Abstract
Furin is a ubiquitously expressed proprotein convertase (PC) that plays a vital role in numerous disease processes including cancer metastasis, bacterial toxin activation (e.g. anthrax and Pseudomonas), and viral propagation (e.g. avian influenza and human immunodeficiency virus). To identify small molecule inhibitors of furin and related processing enzymes, we performed high-throughput screens of chemical diversity libraries utilizing both enzyme-based and cell-based assays. The screens identified partially overlapping sets of compounds that were further characterized for affinity, mechanism, and efficacy in additional cellular processing assays. Dicoumarols were identified as a class of compounds that inhibited furin non-competitively and reversibly with Ki values in the micromolar range. These compounds inhibited furin/furin-like activity both at the cell surface (protecting against anthrax toxin) and in the secretory pathway (blocking processing of the metastasis factor membrane-type 1 matrix metalloproteinase/MT1-MMP) at concentrations close to Ki values. Compounds tested exhibited distinct patterns of inhibition of other furin-family PCs (rat PACE4, human PC5/6 and human PC7), showing that dicoumarol derivatives might be developed as either generic or selective inhibitors of the PCs. The extensive clinical use, high bioavailability and relatively low toxicity of dicoumarols suggests that the dicoumarol structure will be a good starting point for development of drug-like inhibitors of furin and other PCs that can act both intracellularly and at the cell surface.
Furin, is a subtilisin-related serine protease and member of the proprotein convertase (PCs)4 family that functions within the secretory and endocytic pathways and at the cell surface, cleaving proproteins at clusters of basic residues, typically of the form RX(K/R)R↓ (for reviews see Refs. 1–3). The specificity of furin and its yeast homologue Kex2 correlate well with the three-dimensional structures of their catalytic domains (4, 5). Ubiquitously expressed, furin has numerous known or suspected physiological substrates that include growth factors, receptors, coagulation proteins, plasma proteins (e.g. pro-von Willebrand factor), extracellular matrix components, and protease precursors (e.g. matrix metalloproteases) (2). Although the homozygous furin knock-out mouse exhibits embryonic lethality (6), analysis of liver-specific ablation suggests functional overlap with other PCs, such as PACE4, PC5/6, and PC7, that are also widely expressed and act in the constitutive secretory pathway (7). Furin activity contributes to numerous chronic pathological conditions, including Alzheimer disease (8), other non-Alzheimer cerebral amyloidoses (9), osteoarthritis (10), atherosclerosis (11), and tumor progression and malignancy (12). Moreover, activation by host cells of bacterial toxins such as anthrax toxin, Pseudomonas exotoxin A, diphtheria toxin (13), Shiga toxin (14), and Bordetella dermonecrotic toxin (15), requires cleavage by furin or other PCs. Furin or furin-like cleavage of viral envelope glycoproteins is necessary for propagation of many lipid-enveloped viral pathogens including H5N1 avian influenza (16), human immunodeficiency virus-1 (17), ebola (18), measles (19), cytomegalovirus (20), and flaviviruses (21). Even non-enveloped viruses, such as human papillomavirus, can require furin-type processing for entry into the cyotsol after endocytosis (22).
The multiple roles for furin in human pathophysiology have made it a target of interest for development of therapeutic agents. Numerous protein- and peptide-based furin inhibitors have been devised (23). For the most part, these are not drug-like and their use as pharmaceutical agents is hampered by large size, instability, toxicity, and/or low cell permeability. Recently, 2,5-dideoxystreptamine derivatives have shown promise (24), although these molecules have yet to be examined for inhibition of intracellular processing. Important pathophysiological roles exist for furin at the cell surface, such as in the processing of anthrax protective antigen. However, maturation of other bacterial toxins, viral envelope glycoproteins, and metalloprotease precursors such as membrane-type 1 matrix metalloproteinase (MT1-MMP), a matrix metalloprotease whose activity contributes directly to degradation of extracellular matrix components and is important for angiogenesis, tumor invasion, and metastasis (25), require processing by furin in the trans Golgi network and endosomal compartments (2, 26).
Here we report identification of drug-like small molecule inhibitors through simultaneous high-throughput screening (HTS) of chemical diversity libraries with both enzyme-based and cell-based assays for furin and furin-like activities. A preliminary report of the cell-based assay has been published elsewhere (27). Combining the results of the enzymatic screen with the cellular screen allowed identification of small molecule lead compounds with the desired properties of high affinity, high cell permeability, and low toxicity. Dicoumarols, which have an extensive pharmacological history (28), were identified in this study as a family of compounds that inhibited furin reversibly and non-competitively, also inhibited rat PACE4 (rPACE4), human PC5/6 (hPC5/6), and hPC7 and blocked both extracellular maturation of anthrax protective antigen (PA) and intracellular processing of MT1-MMP and other substrates.
EXPERIMENTAL PROCEDURES
Enzymes and Reagents
Secreted, soluble human furin (ssfurin) (29, 30) and hPC5/6, rPACE4, and human hPC7 (31) were expressed and purified as described. Decanoyl-Arg-Val-Lys-Arg-chloromethylketone (decRVKR-CMK), N-tert-butoxycarbonyl-Arg-Val-Arg-Arg-methylcoumarin amide (boc-RVRR-MCA), Ac-Arg-Val-Arg-Arg-pNA (Ac-RVRR-pNA), and boc-Ile-Glu-Gly-Arg-MCA (boc-IEGR-MCA) were purchased from Bachem (King of Prussia, PA). Human α-thrombin and compounds DC1, DC2, DC4, and DC6 were from Sigma. Compounds DC3, DC5, DC7, DC8, DC9, B5, B8, and B9 were from ChemDiv (San Diego, CA). Compounds B1, B2, B6, and B7 were from Chembridge (San Diego, CA). Compounds B3 (CCG8294) (27) and B10 were synthesized, purified, and characterized as described (32). All compounds were dissolved in dimethyl sulfoxide. B3 was dissolved from powder just prior to use. Plasmid pEF-GRAPfurin was as described (27). All HTS materials, including compound libraries (Chembridge DIVERSet, 10,000 compounds; ChemDiv, 20,000 compounds), were supplied by the Center for Chemical Genomics (Life Sciences Institute, University of Michigan). The ToxCount cell viability assay was from Active Motif (Carlsbad, CA). Recombinant PA83 and lethal factor were as described (33).
Enzymatic and Cellular HTS/Dose-response Assay
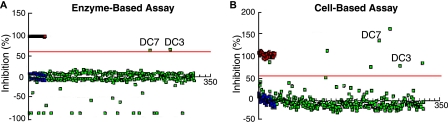
Enzyme solution (15 μl), containing ssfurin (11 nm), 20 mm Na/Mes, pH 7, 1 mm CaCl2, and 0.01% Triton X-100 buffer, or buffer alone (15 μl) was delivered using the Multi-drop 384 (Thermo Labsystems, Waltham, MA) into 384-well plates. Then ∼300 nl of each compound (5–15 μm) was delivered into individual wells (except control wells) using the Biomek FX pin tool (Biomek FX, Beckman, Fullerton, CA). After a 15-min incubation, 5 μl of boc-RVRR-MCA (2 μm) were added to all wells except the controls. The substrate in buffer without enzyme or the standard reaction containing 20 μm dec-RVKR-CMK served as positive controls. After incubation (45 min), residual enzyme activities were monitored by the fluorescence intensity of 7-amino-4-methyl coumarin (AMC) (λex 380 nm, λem 470 nm) using a PHERAstar high-throughput microplate reader (BMGLabtech, Chicago, IL). All plates were bar-coded for identification and linked to compounds from stock plates. Assay quality was determined by calculating mean, standard deviation, coefficient of variation, and Z′ values for each plate. Plates for which Z′ > 0.5 were accepted (34). Plates with Z′ < 0.5 were repeated. Compounds with >60% ssfurin inhibition were chosen as positive hits. Compounds exhibiting >50% inhibition of other targets in the data base of the Center for Chemical Genomics were eliminated. Dose-response analysis was conducted to validate hits. The enzyme-based assay monitored AMC fluorescence. Approximately 1500 compounds fluorescent in the AMC range were ignored as they gave apparent enzyme activities greater than 100%. Examples of enzyme-based and cell-based assays for a sample compound plate are shown in Fig. 1. Note that DC3 and DC7 appear as clear hits for the enzymatic assay, enabling them to be easily identified in the cell-based assay.
FIGURE 1.
Representative enzyme-based and cell-based HTS assays performed on an identical set of compounds. The data shown represent enzymatic and cell-based furin inhibition assays on one of 100 384-well plates containing library compounds. A, the enzyme-based assay identified DC3 and DC7 as hits that inhibited furin activity >60% (red line). Green squares represent AMC fluorescence released by furin cleavage in the presence of library compounds. Red squares represent positive controls, which correspond to signals from reactions inhibited by 20 μm dec-RVKR-CMK or containing substrate boc-RVRR-MCA without enzyme added. Blue squares represent negative controls, in which furin was incubated with the substrate in the absence of any inhibitor. About 2–5% of compounds on any given plate exhibited fluorescence that interfered with measurement of AMC fluorescence and resulted in apparent activity >100%; these are represented by green squares in the negative range. B, the cell-based assay identified these DC2 and DC7 and five additional potential hits as compounds that inhibited GRAP processing >55% (red line). Positive (red squares) and negative (blue squares) controls in the cell-based assay correspond to processing inhibited by 20 μm dec-RVKR-CMK and to complete processing of GRAP, respectively (27). Apparent inhibition greater than 100% is due to cytotoxicity of compounds or, in the case of DC7, inhibition exceeding that seen with the positive control.
For dose-response assays, compounds were transferred from stock plates (3–20 mm in dimethyl sulfoxide) into plates containing ssfurin and furin buffer and serial dilution was performed to give final concentrations of 33 μm to 33 pm in 30 μl (final dimethyl sulfoxide concentration <2%). After 15 min incubation, boc-RVRR-MCA (2 μm) was added. After 45 min incubation, residual enzyme activities were monitored by AMC fluorescence intensity. Dose-response analysis was performed using PRISM software (GraphPad, San Diego, CA). Cellular HTS and dose-response assays were performed as previously described (27). Briefly, these assays depended on inhibition of cleavage of a fusion protein, GRAPfurin, expressed in Chinese hamster ovary (CHO) cells. GRAPfurin consists of alkaline phosphatase connected to the cytosolic and transmembrane domains of β-secretase (BACE) through a 10-amino acid furin recognition sequence derived from Stromolysin 3 (27). Furin cleavage results in release of alkaline phosphatase, which can be assayed in the conditioned medium.
Cell Culture and Transfections
CHO cells were maintained in Ham's F-12 media with 10% fetal bovine serum, 1 μm minimal essential medium non-essential amino acids (Invitrogen), 1% l-glutamine, 100 μg/ml penicillin, and 100 μg/ml streptomycin (P/S/G) (Invitrogen). Cells were transfected with plasmid using FuGENE 6 (Roche). J774A.1 murine macrophage cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). Cells were grown at 37 °C with 5% CO2.
Inhibitor Affinities, Mechanisms, and Enzyme Selectivities
ssfurin was active-site titrated as described (30). Initial assays were performed in furin assay buffer (20 mm Na/MES, pH 7.0, 1 mm CaCl2, 0.1% (v/v) Triton-X-100) (29) by preincubating enzyme (2.0 nm) with inhibitors at a range of concentrations for 30 min at room temperature, then adding substrate boc-RVRR-MCA (2 to 4 μm) and measuring its rate of hydrolysis (residual enzyme activity), which was linear for 15–30 min. AMC fluorescence was measured using an fmax 96-well fluorimeter (Molecular Devices). IC50 values were derived using a Dixon plot (35). Analysis of these data using Eadie-Hoftsee plots suggested that the dicoumarols and compound B3 were non-competitive, whereas compounds B1, B2, and B5–9 were competitive inhibitors (data not shown). In the case of dicoumarols, initial inhibition assays were done by preincubating enzyme with inhibitors because inhibitors appeared to exhibit slow binding. However, this was an artifact due to inhibitor aggregation, which occurred reversibly after dilution into aqueous solution from concentrated stock solutions in dimethyl sulfoxide. When inhibitors were diluted into assay buffer and preincubated for ≥60 min prior to addition of enzyme and substrate, no slow-binding characteristics were seen. Dicoumarols also exhibited complex formation with the AMC substrate boc-RVRR-MCA when the substrate was present at higher concentrations (at or above the Km), making accurate determination of Ki values difficult. A similar phenomenon was described previously (36). The dicoumarols were also fluorescent, which required correction. To circumvent these problems, we performed more accurate inhibition assays using the chromogenic substrate Ac-RVRR-pNA, which permitted use of higher substrate concentrations at high inhibitor concentrations. The Km of ssfurin for this substrate was determined to be 12.3 μm. p-Nitroaniline release was monitored at 410 nm using a TECAN Safire2, microplate reader in 96-well format. Dicoumarol compounds were diluted into assay buffer and incubated for 2 h. Enzyme and substrate were then added together, and inhibition curves (linear) were measured for 30 min. Substrate concentrations were varied from 4 to 80 μm and inhibitors were assayed at concentrations that ranged, roughly, from 0.1 Ki to 3 Ki. Assays were performed in triplicate and repeated at least 3 times. To determine inhibition mechanisms and Ki values, residual enzyme activity was plotted versus inhibitor concentration, generating a series of curves that differed by substrate concentration. The plots were fitted using untransformed equations for competitive, non-competitive, or uncompetitive inhibition using KaleidaGraph (version 4.0). This method was used to determine Ki values and the inhibitory mechanism for all dicoumarol inhibitors and compounds B2 and B3. Ki values determined in this way were nearly identical to IC50 values obtained previously from Dixon plots.
To examine selectivity, inhibition of rPACE4, hPC5/6, and hPC7 was examined in furin assay buffer with furin substrate (boc-RVRR-MCA). Human α-thrombin was assayed in furin assay buffer using boc-IEGR-MCA as a substrate.
Intracellular Processing Assays
To assay processing of the intracellular substrate CPA95 using transient transfection, cells were plated in 6-well dishes, incubated overnight, then transfected with the CPA95 expression plasmid. CPA95 is a derivative of rat carboxypeptidase A1 in which the trypsin cleavage site at Arg-95 (FQAR95Q) has been altered to create a consensus recognition sequence for furin (RQKR95Q) (37).
At 36 h post-transfection, medium was replaced with Opti-MEM (Invitrogen) containing compound or vehicle. After 2 h, medium was discarded and replaced with fresh Opti-MEM containing compound or vehicle. After overnight incubation, conditioned medium and cells were harvested and analyzed by Western blotting.
Anthrax Toxin Killing Assays
Anthrax toxin killing assays were performed as described (33) except as noted below. J774A.1 murine macrophage cells were washed once with modified Ringer's buffer (RB*) (38) and overlaid with RB* or RB* containing compounds to be tested at various concentrations. Anthrax toxin (AT) (recombinant PA83 (12 nm) and recombinant lethal factor (1.2 nm)) was then added, cells were incubated for 2.5 h and live versus dead cells were determined using the ToxCount cell viability assay (Active Motif, Carlsbad, CA). Cells were photographed using a Zeiss Axiovert fluorescence microscope and numbers of green (live) and red (dead) cells (600 total) were counted to calculate percent protection.
Western Blot Analysis of Intracellular Processing
Western blotting was performed as described (39). Protein concentration was determined using the detergent-compatible protein assay kit from Bio-Rad. Carboxypeptidase A was detected using rabbit polyclonal antibody AB1213 (Chemicon, Temecula, CA). solMT1-MMP expression was detected using rabbit anti-MT1-MMP antibody (Chemicon). HA-MT1-MMP expression was detected using a mouse monoclonal antibody to HA (Covanence, Princeton, NJ). Blots were incubated with appropriate horseradish peroxidase-conjugated secondary antibody followed by detection with chemiluminescent horseradish peroxidase substrate (Pierce) or ECL Plus (GE Healthcare). Anthrax PA processing was quantified using a Typhoon Trio (GE Healthcare).
Intracellular IC50 Determination and MT1-MMP Processing Quantification
For IC50 determination, Western blots were scanned and processed using a GE Healthcare Storm 860 phosphorimager and ImageQuant software (GE Healthcare) or films scanned and quantified by NIH ImageJ software. For MT1-MMP processing quantification, the films were scanned and quantified using NIH ImageJ software. The intensity of the processed band was divided by the intensity of the total (processed + unprocessed) bands for the control lane to obtain the percentage processed. The percentage processed in the untreated control represents 0% inhibition. Percentage processed in the presence of compounds were normalized to untreated controls. IC50 values were expressed as mean percent inhibition and plotted against compound concentration. IC50 values were defined as the concentration of compound required to reduce CPA95 processing by 50% relative to control. Assays were repeated in triplicate (error ± S.E.).
RESULTS
HTS of Small Molecule Libraries Using Enzymatic and Cellular Assays
With the goal of identifying compounds that inhibited furin and were bioavailable within the Golgi compartment, we utilized two independent screens, one that utilized the purified enzyme and a second that utilized a live cell assay for furin and furin-like activities. The enzymatic assay measured cleavage of a fluorogenic peptide substrate by purified secreted, ssfurin (29, 30). The cellular assay utilized CHO cells expressing a trans Golgi network-localized furin substrate, GRAP, whose cleavage by furin or other PCs results in secretion of soluble alkaline phosphatase (27). Details of the screens are found under “Experimental Procedures.” Both assays were used to screen commercial small molecule diversity libraries from ChemDiv and Chembridge (∼30,000 compounds), with positive hits defined by inhibition of >60%. Results of the screens were analyzed further to eliminate false-positives, toxic compounds, and nonspecific inhibitors. Inhibitory activities were confirmed by dose-response assays, resulting in selection of 12 compounds that were characterized in detail. This set was supplemented with 2 compounds (B5 and B9) identified by screening against the yeast furin homologue, Kex2, and with structural analogs (DC1, DC2, DC4, DC6, and B10) of compounds identified in the screen.
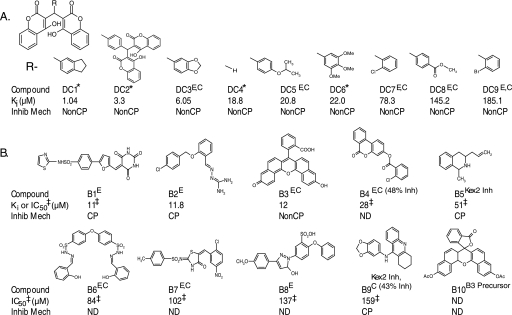
Five of the compounds derived from the HTS (DC3, DC5, and DC7–9) were derivatives of dicoumarol (DC4) (Fig. 2A). Dicoumarol itself and several other dicoumarol derivatives not identified in the screen (DC1, DC2, and DC6) were obtained commercially and also found to inhibit ssfurin.
FIGURE 2.
Small molecule inhibitors of furin. A, dicoumarol-related furin inhibitors; DC4 is dicoumarol. B, non-dicoumarol furin inhibitors. Compound, designation used in paper is given (DC1, etc.); E, compound identified in enzymatic HTS; C, compound identified in cell-based HTS; Kex2 Inh, compound identified in Kex2 HTS. Percent inhibition for the cellular assay is indicated when less than 60%. Asterisk refers to compounds purchased from Sigma. Ki or IC50 (μm), values determined with ssfurin as described under “Experimental Procedures.” IC50 values are indicated by a double dagger; all other values are Ki. For clarity, individual errors are not shown, but in all cases S.E. for Ki values was <10.3%. Inhib Mech (inhibitory mechanism); NonCP, noncompetitive; CP, competitive; ND, not determined.
Affinities and Mechanisms of Inhibition
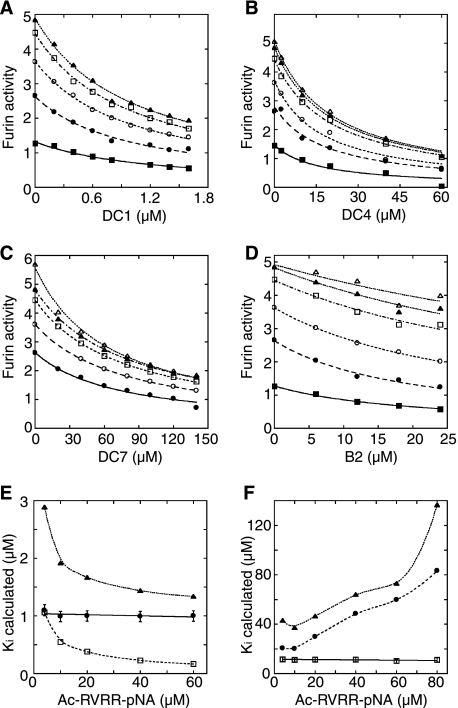
Initially, affinities of all of the inhibitors shown in Fig. 2 were characterized by determining IC50 values assuming a simple binding mechanism with boc-RVRR-AMC as a substrate (see “Experimental Procedures”). Preliminary analysis of these data by Eadie-Hoftstee plots suggested that all of the dicoumarols and B3 inhibited by a non-competitive mechanism and that the basic compounds (B1, B2, B5, and B9) were competitive inhibitors (data not shown). The dicoumarol compounds DC1–9 and compounds B2 and B3 were then characterized more carefully to determine accurate Ki values using Ac-RVRR-pNA as a substrate to avoid problems with inhibitor fluorescence and inhibitor-substrate aggregation (see “Experimental Procedures”). These data were fit to untransformed equations for competitive, non-competitive, and uncompetitive inhibition. Fig. 3, A–D, shows examples of inhibition data fit assuming a non-competitive mechanism for DC1, DC4, and DC7 (panels A–C) and a competitive mechanism for B2 (panel D). Fig. 3, E and F, show plots of Ki determined assuming competitive, non-competitive, or uncompetitive inhibition for DC1 and B2, respectively. As seen in Fig. 3E, only the non-competitive inhibition model gave a consistent Ki value for DC1 (and for the other dicoumarols and B3, data not shown). As seen in Fig. 3F, only the competitive inhibition model gave a consistent Ki value for B2. We conclude that the dicoumarol compounds inhibit by a purely non-competitive mechanism. B2 is clearly competitive. The other basic compounds appear also to be competitive, but were not characterized in greater detail.
FIGURE 3.
Representative example of Ki and inhibition mechanism determinations shown in the enzyme activity versus inhibitor concentration [I] at fixed concentrations of ac-RVRR-pNA [S]; from bottom: 4 μm (filled square), 10 μm (filled circle), 20 μm (empty circle), 40 μm (empty square), 60 μm (filled triangle), and 80 μm (empty triangle). A, noncompetitive fit for DC1; B, noncompetitive fit for DC4; C, noncompetitive fit for DC7; D, competitive fit for B2. Dicoumarol compounds were preincubated into assay buffer for 2 h, and inhibition reactions were started by addition of active site-titrated furin (33 nm) and ac-RVRR-pNA simultaneously in 96-well format. The enzyme inhibition by B2 were performed without the preincubation required for dicoumarols. Ki values determined are listed in Fig. 2. E and F, plots of Ki values versus substrate concentrations for DC1 (E) and B2 (F) calculated from fitting raw inhibition data to untransformed equations for competitive (squares), non-competitive (circles), and uncompetitive (triangles) inhibition. Only the noncompetitive fit yielded a consistent Ki (average of 1.04 ± 0.09 μm for the varied substrate concentrations) for DC1; whereas only the competitive fit yielded a consistent Ki (average of 11.8 ± 1.2 μm for the varied substrate concentrations) for B2. In each case, fits to other models (competitive and uncompetitive for DC1; noncompetitive and uncompetitive for B2) yielded average Ki values with S.E. of >50%.
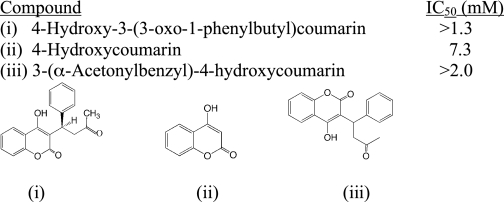
The dicoumarol compounds exhibited Ki values for ssfurin that ranged from 1.0 (DC1) to 185 μm (DC9) (Fig. 2A). 4-Hydroxycoumarin itself, warfarin, and 3-(α-acetonyl-benzyl)-4-(hydroxycoumarin) only exhibited inhibition in the millimolar range (Table 1), suggesting that the dicoumarol structure itself is crucial for affinity. The non-dicoumarol compounds (Fig. 2B) exhibited Ki (or IC50) values ranging from 11 (B1) to 159 μm (B9).
TABLE 1.
IC50values of ssfurin for simple 4-hydroxycoumarin derivatives
IC50values were determined as described under “Experimental Procedures.” Mean ± S.E. for each determination was ≤10.3%. 4-Hydroxy-3-(3-oxo-1-phenylbutylcoumarin is warfarin.
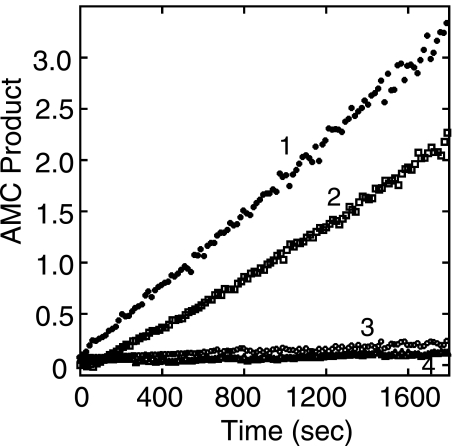
Inhibition of ssfurin by dicoumarol compounds was reversible (Fig. 4). When ssfurin was preincubated with DC4 at 10 × Ki in the absence of substrate and then diluted 10-fold into buffer containing substrate, activity was slightly in excess of that expected (50%) for full reversibility (Fig. 4, curve 2). No such recovery of activity was observed when enzyme was diluted into a reaction containing DC4 at 10 × Ki (Fig. 4, curve 3). Incubation of ssfurin with DC3 and DC9 for up to 2 h also showed no evidence of irreversible inactivation (data not shown).
FIGURE 4.
Reversible inhibition by dicoumarol DC4. ssfurin (11 nm) was preincubated with 188 μm DC4 (10 × Ki) for 15 min at 22 °C, then diluted 10-fold into ssfurin assay buffer containing boc-RVRR-MCA (2 μm) that either lacked inhibitor (line 2) or contained 188 μm DC4 (line 4). As controls, ssfurin (11 nm) was preincubated without DC4 and diluted 10-fold into buffer containing substrate either lacking inhibitor (line 1) or containing DC4 to give a final concentration of 188 μm (line 3). Average S.E., 6.5% (n = 3). Relative slopes of linear portions of plots are: line 1, 1.00; line 2, 0.71; line 3, 0.025; line 4, 0.043.
Inhibition of Extracellular Processing of Anthrax Protective Antigen
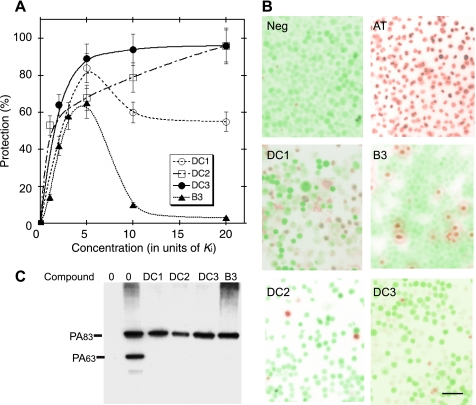
Furin and furin-like proteases act at the cell surface to process the precursor form of anthrax protective antigen (PA), PA83, to the mature form, PA63, which forms the heptameric prepore that binds and facilitates translocation of anthrax lethal factor and edema factor (40). Inhibition of furin activity protects against anthrax toxicity caused by PA and lethal factor (“AT”) (33, 41). As previously described (33), addition of recombinant PA (12 nm) + lethal factor (1.2 nm) killed >99% of J774A.1 murine macrophages within 2.5 h of incubation (Fig. 5B, AT), whereas control cells exhibited no lysis (Fig. 4B, “Neg”). Compounds DC1, DC2, DC3, and B3 all protected macrophages from AT-mediated cell death, and exhibited distinct titration curves (Fig. 5A). DC1 and B3 exhibited optimal protection at 5 × Ki, showing toxicity at higher concentrations. In contrast, DC2 and DC3 exhibited optimal protection at 20 × Ki, with no significant toxicity. Fig. 5B shows representative images of cells treated with AT in the presence of optimal concentrations of the four compounds. In this experiment, treatment of cells with 4.9 μm DC1, 60 μm B3, 66 μm DC2, or 120 μm DC3 protected 83.5, 65, 96.2, or 95.5% of cells, respectively, from AT-dependent killing.
FIGURE 5.
Protection of macrophages from anthrax-toxin mediated killing. A, J774A.1 murine macrophages were incubated with AT and increasing concentrations of compounds DC1, DC2, DC3, or B3 (n = 3). Average S.E., 9.5%, DC1; 10.0%, DC2; 9.0%, DC3; 12.5%, B3. B, representative images of macrophage cells exposed to vehicle (Neg) or anthrax toxin (AT) and AT-exposed cells treated with 5 μm DC1 (5 × Ki), 60 μm B3 (5 × Ki), 66 μm DC2 (20 × Ki), or 120 μm DC3 (5 × Ki) (n = 3). Live cells appear green and lysed cells appear red. Scale bar is 50 μm. C, representative Western blot analysis of PA83 processing on macrophage cell surface. J774A.1 murine macrophages (10-cm plates) were washed twice with RB* and overlaid with RB* or RB* containing DC1 (5 μm), DC2 (12.5 μm), DC3 (24 μm), or B3 (24 μm). Anthrax PA83 (12 nm) was added and cells were incubated for 2.5 h at 37 °C. Cell lysates were probed with anti-PA antibody as described (33).
To confirm that protection was due to inhibition of PA cleavage, we tested each ability of the compound to inhibit processing of PA83 to PA63. When macrophage cultures were incubated with 12 nm PA83 for 2.5 h and cell extracts were analyzed by immunoblotting with anti-PA antibody, ∼40% of cell-associated PA83 was processed to PA63 (Fig. 5C). When cells were treated with DC1, DC2, or DC3 at concentrations equivalent to 5 × Ki or B3 at concentrations equivalent to 2 × Ki, no significant cleavage of PA83 was observed (Fig. 5C).
Inhibition of Intracellular Processing
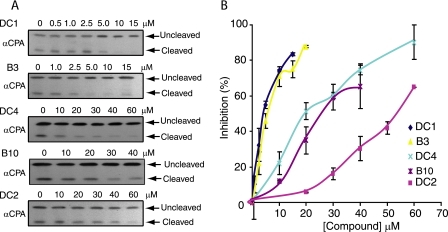
To examine inhibition of intracellular processing reactions catalyzed by furin or other PCs, we employed live cell assays using the engineered furin substrate CPA95 (37). CPA95 is a secreted substrate engineered to contain a furin cleavage motif at residue 95 (see “Experimental Procedures”). CPA95 is a secreted substrate engineered to contain a furin cleavage motif at residue 95. Expression of CPA95 in transfected CHO cells resulted in the appearance of both unprocessed (43 kDa) and processed (32 kDa) forms of the protein in the extracellular medium. Treatment of these cells with specific inhibitory compounds resulted in a dose-dependent decrease in the processed form and increase in the unprocessed form (Fig. 6A). The compounds tested showed a range of inhibitory capacity with DC1, B3, DC4, B10, and DC2 exhibiting IC50 values of ∼4, ∼7, ∼20, ∼25, and ∼55 μm, respectively (Fig. 6B). The same compounds tested in Fig. 6 also inhibited intracellular processing of pro-von Willebrand factor (data not shown).
FIGURE 6.
IC50 determination for small molecule inhibitors. A, representative Western blots, using anti-CPA antibody (αCPA), of conditioned media of CHO cells transfected with the CPA95 expression plasmid (37) and treated with vehicle alone or with increasing concentrations of DC1, B3, DC4, B10, or DC2. B, data from blots quantified as described under “Experimental Procedures” and plotted as percent inhibition versus compound concentration (n = 3). (Values are mean ± S.E.).
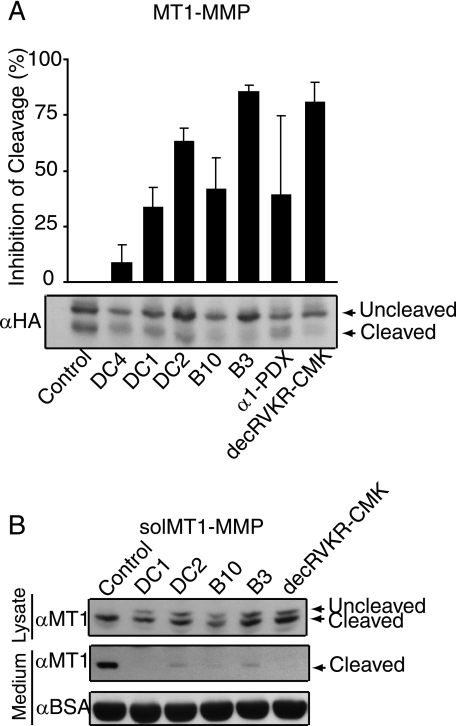
Inhibition of Intracellular Maturation of MT1-MMP
Inhibition of furin/PC-dependent maturation of MT1-MMP offers a potentially important new avenue to impede tumor invasiveness and metastasis (25). To determine whether compounds identified in this study could inhibit processing of MT1-MMP, CHO cells were transfected with a plasmid encoding HA epitope-tagged MT1-MMP. In the absence of furin inhibitors, transfection of MT1-MMP in cells resulted in the appearance of the precursor (∼63 kDa) and mature (∼60 kDa) forms of MT1-MMP as shown by Western blot analysis using an HA-specific antibody (Fig. 7A, bottom). Addition of DC4, DC1, DC2, B10, and B3 resulted in marked reduction in the appearance of mature MT1-MMP. Inhibition of processing, relative to control, was quantified as and expressed as a percentage (Fig. 7A, top). Treatment of these cells with compounds inhibited MT1-MMP processing. An 8.8% reduction was observed with 40 μm DC4, 34% with 10 μm DC1, 63% with 50 μm DC2, 42% with 40 μm B10, and 86% with 15 μm B3. In this experiment, the well characterized furin inhibitors decRVKR-CMK (20 μm) and α1-PDX (42), the latter introduced by cotransfection, inhibited MT1-MMP processing by 40 and 81%, respectively (Fig. 7A, top). To confirm the inhibitory effects of these compounds on furin/PC-mediated MT1-MMP cleavage, we transfected CHO cells with a plasmid encoding soluble MT1-MMP (solMT1-MMP; i.e. MT1-MMP lacking the transmembrane domain) (43), which is completely processed to the mature form by endogenous furin (26). As shown by Western blot analysis using an MT1-MMP antibody, processing of solMT1-MMP by furin/PCs in CHO cells resulted in appearance of the mature (∼50 kDa) form of solMT1-MMP in cell lysates and extracellular medium (Fig. 7B, lane 1). Addition of 10 μm DC1, 50 μm DC2, 40 μm B10, or 15 μm B3 resulted in a reduction of processed solMT1-MMP in cell lysates comparable with that seen with 20 μm decRVKR-CMK (Fig. 7B), with no significant mature solMT1-MMP released to the medium.
FIGURE 7.
Inhibition of furin-mediated cleavage of MT1-MMP. A, top, data from blots quantified as described under “Experimental Procedures” and plotted as percent inhibition compared with control (n = 3). Values are mean ± S.D.. Bottom, representative Western blot, probed with anti-HA antibody, of cellular lysates of CHO cells transfected with MT1-MMP-HA plasmid and treated with vehicle, DC4 (40 μm), DC1 (10 μm), DC2 (50 μm), B10 (40 μm), and B3 (15 μm). DecRVKR-CMK (20 μm) and cotransfection with α1-PDX served as positive controls. B, representative Western blots, probed with MT1-MMP antibody, of lysates and conditioned medium of CHO cells transfected with solMT1-MMP treated with vehicle, DC1 (10 μm), DC2 (50 μm), B10 (40 μm), and B3 (15 μm). Blot of conditioned medium was also probed with bovine serum albumin antibody (BSA) as a loading control. DecRVKR-CMK (20 μm) served as a positive control.
Inhibition of rPACE4, hPC5/6, and hPC7
A subset of compounds was tested against purified catalytic domains of rPACE4, hPC5/6, and hPC7, other members of the PC family that, like furin, function in the constitutive secretory pathway, the endocytic pathway, and/or the cell surface and exhibit broad tissue distributions. Ki values are shown in Table 2. Although all compounds tested inhibited all four enzymes (ssfurin, rPACE4, hPC5/6, and hPC7), each compound exhibited a unique pattern of inhibitory potency. DC1 and DC3 exhibited similar Ki values for ssfurin, rPACE4, and hPC5/6 but showed significantly lower affinity for hPC7. DC2 inhibited all four PCs with similar efficacy. B3 had its strongest inhibitory activity against hPC7, exhibited similar inhibitory activities toward ssfurin and rPACE4, and was less effective with hPC5/6. All four compounds also inhibited human α-thrombin to some degree.
TABLE 2.
Ki or IC50values for inhibition of rPACE4, hPC5/6, hPC7, and α-thrombin by small molecule inhibitors
Ki and IC50 values were determined as described under “Experimental Procedures.” S.E. for each determination was ≤10.3%.
| Enzyme | Compound |
|||
|---|---|---|---|---|
| DC1 | DC2 | DC3 | B3 | |
| μm | ||||
| ssfurina | 1.0 | 3.3 | 6.1 | 12 |
| rPACE4 | 13 | 8.3 | 14 | 12 |
| hPC5/6 | 4.0 | 3.0 | 16 | 76 |
| hPC7 | 79 | 12 | 91 | 4.5 |
| α-Thrombin | 21 | 60 | 15 | 63 |
a Data fromFig. 2 for comparison.
Toxicity of Furin Inhibitors under Conditions of Tests of Inhibition of Processing
Cytotoxicity was examined by incubating J774A.1 murine macrophages with compounds for 2.5 to 8 h (Table 3). Dicoumarol derivatives DC1, DC2, DC3, and DC4 showed no significant toxicity when incubated with macrophages at 20 × Ki for 8 h (Table 3). B3 showed some toxicity when incubated with macrophages at 2 × Ki.
TABLE 3.
Toxicity of small molecule inhibitors in cell culture under conditions of tests of inhibition of processing
J774A.1 murine mac ro phages were incubated in RB* with DC1 (20 ×Ki), DC2 (20 ×Ki), DC3 (20 ×Ki), DC4 (20 ×Ki), or B3 (2 ×Ki) for times as shown. Toxicity was determined as described under “Anthrax Toxin Killing Assays” under “Experimental Procedures.” Cells in triplicate wells (∼600 cells per well) were counted for each time point. Average S.E.: 8.3% for DC1, 7.6% for DC2, 7.2% for DC3, 10.3% for DC4, and 13.9% for B3.
| Time | Compound |
||||
|---|---|---|---|---|---|
| DC1 | DC2 | DC3 | DC4 | B3 | |
| h | |||||
| 0 | 100 | 100 | 100 | 100 | 100 |
| 2.5 | 98 | 96 | 97 | 96 | 85 |
| 6 | 95 | 96 | 98 | 95 | 80 |
| 8 | 86 | 83 | 90 | 94 | 60 |
DISCUSSION
Proprotein processing reactions provide potential targets of drug intervention for both chronic and infectious human disease (44). Because a large number of processing reactions are catalyzed by PCs, these enzymes have emerged as important potential drug targets (23). Drugs that are not cell-permeable may be effective in the case of extracellular processing events, such as maturation of anthrax protective antigen. More generally, however, small molecules with high bioavailability and cell permeability and exhibiting low toxicity are required to inhibit processing in intracellular compartments. Furin, PACE4, PC5/6, and PC7 all function in the constitutive secretory pathway, in the endocytic pathway, and/or at the cell surface and exhibit broad tissue distributions (2). Because of their similar patterns of substrate recognition and at least partial overlap in expression, it is currently unclear whether the goal of drug development targeting pathophysiological effects of furin or furin-like processing should be directed toward inhibitors exhibiting a high degree of discrimination between PC family members or toward more generic inhibitors that can block activities of all the PCs.
Here we report the results of a high-throughput screen for small molecule inhibitors of furin/PCs that relied on simultaneous use of enzymatic and cellular assays. Use of the cell-based assay identified compounds that inhibited intracellular processing catalyzed by furin alone or by furin along with other PCs while eliminating compounds with substantial toxicity. Use of the enzymatic assay identified compounds that directly inhibited ssfurin. The combination of assays allowed us to focus on furin inhibitors that had high promise for intracellular inhibition. The combined screens identified a set of competitive inhibitors that were largely unrelated structurally and a set of non-competitive inhibitors, mostly from one structural family. Many of the competitive inhibitors were basic compounds that performed poorly due to higher toxicity or lower cell permeability when further cell-based assays were employed. We focused our efforts instead on the non-competitive inhibitors, all of which were structurally related or analogous to dicoumarol. Identification of these compounds as furin inhibitors is significant for several reasons. First, to our knowledge, no other non-competitive furin inhibitors have been described. Second, inhibition by several of these compounds (DC3, DC4, and DC9) was reversible, indicating that these molecules do not undergo nonspecific covalent reactions with ssfurin. Third, dicoumarols have an extensive history of use as pharmacological agents that makes them an attractive starting point for drug development.
Dicoumarol (DC4) has been used clinically as an anticoagulant with vitamin K antagonist activity similar to that of warfarin (45–47). A large number of dicoumarol derivatives have been synthesized and assessed for anticoagulant activity (48). The use of dicoumarol as an oral anticoagulent is indicative of high bioavailability, low toxicity, and high cell permeability. Indeed, several dicoumarols examined here exhibited high cell permeability as the IC50 values for inhibition of intracellular processing (Fig. 6B) were quite close to the Ki values measured with purified ssfurin. Although the furin inhibitors identified in this screen exhibited Ki values in the micromolar range, their effective concentrations for inhibition of both cell surface and intracellular processing were also in the low micromolar range. Inhibition of intracellular processing reactions by other characterized furin inhibitors, including engineered proteins (20, 49) and reactive peptides (50, 51), has required inhibitor concentrations ranging from ∼1,000 to 10,000 × Ki. In contrast, IC50 values for inhibition of intracellular processing by DC1, DC2, and DC4 were ∼4 × Ki, 17 × Ki, and 1 × Ki, respectively (Figs. 2A and 6B). Moreover, IC50 values for previously characterized furin inhibitors when assayed in cell-surface processing reactions have also been high in comparison to Ki values: 210 × Ki for RRD-eglin (33), 120 × Ki for Ac-Arg-Glu-Lys-boroArg (33), 2800 × Ki for D9R (52), and 190–360 × Ki for 2,5-dideoxystrepatmine derivatives (24). In contrast, IC50 values for inhibition of intracellular processing by DC1, DC2, and DC3 were <5 × Ki, 1 × Ki, and ∼2 × Ki, respectively (Figs. 2A and 6B). This remarkable efficiency of inhibition may be related to the ability of these compounds to inhibit both the intracellular and surface pools of processing enzymes.
Although the therapeutic target of dicoumarols when used as anticoagulants is presumptively vitamin K epoxide reductase (53), another enzyme, NAD(P)H quinone oxidoreductase (DT diaphorase), has also been shown to be a high-affinity target (54). In addition, dicoumarol has been reported to inhibit gap junction formation in cultured cells (55) and ADP-ribosylation of CtBP3/BARS, resulting in alterations in Golgi morphology and function (56). These effects may or may not influence the toxicity of dicoumarol but have not precluded clinical use.
The PCs represent a new set of physiological targets for inhibition by dicoumarol and its derivatives. Because of the functional redundancy of the PCs, it is difficult to ascertain whether inhibition of processing in the individual cellular assays (processing of anthrax PA, CPA95, GRAPfurin, von Willebrand factor, or MT1-MMP) is due solely to inhibition of furin, inhibition of other PC family members or both. Nevertheless, our results suggest both that single compounds can inhibit multiple members of the PC family (Table 2) and that selectivity between enzymes can be achieved through modification of the dicoumarol structure (Fig. 2A).
For several of the compounds tested, concentrations required for inhibition in cellular assays are similar to the therapeutic doses of dicoumarol, suggesting that affinities may be close to that needed for drug models. Nevertheless, because dicoumarols inhibit multiple target enzymes, optimization of compounds will be necessary. Dicoumarol represents a good platform for structural modification that might yield compounds with increased affinity for furin and other PCs. Increasing selectivity, both for individual PCs and for PCs versus other targets (e.g. vitamin K epoxide reductase and NAD(P)H quinone oxidoreductase) should further reduce off-target effects. Production of higher affinity derivatives may also assist in identifying the dicoumarol binding site within furin and yield information regarding the molecular mechanism of non-competitive inhibition.
Acknowledgments
We thank S. Weiss, S. Zucker, and J. Cao for plasmids and J. Swanson for PA and lethal factor. We thank R. Neubig and S. Decker and the Center for Chemical Genomics, Life Sciences Institute, University of Michigan, for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health NCI Grants P01CA85878, R24CA83099, and P50CA093990 (to A. R. and B. D. R.) and GM39697 and GM50915 (to R. S. F.). This work was also supported by Canadian Institutes of Health Research grants (to R. D.).
- PC
- proprotein convertase
- MT1-MMP
- membrane-type 1 matrix metalloproteinase
- HTS
- high-throughput screening
- r
- rat
- h
- human
- ssfurin
- secreted, soluble human furin
- decRVKR-CMK
- decanoyl-Arg-Val-Lys-Arg-chloromethylketone
- boc-RVRR-MCA
- N-tert-butoxycarbonyl-Arg-Val-Arg-Arg-methylcoumarin amide
- boc-IEGR-MCA
- boc-Ile-Glu-Gly-Arg-MCA
- AMC
- 7-amino-4-methyl coumarin
- pNA
- para-nitroanilide
- ac-RVRR-pNA
- acetyl-Arg-Val-Arg-Arg-pNA
- CHO
- Chinese hamster ovary
- RB*
- modified Ringer's buffer
- PA
- anthrax protective antigen
- AT
- anthrax toxin
- Mes
- 4-morpholineethanesulfonic acid
- HA
- hemagglutinin.
REFERENCES
- 1.Rockwell N. C., Krysan D. J., Komiyama T., Fuller R. S. ( 2002) Chem. Rev. 102, 4525– 4548 [DOI] [PubMed] [Google Scholar]
- 2.Thomas G. ( 2002) Nat. Rev. Mol. Cell Biol. 3, 753– 766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scamuffa N., Calvo F., Chretien M., Seidah N. G., Khatib A. M. ( 2006) FASEB J. 20, 1954– 1963 [DOI] [PubMed] [Google Scholar]
- 4.Henrich S., Cameron A., Bourenkov G. P., Kiefersauer R., Huber R., Lindberg I., Bode W., Than M. E. ( 2003) Nat. Struct. Biol. 10, 520– 526 [DOI] [PubMed] [Google Scholar]
- 5.Holyoak T., Wilson M. A., Fenn T. D., Kettner C. A., Petsko G. A., Fuller R. S., Ringe D. ( 2003) Biochemistry 42, 6709– 6718 [DOI] [PubMed] [Google Scholar]
- 6.Roebroek A. J., Umans L., Pauli I. G., Robertson E. J., van Leuven F., Van de Ven W. J., Constam D. B. ( 1998) Development 125, 4863– 4876 [DOI] [PubMed] [Google Scholar]
- 7.Roebroek A. J., Taylor N. A., Louagie E., Pauli I., Smeijers L., Snellinx A., Lauwers A., Van de Ven W. J., Hartmann D., Creemers J. W. ( 2004) J. Biol. Chem. 279, 53442– 53450 [DOI] [PubMed] [Google Scholar]
- 8.Bennett B. D., Denis P., Haniu M., Teplow D. B., Kahn S., Louis J. C., Citron M., Vassar R. ( 2000) J. Biol. Chem. 275, 37712– 37717 [DOI] [PubMed] [Google Scholar]
- 9.Rostagno A., Tomidokoro Y., Lashley T., Ng D., Plant G., Holton J., Frangione B., Revesz T., Ghiso J. ( 2005) Cell Mol. Life Sci. 62, 1814– 1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arner E. C. ( 2002) Curr. Opin. Pharmacol. 2, 322– 329 [DOI] [PubMed] [Google Scholar]
- 11.Stawowy P., Fleck E. ( 2005) J. Mol. Med. 83, 865– 875 [DOI] [PubMed] [Google Scholar]
- 12.Bassi D. E., Fu J., Lopez de Cicco R., Klein-Szanto A. J. ( 2005) Mol. Carcinog. 44, 151– 161 [DOI] [PubMed] [Google Scholar]
- 13.Gordon V. M., Klimpel K. R., Arora N., Henderson M. A., Leppla S. H. ( 1995) Infect. Immun. 63, 82– 87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garred O., van Deurs B., Sandvig K. ( 1995) J. Biol. Chem. 270, 10817– 10821 [DOI] [PubMed] [Google Scholar]
- 15.Matsuzawa T., Fukui A., Kashimoto T., Nagao K., Oka K., Miyake M., Horiguchi Y. ( 2004) J. Biol. Chem. 279, 2866– 2872 [DOI] [PubMed] [Google Scholar]
- 16.Basak A., Zhong M., Munzer J. S., Chretien M., Seidah N. G. ( 2001) Biochem. J. 353, 537– 545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moulard M., Decroly E. ( 2000) Biochim. Biophys. Acta 1469, 121– 132 [DOI] [PubMed] [Google Scholar]
- 18.Volchkov V. E., Feldmann H., Volchkova V. A., Klenk H. D. ( 1998) Proc. Natl. Acad. Sci. U. S. A. 95, 5762– 5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe M., Hirano A., Stenglein S., Nelson J., Thomas G., Wong T. C. ( 1995) J. Virol. 69, 3206– 3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jean F., Thomas L., Molloy S. S., Liu G., Jarvis M. A., Nelson J. A., Thomas G. ( 2000) Proc. Natl. Acad. Sci. U. S. A. 97, 2864– 2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stadler K., Allison S. L., Schalich J., Heinz F. X. ( 1997) J. Virol. 71, 8475– 8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards R. M., Lowy D. R., Schiller J. T., Day P. M. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 1522– 1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fugere M., Day R. ( 2005) Trends Pharmacol. Sci. 26, 294– 301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao G. S., Cregar L., Wang J., Millis S. Z., Tang C., O'Malley S., Johnson A. T., Sareth S., Larson J., Thomas G. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 19707– 19712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato H., Takino T., Miyamori H. ( 2005) Cancer Sci. 96, 212– 217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yana I., Weiss S. J. ( 2000) Mol. Biol. Cell 11, 2387– 2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coppola J. M., Hamilton C. A., Bhojani M. S., Larsen M. J., Ross B. D., Rehemtulla A. ( 2007) Anal. Biochem. 364, 19– 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borges F., Roleira F., Milhazes N., Santana L., Uriarte E. ( 2005) Curr. Med. Chem. 12, 887– 916 [DOI] [PubMed] [Google Scholar]
- 29.Komiyama T., Fuller R. S. ( 2000) Biochemistry 39, 15156– 15165 [DOI] [PubMed] [Google Scholar]
- 30.Bravo D. A., Gleason J. B., Sanchez R. I., Roth R. A., Fuller R. S. ( 1994) J. Biol. Chem. 269, 25830– 25837 [PubMed] [Google Scholar]
- 31.Fugere M., Limperis P. C., Beaulieu-Audy V., Gagnon F., Lavigne P., Klarskov K., Leduc R., Day R. ( 2002) J. Biol. Chem. 277, 7648– 7656 [DOI] [PubMed] [Google Scholar]
- 32.Coppola J. M., Bhojani M. S., Ross B. D., Rehemtulla A. ( 2008) Neoplasia 10, 363– 370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komiyama T., Swanson J. A., Fuller R. S. ( 2005) Antimicrob. Agents Chemother. 49, 3875– 3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J. H., Chung T. D., Oldenburg K. R. ( 1999) J. Biomol. Screen 4, 67– 73 [DOI] [PubMed] [Google Scholar]
- 35.Dixon M., Webb E. ( 1979) Enzymes, Academic Press, New York [Google Scholar]
- 36.Segel I. H. ( 1993) Enzyme Kinetics, Wiley Interscience, New York [Google Scholar]
- 37.Hamstra D. A., Rehemtulla A. ( 1999) Hum. Gene Ther. 10, 235– 248 [DOI] [PubMed] [Google Scholar]
- 38.Beauregard K. E., Collier R. J., Swanson J. A. ( 2000) Cell. Microbiol. 2, 251– 258 [DOI] [PubMed] [Google Scholar]
- 39.Chen G., Bhojani M. S., Heaford A. C., Chang D. C., Laxman B., Thomas D. G., Griffin L. B., Yu J., Coppola J. M., Giordano T. J., Lin L., Adams D., Orringer M. B., Ross B. D., Beer D. G., Rehemtulla A. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 12507– 12512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collier R. J., Young J. A. ( 2003) Annu. Rev. Cell Dev. Biol. 19, 45– 70 [DOI] [PubMed] [Google Scholar]
- 41.Sarac M. S., Peinado J. R., Leppla S. H., Lindberg I. ( 2004) Infect. Immun. 72, 602– 605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson E. D., Thomas L., Hayflick J. S., Thomas G. ( 1993) J. Biol. Chem. 268, 24887– 24891 [PubMed] [Google Scholar]
- 43.Cao J., Sato H., Takino T., Seiki M. ( 1995) J. Biol. Chem. 270, 801– 805 [DOI] [PubMed] [Google Scholar]
- 44.Fear G., Komarnytsky S., Raskin I. ( 2007) Pharmacol. Ther. 113, 354– 368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lockner D., Paul C. ( 1979) Br. J. Clin. Pharmacol. 8, 59– 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulman S., Granqvist S., Holmstrom M., Carlsson A., Lindmarker P., Nicol P., Eklund S. G., Nordlander S., Larfars G., Leijd B., Linder O., Loogna E. ( 1997) N. Engl. J. Med. 336, 393– 398 [DOI] [PubMed] [Google Scholar]
- 47.McAlister V. ( 2006) Clin. Investig. Med. 29, 373– 377 [PubMed] [Google Scholar]
- 48.Guminska M., Eckstein M. ( 1961) J. Med. Pharm. Chem. 3, 583– 595 [DOI] [PubMed] [Google Scholar]
- 49.Komiyama T., VanderLugt B., Fugere M., Day R., Kaufman R. J., Fuller R. S. ( 2003) Proc. Natl. Acad. Sci. U. S. A. 100, 8205– 8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jean F., Stella K., Thomas L., Liu G., Xiang Y., Reason A. J., Thomas G. ( 1998) Proc. Natl. Acad. Sci. U. S. A. 95, 7293– 7298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hallenberger S., Bosch V., Angliker H., Shaw E., Klenk H. D., Garten W. ( 1992) Nature 360, 358– 361 [DOI] [PubMed] [Google Scholar]
- 52.Kacprzak M. M., Peinado J. R., Than M. E., Appel J., Henrich S., Lipkind G., Houghten R. A., Bode W., Lindberg I. ( 2004) J. Biol. Chem. 279, 36788– 36794 [DOI] [PubMed] [Google Scholar]
- 53.Wallin R., Hutson S. M. ( 2004) Trends Mol. Med. 10, 299– 302 [DOI] [PubMed] [Google Scholar]
- 54.Asher G., Dym O., Tsvetkov P., Adler J., Shaul Y. ( 2006) Biochemistry 45, 6372– 6378 [DOI] [PubMed] [Google Scholar]
- 55.Abdelmohsen K., Stuhlmann D., Daubrawa F., Klotz L. O. ( 2005) Arch. Biochem. Biophys. 434, 241– 247 [DOI] [PubMed] [Google Scholar]
- 56.Mironov A. A., Colanzi A., Polishchuk R. S., Beznoussenko G. V., Mironov A. A., Jr., Fusella A., Di Tullio G., Silletta M. G., Corda D., De Matteis M. A., Luini A. ( 2004) Eur. J. Cell Biol. 83, 263– 279 [DOI] [PubMed] [Google Scholar]