Abstract
LINGO-1 is a component of the tripartite receptor complexes, which act as a convergent mediator of the intracellular signaling in response to myelin-associated inhibitors and lead to collapse of growth cone and inhibition of neurite extension. Although the function of LINGO-1 has been intensively studied, its downstream signaling remains elusive. In the present study, a novel interaction between LINGO-1 and a serine-threonine kinase WNK1 was identified by yeast two-hybrid screen. The interaction was further validated by fluorescence resonance energy transfer and co-immunoprecipitation, and this interaction was intensified by Nogo66 treatment. Morphological evidences showed that WNK1 and LINGO-1 were co-localized in cortical neurons. Furthermore, either suppressing WNK1 expression by RNA interference or overexpression of WNK1-(123–510) attenuated Nogo66-induced inhibition of neurite extension and inhibited the activation of RhoA. Moreover, WNK1 was identified to interact with Rho-GDI1, and this interaction was attenuated by Nogo66 treatment, further indicating its regulatory effect on RhoA activation. Taken together, our results suggest that WNK1 is a novel signaling molecule involved in regulation of LINGO-1 mediated inhibition of neurite extension.
Axons of the adult mammalian central nervous system possess an extremely limited ability to regenerate after injury, largely because of inhibitory components of myelin preventing axon growth (1, 2). Several myelin-associated inhibitors have been identified, including myelin-associated glycoprotein (3–5), chondroitin sulfate proteoglycans (6), oligodendrocyte myelin glycoprotein (7), and Nogo (8–10). Myelin-associated glycoprotein, oligodendrocyte myelin glycoprotein, and Nogo bind to the Nogo-66 receptor (NgR)3 and exert their actions through a tripartite receptor complex NgR/LINGO-1/p75NTR (11) or NgR/LINGO-1/TROY (12, 13).
LINGO-1 is a transmembrane protein that contains a leucine-rich repeat, an immunoglobulin domain, and a short intracellular tail (11). LINGO-1 functions as an essential component of the NgR complexes that mediate the activity of myelin inhibitors to regulate central nervous system axon growth (11, 14). In neurons, the NgR complexes activate RhoA in the presence of myelin inhibitors, which lead to growth cone collapse and neurite extension inhibition (11). Attenuation of LINGO-1 function is able to overcome the myelin inhibitory activity in the spinal cord that prevents axonal regeneration after lesion in rats (15). Besides, it has been reported that LINGO-1 is also expressed in oligodendrocytes, where it negatively regulates oligodendrocyte differentiation and axon myelination (16). Inhibition of LINGO-1 promotes spinal cord remyelination in an experimental model of autoimmune encephalitis (17). Moreover, inhibition of LINGO-1 has been shown to enhance survival, structure, and function of dopaminergic neurons in Parkinson disease models (18). Although the function of LINGO-1 has been intensively studied, much less is known about its downstream signaling.
To gain insight into the mechanisms by which LINGO-1 functions, it is of considerable importance to identify new binding partners of LINGO-1. Therefore, using the intracellular domain of LINGO-1 as bait, we employed yeast two-hybrid screening on a brain cDNA library and identified several candidates that interact with LINGO-1, one of which is the protein kinase WNK1.
WNKs (with no lysine [K]) are a distinct subfamily of serine-threonine kinases, which are characterized by a unique placement of the lysine that is involved in binding ATP and catalyzing phosphoryl transfer (19). Thus far, WNKs are known composed of four members, WNK1, WNK2, WNK3, and WNK4. Mutations in the serine-threonine kinases WNK1 and WNK4 cause a Mendelian disease PAHII, featuring hypertension and hyperkalemia (20, 21), and their roles in the regulation of electrolyte flux in the kidney have been well established (22). Recently, other important features of WNKs are beginning to be understood. WNKs have also been proposed functioning in a number of non-transport processes, including cell growth, differentiation, and apoptosis (23–26). Although WNK1 has been shown to be expressed in brain (27, 28), little is known about its function in the nervous system until recently; mutations of a nervous system-specific exon of the WNK1 gene were found to cause Hereditary sensory and autonomic neuropathy type II (HSANII) (29). In this study WNK1 was demonstrated to interact with LINGO-1 and regulate Nogo-induced inhibition of neurite extension.
EXPERIMENTAL PROCEDURES
Plasmid Construct
The full-length LINGO-1 was generated by PCR from a human brain cDNA library and inserted into the YFP or GFP fusion vector pEYFP-N1 or pEGFP-N1 to generate LINGO-1-YFP or LINGO-1-GFP. The cDNA for the intracellular domain (amino acids 580–620) of the human LINGO-1(LINGO-1ICD) was subcloned in-frame into the GAL4 fusion vector pGBKT7 (Clontech). The cDNA for WNK1-(123–510) was subcloned into the pECFP-N1 vector to generate WNK1-(123–510)-CFP. Full-length human WNK1 construct EGFP-C2-hWNK1 and rat WNK1 construct pCMV5-Myc-rWNK1 were generous gifts from Prof. Peter Jordan and Prof. Melanin H. Cobb, respectively. The pCDNA-Myc-Rho-GDI1 was kindly provided by Prof. Lan Bao. All of the constructs were fully sequenced.
Yeast Two-hybrid Screening
Yeast two-hybrid screening was performed according to the manufacturer's protocols (BD Biosciences Clontech). The Saccharomyces cerevisiae strain AH109 was cultured in YPDA (1% w/v yeast extract, 2% w/v peptone, 2% w/v dextrose, 0.003% adenine) and transformed with the bait plasmid pGBKT7-LINGO-1ICD and mated with Y187 pretransformed with the library plasmids. At least 4.5 × 106 individual library clones were screened based on the ability of the transformed yeast to grow on synthetic defined medium deficient in tryptophan, leucine, histidine, and adenine (SD/−Trp/−Leu/−His/−Ade plates) and express β-galactosidase. Potential positive clones were selected, and prey plasmids containing library cDNA inserts were isolated and shuttled into Escherichia coli KC8 cells. After colonies bearing the same activating domain/library plasmid were eliminated by PCR, positive colonies were further confirmed by testing pACT2-cDNA against GAL4 to eliminate false positives and then sequenced. The co-transformants were further analyzed for β-galactosidase expression by filter assay as described before (30, 31). During the analysis, cotransformants of the prey vector pACT2 and bait vector pGBKT7 vector were set as the negative control, and yeast mating diploids containing murine p53 and SV40 large T antigen were used as the positive control.
FRET Measurements with Three-channel Microscopy
PC12 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 5% fetal bovine serum and 10% heat-inactivated horse serum (HyClone) in a humidified 5% CO2 atmosphere at 37 °C. For transient transfection, cells were plated onto 8-mm2 glass coverslips as a monolayer in 12-well plates coated with poly-l-lysine and treated with NGF (100 ng/ml) for 48 h and then transfected with equal amount of the pEYFP-N1-LINGO-1 and pECFP-N1-WNK1-(123–510) by using Lipofectamine2000 (Invitrogen). After a 4-h incubation, the medium containing transfection reagents was replaced with fresh medium with NGF (100 ng/ml) supplement. Thirty-six hours later 100 nm GST-Nogo66 or GST was added to the cells for 3 h and washed twice with phosphate-buffered saline, and then images were taken with an Olympus IX71 inverted microscope equipped with a ×40 objective lens, cooled charge-coupled device cascade 512FCCD camera (Roper Scientific, Inc.), dual filter wheels, and a xenon 85-watt light source, all controlled by Image-Pro®plus Version 5.0.1 software (Media Cybernetics).
The quantitative FRET measurement has been described previously (32, 33). Briefly, images were acquired sequentially through YFP, CFP, and FRET filter channels, and the filter sets used were YFP (excitation, 500/25 nm; emission, 545/35 nm), CFP (excitation, 440/21 nm; emission, 480/30 nm), and FRET (excitation, 440/21 nm; emission, 535/26 nm). Corrected FRET (FRETC) was calculated on a pixel-by-pixel basis for the entire image using the equation FRETC = FRET − (a × YFP) − (b × CFP), where FRET, CFP, and YFP correspond to background-subtracted images of cells co-expressing CFP and YFP fusion proteins acquired through the FRET, CFP, and YFP channels, respectively, a and b are the fractions of bleed-through of YFP and CFP fluorescence through the FRET filter channel, here quantified as 0.081 and 0.753, respectively. As for the statistical chart, we selected FRET ratio (FR) (34, 35) to quantify the FRET signal. In our system, the formula is FR = (FRET − 0.753 × CFP)/0.081 × YFP.
Immunofluorescence Staining
Cells cultured on coverslips or tissue slices from P0 rat brain were washed in phosphate-buffered saline, then fixed for 30 min with 4% paraformaldehyde at room temperature. The fixed samples were permeabilized with 0.1% Triton X-100 for 30 min, subsequently blocked with 1% bovine serum albumin in phosphate-buffered saline, incubated at 4 °C overnight with primary antibody (rabbit-anti-LINGO-1, 1:100, Upstate; goat-anti-LINGO-1, 1:100, Santa Cruz; rabbit-anti-WNK1, 1:100, Alpha Diagnostic; mouse-anti-Tuj1, 1:100, Chemicon, Temecula, CA), and detected by species-specific fluorescein isothiocyanate- or rhodamine- conjugated secondary antibodies (1:100, Santa Cruz). Fluorescence images were taken with a Leica SP5 confocal microscope.
Inhibition of WNK1 Expression by RNA Interference
Rat WNK1 DNA sequences were selected for designing candidate small hairpin RNAs. The siRNA sequences are: WNK1si-1, GCAACAGGATGATATCGAA; WNK1si-2, GCCAGAGCCTAATGGAATT. These WNK1si sequences were constructed into pSUPER vector to generate shRNAs. The WNK1si constructs were transfected into PC12 cells, and cells were subjected to reverse transcription-PCR or Western blot 48 h later. For qualitative expression analysis, the High Fidelity PCR system (Roche Applied Science) was used with the following primers: for WNK1, 5′-GCCATTTCAATACCAAGCC-3′/5′-ACTCCACTGAGTGCCGAAT-3′; for GAPDH, 5′-ATCACTGCCACCCAGAAGAC-3′/5′-ATGAGGTCCACCACCCTGTT-3′. In Western blot and morphology analysis, the control was the pSUPER vector without inserts, and the control siRNA (control si) in reverse transcription-PCR analysis was another unrelated siRNA oligonucleotide ligated into the pSUPER vector.
Immunoprecipitation and Immunoblotting
Tissue homogenates and cell lysates were prepared as described previously (31) and clarified by centrifugation at 11,200 × g for 20 min at 4 °C. An equal amount (300–500 μl) of supernatants was incubated with 5 μl of the corresponding antibodies for 2 h at 4 °C. Protein G-agarose beads (Roche Applied Science) were then added for another 12 h rotation at 4 °C; immunoprecipitated samples were then washed 3 times with lysis buffer, boiled 3–5 min in sample-loading buffer, and then subjected to SDS-PAGE, immunoblotted, and visualized with enhanced chemiluminescence (ECL, Pierce). The following antibodies were used: rabbit anti-LINGO-1 (1:500, Upstate), rabbit anti-WNK1 (1:500, Alpha Diagnostics), monoclonal mouse anti-Rho-GDIα (1:1000, Santa Cruz), monoclonal mouse anti-Myc (1:1000, Santa Cruz), rabbit anti-GFP (1:1000, Santa Cruz), horseradish peroxidase (HRP)-conjugated anti-GAPDH (1:10000, Kangcheng), and HRP-conjugated secondary antibody (1:10000, Santa Cruz).
RhoA Activity Assay
Active RhoA was determined with the GST-Rhotekin binding domain as described previously (36). Briefly, cells were treated with 100 nm GST-Nogo66 or GST for 60 min then washed with ice-cold phosphate-buffered saline and lysed in RIPA (50 mm Tris, pH 7.2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 500 mm NaCl, 10 mm MgCl2, plus protease inhibitors). Clarified cell lysates were incubated with GST-Rhotekin binding domain (20 μg) bound to beads at 4 °C for 60 min. The beads were then washed 4 times in buffer B (Tris buffer containing 1% Triton X-100, 150 mm NaCl, 10 mm MgCl2, plus protease inhibitors) at 4 °C. Bound RhoA proteins were detected by Western blot using a monoclonal antibody against RhoA (1:1000, Santa Cruz). The amount of GTP-bound RhoA was normalized to the total amount of RhoA in cell lysates as previously described (36).
Neurite Outgrowth Assay
PC12 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 5% fetal bovine serum and 10% heat-inactivated horse serum (HyClone) in a 37 °C and 5% CO2 incubator. Cells were seeded on a 12-well plate coated with poly-l-lysine 24 h before transfection using Lipofectamine2000. After a 4-h incubation with the transfection medium with NGF (100 ng/ml) supplement, 100 nm GST-Nogo66 or GST was added concurrently. Neurite outgrowth assay of PC12 cells was performed 72 h after transfection as previously described (31), and the cells possessing one or more neurites of a length more than twice the diameter of the cell body were scored as differentiated cells. The data were analyzed by Student's t test, and each value was the mean ± S.D. sampled from three independent experiments.
Cortical neurons were isolated from P0 Sprague-Dawley rats as previously described (37) and subjected to electro-transfection with a Nuclei transfector (AMAXA) according to the instructions. Three hours later the cultures were replaced with fresh Neurobasal-A medium with 2% B27 supplement (Invitrogen), and at the same time 100 nm GST-Nogo66 or GST were added. Forty-eight hours after transfection cells were observed under a fluorescent microscope (Olympus, excitation 454 nm), and the length of the longest neurite of GFP-positive neurons was quantified by MetaMorph image analysis software. The data were analyzed by Student's t test, and each value was the mean ± S.D.
RESULTS
WNK1 Is Identified as a LINGO-1-binding Protein in a Yeast Two-hybrid Screening
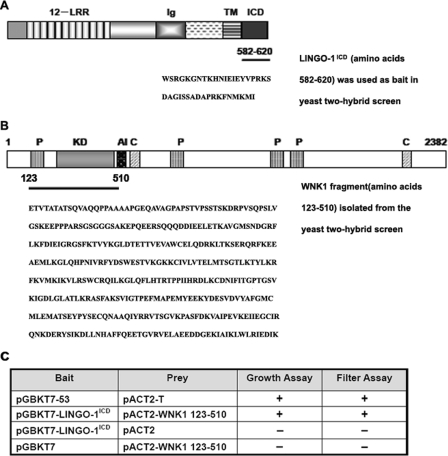
In an attempt to identify intracellular proteins involved in the regulation of LINGO-1 signaling, a yeast two-hybrid screening was performed with LINGO-1ICD, the intracellular domain of LINGO-1 (amino acids residues 582–620), as bait (Fig. 1A). In the screen 317 potential positive clones were isolated, and of these, 56 clones scored positive based on the β-galactosidase filter assay. False positives were further eliminated by testing autonomous activation, and 21 clones remained positive and were subjected to sequencing. One of the positive clones was identified harboring a 1164-bp cDNA fragment that corresponds to amino acid residues 123–510 of protein kinase WNK1 (38). The region includes the entire kinase domain (amino acid 221–479) plus part of the flanking sequences as shown in Fig. 1B. The interaction between LINGO-1-(582–620) and WNK1-(123–510) was further confirmed by cotransformation analysis, according to nutrition selection growth assay as well as β-galactosidase filter assay. In contrast, neither the combination of pGBKT7-LINGO-1ICD with the prey vector nor of pACT2-WNK1-(123–510) with the bait vector can initiate the reporter genes (Fig. 1C). Thus, WNK1 interacts with LINGO-1 in yeast two-hybrid screening system.
FIGURE 1.
WNK1 interacts with the cytoplasmic domain of LINGO-1 in a two-hybrid screen. A, schematic representation of the LINGO-1. Ig, immunoglobulin domain; TM, transmembrane domain; ICD, intracellular domain (amino acid 582–620), underlined as bait in the screening. LRR, leucine-rich repeat. B, the human WNK1 clone (amino acid 123–510, underlined) isolated from the two-hybrid screen. P, proline-rich regions; AI, autoinhibitory domain; KD, kinase domain; C, coiled-coil regions. C, prey and bait were co-transformed into AH109 yeast for autotrophic growth assay and filter assay to analyze the specificity of the interaction between LINGO-1ICD and WNK1-(123–510) in the two-hybrid system.
WNK1 Interacts Directly with LINGO-1 in Living Cells
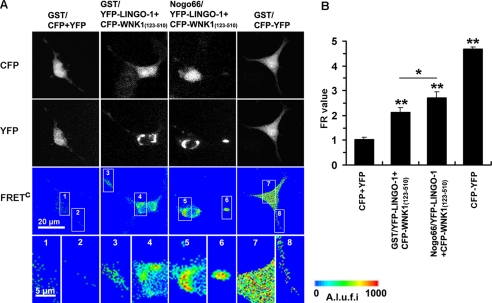
We next used a biophysical approach based on FRET to analyze the direct interaction between WNK1 and LINGO-1 in living cells. Full-length LINGO-1 and WNK1-(123–510) were cloned in the pEYFP-N1 and pECFP-N1 vector, respectively. To measure steady-state FRET, PC12 cells co-transfected with plasmids encoding CFP/YFP fusion proteins and subjected to GST or GST-Nogo66 treatment were imaged through CFP, YFP, and FRET filter channels. As shown in Fig. 2, cells co-expressing CFP and YFP, which served as a negative control, showed no FRET, with FR = 1.04 ± 0.07 (n = 51). On the other hand, cells co-expressing the CFP-YFP concatamer, a positive control for FRET, showed a significant increase in FR (FR = 4.68 ± 0.09, n = 55). Cells co-expressing LINGO-1-YFP with WNK1-(123–510)-CFP produced significant FRET signals in GST treatment, with FR = 2.13 ± 0.17 (n = 30), whereas the GST-Nogo66-treated group produced even greater FRET signals, with FR = 2.72 ± 0.23 (n = 34). The FRET signals could be observed in the cell body and at the terminus of neurites as visualized in images of higher magnification (Fig. 2A). These results suggested that fluorescent protein-tagged LINGO-1 and WNK1-(123–510) are able to interact directly in living PC12 cells, and the interaction can be intensified by Nogo treatment.
FIGURE 2.
FRET imaging to visualize interaction of LINGO-1 with WNK1-(123–510) in living cells. A, the indicated CFP and YFP fusion proteins were expressed in PC12 cells. After a 3-h treatment with GST-Nogo66 (100 nm) or GST (100 nm), images were taken by the CFP channel (upper lane), YFP channel (middle lane), and the FRET channel. FRETC was calculated as described under “Experimental Procedures” and is presented as pseudocolor (lower lane). A.l.u.f.i, arbitrary linear units of fluorescence intensity. Bar, 20 μm. B, FR values obtained from individual COS-7 cells expressing the indicated fusing proteins were calculated. Data shown are the mean ± S.D. from three independent experiments. p < 0.05 (*) and p < 0.001 (**) by Student's t test versus PC12 cells co-transfected with CFP and YFP (negative control) or as indicated.
WNK1 Forms a Co-precipitable Protein Complex with LINGO-1 in Brain Tissues and Cell Lysates
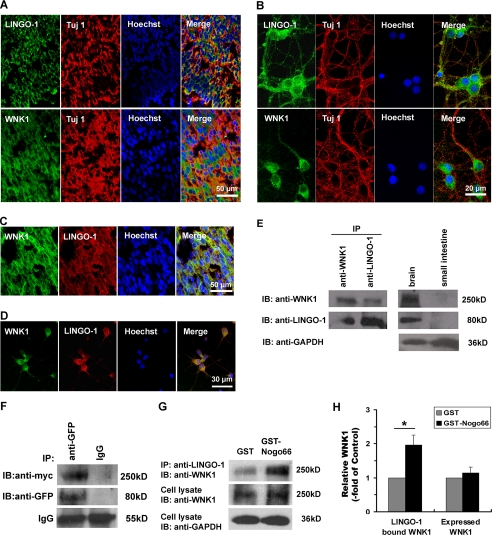
WNK1 has been detected in brain lysate by immunoblotting as previous report (27, 38). To examine the cellular localization and expression patterns of WNK1, we carried out immunohistochemistry analysis on rat brain slices and primary cultured neurons. As shown in Fig. 3, A and B, in the cortex slices and primary cultured neurons, both LINGO-1 and WNK1 were highly expressed in neurons which were marked by anti-Tuj1 in antibody staining. Furthermore, immunostaining of WNK1 along with LINGO-1 on P0 rat cortex slices showed they could colocalize in the same cells (Fig. 3C). To further examine the location of these proteins, primary cultured cortical neurons were co-stained with antibodies against WNK1 and LINGO-1. As shown in Fig. 3D, LINGO-1 was predominantly located on membrane, whereas WNK1 seemed more diffusely distributed. Our result suggested LINGO-1 and WNK1 could colocalize in the same cells in brain slices and primary cultured cortical neurons.
FIGURE 3.
WNK1 forms a co-precipitable protein complex with LINGO-1 both in vivo and in vitro. A–D, IHC staining to show WNK1 and LINGO-1 are colocalized in rat cortical neurons. A, IHC staining of LINGO-1, WNK1, and β-III tubulin in P0 rat brain cortex sections. Green, stained with anti-LINGO-1 or anti-WNK1. Red, stained with anti-Tuj 1 antibody. Bar, 50 μm. B, IHC staining of LINGO-1, WNK1, and Tuj 1 in primary cultured rat cortical neurons; scale bar, 20 μm. C and D, IHC staining of LINGO-1 and WNK1 in P0 rat brain cortex sections (C) or primary cultured rat cortical neurons (D). Green, stained with anti-WNK1; red, stained with anti-LINGO-1. Scale bars: C, 50 μm; D, 30 μm. E, immunoprecipitation (IP) shows endogenous WNK1 interacts with LINGO-1 in rat brain tissues. Protein expressions in brain and small intestine tissues were set as positive and negative controls, and GAPDH was were used to normalize the inputs. IB, immunoblot. F, immunoprecipitation shows Myc-tagged full-length WNK1 and GFP-tagged LINGO-1 interacts in COS-7 cells. The rabbit IgG was used as the immunoprecipitation control, and the heavy chain of IgG is visualized for input control. G, co-immunoprecipitation of WNK1 with LINGO-1 in primary cultured cortical neurons. The interaction between endogenous LINGO-1 and WNK1 was intensified by Nogo66 treatment. H, quantification of LINGO-1 bound WNK1 or endogenous expressed WNK1. The blots of WNK1 were normalized with GAPDH levels in cell lysates and then presented as -fold of GST-treated controls. Data shown are the mean ± S.D. from three independent experiments. *, p < 0.01 by Student's t test.
To examine whether LINGO-1 and WNK1 form a natural complex in mammalian brain tissues, we performed immunoprecipitation assays. As shown in Fig. 3E, P1 rat brain tissue lysates were immunoprecipitated with anti-WNK1 or anti-LINGO-1, then immunoblotted with anti-LINGO-1 and anti-WNK1 separately. Both LINGO-1 and WNK1 were detected in immunoprecipitates of anti-WNK1 or anti-LINGO-1. As a control, WNK1 and LINGO-1 were detected in brain tissue lysates but not in small intestine tissue lysates. Thus, endogenous LINGO-1 and WNK1 form a co-precipitable protein complex in brain tissues.
To further validate the interaction between LINGO-1 and WNK1, GFP-tagged LINGO-1 and Myc-tagged full-length rat WNK1 were co-transfected into COS-7 cells then subjected to immunoprecipitation with anti-GFP or control IgG. As shown in Fig. 3F, Myc-tagged WNK1 was co-immunoprecipitated by anti-GFP, but not by the control IgG, further suggesting the specific association between LINGO-1 and WNK1.
To examine whether the combination between the endogenous protein responses to Nogo66 stimulation, primary cultured cortical neurons were treated with GST-Nogo66 (100 nm) or GST (100 nm) for 48 h and then subjected to immunoprecipitation with anti-LINGO-1 and further blotted with anti-WNK1 with an equal proportion of the cell lysate set in parallel for input control. As shown in Fig. 3G, the combination between endogenous LINGO-1 and WNK1 was significantly intensified after Nogo66 treatment, whereas the endogenous expression of WNK1 was not altered. The quantitative analysis was shown in Fig. 3H. When normalized with GAPDH and standardized by the control (GST-treated group), LINGO-1-bound WNK1 increased by approximate 1-fold. Therefore, WNK-1 forms a co-precipitable protein complex with LINGO-1 both in vivo and in vitro, whereas Nogo66 treatment intensifies the endogenous combination.
WNK1 Mediates the Inhibitory Effect of Nogo on Neurite Outgrowth in PC12 Cells
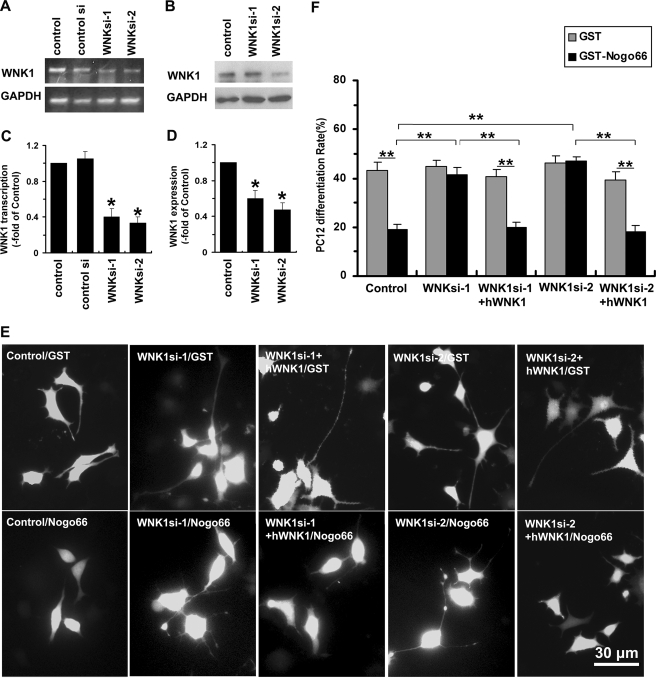
To identify a possible physiological role of WNK1, the RNAi approach was employed. Two different siRNA sequences were selected and constructed into pSUPER vector, referred as WNK1si-1 or WNKsi-2. PC12 cells were transfected with either pSUPER/shWNK1 (WNK1si-1 or WNKsi-2), pSUPER/scramble (Control si), or pSUPER vector (Control) and subjected to reverse transcription-PCR or Western blot 72 h later. We observed that the level of WNK1 mRNA was significantly suppressed in cells transfected with pSUPER/shWNK1, but it was not affected in the scramble control (Fig. 4, A and C). Western blot analysis also confirmed that each pSUPER/shWNK1 strongly suppressed WNK1 protein levels (Fig. 4, B and D). These results indicated that promoter-driven endogenous expression of shWNK1 was able to mediate RNAi of WNK1 in PC12 cells. We further performed neurite outgrowth assay in PC12 cells. Following transfection, all of the PC12 cells were treated with 100 ng/ml NGF for 3 days accompanied with GST-Nogo66 (100 nm) or GST (100 nm). As expected, in the group transfected with pSUPER vector, which was set as a control, cell differentiation rate was significantly decreased in GST-Nogo66 treated PC12 cells (19.2 ± 2.1%, n = 213) compared with the GST-treated cells (43.3 ± 3.2%, n = 221). In contrast, when pSUPER/shWNK1 was transfected, cell differentiation rates were significantly increased, and there was no difference between GST (WNK1si-1, 45.3 ± 2.5%, n = 211; WNK1si-2: 46.1 ± 2.8%, n = 243) and GST-Nogo66 treatment (WNK1si-1, 42.1 ± 2.9%, n = 208; WNK1si-2, 46.9 ± 1.9%, n = 239) (Fig. 4, E and F), suggesting that knockdown of endogenous WNK1 abrogates Nogo inhibition in PC12 cell differentiation. Furthermore, when a human full-length WNK1, which is resistant to the RNAi approach, was co-transfected with either of the two pSUPER/shWNK1s, the effect of WNK1 RNAi on PC12 differentiation in the presence of Nogo66 was abolished (WNK1si-1+hWNK1, GST, 40.5 ± 3%, n = 295; GST-Nogo66, 20.1 ± 2.1%, n = 265; WNK1si-2+hWNK1, GST, 39.5 ± 3.3%, n = 287; GST-Nogo66, 18.1 ± 2.5%, n = 245), further indicating the specificity of the WNK1 RNAi. Therefore, endogenous WNK1 mediates the inhibitory effect of Nogo on neurite outgrowth in PC12 cells.
FIGURE 4.
Knockdown of WNK1 eliminates Nogo-induced inhibition of neurite outgrowth in PC12. A, reverse transcription-PCR analysis of WNK1 mRNA expression from PC12 cells transfected with pSUPER/WNK1shRNAs (WNK1si-1, WNK1si-2), pSUPER/scramble (Control si), or pSUPER vector (Control). GAPDH was used as an internal standard. B, WNK1 expression level was also determined by Western blot. C and D, quantification of A and B, respectively, presented as -fold of control. Data shown are the mean ± S.D. from three independent experiments. *, p < 0.01 by Student's t test. E, PC12 cells transfected with pSUPER/WNK1shRNAs, pSUPER vector, or pSUPER/WNK1shRNAs along with the full-length human WNK1 construct were treated with 100 nm GST or GST-Nogo66 in the presence of NGF (100 ng/ml) for 72 h and then visualized using fluorescence microscopy. F, quantification of differentiated cells. Values are the mean ± S.D. **, p < 0.001 by Student's t test compared as indicated.
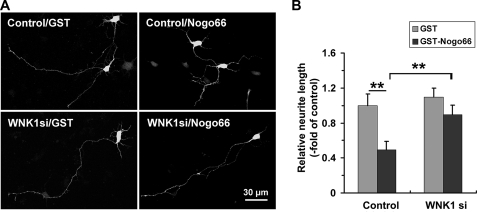
Suppressing WNK1 Promotes Neurite Extension and Abrogates the Inhibitory Response to Nogo in Cortical Neurons
To examine the function of WNK1 in neurons, cortical neurons from embryonic day 18 (E18) rats were cultured and transfected with the most effective pSUPER/shWNK1 (WNK1si-2) or pSUPER (control). As shown in Fig. 5A, in pSUPER-transfected neurons (GFP-positive), as expected, neurite extension was significantly decreased in GST-Nogo66-treated cells compared with the GST-treated cells. In contrast, pSUPER/shWNK1-transfected neurons showed diminished responses to GST-Nogo66, as is evidenced by the presence of longer neurites than those observed in control cells transfected with pSUPER (Fig. 5B). These results suggested that WNK1 is involved in mediating Nogo inhibitory effect on neurite extension in cortical neurons.
FIGURE 5.
Suppressing WNK1 expression promotes neurite extension in primary cultured cortical neurons. A, neurite extension of pSUPER/WNK1shRNA (WNK1si)- or pSUPER vector (Control)-transfected cortical neurons in the presence of GST or GST-Nogo66 visualized by confocal microscope. Scale bar, 30 μm. B, the longest neurite length of the transfected neurons was quantified, presented as -fold of control. Values are the mean ± S.D. **, p < 0.001 by Student's t test compared as indicated.
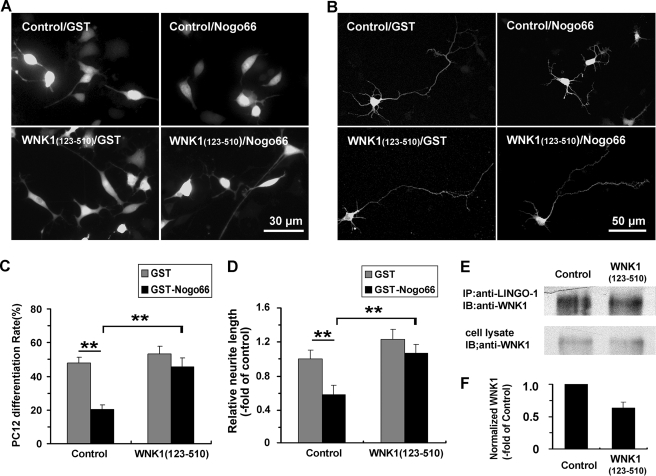
Overexpressing WNK1-(123–510) Attenuates Nogo-induced Inhibition of Neurite Extension
To test the effect of overexpressing WNK1-(123–510), PC12 cells and primary cultured cortical neurons were transfected with pEYFP-N1-WNK1-(123–510) construct or pEYFP-N1 vector as a control, then cells were treated with NGF as well as GST-Nogo66 or GST as previously described. In the group transfected with pEYFP-N1 vector, the PC12 cell differentiation rate was significantly decreased in GST-Nogo66-treated PC12 cells (20.1 ± 2.9%, n = 131) compared with the GST-treated cells (47.9 ± 3.5%, n = 125). When pEYFP-N1-WNK1-(123–510) was transfected, the cell differentiation rates in the GST- and GST-Nogo66-treated group were 53.3 ± 4.5% (n = 137) and 45.7 ± 5.1% (n = 122), respectively, which were significantly increased compared with the group transfected with pEYFP-N1 vector (Fig. 6, A and C). In primary cultured cortical neurons, pEYFP-N1-WNK1-(123–510)-transfected neurons also showed diminished responses to GST-Nogo66, as is evidenced by the presence of longer neurites than those observed in control cells transfected with pEYFP-N1 vector (Fig. 6, B and D). These results suggest that overexpressing WNK1-(123–510) could inhibit Nogo-induced inhibition of neurite extension instead of strengthening it. To explore the underlying mechanism, we further examine whether overexpressing WNK1-(123–510) could affect the interaction between endogenous WNK1 and LINGO-1. PC12 cells were transfected with pEYFP-N1-WNK1-(123–510) or pEYFP-N1 vector. After culturing with GST-Nogo66 (100 nm) for 2 days, cells were lysed and immunoprecipitated, whereas an equal portion of cell lysate was set aside for quantity control. As shown in Fig. 6, E and F, overexpression of WNK1-(123–510) was found to significantly attenuate endogenous WNK1 interaction with LINGO-1.
FIGURE 6.
Overexpressing WNK1-(123–510) attenuates Nogo-induced inhibition of neurite extension and inhibits the interaction between endogenous WNK1 and LINGO-1. A, PC12 cells transfected with pEYFP-N1 vector (Control) or pEYFP-N1-WNK1-(123–510) were treated with 100 nm GST or GST-Nogo66 in the presence of NGF (100 ng/ml) for 72 h then visualized using fluorescence microscopy. Scale bar, 30 μm. B, neurite extension of the transfected cortical neurons in the presence of GST or GST-Nogo66 (100 nm), visualized by confocal microscope. Scale bar, 50 μm. C and D, neuronal differentiation rate of transfected PC12 cells and the longest neurite length of transfected cortical neurons were analyzed. Values are the mean ± S.D. **, p < 0.001 by Student's t test compared as indicated. E, PC12 cells were transfected with pEYFP-N1 (Control) or pEYFP-N1-WNK1-(123–510) and treated with GST-Nogo66 (100 nm) for 48 h. The cell lysates were immunoprecipitated (IP) with anti-LINGO-1 and blotted (IB) with anti-WNK1, and an equal portion of the cell lysate was set as an endogenous expression control. F, quantification of E was presented as -fold of control. Data shown are the mean ± S.D. of three independent experiments. *, p < 0.01 by Student's t test.
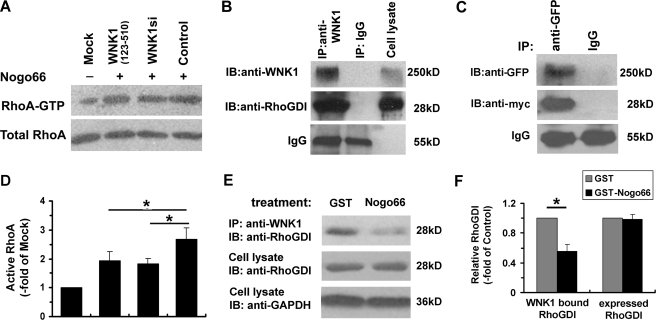
WNK1 Is Involved in Regulation of Nogo-induced RhoA Activation
LINGO-1 has been found to mediate Nogo66 inhibitory signaling by activation of RhoA (11, 39). To investigate the involvement of WNK1 in the Nogo-LINGO-1-RhoA-signaling pathway, PC12 cells were transfected with pSUPER/shWNK1 (WNK1si), pSUPER vector (Control), or pEYFP-N1-WNK1-(123–510); no transfection served as the mock control. After treatment with GST-Nogo66 (100 nm) or GST(100 nm) for 1 h, the activation of RhoA by Nogo66 in these PC12 cells were tested by using GST-fused Rho binding domain of Rhotekin to precipitate GTP-bound RhoA from the cell lysates (36). As shown in Fig. 7, A and D, Nogo66 treatment greatly elevated the level of active RhoA in PC12 cells, which was consistent with the previous report (39), whereas knockdown of WNK1 by RNAi could dramatically decrease the elevation of RhoA activity by Nogo treatment. Furthermore, overexpression of WNK1-(123–510) also attenuated the RhoA activation, which was consistent with our previous results in neurite outgrowth analysis. These results indicate that WNK1 is involved in regulation of Nogo-induced RhoA activation.
FIGURE 7.
WNK1 is involved in regulation of Nogo-induced RhoA activation. A, PC12 cells transfected with pEYFP-WNK1-(123–510), pSUPER/WNK1shRNA (WNK1si), or pSUPER vector (Control) were treated with GST Nogo66, and non-transfected PC12 without Nogo66 treatment were set as Mock control. The cell lysates were affinity-precipitated with GST-rhotekin binding domain to detect GTP-bound Rho as mentioned under “Experimental Procedures,” and total Rho in cell lysate was set aside as inputs. Quantification shows in D, where GTP Rho levels were normalized to total Rho protein levels in the cell lysate and presented as -fold Mock control. B, co-immunoprecipitation (IP) of WNK1 with Rho-GDI was performed using lysates prepared from P1 rat brain tissue using rabbit IgG for control, and cell lysate was set in parallel at the same time. IB, immunoblot. C, immunoprecipitation shows full-length WNK1 and Rho-GDI interacts in COS-7 cells co-transfected with Myc-tagged Rho-GDI and GFP-tagged full-length human WNK1. The rabbit IgG was used as the immunoprecipitation control, and the heavy chain of IgG is visualized for input control. E. Co-immunoprecipitation of WNK1 with Rho-GDI in primary cultured cortical neurons treated with GST-Nogo66 or GST. F, quantification of WNK1-bound Rho-GDI or endogenous expressed Rho-GDI. The blots of Rho-GDI were first normalized with GAPDH levels in cell lysates and then presented as -fold of GST-treated controls. Data shown are the mean ± S.D. from three independent experiments. *, p < 0.01 by Student's t test.
Rho guanine nucleotide dissociation inhibitors (Rho-GDIs) were considered as central regulatory molecules in Rho GTPase activation, and it had been reported that stimulation with Nogo would lead to release Rho from Rho-GDI (40). To test whether WNK1 could associate with Rho-GDI1, P1 rat brain tissue extracts were prepared and immunoprecipitated with anti-WNK1 or control IgG. As shown in Fig. 7B, Rho-GDI was co-immunoprecipitated by anti-WNK1, but not by the control IgG, indicating the specific association between endogenous Rho-GDI and WNK1. Meanwhile, this association was also confirmed in the in vitro mammalian expressed system. As shown in Fig. 7C, COS-7 cells co-transfected with Myc-tagged Rho-GDI and GFP-tagged full-length human WNK1 were subjected to immunoprecipitation with anti-GFP or control IgG. As shown in Fig. 3F, Myc-tagged Rho-GDI was immunoprecipitated by anti-GFP, but not by the control IgG, further suggesting the specific interaction between Rho-GDI and WNK1.
To further investigate whether the association between Rho-GDI and WNK1 was affected by Nogo66 treatment, primary cultured cortical neurons were treated with GST-Nogo66 (100 nm) or GST (100 nm) and then subjected to immunoprecipitation. As shown in Fig. 7, E and F, the interaction between endogenous Rho-GDI and WNK1 was significantly reduced after Nogo66 treatment, whereas the endogenous expression of Rho-GDI was not affected. Taken together, these results indicate that WNK1 may mediate regulation of Nogo-induced RhoA activation via interaction with Rho-GDI.
DISCUSSION
Although many axon growth inhibitors have been identified in myelin, myelin-associated glycoprotein, oligodendrocyte myelin glycoprotein, and Nogo account for most of the central nervous system myelin inhibitory activities (7, 41). They share a convergence in mediating inhibitory signaling via the triple receptor complex NgR/LINGO-1/p75 (11) or NgR/LINGO-1/Troy (12, 13), in which NgR1 mainly functions as a binding partner for ligands, whereas the other transmembrane receptors are responsible for signaling transduction. The p75 neurotrophin receptor, a member of the tumor necrosis factor receptor family (42), has been well documented to serve as a component of several discrete cell surface signaling platforms that perform diverse functions not limited to NgR1 pathway (43, 44). Also, as an orphan tumor necrosis factor receptor, Troy exhibits a similar function and is considered as a substitute of p75 (12, 13). In contrast, LINGO-1, selectively expressed in the central nervous system, is a member of an emerging group of central nervous system-enriched, type I surface proteins with an ectodomain-containing leucine-rich repeats and motifs found in cell adhesion molecules, which are considered either signaling intracellularly through their cytoplasmic domains or by engagement of other transmembrane proteins in a signaling complex (45). LINGO-1 acts as an adaptor that connects NgR with p75NTR or TROY by forming the tripartite receptor complex (11, 45, 46). Interestingly, LINGO-1 seems not always to coexist with NgR and p75 (or Troy) (47), indicating additional signaling of LINGO-1 independent with NgR and p75 (or Troy). The cytoplasmic domain of LINGO-1 contains a canonical EGFR-like tyrosine phosphorylation site (11). A recent study proposed the intracellular domain of LINGO-1 interacts with the post-mitotic neuronal-specific zinc finger protein Myt1l, suggesting that LINGO-1 may regulate Myt1l transcription factor activity by affecting its subcellular localization (47). In the present study we employed a yeast two-hybrid screen by using the intracellular domain of LINGO-1 as bait and identified the protein kinase WNK1 as a LINGO-1 binding candidate. In addition, the interaction between WNK1 and LINGO-1 was validated in living cells by FRET and in brain tissue lysates or an in vitro mammalian expressing system by co-immunoprecipitation, demonstrating WNK1 as a novel LINGO-1 binding partner. Furthermore, Nogo66 treatment was found to intensify this combination, indicating WNK1 may play a role in LINGO-1-mediated Nogo66 signaling.
The functions of WNK1 on ion homeostasis and transportation, which contribute greatly to the regulation of blood pressure, have been well studied (Refs. 48–55; for review, see Refs. 22, 56, and 57). It has been reported that WNK1 is expressed widely including in brain (27, 28, 38, 58). Recently, a more detailed profile of WNK1 expression in both the central nervous system and peripheral neural system was examined (29). Moreover, a nervous system-specific exon of the WNK1 gene, also known as a single-exon open reading frame hereditary sensory neuropathy type II (HSN2), mutations of which were originally identified in affected families, were found to cause hereditary sensory and autonomic neuropathy type II (HSANII) (29). In the present study we showed WNK1 was highly expressed in the cortex, particularly within the same neuron with LINGO-1. Suppression of WNK1 was found to promote neurite extension and abrogate the inhibitory response to Nogo66 in PC12 cells and cortical neurons. More importantly, overexpression of WNK1-(123–510), which binds to LINGO-1, also came out to attenuate Nogo-induced inhibition of neurite extension in cortical neurons. Taken together, these results strongly suggest that WNK1 is involved in regulating Nogo signaling during neuronal differentiation.
As shown in Figs. 4 and 5, attenuation of WNK1 by RNAi would also, to some extent promote PC12 differentiation and cortical neuron extension in the absence of Nogo. The question remained of whether PC12 cells and cortical neurons constitutively secrete Nogo or related inhibitory molecules to the medium or WNK1 itself is a negative regulator of neurite outgrowth independent of Nogo action. To examine this, we performed immunocytochemistry (IHC) staining on both PC12 cells and cortical neurons with anti-NogoA and anti-NgR antibodies. As shown in supplemental Fig. 1, both NogoA and NgR are expressed on cortical neurons and PC12 cells. It is possible that Nogo or other related inhibitory molecules would secret into medium, which might be the source of the “background activation” of LINGO-1 without adding exogenous Nogo. On the other hand, WNK1 itself may not serve as a negative regulator of neurite outgrowth independent of Nogo action, as overexpression of full-length WNK1 in PC12 cells did not affect NGF-induced PC12 differentiation either in the presence of GST or GST-Nogo66 (supplemental Fig. 2). From another point of view, it seems that endogenous WNK1 is adequate or enough to regulate LINGO-1 signaling activated by Nogo in PC12 cells.
WNK1 was first identified as the prototype of the WNK family of serine-threonine kinases, characterized with the unique substitution of cysteine for lysine at catalytic domain (27). Besides a serine-threonine kinase domain, there are three potential coiled-coil and four proline-rich regions, a glutamine-rich region, two serine-rich regions, and multiple SH3 domain binding motifs as well as a potential bipartite nuclear localization signal, which may contribute to their discrete functional states that impart different effects on downstream effectors (38). It has been reported that WNK family members function via both kinase-dependent and kinase-independent mechanisms. For example, WNK1 acts upstream in the ERK5 3-mitogen-activated protein kinase (MAPK) pathway and can phosphorylate the MAPK kinase kinases MEKK2 and MEKK3 (49, 59). On the other hand, independent of its kinase domain, WNK1 is a negative regulator of insulin-stimulated cell proliferation (50). In the present study overexpression of WNK1-(123–510), which keeps a constitutive serine-threonine kinase activity (60), did not strengthen Nogo-induced inhibition of neurite extension and RhoA activation but came to attenuate them. Our results further showed that overexpression WNK1-(123–510) inhibited the interaction of endogenous WNK1 with LINGO-1, indicating that the serine-threonine kinase domain may serve as a binding platform, and the kinase activity may be unnecessary to mediate LINGO-1 signaling.
Nogo has been reported to activate the small GTPase RhoA in a LINGO-1-dependent manner to exert its inhibitory activity on neurite extension (11, 39). In this study knockdown of WNK1 by RNAi was found to attenuate Nogo-induced RhoA activation, suggesting a regulating role of WNK1 on Nogo-induced RhoA activation. RhoA acts as binary switches, which cycle between the GDP-bound inactive state and the GTP-bound active conformation (61). The GDIs are pivotal regulators for Rho GTPase function, as they control the access of Rho GTPases to regulatory guanine nucleotide exchange factors and GTPase-activating proteins to effector targets and to membranes where such effectors reside (62–64). Rho-GDI1 is ubiquitously expressed and has the binding specificity for Rho family GTPases (RhoA, Rac, Cdc42), which play an essential role in the control of cellular morphology, for example, neurite extension (65, 66). Previous work has shown that the association of p75 with Rho-GDI was enhanced by Nogo, and p75 has an ability to release RhoA from Rho-GDI (40), which demonstrated that activation of RhoA by Nogo through p75 may be attributable, at least partly, to Rho-GDI displacement. A recent study further reported that Kalirin9, a dual RhoGEF, binds p75 directly and regulates p75-Nogo receptor-dependent RhoA activation in response to myelin-associated glycoprotein (67). In line with this finding, we found WNK1 associated with Rho-GDI and Nogo66 treatment would promote Rho-GDI dissociation from WNK1. It is possible that the interaction of LINGO-1 with WNK1 may bring Rho-GDI-RhoA close to p75-bound guanine nucleotide exchange factors, such as Kalirin9, thereby facilitating RhoA activation, although other guanine nucleotide exchange regulatory factors may also be cooperative in this process. The precise mechanism underlying the regulation of RhoA activation by LINGO-1-WNK1 interaction remains to be investigated.
In summary, WNK1 was identified to interact with LINGO-1 and regulate Nogo-induced inhibition of neurite extension. These results may shed new light on both LINGO-1 and WNK1 signaling in neuronal differentiation.
Supplementary Material
Acknowledgments
We are grateful to Prof. Xiao-bin Yuan for generously providing pEYFP-N1, pECFP-N1, and pSUPER vectors, Prof. Lan Bao for generously providing pCDNA-Myc-Rho-GDI1, Prof. Melanine H. Cobb for generously providing full-length rat WNK1 construct pCMV5-Myc-rWNK1, and Prof. Peter Jordan for generously providing EGFP-C2-hWNK1.
This work was supported by National Key Basic Research Program 2006CB500702, Ministry of Science and Technology of China Grant 2007CB947100, National Natural Science Foundation Grants 30500151, 30530240, and 30770657, National 863 Program 2006AA02A114, Program for Changjiang Scholars and Innovative Research Team in University IRT0528, and Shanghai Metropolitan Fund for Research and Development Grant 07DJ14005.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- NgR
- Nogo-66 receptor
- LINGO-1
- leucine-rich repeat and Ig domain-containing, Nogo receptor-interacting protein
- WNK1
- with no lysine [K]
- Rho-GDI
- GDP dissociation inhibitors of Rho-GTPases
- FRET
- fluorescence resonance energy transfer
- FR
- FRET ratio
- IHC
- immunocytochemistry
- YFP
- yellow fluorescent protein
- GFP
- green fluorescent protein
- NGF
- nerve growth factor
- GST
- glutathione S-transferase
- shRNA
- short hairpin RNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- siRNA
- small interfering RNA.
REFERENCES
- 1.Qiu J., Cai D., Filbin M. T. ( 2000) Glia 29, 166– 174 [PubMed] [Google Scholar]
- 2.Fournier A. E., GrandPre T., Strittmatter S. M. ( 2001) Nature 409, 341– 346 [DOI] [PubMed] [Google Scholar]
- 3.McKerracher L., David S., Jackson D. L., Kottis V., Dunn R. J., Braun P. E. ( 1994) Neuron 13, 805– 811 [DOI] [PubMed] [Google Scholar]
- 4.Mukhopadhyay G., Doherty P., Walsh F. S., Crocker P. R., Filbin M. T. ( 1994) Neuron 13, 757– 767 [DOI] [PubMed] [Google Scholar]
- 5.Liu B. P., Fournier A., GrandPré T., Strittmatter S. M. ( 2002) Science 297, 1190– 1193 [DOI] [PubMed] [Google Scholar]
- 6.Niederöst B. P., Zimmermann D. R., Schwab M. E., Bandtlow C. E. ( 1999) J. Neurosci. 19, 8979– 8989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K. C., Koprivica V., Kim J. A., Sivasankaran R., Guo Y., Neve R. L., He Z. ( 2002) Nature 417, 941– 944 [DOI] [PubMed] [Google Scholar]
- 8.Chen M. S., Huber A. B., van der Haar M. E., Frank M., Schnell L., Spillmann A. A., Christ F., Schwab M. E. ( 2000) Nature 403, 434– 439 [DOI] [PubMed] [Google Scholar]
- 9.GrandPré T., Nakamura F., Vartanian T., Strittmatter S. M. ( 2000) Nature 403, 439– 444 [DOI] [PubMed] [Google Scholar]
- 10.Prinjha R., Moore S. E., Vinson M., Blake S., Morrow R., Christie G., Michalovich D., Simmons D. L., Walsh F. S. ( 2000) Nature 403, 383– 384 [DOI] [PubMed] [Google Scholar]
- 11.Mi S., Lee X., Shao Z., Thill G., Ji B., Relton J., Levesque M., Allaire N., Perrin S., Sands B., Crowell T., Cate R. L., McCoy J. M., Pepinsky R. B. ( 2004) Nat. Neurosci. 7, 221– 228 [DOI] [PubMed] [Google Scholar]
- 12.Park J. B., Yiu G., Kaneko S., Wang J., Chang J., He X. L., Garcia K. C., He Z. ( 2005) Neuron 45, 345– 351 [DOI] [PubMed] [Google Scholar]
- 13.Shao Z., Browning J. L., Lee X., Scott M. L., Shulga-Morskaya S., Allaire N., Thill G., Levesque M., Sah D., McCoy J. M., Murray B., Jung V., Pepinsky R. B., Mi S. ( 2005) Neuron 45, 353– 359 [DOI] [PubMed] [Google Scholar]
- 14.Carim-Todd L., Escarceller M., Estivill X., Sumoy L. ( 2003) Eur. J. Neurosci. 18, 3167– 3182 [DOI] [PubMed] [Google Scholar]
- 15.Ji B., Li M., Wu W. T., Yick L. W., Lee X., Shao Z., Wang J., So K. F., McCoy J. M., Pepinsky R. B., Mi S., Relton J. K. ( 2006) Mol. Cell. Neurosci. 33, 311– 320 [DOI] [PubMed] [Google Scholar]
- 16.Mi S., Miller R. H., Lee X., Scott M. L., Shulag-Morskaya S., Shao Z., Chang J., Thill G., Levesque M., Zhang M., Hession C., Sah D., Trapp B., He Z., Jung V., McCoy J. M., Pepinsky R. B. ( 2005) Nat. Neurosci. 8, 745– 751 [DOI] [PubMed] [Google Scholar]
- 17.Mi S., Hu B., Hahm K., Luo Y., Kam Hui E. S., Yuan Q., Wong W. M., Wang L., Su H., Chu T. H., Guo J., Zhang W., So K. F., Pepinsky B., Shao Z., Graff C., Garber E., Jung V., Wu E. X., Wu W. ( 2007) Nat. Med. 13, 1228– 1233 [DOI] [PubMed] [Google Scholar]
- 18.Inoue H., Lin L., Lee X., Shao Z., Mendes S., Snodgrass-Belt P., Sweigard H., Engber T., Pepinsky B., Yang L., Beal M. F., Mi S., Isacson O. ( 2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14430– 14435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanks S. K., Quinn A. M., Hunter T. ( 1988) Science 241, 42– 52 [DOI] [PubMed] [Google Scholar]
- 20.Wilson F. H., Disse-Nicodème S., Choate K. A., Ishikawa K., Nelson-Williams C., Desitter I., Gunel M., Milford D. V., Lipkin G. W., Achard J. M., Feely M. P., Dussol B., Berland Y., Unwin R. J., Mayan H., Simon D. B., Farfel Z., Jeunemaitre X., Lifton R. P. ( 2001) Science 293, 1107– 1112 [DOI] [PubMed] [Google Scholar]
- 21.Golbang A. P., Murthy M., Hamad A., Liu C. H., Cope G., Van't Hoff W., Cuthbert A., O'Shaughnessy K. M. ( 2005) Hypertension 46, 295– 300 [DOI] [PubMed] [Google Scholar]
- 22.Kahle K. T., Ring A. M., Lifton R. P. ( 2008) Annu. Rev. Physiol. 70, 329– 355 [DOI] [PubMed] [Google Scholar]
- 23.Stephens P., Edkins S., Davies H., Greenman C., Cox C., Hunter C., Bignell G., Teague J., Smith R., Stevens C., O'Meara S., Parker A., Tarpey P., Avis T., Barthorpe A., Brackenbury L., Buck G., Butler A., Clements J., Cole J., Dicks E., Edwards K., Forbes S., Gorton M., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jones D., Kosmidou V., Laman R., Lugg R., Menzies A., Perry J., Petty R., Raine K., Shepherd R., Small A., Solomon H., Stephens Y., Tofts C., Varian J., Webb A., West S., Widaa S., Yates A., Brasseur F., Cooper C. S., Flanagan A. M., Green A., Knowles M., Leung S. Y., Looijenga L. H., Malkowicz B., Pierotti M. A., Teh B., Yuen S. T., Nicholson A. G., Lakhani S., Easton D. F., Weber B. L., Stratton M. R., Futreal P. A., Wooster R. ( 2005) Nat. Genet. 37, 590– 592 [DOI] [PubMed] [Google Scholar]
- 24.Veríssimo F., Silva E., Morris J. D., Pepperkok R., Jordan P. ( 2006) Oncogene 25, 4172– 4182 [DOI] [PubMed] [Google Scholar]
- 25.Sun X., Gao L., Yu R. K., Zeng G. ( 2006) J. Neurochem. 99, 1114– 1121 [DOI] [PubMed] [Google Scholar]
- 26.Hong C., Moorefield K. S., Jun P., Aldape K. D., Kharbanda S., Phillips H. S., Costello J. F. ( 2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10974– 10979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu B., English J. M., Wilsbacher J. L., Stippec S., Goldsmith E. J., Cobb M. H. ( 2000) J. Biol. Chem. 275, 16795– 16801 [DOI] [PubMed] [Google Scholar]
- 28.Choate K. A., Kahle K. T., Wilson F. H., Nelson-Williams C., Lifton R. P. ( 2003) Proc. Natl. Acad. Sci. U.S.A. 100, 663– 668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shekarabi M., Girard N., Rivière J. B., Dion P., Houle M., Toulouse A., Lafrenière R. G., Vercauteren F., Hince P., Laganiere J., Rochefort D., Faivre L., Samuels M., Rouleau G. A. ( 2008) J. Clin. Investig. 118, 2496– 2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Yan Z., Farooq A., Liu X., Lu C., Zhou M. M., He C. ( 2004) J. Mol. Biol. 343, 1147– 1155 [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Zhu W., Wang Y. G., Liu X. J., Jiao L., Liu X., Zhang Z. H., Lu C. L., He C. ( 2006) J. Cell Sci. 119, 1666– 1676 [DOI] [PubMed] [Google Scholar]
- 32.Fu G., Wang C., Wang Y. G., Chen Y. Z., He C., Xu Z. Z. ( 2006a) Biochem. Biophys. Res. Commun. 351, 847– 852 [DOI] [PubMed] [Google Scholar]
- 33.Zhang F., Fu G., Wang C., Cao L., Yang H. Y., Wang Y. G., Chen Y. Z., He C. ( 2008) Mol. Imaging Biol., in press [DOI] [PubMed] [Google Scholar]
- 34.Erickson M. G., Alseikhan B. A., Peterson B. Z., Yue D. T. ( 2001) Neuron 31, 973– 985 [DOI] [PubMed] [Google Scholar]
- 35.Erickson M. G., Liang H., Mori M. X., Yue D. T. ( 2003) Neuron 39, 97– 107 [DOI] [PubMed] [Google Scholar]
- 36.Ren X. D., Kiosses W. B., Schwartz M. A. ( 1999) EMBO J. 18, 578– 585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X., He C., Zhang Z., Chen Y. ( 2006) J. Cell Sci. 119, 452– 458 [DOI] [PubMed] [Google Scholar]
- 38.Veríssimo F., Jordan P. ( 2001) Oncogene 20, 5562– 5569 [DOI] [PubMed] [Google Scholar]
- 39.Fournier A. E., Takizawa B. T., Strittmatter S. M. ( 2003) J. Neurosci. 23, 1416– 1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamashita T., Tohyama M. ( 2003) Nat. Neurosci. 6, 461– 467 [DOI] [PubMed] [Google Scholar]
- 41.Barton W. A., Liu B. P., Tzvetkova D., Jeffrey P. D., Fournier A. E., Sah D., Cate R., Strittmatter S. M., Nikolov D. B. ( 2003) EMBO J. 22, 3291– 3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K. C., Kim J. A., Sivasankaran R., Segal R., He Z. ( 2002) Nature 420, 74– 78 [DOI] [PubMed] [Google Scholar]
- 43.Dechant G., Barde Y. A. ( 2002) Nat. Neurosci. 5, 1131– 1136 [DOI] [PubMed] [Google Scholar]
- 44.Barker P. A. ( 2004) Neuron 42, 529– 533 [DOI] [PubMed] [Google Scholar]
- 45.Chen Y., Aulia S., Li L., Tang B. L. ( 2006) Brain Res. Rev. 51, 265– 274 [DOI] [PubMed] [Google Scholar]
- 46.Mosyak L., Wood A., Dwyer B., Buddha M., Johnson M., Aulabaugh A., Zhong X., Presman E., Benard S., Kelleher K., Wilhelm J., Stahl M. L., Kriz R., Gao Y., Cao Z., Ling H. P., Pangalos M. N., Walsh F. S., Somers W. S. ( 2006) J. Biol. Chem. 281, 36378– 36390 [DOI] [PubMed] [Google Scholar]
- 47.Llorens F., Gil V., Iraola S., Carim-Todd L., Martí E., Estivill X., Soriano E., Del Rio J. A., Sumoy L. ( 2008) Dev. Neurobiol. 68, 521– 541 [DOI] [PubMed] [Google Scholar]
- 48.Cope G., Golbang A., O'Shaughnessy K. M. ( 2005) Pharmacol. Ther. 106, 221– 231 [DOI] [PubMed] [Google Scholar]
- 49.Xu B. E., Lee B. H., Min X., Lenertz L., Heise C. J., Stippec S., Goldsmith E. J., Cobb M. H. ( 2005) Cell Res. 15, 6– 10 [DOI] [PubMed] [Google Scholar]
- 50.Xu B. E., Stippec S., Chu P. Y., Lazrak A., Li X. J., Lee B. H., English J. M., Ortega B., Huang C. L., Cobb M. H. ( 2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10315– 10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anselmo A. N., Earnest S., Chen W., Juang Y. C., Kim S. C., Zhao Y., Cobb M. H. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10883– 10888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wade J. B., Fang L., Liu J., Li D., Yang C. L., Subramanya A. R., Maouyo D., Mason A., Ellison D. H., Welling P. A. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8558– 8563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu Y., Subramanya A., Rozansky D., Cohen D. M. ( 2006b) Am. J. Physiol. Renal Physiol. 290, F1305– 1314 [DOI] [PubMed] [Google Scholar]
- 54.Yang C. L., Zhu X., Ellison D. H. ( 2007) J. Clin. Investig. 117, 3403– 3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richardson C., Rafiqi F. H., Karlsson H. K., Moleleki N., Vandewalle A., Campbell D. G., Morrice N. A., Alessi D. R. ( 2008) J. Cell Sci. 121, 675– 684 [DOI] [PubMed] [Google Scholar]
- 56.Kahle K. T., Rinehart J., Ring A., Gimenez I., Gamba G., Hebert S. C., Lifton R. P. ( 2006) Physiology (Bethesda) 21, 326– 335 [DOI] [PubMed] [Google Scholar]
- 57.Xie J., Craig L., Cobb M. H., Huang C. L. ( 2006) Pediatr. Nephrol. 21, 1231– 1236 [DOI] [PubMed] [Google Scholar]
- 58.Delaloy C., Hadchouel J., Imbert-Teboul M., Clemessy M., Houot A. M., Jeunemaitre X. ( 2006) Am. J. Pathol. 169, 105– 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu B. E., Stippec S., Lenertz L., Lee B. H., Zhang W., Lee Y. K., Cobb M. H. ( 2004) J. Biol. Chem. 279, 7826– 7831 [DOI] [PubMed] [Google Scholar]
- 60.Min X., Lee B. H., Cobb M. H., Goldsmith E. J. ( 2004) Structure 12, 1303– 1311 [DOI] [PubMed] [Google Scholar]
- 61.Schmidt A., Hall A. ( 2002) Genes Dev. 16, 1587– 1609 [DOI] [PubMed] [Google Scholar]
- 62.Bokoch G. M., Bohl B. P., Chuang T. H. ( 1994) J. Biol. Chem. 269, 31674– 31679 [PubMed] [Google Scholar]
- 63.Olofsson B. ( 1999) Cell. Signal. 11, 545– 554 [DOI] [PubMed] [Google Scholar]
- 64.Zalcman G., Dorseuil O., Garcia-Ranea J. A., Gacon G., Camonis J. ( 1999) Prog. Mol. Subcell. Biol. 22, 85– 113 [DOI] [PubMed] [Google Scholar]
- 65.Lehmann M., Fournier A., Selles-Navarro I., Dergham P., Sebok A., Leclerc N., Tigyi G., McKerracher L. ( 1999) J. Neurosci. 19, 7537– 7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niederöst B., Oertle T., Fritsche J., McKinney R. A., Bandtlow C. E. ( 2002) J. Neurosci. 22, 10368– 10376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harrington A. W., Li Q. M., Tep C., Park J. B., He Z., Yoon S. O. ( 2008) J. Biol. Chem. 283, 24690– 24697 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.