figs. 1 and 2 and supplemental Table 1). Variants with single Ala substitutions throughout the hand of the D. melanogaster tRNase Z FA were constructed, expressed, and used in repeated processing experiments to obtain the kinetic parameters kcat, Km, and kcat/Km (as in supplemental Fig. 1 and columns to the left in supplemental Table 1). To control for day-to-day variations, each variant processing experiment was accompanied by a wild type tRNase Z processing experiment. The variant relative to wild type values (three right-most columns of supplemental Table 1) were calculated from the variant and wild type experiments performed at the same time, and therefore do not coincide with results calculated from the mean WT values (top row in supplemental Table 1).
Increases in Km and decreases in kcat were observed with the Ala substitutions throughout the FA hand. Taking into consideration the S.E. (± values in supplemental Table 1), only >3.5× increases in Km relative to WT and decreases of >5× in kcat relative to WT are highlighted (blue and red, respectively, in Fig. 2, cf. supplemental Table 1 and supplemental Fig. 2). Supplemental Fig. 2, prepared by suppressing the larger effects of the ΔFA hand and L187A variants, more clearly illustrates the moderate increases in Km (3.5–14×) with Ala substitutions. A cluster of increases in Km relative to WT is observed throughout the GP (α4–α5) loop, most strongly at Gly200 (14× WT), sprinkled through α5 (Leu203, Leu206, Lys207, and Gly209) and with one residue in β10 (Ile212). The reductions in kcat are centered on the β10–β11 loop (Leu214-Lys218) and at three other positions (Gly200, Leu206, and Lys207).
DISCUSSION
A Discrete Region of tRNase Z, the FA Hand
This study refines results from earlier studies of the flexible arm using kinetic analysis of a hand deletion and an Ala scan throughout the hand. The FA has been reported previously to be indispensable or, in contrast, dispensable for pre-tRNA processing (21, 31). Results with tRNase Z ΔFA establish that the hand of the FA contributes almost two orders of magnitude to the base Km for wild type tRNase Z of 3.4 × 10−8 m. The ability to assign a value to the contribution made by the FA to substrate binding is due to care taken with kinetic analysis and the quality of baculovirus-expressed tRNase Z. No changes in the cleavage site were observed, unlike in Ref. 31.
The Ala scan through the hand of the FA led to discovery of clusters of residues in which substitutions principally affect Km or kcat (the GP and β10–β11 loops, respectively) and of a single residue (Leu187) at the ascending stalk/hand (α4) boundary in which a substitution strikingly increases Km. These results were obtained by careful kinetic analysis and the substantial effort of a single residue Ala scan compared with punctate mutagenesis strategies.
Multiple sequence alignments show three classes of FA (21): the tRNase ZL FA and a slightly shorter FA of the bacterial tRNase Z class align well (Fig. 1A); an atypically short FA exemplified by Arabidopsis thaliana TRZ1 and T. maritima tRNase Z lacks the bulky hydrophobic at the ascending stalk/hand boundary, and the GP loop and is instead characterized by a cluster of 4–5 basic residues. Curiously, both tRNase Z FAs that have been subjected previously to mutagenesis analysis are from this latter category (31, 34), although the original FA deletion reported to be indispensable was from the more typical E. coli tRNase Z (21). Significance of L187A and the GP loop for substrate binding would therefore not have been detected in the latter studies. The results presented here, in light of previous reports, suggest that there are at least two general mechanisms by which the FA recognizes tRNA, one primarily based on interactions of complementary surfaces and the other based on electrostatic contacts.
Ala Substitutions That Produce Greatest Increases in Km
Substitutions in the hand of the FA that produce the greatest increases in Km (L187A and Gly200) are in conserved residues that were suggested by the co-crystal structure to contact substrate (blue asterisks in Fig. 1, C and D and supplemental Fig. 3) (20). Interestingly, neither of these side chains has potential for H-bonding or electrostatic interactions with nucleoside bases or the polynucleotide backbone. More likely, contacts are based principally on shapes and paths of the respective backbones and through stacking interactions.
Leu187 probably functions as a primary contact with substrate (20), and judging from its position at the ascending stalk-α4 boundary, could also maintain the structure, position, or orientation of the hand of the FA relative to the stalk and body of the enzyme, thereby influencing ability of the entire FA to bind substrate. The L187A substitution could increase flexibility at this boundary, enabling a stable interaction between the hand of the FA and the body of tRNase Z (arrows in supplemental Fig. 3A) so that the hand would no longer bind tRNA.
The bulky hydrophobic Leu187 side chain is in the interior of the FA hand in B. subtilis tRNase Z (supplemental Fig. 3B). The structure of the FA hand could be maintained by a combination of internal hydrophobicity and van der Waals contacts. According to this hypothesis (supplemental Fig. 3C, cf. Fig. 3B), the L187A substitution would allow the hand to collapse due to a net reduction in internal hydrophobicity and van der Waals attractions, explaining the large effect on Km.
A backbone amino group at Gly200 could electrostatically contact a backbone phosphate on the tRNA (supplemental Fig. 3D) (20); Ala substitution at that position might alter the path of the polypeptide or interfere sterically with its position to prevent this backbone contact. The path of the polypeptide chain in this region of the GP (α4–α5) loop runs parallel to the plane of the Gly19-Cys56 base pair, suggesting a stacking interaction (Fig. 2D and supplemental Fig. 3D). The increased Km observed with substitutions in the GP loop may informatively identify a broad contact surface. Processing kinetic analysis reports the Km effects on a dynamic ES complex, whereas the co-crystal was a static complex of enzyme with product; thus, the whole GP loop could directly contact the elbow of the substrate during catalysis. On the other hand, effects of these substitutions could be locally propagated along the path of the polypeptide consistent with a single contact corresponding to Gly200, as suggested previously (20).
Contact Positions in Elbow of Pre-tRNA Substrate
The sequence of several nucleotides in the elbow of tRNA are conserved in canonical tRNAs. (The Gly18-Gly19/Ψ55-Cys56 D/T loop tertiary base pairs that define the elbow and the Thr54-Ala58 pairing across the T loop that defines the U-turn.) These conserved features and the characteristic length of the coaxially stacked acceptor and T stems present potential characteristics for recognition by tRNase Z and other enzymes in the tRNA maturation pathway that distinguish tRNAs from other RNA molecules without discriminating between individual tRNAs (36–38). Moderate effects on Km of human tRNase ZL were observed with substitutions in the D and T loops of the substrate (28), consistent with a function in tRNase Z recognition for these structural elements. Importance of the elbow and T loop for tRNase Z reaction has been questioned (39), but this and similar studies were performed with a much higher enzyme concentration than in the present study.
Effects of Substitutions on kcat
The FA appears to be principally concerned with substrate recognition and binding, leading one to anticipate greater effects on Km than on kcat. Deletion of the entire FA hand (ΔFA) and one of the Ala substitutions (L187A) cause a large increase in Km with little decrease in kcat, which is consistent with this binding model. Most of the FA hand substitutions have a kcat relative to WT below 1, however, most clearly illustrated by a cluster of low kcat relative to WT values centered on the β10–β11 loop. One side of the β10-loop-β11 region faces α4-GP loop-α5, and the other side faces the solvent; thus, the region in which effects were observed does not obviously contact the substrate. A change in orientation of the FA hand could allow FA-tRNA binding to persist, but with the tRNA pointed away from the body of tRNase Z and the active site. kcat effects could also be explained by changes in protein-protein contacts or in protein folding and stability.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Drupattie Dial, Victoria Edwards, Shay Karkashon, and Asif Rizwan. We also acknowledge Kevin Ryan (City College) and Liang Tong (Columbia University) for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants S06GM08153, R15CA120072, and SC3GM084764. This work was also supported by grants from the Professional Staff Congress-City University of New York.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- FA
- flexible arm
- nt
- nucleotide
- WT
- wild type
- tRNase ZL
- long form of tRNase Z
- tRNase ZS
- short form of tRNase Z
- GP loop
- glycine/proline-rich loop.
REFERENCES
- 1.Aebi M., Kirchner G., Chen J. Y., Vijayraghavan U., Jacobson A., Martin N. C., Abelson J. ( 1990) J. Biol. Chem. 265, 16216– 16220 [PubMed] [Google Scholar]
- 2.Tavtigian S. V., Simard J., Teng D. H., Abtin V., Baumgard M., Beck A., Camp N. J., Carillo A. R., Chen Y., Dayananth P., Desrochers M., Dumont M., Farnham J. M., Frank D., Frye C., Ghaffari S., Gupte J. S., Hu R., Iliev D., Janecki T., Kort E. N., Laity K. E., Leavitt A., Leblanc G., McArthur-Morrison J., Pederson A., Penn B., Peterson K. T., Reid J. E., Richards S., Schroeder M., Smith R., Snyder S. C., Swedlund B., Swensen J., Thomas A., Tranchant M., Woodland A. M., Labrie F., Skolnick M. H., Neuhausen S., Rommens J., Cannon-Albright L. A. ( 2001) Nat. Genet. 27, 172– 180 [DOI] [PubMed] [Google Scholar]
- 3.Chen Y., Beck A., Davenport C., Chen Y., Shattuck D., Tavtigian S. V. ( 2005) BMC Mol. Biol. 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nashimoto M. ( 1997) Nucleic Acids Res. 25, 1148– 1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohan A., Whyte S., Wang X., Nashimoto M., Levinger L. ( 1999) RNA 5, 245– 256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pellegrini O., Nezzar J., Marchfelder A., Putzer H., Condon C. ( 2003) EMBO J. 22, 4534– 4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zareen N., Hopkinson A., Levinger L. ( 2006) RNA 12, 1104– 1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiffer S., Rösch S., Marchfelder A. ( 2003) Biol. Chem. 384, 333– 342 [DOI] [PubMed] [Google Scholar]
- 9.Korver W., Guevara C., Chen Y., Neuteboom S., Bookstein R., Tavtigian S., Lees E. ( 2003) Int. J. Cancer 104, 283– 288 [DOI] [PubMed] [Google Scholar]
- 10.Takaku H., Minagawa A., Takagi M., Nashimoto M. ( 2004) Nucleic Acids Res. 32, 4429– 4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith M. M., Levitan D. J. ( 2004) Dev. Biol. 266, 151– 160 [DOI] [PubMed] [Google Scholar]
- 12.Hölzle A., Fischer S., Heyer R., Schütz S., Zacharias M., Walther P., Allers T., Marchfelder A. ( 2008) RNA 14, 928– 937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominski Z. ( 2007) Crit. Rev. Biochem. Mol. Biol. 42, 67– 93 [DOI] [PubMed] [Google Scholar]
- 14.Vogel A., Schilling O., Meyer-Klaucke W. ( 2004) Biochemistry 43, 10379– 10386 [DOI] [PubMed] [Google Scholar]
- 15.de la Sierra-Gallay I. L., Pellegrini O., Condon C. ( 2005) Nature 433, 657– 661 [DOI] [PubMed] [Google Scholar]
- 16.Mandel C. R., Kaneko S., Zhang H., Gebauer D., Vethantham V., Manley J. L., Tong L. ( 2006) Nature 444, 953– 956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karkashon S., Hopkinson A., Levinger L. ( 2007) Biochemistry 46, 9380– 9387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominski Z., Yang X. C., Marzluff W. F. ( 2005) Cell 123, 37– 48 [DOI] [PubMed] [Google Scholar]
- 19.Zareen N., Yan H., Hopkinson A., Levinger L. ( 2005) J. Mol. Biol. 350, 189– 199 [DOI] [PubMed] [Google Scholar]
- 20.de la Sierra-Gallay I. L., Mathy N., Pellegrini O., Condon C. ( 2006) Nat. Struct. Mol. Biol. 13, 376– 377 [DOI] [PubMed] [Google Scholar]
- 21.Schilling O., Späth B., Kostelecky B., Marchfelder A., Meyer-Klacke W., Vogel A. ( 2005) J. Biol. Chem. 280, 17857– 17862 [DOI] [PubMed] [Google Scholar]
- 22.Schiffer S., Rösch S., Marchfelder A. ( 2002) EMBO J. 21, 2769– 2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takaku H., Minagawa A., Takagi M., Nashimoto M. ( 2003) Nucleic Acids Res. 31, 2272– 2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan H., Zareen N., Levinger L. ( 2006) J. Biol. Chem. 281, 3926– 3935 [DOI] [PubMed] [Google Scholar]
- 25.Redko Y., Li de Lasierra-Gallay I., Condon C. ( 2007) Nat. Rev. Microbiol. 5, 278– 286 [DOI] [PubMed] [Google Scholar]
- 26.Ishii R., Minagawa A., Takaku H., Takagi M., Nashimoto M., Yokoyama S. ( 2005) J. Biol. Chem. 280, 14138– 14144 [DOI] [PubMed] [Google Scholar]
- 27.Kostelecky B., Pohl E., Vogel A., Schilling O., Meyer-Klaucke W. ( 2006) J. Bacteriol. 188, 1607– 1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopkinson A., Levinger L. ( 2008) RNA Biol. 5, 104– 111 [DOI] [PubMed] [Google Scholar]
- 29.Minagawa A., Takaku H., Takagi M., Nashimoto M. ( 2004) J. Biol. Chem. 279, 15688– 15697 [DOI] [PubMed] [Google Scholar]
- 30.Ishii R., Minagawa A., Takaku H., Takagi M., Nashimoto M., Yokoyama S. ( 2007) Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 63, 637– 641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minagawa A., Ishii R., Takaku H., Yokoyama S., Nashimoto M. ( 2008) J. Mol. Biol. 381, 289– 299 [DOI] [PubMed] [Google Scholar]
- 32.Dubrovsky E. B., Dubrovskaya V. A., Levinger L., Schiffer S., Marchfelder A. ( 2004) Nucleic Acids Res. 32, 255– 262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fechter P., Rudinger J., Giegé R., Théobald-Dietrich A. ( 1998) FEBS Lett. 436, 99– 103 [DOI] [PubMed] [Google Scholar]
- 34.Späth B., Kirchner S., Vogel A., Schubert S., Meinlschmidt P., Aymanns S., Nezzar J., Marchfelder A. ( 2005) J. Biol. Chem. 280, 35440– 35447 [DOI] [PubMed] [Google Scholar]
- 35.DeLano W. L. ( 2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 36.McClain W. H., Guerrier-Takada C., Altman S. ( 1987) Science 238, 527– 530 [DOI] [PubMed] [Google Scholar]
- 37.Levinger L., Bourne R., Kolla S., Cylin E., Russell K., Wang X., Mohan A. ( 1998) J. Biol. Chem. 273, 1015– 1025 [DOI] [PubMed] [Google Scholar]
- 38.Shi P. Y., Weiner A. M., Maizels N. ( 1998) RNA 4, 276– 284 [PMC free article] [PubMed] [Google Scholar]
- 39.Shibata H. S., Takaku H., Takagi M., Nashimoto M. ( 2005) J. Biol. Chem. 280, 22326– 22334 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.