Abstract
The presence of histone H3 lysine 36 methylation (H3K36me) correlates with actively transcribed genes. In yeast, histone H3K36me mediated by KMT3 (also known as Set2) recruits a histone deacetylase complex, Rpd3s, to ensure the fidelity of transcription initiation. We report the purification of human KMT3a (also known as HYPB or hSet2) complex and the identification of a novel, higher eukaryotic specific subunit, heterogeneous nuclear ribonucleoprotein L (HnRNP-L). Interestingly, although KMT3a has intrinsic activity in vitro, HnRNP-L is essential in vivo. Moreover, KMT3a generates mono-, di-, and trimethylated products in vitro, but RNA interference against KMT3a or HnRNP-L down-regulates exclusively the H3K36me3 mark in vivo.
In the last two decades, histone modifications have been recognized as being pivotal to nearly all DNA templated processes in eukaryotes (for reviews, see Refs. 1–5). Eukaryotic transcription is highly influenced by various histone modification events. Besides the well studied marks of histone acetylation, several histone marks have been linked to active transcription: mono-ubiquitination of histone H2BK123 (Lys-120 in vertebrates) and methylation of histones H3K4, H3K36, and H3K79 (for reviews, see Refs. 6–9).
Yeast KMT3, the first enzyme identified as mediating H3K36 methylation (10), is linked to transcription elongation via its interaction with the Ser-2-phosphorylated C-terminal domain of RNA polymerase II (11–14) through its C-terminal SRI domain (15). KMT3-mediated H3K36 methylation (H3K36me)4 serves as a docking site to recruit a histone deacetylase complex, Rpd3s, which in turn deacetylates the nucleosomes residing at the coding regions to ensure the fidelity of transcription initiation (16–19). On the other hand, H3K36 methylation can also recruit the NuA3 histone acetyltransferase complex through its PhD finger domain-containing subunit Nto1 (20, 21). The means by which H3K36me-mediated recruitment of histone deacetylase and histone acetyltransferase activities is balanced remains an interesting question. However, NuA3 interacts with the Set1 methyltransferase, and NuA3 function depends on both Set1 and its substrate H3K4 (20). Therefore a possibility is that these histone deacetylase and histone acetyltransferase complexes are differentially recruited depending on the co-presence of H3K4me and H3K36me within the same or neighboring nucleosomes.
In higher eukaryotes, H3K36 methylation is also linked with active transcription (22–24). Interestingly, reduced levels of H3K36 methylation lead to subsequent reductions in H4K16 acetylation (23), which further impacts dosage compensation in Drosophila (25). In mammals, methylation of H3K36 is also mediated by the NSD family of enzymes (26),5 and NSD family HKMTs share high sequence similarity with yeast KMT3 only at the Set domain, but not other regions. In contrast, KMT3a (also known as HYPB or hSet2) is the mammalian orthologue of yeast KMT3, which shares high sequence similarity with yeast KMT3a at the Set domain, as well as at the WW and SRI domains. KMT3a specifically methylates H3K36 (27). Interestingly, unlike the yeast KMT3, knockdown of KMT3a results in the reduction of H3K36me3 exclusively, without affecting the other H3K36 methylation states (me1 and me2) in mammals (28) as well as in Drosophila (23).
Here we report the purification of human KMT3a complex and the identification of a novel, higher eukaryotic specific subunit, heterogeneous nuclear ribonucleoprotein L (HnRNP-L). Interestingly, although KMT3a has intrinsic activity in vitro, RNAi against HnRNP-L leads to the reduction of H3K36me3 in vivo, without affecting KMT3a expression levels.
HnRNP-L is a member of a large family of RRM domain-containing RNA-binding proteins. It has been implicated in various RNA-related processes, particularly in mediating selective exon inclusion during alternative splicing (29–32) and facilitating polyadenylation (32, 33). These so-called “post-transcriptional” events are known to be “co-transcriptional” (34, 35) and occur proximal to the chromatin templates. Our finding that HnRNP-L plays a role in regulating chromatin modification suggests the existence of an elaborative cross-talk between the chromatin template and co-transcriptional pre-mRNA processing.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies against modified histones were purchased from Millipore. Antibody against H3 was kindly provided by Dr. Xingwang Deng, from the National Institute of Biological Sciences, Beijing. This antibody is a rabbit polyclonal generated with recombinant full-length H3 as immunogen, which recognize H3 regardless of its modification status. Antibody against HnRNP-L was a kind gift from Dr. Gideon Dreyfuss from the Howard Hughes Medical Institute, Dept. of Biochemistry and Biophysics, University of Pennsylvania School of Medicine, Philadelphia.
Stable Cell Lines
pFLAG-KMT3a-C and pFLAG-HnRNP-L were stably transfected into HEK293 cells to generate the FLAG-KMT3a-C and FLAG-HnRNP-L stable cell lines, respectively. Both plasmids were based on pCMV4-FLAG from Sigma, with a FLAG tag fused at the N terminus, under the control of CMV promoter.
Affinity Purification of the Human KMT3a Complex
M2 anti-FLAG-agarose (Sigma) was equilibrated with the same buffer used in nuclear extract preparation (20 mm Tris-HCl (pH 7.9), 1.5 mm MgCl2, 0.42 m NaCl, and 0.2 mm phenylmethylsulfonyl fluoride) and then incubated overnight at 4 °C with nuclear extract derived from the stable cell lines. The resin was washed with excess amounts of buffer containing 20 mm Tris-HCl (pH 7.9), 1.5 mm MgCl2, 0.2 mm phenylmethylsulfonyl fluoride, 0.5 m KCl, and proteins eluted with the same buffer supplemented with 0.1 mg/ml FLAG peptide.
Anti-FLAG affinity-purified materials were further fractionated on a 2.4-ml Superose 6 gel filtration column at 4 °C, with buffer containing 20 mm Tris-HCl (pH 7.9), 0.2 mm phenylmethylsulfonyl fluoride, and 0.5 m KCl.
Protein Identification
Gel-resolved proteins were digested with trypsin and fractionated, and resulting peptide pools were analyzed by matrix-assisted laser-desorption/ionization (MALDI) reflectron time-of-flight (TOF) MS. Mass spectrometric sequencing of selected peptides was done by MALDI-TOF/TOF (MS/MS) analysis on the same prepared samples. Any identification thus obtained was verified by comparing the computer-generated fragment ion series of the predicted tryptic peptide with the experimental MS/MS data.
RNase Treatment and Co-immunoprecipitation
200 μg of nuclear extracts derived from HEK293 cells stably expressing FLAG-KMT3a-C were incubated with excess amounts (100 μg) of RNaseA at 37 °C for 2 h. The extracts were then subjected to co-immunoprecipitation using antibody against HnRNP-L in parallel with its mock treated control extracts.
Co-immunoprecipitation between KMT3a and HnRNP-L was performed under stringent conditions by washing with buffer containing 20 mm Tris-HCl (pH 7.9), 1.5 mm MgCl2, 0.2 mm phenylmethylsulfonyl fluoride, 0.5 m KCl, and 0.1% Nonidet P-40.
HKMT Assay
A 30 μl of reaction mixture containing S-[methyl-3H]adenosylmethionine (PerkinElmer Life Sciences), recombinant oligonucleosomes and enzymes in HKMT assay buffer (50 mm Tris, pH 8.5, 20 mm KCl, 10 mm MgCl2, 10 mm dithiothreitol, and 250 mm sucrose) was incubated for 1 h at 30 °C. The reaction products were separated by 13% SDS-PAGE and then subjected to autoradiography.
RNAi Knockdown of KMT3a and HnRNP-L
HeLa cells treated with HnRNP-L siRNA were analyzed 72 h after transfection. The sequences of chemically synthesized siRNA for HnRNP-L transient knockdown experiments were adopted from Hung et al. (32). The upper strand sequences were as follows: HnRNP-L, 1) 5′-GAAUGGAGUUCAGGCGAUGTT-3′; 2) 5′-CUACGAUGACCCGCACAAATT-3′; and Scrambled, 5′-UUCUCCGAACGUGUCACGUTT-3′. Lentivirus short hairpin RNA vectors for establishing stable RNAi cell lines for KMT3a were purchased from Sigma.
Immunofluorescence
Immunofluorescence were performed with HEK293 cells transiently transfected with siRNA against HnRNP-L (pair 1). Cells were stained 72 h after transfection.
RESULTS
HnRNP-L Is a Higher Eukaryotic Specific Subunit of Human KMT3a Complex
Yeast KMT3 interacts with RNA polymerase II (11–14), and its purification under stringent conditions did not reveal any other co-purifying species present stoichiometrically (10). Given our previous experiences with protein complexes in higher eukaryotes and their complexity in that they often differ from their yeast counterparts (36, 37), we attempted to purify the human KMT3a complex to address its function in mammals.
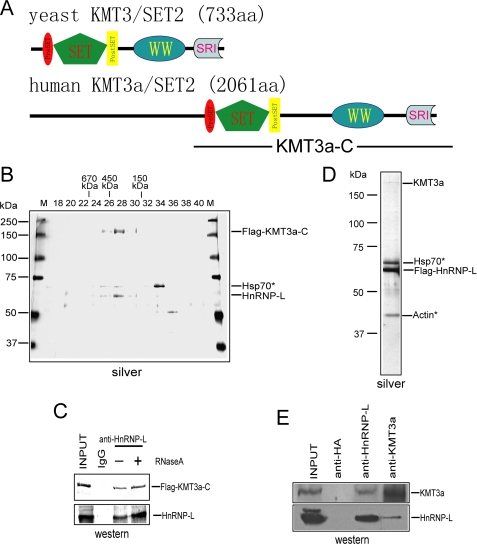
The closest homologue of yeast KMT3 in Homo sapiens is KMT3a, which has highly conserved domains and additional sequences of unknown function (Fig. 1A). Due to the failure of multiple attempts to stably express tagged full-length KMT3a in mammalian cells, we established an HEK293 stable cell line that expresses a FLAG-tagged version of the C-terminal half of KMT3a (FLAG-KMT3a-C, Fig. 1A). Nuclear extracts derived from this stable cell line were subjected to affinity purification using M2 anti-FLAG resin under stringent conditions (for details see “Experimental Procedures”), followed by Superose 6 gel filtration. Interestingly, FLAG-KMT3a-C co-eluted with a 60-kDa protein forming a complex with an apparent mass of ∼300 kDa. MALDI-TOF mass spectrometry analysis identified the co-eluting protein as HnRNP-L (Fig. 1B). HnRNP-L is an RNA-binding protein with three RNA recognition motifs, therefore it was possible that the apparent association between KMT3a and HnRNP-L was indirect, being mediated by their co-binding an RNA molecule. However, treatment of the nuclear extracts with excess amounts of RNaseA at 37 °C prior to affinity purification did not abolish the association between KMT3a and HnRNP-L (Fig. 1C), indicating the association is likely due to direct protein-protein interaction.
FIGURE 1.
HnRNP-L is a novel subunit of the human KMT3a complex. A, domain structure of human KMT3a and yeast KMT3. B, silver staining indicates co-elution of HnRNP-L with FLAG -KMT3a-C. Fractions were from a 2-ml Smart Superose 6 gel filtration column. The input of this column is affinity-purified KMT3a from cells stably expressing FLAG-KMT3a-C. Fraction numbers are shown at the top of the panel. C, interaction between KMT3a and HnRNP-L is independent of RNase treatment. D, endogenous, full-length KMT3a co-purifies with FLAG-HnRNP-L in nuclear extracts from stable cells expressing FLAG-HnRNP-L. E, reciprocal co-immunoprecipitation experiments demonstrate interactions between endogenous KMT3a and HnRNP-L in HeLa cells.
To further scrutinize the above finding as well as to obtain a complex containing full-length KMT3a, we established a stable cell line expressing FLAG-tagged HnRNP-L. Reciprocal affinity purification with nuclear exacts from these cells indeed co-purified the endogenous full-length KMT3a (Fig. 1D), which was confirmed by Western blot (data not shown, see also Fig. 1E) and HKMT activity assays (see Fig. 3, A and B).
FIGURE 3.
KMT3a HKMT activity. A, KMT3a complex purified from HEK293 cells stably expressing FLAG-HnRNP-L is a nucleosomal HKMT that does not use core histones as substrates. B, KMT3a complex is specific for H3K36. C, mass spectrometric analysis showing that KMT3a has mono-, di-, and trimethylation activities in vitro.
We next performed co-immunoprecipitation experiments using HeLa cell nuclear extracts and antibodies against KMT3a and HnRNP-L to test for their interaction under endogenous conditions. Antibodies against KMT3a co-immunoprecipitated the endogenous HnRNP-L (Fig. 1E, lower panel). In a reciprocal experiment, antibodies against HnRNP-L co-immunoprecipitated the endogenous KMT3a (Fig. 1E, upper panel). In control experiments, antibody against HA was ineffectual (Fig. 1E).
Interaction Domain Mapping
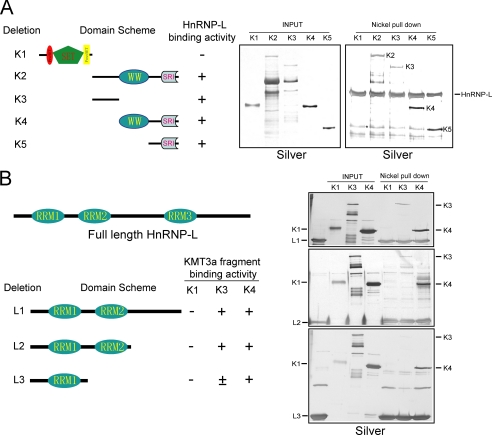
To map the regions of KMT3a responsible for its interaction with HnRNP-L, several GST-tagged KMT3a deletion mutants (Fig. 2A, left panel) were expressed and purified from Escherichia coli (Fig. 2A, middle panel), then incubated with recombinant His-tagged HnRNP-L purified from E. coli, and subjected to the nickel-agarose pulldown assay. Silver staining of the eluates demonstrated that two regions of KMT3a could interact with HnRNP-L independently. One region encompasses the linker sequence between the WW domain and the SET domain, and another region consists of the SRI domain (Fig. 2A, right panel). Nickel-agarose alone did not pull down any of the deletion mutants in control experiments (data not shown). In addition, the KMT3a SET domain failed to interact with HnRNP-L (Fig. 2A, right panel).
FIGURE 2.
Interaction domain mapping. A, left, schematic representation of GST-tagged KMT3a deletion mutants used for the interaction assay. Middle, silver stain of purified recombinant GST-KMT3a deletion mutants. Right, results from the nickel agarose pulldown assay using His-HnRNP-L. B, left, schematic representation of His-tagged HnRNP-L deletion mutants. Right, results from nickel pulldown assays using deletion mutants of His-tagged HnRNP-L and of GST-KMT3a, as indicated.
To map the domains on HnRNP-L responsible for its interaction with KMT3a, several His-tagged HnRNP-L deletion mutants (Fig. 2B, left panel) were expressed and purified from E. coli, then incubated with the recombinant GST-tagged KMT3a deletion mutants and subjected to the nickel-agarose pulldown assay. Silver staining of the eluates demonstrated that two regions of KMT3a could interact with HnRNP-L independently. The region containing the first RRM domain of HnRNP-L was sufficient to interact with the most C-terminal part of KMT3a (deletion K4) (Fig. 2B, bottom of right panel), whereas the region encompassing the second RRM domain could stabilize the interaction by binding to the KMT3a linker sequence between the SET and WW domains (Fig. 2B, top and middle of right panel). These studies collectively demonstrate the domains of each protein require for interaction, and further reinforce our purification studies. None of the interactions described above were sensitive to RNase treatment (data not shown).
Substrate and Reaction Specificity of Human KMT3a Complex
We then performed the histone methyltransferase assay using the human KMT3a complex purified from HEK293 cells stably expressing FLAG-HnRNP-L. The results showed it to be a nucleosomal specific HKMT, exhibiting barely detectable activity on core histones (Fig. 3A). In addition, its activity was specific for H3K36, because the K36A substitution mutant was completely defective as a substrate, while other K→A substitution mutants of histone H3 were successfully methylated (Fig. 3B). This result is consistent with that reported for the substrate specificity of the recombinant KMT3a SET domain (27).
Mass spectrometric analysis of the products formed in the in vitro HKMT assay demonstrated that KMT3a is capable of methylating H3K36 to mono-, di-, and trimethylated status (Fig. 3C). This is in contrast to that observed in vivo. RNAi against KMT3a exclusively reduced the H3K36me3 mark in vivo, without affecting the mono- and dimethylated status either in mammals (supplemental Fig. S1) (28) or in Drosophila (23).
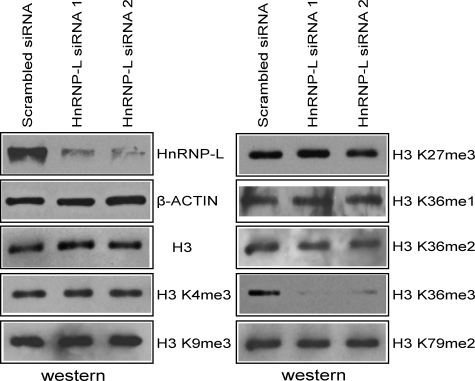
The Level of Histone H3K36me3 Is Dependent on KMT3a and HnRNP-L in Vivo
To further understand the functional importance of the KMT3a complex in vivo, especially with regard to its newly identified HnRNP-L subunit, we investigated the levels of various histone modifications in cells after HnRNP-L knockdown. Two different siRNA pairs successfully achieved HnRNP-L knockdown in HeLa cells (Fig. 4). Nuclear lysates were then analyzed by Western blot using various antibodies against histone proteins and specific histone modifications. In cells treated with either of the HnRNP-L siRNA pairs, we observed significantly lower H3K36me3 levels, but no apparent change in the levels of H3K36me1/2 or the other modifications tested (Fig. 4).
FIGURE 4.
RNAi against HnRNP-L down-regulates H3K36me3 levels. Western analysis showing specific reduction in HnRNP-L and H3K36me3 levels in HeLa cells treated with two different siRNA pairs against HnRNP-L. A pair of siRNA with scrambled sequences was used as control.
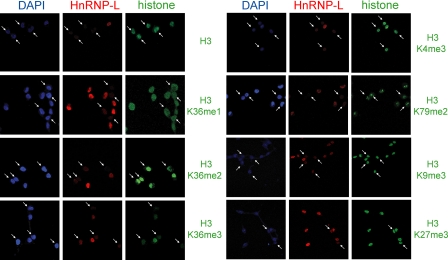
To validate these results, we also performed immunofluorescence studies using HEK293 cells transiently transfected with siRNA targeting HnRNP-L and co-staining with antibodies against HnRNP-L and either histone H3 or modified histone H3. Fig. 5 demonstrates the essential role of HnRNP-L in maintaining H3K36me3 levels in vivo. Consistent with the Western results shown in Fig. 4, none of the other modifications tested, including H3K36me1/2, were affected.
FIGURE 5.
Immunofluorescence indicates that reduction in H3K36me3 levels correlates with reduced HnRNP-L levels in HEK293 cells transiently transfected with siRNA against HnRNP-L. siRNA-treated cells were co-stained with antibody against HnRNP-L and either histone proteins or histones containing specific modifications. Cells exhibiting reduced levels of HnRNP-L are marked by arrows. Reduced HnRNP-L levels down-regulate H3K36me3, but were ineffectual with respect to H3K36me1/2, H3K4me3, H3K9me3, H3K27me3, H3K79me2, and histone H3.
HnRNP-L Does Not Stimulate KMT3a Activity in Vitro
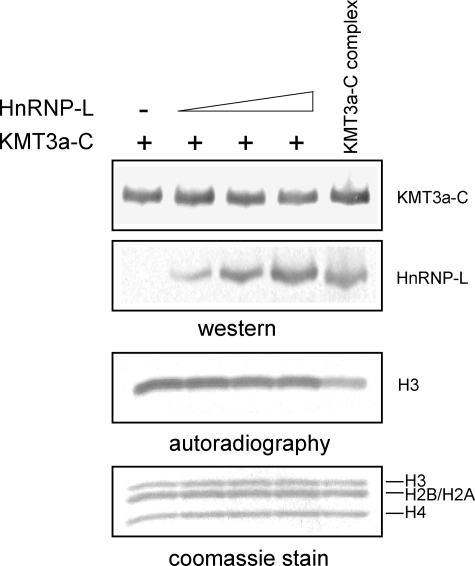
Because HnRNP-L knockdown had the same effect on histone H3K36 trimethylation as previously reported for KMT3a knockdown (supplemental Fig. S1) (28), we next examined if HnRNP-L could stimulate the HKMT activity of KMT3a biochemically. To address this, reconstitution experiments were performed using recombinant KMT3a-C and HnRNP-L, expressed from baculovirus independently or together. Co-expression of the two recombinant proteins displayed similar activity as KMT3a-C alone. In addition, titrating in recombinant HnRNP-L was ineffectual with respect to KMT3a-C activity (Fig. 6). Thus, HnRNP-L does not appear to up-regulate KMT3a activity under these conditions. Similar results were observed in the presence of synthetic RNA (data not shown).
FIGURE 6.
Addition of HnRNP-L does not stimulate KMT3a activity in vitro..
DISCUSSION
In this report, we presented the isolation, biochemical characterization, and functional analysis of the human KMT3a complex, which is composed of KMT3a and a higher eukaryotic specific subunit HnRNP-L. Although KMT3a has intrinsic activity in vitro (27) (Figs. 3 and 6), HnRNP-L is required for its activity in vivo (Figs. 4 and 5). Moreover, KMT3a generates mono-, di-, and trimethylated products in vitro (Fig. 3C), but RNAi against KMT3a (supplemental Fig. S1) (23, 28) or HnRNP-L (Figs. 4 and 5) down-regulates exclusively the H3K36me3 mark in vivo, suggesting other H3K36-specific HKMTs might be responsible for H3K36 mono- and dimethylation in vivo. Indeed our unpublished studies demonstrate that NSD1, NSD2, and NSD3 dimethylates H3K36.5
How Does HnRNP-L Contribute to the HKMT Activity of KMT3a?
The exact mechanism by which HnRNP-L contributes to the HKMT activity of KMT3a is presently an open question. One possibility is that HnRNP-L plays a role in targeting the KMT3a complex to certain regions in chromatin. This is plausible given that HnRNP-L is an RNA-binding protein with certain sequence context preferences (29, 38). It is an intriguing possibility that selective binding toward certain RNA sequences could enrich the local concentration of the KMT3a complex at particular chromatin region(s), giving rise to hot spots of H3K36me3. Because HnRNP-L is a higher eukaryotic specific subunit of the KMT3a complex, such H3K36me3 hot spots are unlikely to exist in yeast. Additional high resolution chromatin immunoprecipitation experiments in higher eukaryotes might be necessary to prove or disprove the existence of such hot spots. Another intriguing possibility is that HnRNP-L may bind to the nascent RNA, thus directing KMT3a to establish the H3K36me3 mark on the template chromatin, which might in turn recruit factors involved in co-transcriptional RNA processing (see detailed discussions below).
Are There Any Functions of H3K36me3 Specific to Higher Eukaryotes?
That higher eukaryotes express a subunit that specifically regulates HKMT trimethylase activity toward H3K36 begs the question of whether there are higher eukaryotic specific functions of H3K36me3 yet to be appreciated. Because HnRNP-L is an RNA-binding protein involved in multiple pre-mRNA processing events (29–33), if there are higher eukaryotic specific functions of H3K36me3, they are likely linked to co-transcriptional pre-mRNA processing such as splicing (or alternative splicing) and polyadenylation.
Splicing is largely a higher eukaryotic specific event, because most yeast genes do not have intron(s). In contrast, human exons are often dispersed and spaced apart by huge introns. Although multiple mechanisms (for reviews, see Refs. 39, 40) exist for correctly selecting these small exons out of the bulky introns, it is certainly possible that other regulatory mechanisms are involved. One attractive hypothesis is that the chromatin templates of exons in most, or alternatively spliced genes are marked by specific histone modification(s) that recruit the splicing machinery thereby elevating its local concentration to the sites of the splicing substrates. Of note, H3K4me3, a mark for the start site of transcription, could recruit CHD1 that in turn recruits U2 and stimulates the rate of splicing in vivo (41). However, as of now, chromatin modification(s) specifically marking the exons or exon-intron junctions have not been reported. Yet H3K36me3 would be a good candidate in this scenario as HnRNP-L is a factor involved in alternative splicing (29–32) and, as shown here, is also a subunit of a chromatin-modifying enzyme complex essential for its activity. In support of this hypothesis that the splicing machinery could recognize the H3K36me3 mark, a recent report demonstrated that H3K36me3 is specifically enriched at the intron-exon boundaries (42).
Another post-transcriptional process to be considered is polyadenylation. Polyadenylation sites consist of a highly conserved sequence motif AAUAAA on the pre-mRNA (43, 44). Such a sequence could occur once every 4096 bp by chance. Although this is unlikely to be an issue in the case of yeast, it could pose a huge challenge to the integrity of polyadenylation in mammals that often produce pre-mRNA products of over 200 kb, each potentially containing over 50 such AAUAAA motifs. Thus higher eukaryotes might require additional mechanisms to ensure correct transcription termination and polyadenylation. This might entail that a gene, encoding a nascent mRNA that has been successfully cleaved by the polyadenylation machinery, would be marked with certain histone modification(s) to facilitate transcription termination. Of note, a histone modification (H3K4me3) marking the transcription start site (45, 46) has been reported to recruit RNA polymerase II general transcription factor D (TFIID) (47). In this case, productive transcription sets the mark at the initiation site, which in turn stabilizes the recruited transcription machinery, safeguarding the proper initiation of subsequent transcription. H3K36me3 serves as an interesting candidate to mark the termination site not only because it has a higher eukaryotic-specific regulator, but also its levels tend to peak at the 3′-end of genes (22).
Supplementary Material
Acknowledgments
We are grateful to Dr. Gideon Dreyfuss from the Howard Hughes Medical Institute, Dept. of Biochemistry and Biophysics, University of Pennsylvania School of Medicine, Philadelphia for antibodies against HnRNP-L. We truly appreciate Dr. Bindereif from the Institute of Biochemistry, Justus-Liebig-University of Giessen, Germany for providing human HnRNP-L cDNA. We thank Dr. Xingwang Deng from the National Institute of Biological Sciences, Beijing for providing antibodies against histone H3. We are grateful to Dr. L. D. Vales for valuable comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM37120. This work was also supported by the NCI Cancer Center Support Grant P30 CA08748 (to P. T.), by the Howard Hughes Medical Institute (to D. R.), and by the Chinese Ministry of Science and Technology 863 Project 2007AA02Z1A6 (to B. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Y. Li and D. Reinberg, unpublished observations.
- H3K36me
- H3K36 methylation
- HnRNP-L
- heterogeneous nuclear ribonucleoprotein L
- RNAi
- RNA interference
- CMV
- cytomegalovirus
- MALDI
- matrix-assisted laser-desorption/ionization
- TOF
- time-of-flight
- MS
- mass spectrometry
- MS/MS
- tandem MS
- siRNA
- small interference RNA
- GST
- glutathione S-transferase.
REFERENCES
- 1.Jenuwein T., Allis C. D. ( 2001) Science 293, 1074– 1080 [DOI] [PubMed] [Google Scholar]
- 2.Margueron R., Trojer P., Reinberg D. ( 2005) Curr. Opin. Genet. Dev. 15, 163– 176 [DOI] [PubMed] [Google Scholar]
- 3.Martin C., Zhang Y. ( 2005) Nat. Rev. Mol. Cell Biol. 6, 838– 849 [DOI] [PubMed] [Google Scholar]
- 4.Li B., Carey M., Workman J. L. ( 2007) Cell 128, 707– 719 [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. ( 2007) Cell 128, 693– 705 [DOI] [PubMed] [Google Scholar]
- 6.Hampsey M., Reinberg D. ( 2003) Cell 113, 429– 432 [DOI] [PubMed] [Google Scholar]
- 7.Shilatifard A. ( 2006) Annu. Rev. Biochem. 75, 243– 269 [DOI] [PubMed] [Google Scholar]
- 8.Berger S. L. ( 2007) Nature 447, 407– 412 [DOI] [PubMed] [Google Scholar]
- 9.Weake V. M., Workman J. L. ( 2008) Mol. Cell 29, 653– 663 [DOI] [PubMed] [Google Scholar]
- 10.Strahl B. D., Grant P. A., Briggs S. D., Sun Z. W., Bone J. R., Caldwell J. A., Mollah S., Cook R. G., Shabanowitz J., Hunt D. F., Allis C. D. ( 2002) Mol. Cell Biol. 22, 1298– 1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J., Moazed D., Gygi S. P. ( 2002) J. Biol. Chem. 277, 49383– 49388 [DOI] [PubMed] [Google Scholar]
- 12.Li B., Howe L., Anderson S., Yates J. R., 3rd, Workman J. L. ( 2003) J. Biol. Chem. 278, 8897– 8903 [DOI] [PubMed] [Google Scholar]
- 13.Xiao T., Hall H., Kizer K. O., Shibata Y., Hall M. C., Borchers C. H., Strahl B. D. ( 2003) Genes Dev. 17, 654– 663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogan N. J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D. P., Beattie B. K., Emili A., Boone C., Shilatifard A., Buratowski S., Greenblatt J. ( 2003) Mol. Cell Biol. 23, 4207– 4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kizer K. O., Phatnani H. P., Shibata Y., Hall H., Greenleaf A. L., Strahl B. D. ( 2005) Mol. Cell Biol. 25, 3305– 3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. ( 2005) Cell 123, 581– 592 [DOI] [PubMed] [Google Scholar]
- 17.Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., Collins S. R., Schuldiner M., Chin K., Punna T., Thompson N. J., Boone C., Emili A., Weissman J. S., Hughes T. R., Strahl B. D., Grunstein M., Greenblatt J. F., Buratowski S., Krogan N. J. ( 2005) Cell 123, 593– 605 [DOI] [PubMed] [Google Scholar]
- 18.Joshi A. A., Struhl K. ( 2005) Mol. Cell 20, 971– 978 [DOI] [PubMed] [Google Scholar]
- 19.Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J. L. ( 2007) Science 316, 1050– 1054 [DOI] [PubMed] [Google Scholar]
- 20.Martin D. G., Grimes D. E., Baetz K., Howe L. ( 2006) Mol. Cell Biol. 26, 3018– 3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi X., Kachirskaia I., Walter K. L., Kuo J. H., Lake A., Davrazou F., Chan S. M., Martin D. G., Fingerman I. M., Briggs S. D., Howe L., Utz P. J., Kutateladze T. G., Lugovskoy A. A., Bedford M. T., Gozani O. ( 2007) J. Biol. Chem. 282, 2450– 2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannister A. J., Schneider R., Myers F. A., Thorne A. W., Crane-Robinson C., Kouzarides T. ( 2005) J. Biol. Chem. 280, 17732– 17736 [DOI] [PubMed] [Google Scholar]
- 23.Bell O., Wirbelauer C., Hild M., Scharf A. N., Schwaiger M., MacAlpine D. M., Zilbermann F., van Leeuwen F., Bell S. P., Imhof A., Garza D., Peters A. H., Schübeler D. ( 2007) EMBO J. 26, 4974– 4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L., Zhao Z., Dong A., Soubigou-Taconnat L., Renou J. P., Steinmetz A., Shen W. H. ( 2008) Mol. Cell Biol. 28, 1348– 1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell O., Conrad T., Kind J., Wirbelauer C., Akhtar A., Schübeler D. ( 2008) Mol. Cell Biol. 28, 3401– 3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rayasam G. V., Wendling O., Angrand P. O., Mark M., Niederreither K., Song L., Lerouge T., Hager G. L., Chambon P., Losson R. ( 2003) EMBO J. 22, 3153– 3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun X. J., Wei J., Wu X. Y., Hu M., Wang L., Wang H. H., Zhang Q. H., Chen S. J., Huang Q. H., Chen Z. ( 2005) J. Biol. Chem. 280, 35261– 35271 [DOI] [PubMed] [Google Scholar]
- 28.Edmunds J. W., Mahadevan L. C., Clayton A. L. ( 2008) EMBO J. 27, 406– 420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hui J., Stangl K., Lane W. S., Bindereif A. ( 2003) Nat. Struct. Biol. 10, 33– 37 [DOI] [PubMed] [Google Scholar]
- 30.Rothrock C. R., House A. E., Lynch K. W. ( 2005) EMBO J. 24, 2792– 2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melton A. A., Jackson J., Wang J., Lynch K. W. ( 2007) Mol. Cell Biol. 27, 6972– 6984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung L. H., Heiner M., Hui J., Schreiner S., Benes V., Bindereif A. ( 2008) RNA 14, 284– 296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guang S., Felthauser A. M., Mertz J. E. ( 2005) Mol. Cell Biol. 25, 6303– 6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orphanides G., Reinberg D. ( 2002) Cell 108, 439– 451 [DOI] [PubMed] [Google Scholar]
- 35.Maniatis T., Reed R. ( 2002) Nature 416, 499– 506 [DOI] [PubMed] [Google Scholar]
- 36.Zhu B., Mandal S. S., Pham A. D., Zheng Y., Erdjument-Bromage H., Batra S. K., Tempst P., Reinberg D. ( 2005) Genes Dev. 19, 1668– 1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu B., Zheng Y., Pham A. D., Mandal S. S., Erdjument-Bromage H., Tempst P., Reinberg D. ( 2005) Mol. Cell 20, 601– 611 [DOI] [PubMed] [Google Scholar]
- 38.Liu X., Mertz J. E. ( 1995) Genes Dev. 9, 1766– 1780 [DOI] [PubMed] [Google Scholar]
- 39.Black D. L. ( 2003) Annu. Rev. Biochem. 72, 291– 336 [DOI] [PubMed] [Google Scholar]
- 40.Konarska M. M., Query C. C. ( 2005) Genes Dev. 19, 2255– 2260 [DOI] [PubMed] [Google Scholar]
- 41.Sims R. J., 3rd, Millhouse S., Chen C. F., Lewis B. A., Erdjument-Bromage H., Tempst P., Manley J. L., Reinberg D. ( 2007) Mol. Cell 28, 665– 676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolasinska-Zwierz P., Down T., Latorre I., Liu T., Liu X. S., Ahringer J. ( 2009) Nat. Genet. 41, 376– 381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiddes J. C., Goodman H. M. ( 1980) Nature 286, 684– 687 [DOI] [PubMed] [Google Scholar]
- 44.Fitzgerald M., Shenk T. ( 1981) Cell 24, 251– 260 [DOI] [PubMed] [Google Scholar]
- 45.Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. ( 2002) Nature 419, 407– 411 [DOI] [PubMed] [Google Scholar]
- 46.Schneider R., Bannister A. J., Myers F. A., Thorne A. W., Crane-Robinson C., Kouzarides T. ( 2004) Nat. Cell Biol. 6, 73– 77 [DOI] [PubMed] [Google Scholar]
- 47.Vermeulen M., Mulder K. W., Denissov S., Pijnappel W. W., van Schaik F. M., Varier R. A., Baltissen M. P., Stunnenberg H. G., Mann M., Timmers H. T. ( 2007) Cell 131, 58– 69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.