Abstract
Proinflammatory responses induced by Plasmodium falciparum glycosylphosphatidylinositols (GPIs) are thought to be involved in malaria pathogenesis. In this study, we investigated the role of MAPK-activated protein kinase 2 (MK2) in the regulation of tumor necrosis factor-α (TNF-α) and interleukin (IL)-12, two of the major inflammatory cytokines produced by macrophages stimulated with GPIs. We show that MK2 differentially regulates the GPI-induced production of TNF-α and IL-12. Although TNF-α production was markedly decreased, IL-12 expression was increased by 2–3-fold in GPI-stimulated MK2−/− macrophages compared with wild type (WT) cells. MK2−/− macrophages produced markedly decreased levels of TNF-α than WT macrophages mainly because of lower mRNA stability and translation. In the case of IL-12, mRNA was substantially higher in MK2−/− macrophages than WT. This enhanced production is due to increased NF-κB binding to the gene promoter, a markedly lower level expression of the transcriptional repressor factor c-Maf, and a decreased binding of GAP-12 to the gene promoter in MK2−/− macrophages. Thus, our data demonstrate for the first time the role of MK2 in the transcriptional regulation of IL-12. Using the protein kinase inhibitors SB203580 and U0126, we also show that the ERK and p38 pathways regulate TNF-α and IL-12 production, and that both inhibitors can reduce phosphorylation of MK2 in response to GPIs and other toll-like receptor ligands. These results may have important implications for developing therapeutics for malaria and other infectious diseases.
Malaria, caused by Plasmodium species of protozoan parasites, is a major public health and economic burden in many parts of the world. An estimated 300–500 million people suffer from infection, and 1–2 million die of severe malaria annually (1–4). Among the different species of parasites that infect humans, Plasmodium falciparum causes the most fatal forms of malaria and is responsible for most deaths (3, 4). Severe malaria is associated with a broad spectrum of systemic as well as single and multiple organ pathologies, including periodic and intense fever and chills, shock, severe anemia, metabolic acidosis, hypoglycemia, renal failure, jaundice, acute respiratory distress, convulsion, seizures, and coma. Although the molecular mechanisms involved in malaria pathogenesis are highly complex and multifactorial, accumulating evidence indicates that dysregulated innate immune responses play important roles in the pathology of severe malaria (4).
During malaria infection, like in most other pathogenic infections, the innate immune system responds early on by producing high levels of proinflammatory cytokines such as TNF-α,3 IFN-γ, IL-12, IL-6, IL-1, and NO (5–8). In the absence of prior immunity, these proinflammatory mediators function as the first line of defense against parasites and are crucial for controlling infection; otherwise, parasites grow rapidly and overwhelm the host, causing severe illness and fatality. The inflammatory mediators exert toxic effects on parasites by initiating a variety of effector mechanisms, such as cytotoxicity by free radicals, phagocytosis, complement activation, and cell and antibody-mediated adaptive immune responses (5, 9, 11–13). For example, IFN-γ is a potent immunostimulatory cytokine that primes macrophages for the efficient production of cytokines, including TNF-α, IL-12, IL-6, and reactive oxygen and nitrogen free radicals. IL-12, another potent immunostimulatory cytokine, induces IFN-γ secretion by NK cells and also modulates cell-mediated and humoral responses. TNF-α, IFN-γ, and IL-12 can activate macrophages to produce oxygen and nitrogen free radicals for killing parasites by cytotoxic effects (11).
Proinflammatory responses are harmful to the host if they are not appropriately regulated and continue to be overproduced (4, 5). Usually, after parasite growth is brought under control, proinflammatory responses are down-regulated by the increased expression of anti-inflammatory cytokines. However, the required tight regulation between pro- and anti-inflammatory responses is not always maintained. In some infected individuals, altered/impaired immune responses due to defects in parasite recognition and/or signaling events lead to distinct and varied clinical conditions. Accordingly, a number of studies have shown that prolonged and excessive production of TNF-α, IFN-γ, IL-12, IL-6, IL-1, NO, and other mediators during infection are associated with severe malaria (14–16). Understanding the details of the signaling events that govern pro- and anti-inflammatory responses may offer targets for developing novel drugs or immunotherapeutics. However, very little is known about the myriad signaling events involved in innate immune responses to malaria parasites.
Glycosylphosphatidylinositols (GPIs) of P. falciparum have been recognized as the major factors involved in the production of proinflammatory mediators, thereby contributing to malaria pathogenesis (17, 18). Recent studies have shown that GPIs activate macrophages mainly through TLR2/TLR1-dependent signaling, leading to the activation of ERK, p38, and JNK MAPK and NF-κB pathways and the production of cytokines (19). The MAPK pathways and NF-κBs differentially contribute to the expression of various proinflammatory mediators (20). For example, in macrophages primed with IFN-γ, inhibition of either ERK or p38 pathway caused only a marginal decrease in TNF-α production in response to malarial GPIs (20). Inhibition of ERK activation resulted in a severalfold increase in IL-12 production, whereas inhibition of p38 activity markedly reduced the IL-12 expression. In a preliminary study using unprimed macrophages, inhibition of ERK activation caused a significant decrease in TNF-α production, whereas that of p38 activity had little or no effect on TNF-α production. In contrast, inhibition of either ERK activation or p38 activity resulted in increased IL-12 production. In this study, we investigated the mechanisms that underlie the differential regulation of TNF-α and IL-12 production using macrophages deficient in MK2, one of the signaling molecules, which has been described to be downstream of p38 (21). Furthermore, we examined the effect of inhibiting ERK and p38 MAPKs in WT macrophages. Our results show that MK2 is targeted by both ERK and p38 and that MK2 directly or indirectly regulates TNF-α protein expression at both transcriptional and post-transcriptional levels. The results also suggest that MK2 regulates the GPI-induced IL-12 production by controlling NF-κB binding to gene promoters and the expression of c-Maf and binding of GAP-12 to GATA sequences of the IL-12 gene promoter.
EXPERIMENTAL PROCEDURES
Materials
SB203580, U0126, and PD98059 were from Calbiochem. Actinomycin D was from Sigma. The anti-peptide and phospho-specific antibodies that specifically recognize ERK1/ERK2, p38, JNK, MK2, IκBα, and β-actin, and HRP-conjugated goat anti-mouse IgGs and goat anti-rabbit IgGs were from Cell Signaling Technology Inc. (Beverly, MA). Rabbit antibodies that are specific to mouse NF-κB p50 (sc-114), NF-κB p65 (sc-372), c-Rel (sc-71), and c-Maf (sc-7866) were from Santa Cruz Biotechnology, Inc. Rabbit anti-mouse IκBζ antibody was produced in the laboratory of Dr. Tatsushi Muta as described previously (22). Kits for the estimation of TNF-α and IL-12 (p40) by ELISA (Duoset ELISA development system) were from R & D Systems, Inc. (Minneapolis, MN). NE-PER nuclear and cytoplasm extraction reagents and LightShift® chemiluminescent EMSA kit were from Pierce. MK2 immunoprecipitation kinase assay kit was from Upstate cell signaling solutions. FBS, DMEM, TRIzol RNA isolation reagent, and PCR primers and DNA probes labeled at the 5′-ends with biotin were from/custom-synthesized by Invitrogen. CpG was from Coley Pharmaceutical (Kanata, Ontario, Canada). Pam3CSK4 was from EMC Microcollections GmbH (Tübingen, Germany). Human blood and serum collected from healthy donors were received from the Blood Bank of the Hershey Medical Center. Endotoxin-free water and buffers were used for all the experimental procedures.
P. falciparum Culturing and Isolation of GPIs
P. falciparum parasites (FCR-3 or 3D7 strain) were cultured using O-positive human erythrocytes in RPMI 1640 medium containing 10–20% human O-positive plasma and 50 μg/ml gentamycin under 90% nitrogen, 5% oxygen, and 5% carbon dioxide atmosphere as described previously (23). Cultures were harvested at 20–30% parasitemia, and parasites were released by treatment with 0.05% saponin and purified by centrifugation on 5% bovine serum albumin in PBS, pH 7.4. Isolation and high pressure liquid chromatography purification of GPIs were performed as described previously (24).
Mice
MK2 knock-out mice (generated in the laboratory of Dr. Gaestel) and wild type control mice (purchased from The Jackson Laboratories) both in a C57BL/6 genetic background were used for this study. The animals were maintained in a germ-free environment, and animal care was in accordance with the Institutional Guidelines of the Pennsylvania State University College of Medicine.
Preparation of L929 Cell Culture Conditioned Medium
Mouse L929 fibroblasts were cultured in DMEM containing 10% FBS, 1% l-glutamine, 1 mm sodium pyruvate, 50 μm 2-mercaptoethanol, nonessential amino acids, and 10 units/ml penicillin/streptomycin (DMEM complete medium) in roller flasks at 37 °C for 4–5 days. Supernatants were collected, centrifuged at 1,300 × g for 20 min, and used as the source of macrophage-colony-stimulating factor.
Preparation and Culturing of Mouse Bone Marrow-derived Macrophages
Bone marrow from the femurs of mice was flushed with DMEM and 10 units/ml penicillin/streptomycin using a syringe-needle. Tissue debris was removed by filtration through a 70-μm strainer, and cells were pelleted by centrifugation at 300 × g. The cell pellets were washed twice with DMEM and penicillin/streptomycin and dispersed in DMEM complete medium (see above) containing 30% L929 cell culture conditioned medium, and cultured at 37 °C for 2 days. The nonadherent cells were removed, and the adherent cells were cultured by changing the medium every 2 days. After 7 days, macrophages were harvested by scraping in incomplete DMEM, suspended in complete DMEM, and used for stimulation with parasite GPIs.
Stimulation of Macrophages with P. falciparum GPIs
Freshly harvested macrophages, prepared as described above, were seeded into 96-well microtiter plates (2.5 × 104 cells/well) and cultured overnight in DMEM complete medium (19). The cells were treated with GPIs coated on gold particles. Cells treated with uncoated gold particles were used as controls. For inhibition studies, inhibitors were added to the culture medium 1 h prior to the addition of GPIs. Culture supernatants were collected at the indicated time points and stored at −70 °C for cytokine measurements.
Analysis of Signaling Proteins
Macrophages in 24-well microtiter plates (0.5 × 106 cells/well) were cultured overnight in DMEM complete medium but containing 0.5% FBS. The cells were stimulated with 200 nm GPIs in DMEM complete medium. At various time points, the supernatant was removed, and the cells were washed with ice-cold PBS and lysed with ice-cold RIPA buffer (50 mm Tris-HCl, 150 mm NaCl, pH 7.2, containing 1% sodium deoxycholate, 0.1% SDS, and 1% Triton X-100). Lysates were centrifuged at 4 °C, and supernatant was used for Western blotting.
Analysis of Gene Transcription by RT-PCR
Macrophages in 24-well plates (1 × 106 cell/well) were cultured overnight in DMEM, 0.5% FBS and then stimulated with 200 nm GPIs in DMEM, 10% FBS. At various time points, culture supernatants were removed and cells washed with ice-cold PBS, pH 7.2. Cells were lysed in TRIzol, total RNA isolated, and used for cDNA preparation for RT-PCR.
Total RNA samples from wild type and gene knock-out mice were reverse-transcribed using random oligonucleotide hexamers. Aliquots of cDNA samples were analyzed for TNF-α, IL-12p40, and β-actin (endogenous control) genes by PCR. The primers used were as follows: TNF-α, forward 5′-CGG TGC CTA TGT CTC AGC CT-3′, reverse 5′-TTG GGC AGA TTG ACC TCA GC-3′; IL-12p40 (25), forward 5′-CAG AAG CTA ACC ATC TCC TGG TTT G-3′, Reverse 5′-CCG GAG TAA TTT GGT GCT TCA CAC-3′; and β-actin, forward 5′-ACC CTA AGG CCA ACC GTG AA-3′, reverse 5′-CCG CTC GTT GCC AAT AGT GA-3′. The conditions used for PCR were as follows: for TNF-α and β-actin, 30 cycles of 94 °C denaturation for 30 s, 50 °C annealing for 30 s, and 72 °C extension for 60 s; for IL-12p40, 40 cycles of 94 °C denaturation for 30 s, 60 °C annealing for 30 s, and 72 °C extension for 60 s.
Real time RT-PCR was performed for 50 cycles using SYBR green master mix kit using 7900 HT real time PCR equipment (Applied Biosystems). The transcript levels were quantified using ΔΔCt method, and values were normalized with respect to β-actin transcript as an internal control. The primers used were same as above used for RT-PCR.
Analysis of mRNA Stability
MK2−/− macrophages and WT macrophages pretreated with SB203580 and untreated WT macrophages were stimulated with 200 nm GPIs for 4 h, and then 10 μg/ml of actinomycin D was added. Thereafter, cells were harvested at different time points, and total RNA was extracted with TRIzol reagent; TNF-α and IL-12p40 mRNA levels were analyzed by RT-PCR and quantitative RT-PCR as described above.
Extraction of Cell Nuclear Proteins and Analysis of NF-κB
The MK2−/−, SB203580-pretreated and control macrophages (5–10 × 106 cells) in T-25 culture flasks were stimulated with 200 nm GPIs. At various time points, cells were harvested by scraping and washed with PBS. The nuclear and cytoplasm fractions were isolated using NE-PER nuclear and cytoplasm extraction reagents (NER and CER) according to the manufacturer's instructions. Briefly, the cell suspensions were centrifuged; CERI solution was added to cell pellets and vortexed, and then CERII solution was added and vortexed. After centrifugation, the supernatant cytoplasmic extracts were removed. Nuclear pellets were suspended in 50 μl of NER solution, vortexed every 10 min, incubated in an ice-bath for 40 min, and supernatant containing nuclear extracts collected. The nuclear extracts were analyzed by Western blotting using antibodies against p50, p65 (Rel-A), c-Rel, c-Maf, and IκBζ.
Cytokine Measurement
TNF-α and IL-12p40 levels in the culture supernatants of macrophages stimulated with GPIs were determined by sandwich ELISA with HRP-conjugated streptavidin and TMB color reagent using 96-well plastic plates, and Duoset ELISA development kit (R & D Systems). After sufficient color development, the reaction stopped by the addition of 1 m sulfuric acid and absorbance at 450 nm was measured with SpectraMax Plus384 plate reader (Molecular Devices). The cytokine concentrations were calculated with reference to standard graphs.
Statistical Analysis
The cytokine data were plotted as mean values ± S.E. Statistical analysis of results was performed by either Student's t test (Fig. 1) or one-way analysis of variance (Fig. 2) using Prism GraphPad 3.0. p values of <0.05 were considered statistically significant.
FIGURE 1.
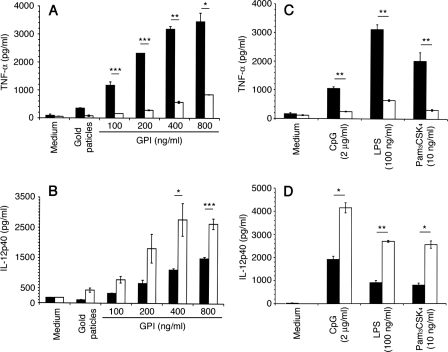
Production of TNF-α and IL-12 by macrophages stimulated with P. falciparum GPIs. WT and MK2−/− macrophages (∼2.5 × 104 cells/well) were plated in 96-well plates. After overnight culturing, the cells were stimulated with P. falciparum GPIs or with control ligands at the indicated concentrations. After 48 h, the culture supernatants were collected and assayed for TNF-α (A and C) and IL-12 (B and D) by ELISA. Black bars, WT cells; open bars, MK2−/− cells. The experiments were performed three times each in duplicate. Means values ± S.E. are plotted. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 2.
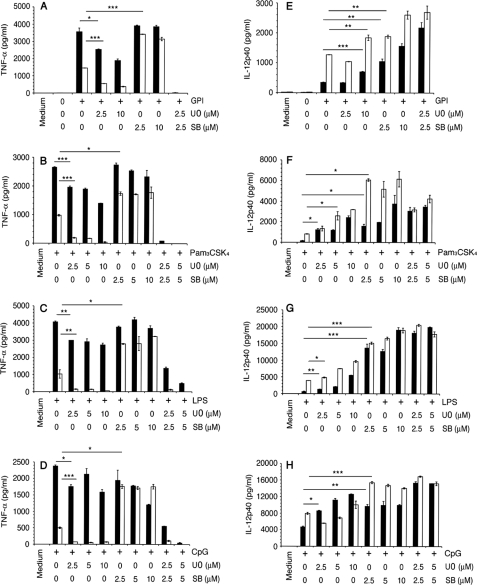
Effect of ERK and p38 MAPK inhibition on the production of TNF-α and IL-12 by macrophages stimulated with P. falciparum GPIs and other TLR ligands. WT and MK2−/− macrophages were plated in 96-well plates (∼2.5 × 104 cells/well). After overnight culturing, the cells were treated with U0126, SB203580, or U0126 plus SB203580 for 1 h and then stimulated with 200 nm GPIs (A and E), 10 ng/ml Pam3CSK4 (B and F), 100 ng/ml LPS (C and G), or 2 μg/ml CpG (D and H). Cells not treated with inhibitors were used as controls. After stimulation for 48 h, TNF-α and IL-12 levels in the culture supernatants were measured by ELISA. Black bars, wild type cells; open bars, MK2−/− cells. For each panel, the ligand used for cell stimulation is indicated. The experiments were performed three times each in duplicate. Means values ± S.E. are plotted. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Western Blotting
The macrophage lysates or nuclear extracts obtained as outlined above were made to 1× with respect to the SDS-PAGE sample buffer containing 2-mercaptoethanol, boiled for 5 min, and electrophoresed on 8 or 10% SDS-polyacrylamide gels. The protein bands in the gels were transferred onto nitrocellulose membranes. The membranes were blocked with 5% (w/v) nonfat dry milk in PBS, pH 7.4, containing 0.1% Tween 20, at room temperature for 1 h, and then incubated with either anti-peptide or phospho-specific antibodies in the above buffer containing 2.5% nonfat dry milk. After washing, the membranes were incubated with HRP-conjugated anti-mouse or anti-rabbit secondary antibodies, treated with chemiluminescent substrate (LumiGLO from Kirkegaard & Perry Laboratories), and exposed to x-ray films.
The primary and secondary antibodies bound to signaling proteins in the membranes were stripped off by incubating at 50 °C for 45 min in 63 mm Tris-HCl, 100 mm 2-mercaptoethanol, 2% SDS, pH 6.8. The blots were treated with either β-actin or nonphosphopeptide-specific antibodies, and the bound antibodies were detected as above.
MK2 in Vitro Kinase Assay
The MK2 in vitro kinase assay was performed using immunoprecipitation kinase assay kit according to the manufacturer's instructions (Upstate cell signaling solutions). Briefly, cells pretreated with inhibitors were stimulated with GPIs and lysed in lysis buffer. The cell lysates were incubated at 4 °C for 1.5 h with rabbit anti-mouse MK2 antibodies immobilized on agarose by gentle rotation. The immunoprecipitates were washed with lysis buffer followed by kinase buffer and incubated with HSP27 and magnesium/ATP mixture at 30 °C for 45 min with vigorous shaking. The reaction mixture was analyzed by Western blotting using mouse anti-phospho-human HSP27 (Ser-78) antibody.
EMSA
WT and MK2−/− macrophages (2 × 106 cells) were plated into 24-well plates and cultured overnight in DMEM containing 10% FBS. Cells in some wells were treated with SB203580 and U0126 for 1 h. Both untreated and inhibitor-treated cells were stimulated with 200 nm GPI, at the indicated time points harvested using trypsin/EDTA in incomplete medium, and washed with cold PBS. The nuclear and cytoplasmic fractions were isolated using NE-PER nuclear and cytoplasmic extraction reagents as described above. To 2 μl of nuclear extracts were added 20 mm HEPES, pH 7.5, 60 mm KCl, 4% Ficoll 400, 2 mm dithiothreitol, 20 μg of purified bovine serum albumin, 2 μg of poly(dI·dC), and 50 fmol of 5′-end biotin-labeled DNA probes in 20 μl at room temperature for 20 min. The reaction mixtures were electophoresed on 6% polyacrylamide gel in 0.5× TBE at 100 V for 1 h. The protein-DNA complexes in gels were transferred to nylon membranes, cross-linked with UV light at 120 mJ/cm2 for 60 s, and the biotinylated protein-DNA bands detected with HRP-conjugated streptavidin using chemiluminescence nucleic acid detection reagents in the EMSA kit according to the manufacturer's instructions. The DNA probes used were as follows: TNF-α promoter κB-binding site AAA CAG GGG GCT TTC CCT CCT CAA; IL-12p40 κB-binding site, CTT CTT AAA ATT CCC CCA GA; and GA-12, CCT CGT TAT TGA TAC ACA CAC AGA GA. Complementary oligonucleotide mixtures were denatured at 90 °C for 1 min, annealed by slow cooling, and then incubated at melting temperatures for 30 min. Annealed DNA probes were purified with QIAquick® nucleotide removal kit from Qiagen. EMSA was also performed after incubating nuclear extracts with anti-p50, anti-p65, and anti-c-Rel antibodies for 30 min on ice prior to the addition of biotin-labeled IL-12p40 DNA probe.
RESULTS
Macrophages stimulated with purified P. falciparum GPIs produce elevated levels of TNF-α, IL-12, IL-6, and nitric oxide through TLR2/TLR1-mediated activation of ERK, p38, JNK, and NF-κB signaling pathways (19, 20). Studies have shown that MK2, which is described to be downstream of p38, plays an important role in regulation of microbial ligand-induced production of cytokines (21). To determine the role of MK2 in P. falciparum GPI-induced production of proinflammatory mediators, bone marrow-derived macrophages from MK2−/− and WT mice were stimulated with GPIs. In parallel, cells were also stimulated with other TLR ligands, LPS (TLR4), Pam3CSK4 (TLR2-TLR1), and CpG (TLR9). All experiments were performed using cells not primed with IFN-γ to avoid the effects of IFN-γ receptor-mediated signaling pathway. ELISA analysis of cell supernatants showed that both WT and MK2−/− macrophages stimulated with GPIs produced TNF-α and IL-12 in a dose-dependent manner (Fig. 1, A and B). However, the production of TNF-α by MK2−/− macrophages was markedly (reduced by ∼80%) lower compared with WT macrophages. In contrast, MK2−/− macrophages produced 2–3-fold higher levels of IL-12 than WT macrophages. Similar results were obtained for macrophages stimulated with Pam3CSK4, LPS, or CpG; in all cases, markedly lower levels of TNF-α and significantly increased levels of IL-12 were produced by MK2−/− macrophages when compared with WT cells (Fig. 1, C and D).
Next, we studied the effect of U0126 (inhibitor of ERK activation) and SB203580 (inhibitor of p38 MAPK activity) on the production of TNF-α and IL-12 in GPI-stimulated WT and MK2−/− macrophages (Fig. 2, A and E). In parallel, we also analyzed the production of TNF-α and IL-12 by cells pretreated with the ERK and p38 MAPK inhibitors and stimulated with the other TLR ligands, LPS, Pam3CSK4, and CpG (Fig. 2, B–D and F–H). Inhibition of ERK activation resulted in ∼50% decrease in TNF-α production by WT macrophages stimulated with GPI or Pam3CSK4 (Fig. 2, A and B). In WT cells stimulated with LPS or CpG, inhibition of ERK activation caused only an ∼20% reduction in TNF-α production (Fig. 2, C and D). In MK2−/− macrophages, however, inhibition of ERK activation could abolish the GPI-induced TNF-α production.
In contrast to the effect of blocking ERK activation, inhibition of p38 activity resulted in either only a marginal decrease or did not affect TNF-α production by WT macrophages stimulated with GPIs, Pam3CSK4, or LPS (Fig. 2, A–C); in the case of CpG, however, substantial decrease was observed (Fig. 2D). Inhibition of p38 activity resulted in the TNF-α production by MK2−/− macrophages at levels similar to that by WT macrophages in response to stimulation with GPIs or the other TLR ligands (Fig. 2, A–D). Inhibition of both ERK activation and p38 activity completely abolished the TNF-α production in both WT and MK2−/− macrophages. The production of IL-12 was substantially increased in both WT and MK2−/− macrophages pretreated with U0126 or SB203580 and simulated with GPIs or other TLR ligands (Fig. 2, E–H). Interestingly, inhibition of both ERK and p38 also resulted in increased production of IL-12 by either cell types stimulated with GPIs or other ligands.
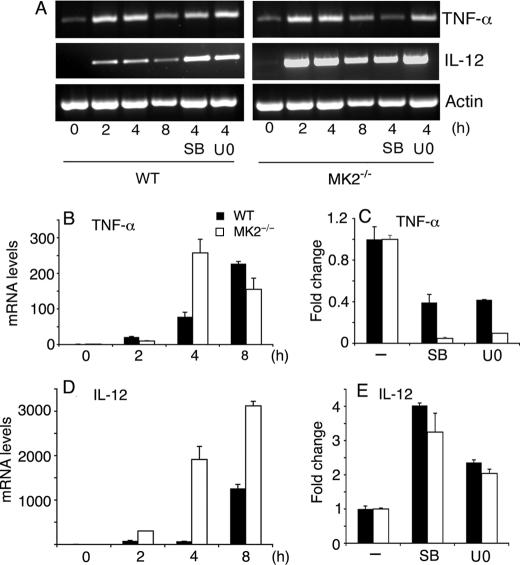
To examine whether the observed altered production of TNF-α and IL-12 by MK2−/− macrophages and in ERK- or p38-inhibited macrophages stimulated with GPIs is because of changes in mRNA levels, we analyzed the kinetics of TNF-α and IL-12 mRNA synthesis. WT and MK2−/− macrophages, untreated or treated with either U0126 or SB203580, were stimulated with GPIs, and steady state mRNA levels at 2, 4, and 8 h after stimulation were determined by RT-PCR and by real time RT-PCR (Fig. 3). The levels of TNF-α mRNA in WT macrophages stimulated with GPIs steadily increased until 8 h, whereas in MK2−/− macrophages, the TNF-α transcript was maximal at 4 h, with a decreased level at 8 h (Fig. 3B). These data could be explained by different kinetics of TNF-α mRNA synthesis or, more likely, by the decreased TNF-α mRNA stability in MK2−/− cells. Inhibition of either ERK or p38 resulted in, respectively, 0.6- and ∼0.9-fold decrease in the level of TNF-α mRNA in WT and MK2−/− macrophages stimulated with GPIs (Fig. 3C).
FIGURE 3.
Analysis of TNF-α and IL-12 mRNA levels in macrophages stimulated with P. falciparum GPIs. WT and MK2−/− macrophages were plated in 24-well microtiter plates (∼1 × 106 cells/well). After overnight culturing, the cells were stimulated with 200 nm P. falciparum GPIs for the indicated time periods. The cells pretreated for 1 h with either 10 μm of U0126 or 10 μm of SB203580 were also stimulated with 200 nm GPIs for 4 h. The cells were harvested; total RNA was isolated, and mRNA levels of TNF-α and IL-12 were analyzed by RT-PCR (A) and by real time RT-PCR (B–E). The mRNA levels in B–E were normalized with respect to endogenous β-actin mRNA. C and E represent relative mRNA levels of cells either untreated or inhibitor pretreated cells stimulated with GPIs for 4 h. The experiments were performed two times each in triplicate. Mean values ± S.E. are plotted. The TNF-α and IL-12 mRNA synthesis was also analyzed by quantitative RT-PCR in WT and MK2−/− macrophages stimulated with 10 ng/ml Pam3CSK4 (data not shown).
In the case of IL-12, mRNA accumulated with faster kinetics and persisted at much higher levels in MK2−/− macrophages than in WT cells (Fig. 3D). Inhibition of ERK activation increased the steady state IL-12 mRNA level by ∼2.5- and ∼2.2-fold in WT and MK2−/− macrophages, respectively, whereas the inhibition of p38 activity caused ∼4- and ∼3.5-fold increase in WT and MK2−/− macrophages, respectively (Fig. 3E). We also determined the mRNA levels in WT and MK2−/− macrophages stimulated with Pam3CSK4. Interestingly, with Pam3CSK4, the TNF-α and IL-12 of mRNA accumulated much faster in both MK2−/− and WT macrophages as compared with stimulation with GPIs (data not shown). For TNF-α, the maximal level of mRNA was observed at 2 h in both types of macrophages. For IL-12, the maximal level of mRNA was at 2–4 h in both cell types. Furthermore, as in the case of stimulation with GPIs, the mRNA levels were substantially higher in Pam3CSK4-stimulated MK2−/− macrophages than in WT macrophages (data not shown).
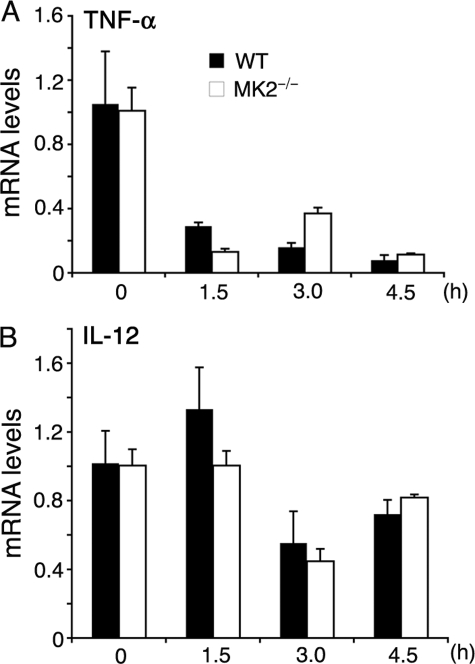
MK2 has been shown to regulate LPS-dependent TNF-α production by controlling mRNA stability through phosphorylation of TTP, an inducible protein that binds to AU-rich elements (AREs) in the 3′-untranslated region of TNF-α mRNA, causing mRNA degradation only when nonphosphorylated (21, 26, 27). The IL-12 mRNA, however, lacks AREs in the 3′-untranslated region (28) and does not appear to be destabilized by TTP. To determine whether this situation is similar in GPI-stimulated macrophages, we measured TNF-α and IL-12 mRNA levels by real time RT-PCR at various time points after stimulating cells with GPIs and stopping transcription with actinomycin D. TNF-α mRNA degraded rapidly in both WT and MK2−/− macrophages (Fig. 4A). However, the initial stability of TNF-α mRNA (t½ = 48 min) was considerably lower in MK2−/− macrophages than that (t½ = 70 min) in WT macrophages. These t½ values closely resemble 51 and 76 min, the values previously reported for mRNA degradation in LPS-stimulated MK2−/− and WT macrophages, respectively (26). Inhibition of either ERK activation or p38 activity caused little or no decrease in TNF-α mRNA stability in GPI-stimulated WT macrophages (data not shown). IL-12 mRNA was relatively stable and comparable in WT and MK2−/− macrophages (t½ = 5.22 and 5.16 h, respectively; Fig. 4B). Similarly, MK2−/− macrophages stimulated with Pam3CSK4 showed very low stability of TNF-α mRNA and relatively stable IL-12 mRNA compared with WT cells (supplemental Fig. S1).
FIGURE 4.
Analysis of TNF-α and IL-12 mRNA stability in P. falciparum GPI-stimulated macrophages. WT and MK2−/− macrophages, plated in 24-well microtiter plates (∼1 × 106 cells/well), were stimulated with 200 nm of P. falciparum GPIs. After 4 h, actinomycin D (10 μg/ml) was added to stop transcription. At the indicated time points after the addition of actinomycin D, cells were harvested, and total RNA was isolated. The levels of TNF-α (A) and IL-12 (B) mRNA were quantified by real time RT-PCR. The mRNA levels were normalized with respect to endogenous β-actin mRNA. The experiments were performed two times each time in triplicate. Mean values ± S.E. are plotted. The TNF-α and IL-12 mRNA stability was also analyzed by quantitative RT-PCR in WT and MK2−/− macrophages stimulated with 10 ng/ml of Pam3CSK4 (supplemental Fig. S1).
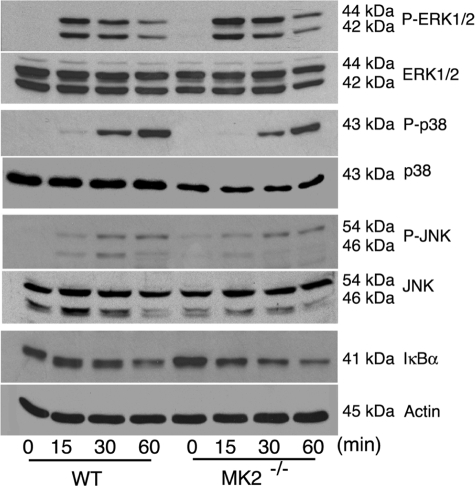
MK2 has been reported to interact with p38 and stabilize the protein (29, 30). We examined whether MK2 deficiency alters the level of activation of p38 or other MAPK and/or NF-κB signaling pathways in response to GPI stimulation. Macrophages from MK2−/− mice and WT mice were stimulated with P. falciparum GPIs, and cell lysates were analyzed for activation of MAPK and NF-κB by Western blotting. The activation of all three MAPKs (ERK1/2, p38, and JNK) as well as the activation of the NF-κB pathway was normal compared with WT cells (Fig. 5). However, the levels of p38 recognized by both anti-p38 and anti-phospho-p38 antibodies were noticeably lower in MK2−/− macrophages than in WT cells. These observations are consistent with the results of previous studies (29).
FIGURE 5.
Activation of MAPK and NF-κB signaling pathways in macrophages stimulated with P. falciparum GPIs. WT and MK2−/− macrophages were plated in 24-well plates (∼5 × 105 cells/well). After overnight culturing, the cells were stimulated with 100 nm GPIs. At the indicated time points, the cells were lysed with 30 μl of RIPA buffer and lysates electrophoresed on 10% SDS-polyacrylamide gels. The proteins bands on gels were transferred onto nitrocellulose membranes, and membranes were treated with anti-peptide antibodies specific to phosphorylated (P) and nonphosphorylated forms of ERK, p38, and JNK MAPKs. The membranes were also separately treated with anti-IκBα and anti-β-actin antibodies. The bound antibodies were detected with HRP-conjugated secondary antibodies (1:2000 dilution) and chemiluminescent substrate system.
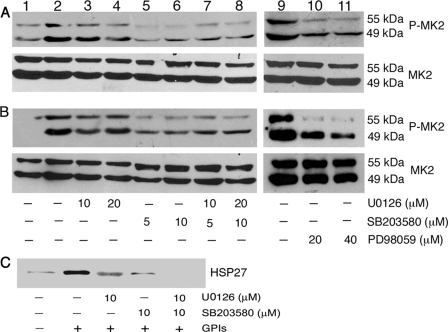
The differential regulation of GPI-induced TNF-α production by p38 and MK2 (see Figs. 1 and 2) suggested the possibility of MK2 being targeted by another kinase, contributing to the production of TNF-α. Although MK2 is thought to be a downstream target of p38, some studies have reported that both p38 and ERK can activate MK2 in human neutrophils and mouse macrophages in response to inflammatory stimulation (31, 32), and it remains unclear whether ERK can also activate MK2 in macrophages in response to microbial stimuli. We analyzed the activation of MK2 by measuring its phosphorylation in GPI-stimulated macrophages pretreated with SB203580 and/or U0126 or PD98059 (another inhibitor of ERK activation). As shown in Fig. 6A, although inhibition of p38 activity by SB203580 substantially decreased the phosphorylation of MK2 in GPI-stimulated macrophages, pretreatment of cells with ERK inhibitor, either with U0126 or PD98059, could also appreciably inhibit MK2 phosphorylation. To determine whether both ERK and p38 can activate MK2 in response to other microbial ligands, we analyzed the effect of ERK and p38 inhibitors on the activation of MK2 in WT macrophages stimulated with Pam3CSK4 and LPS. As in the case of GPIs, both U0126 and SB203580 could inhibit the phosphorylation of MK2 in WT cells stimulated with Pam3CSK4 and LPS (Fig. 6B and data not shown). To confirm the observed involvement of both ERK and p38 in the activation of MK2, we further measured the effect of inhibitors on MK2 activity by determining the phosphorylation of MK2-specific substrate HSP27 in GPI-stimulated WT macrophages pretreated with either U0126 or SB203580. In each case, phosphorylation of HSP27 was significantly inhibited (Fig. 6C). Treatment of macrophages with both U0126 and SB203580 completely inhibited the MK2-dependent phosphorylation of HSP27. These data suggest that both p38 and ERK pathways regulate the activation of MK2 in macrophages stimulated with GPIs and other TLR ligands.
FIGURE 6.
Analysis of MK2 activation/activity in P. falciparum GPI-stimulated macrophages. WT macrophages were plated in 24-well plates (∼5 × 105 cells/well). After overnight culturing, the cells were treated with U0126, SB203580, and U0126 plus SB203580 or PD98059 as indicated for 1 h and stimulated with 200 nm GPIs (A, lanes 2–11), 10 ng/ml Pam3CSK4 (B, lanes 2–11), or 100 ng/ml LPS (data not shown) for 1 h. Lane 1, unstimulated control cells. The cells were lysed with 30 μl of RIPA buffer and lysates electrophoresed on 8% SDS-polyacrylamide gels. The proteins bands on gels were transferred onto nitrocellulose membranes, and membranes were treated with anti-peptide antibodies specific to phosphorylated (P) and nonphosphorylated forms of MK2. The nonphosphorylated MK2 bands served as loading control. The bound antibodies were detected with HRP-conjugated secondary antibodies (1:2000 dilution) and chemiluminescent substrate system. C, WT macrophages (∼5 × 106/flask) were treated with U0126, SB203580, or U0126 plus 10 μm SB203580 for 1 h. Untreated cells were used as controls. The cells were stimulated with 100 nm GPIs for another 1 h, lysed with 500 μl of lysis buffer, and aliquots of lysates were analyzed for MK2 kinase activity with HSP27 as the substrate using MK2 immunoprecipitation kinase assay kit according to the manufacturer's procedure. Phosphorylated HSP27 was analyzed by Western blotting using HSP-Ser-78 phospho-specific antibody (1:1000 dilution).
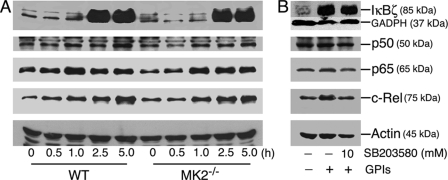
Because IL-12 is a key proinflammatory cytokine thought to be involved in controlling malaria infection and in disease pathogenesis (9), we studied mechanisms that underlie the observed marked increase in IL-12 production by MK2−/− macrophages stimulated with GPIs. NF-κB transcriptional factors are important for the expression of many genes, including TNF-α, and IL-12. Recently, IκBζ has been reported to be critical for the expression of a subset of genes, including IL-12 (33, 34). We analyzed the pattern of activation of various NF-κB transcriptional factors in macrophages treated with GPIs by Western blotting of nuclear extracts prepared from GPI-stimulated cells. The activation and nuclear translocation of p50, p65, and c-Rel NF-κB proteins and nuclear induction of IκBζ were essentially comparable in MK2−/− and WT macrophages (Fig. 7A). Inhibition of p38 with SB203580 prior to stimulation with GPI caused either marginal or no effect on the induction of IκBζ or nuclear translocation of p50, p65, and c-Rel NF-κB proteins (Fig. 7B).
FIGURE 7.
Induction and nuclear translocation of transcription factors in P. falciparum GPI-stimulated macrophages. A, WT and MK2−/− macrophages were plated in T-25 flasks (5–10 × 106 cells/flask). After overnight culturing, the cells were stimulated with 100 nm P. falciparum GPIs. At the indicated time points, the cells were harvested, and nuclear extracts (50 μl total volume) were prepared as described under “Experimental Procedures.” The extracts (20 μl) were electrophoresed on 10% SDS-polyacrylamide gels, and protein bands were transferred onto nitrocellulose membranes and treated with 1 μg/ml anti-IκBζ, anti-p50, anti-p65, anti-c-Rel, or anti-β-actin antibodies. The bound antibodies were detected with HRP-conjugated secondary antibodies (1:2000 dilution) and chemiluminescent substrate system. B, WT macrophages (5–10 × 106 cells/flasks) were treated with 10 μm SB203580 for 1 h. Untreated cells were used as controls. The cells were stimulated with 100 nm P. falciparum GPIs for 3 h and harvested, and nuclear extracts were collected. Aliquots of lysates were analyzed by Western blotting as outlined in A. The experiments were repeated three times, and the results of representative experiments are shown.
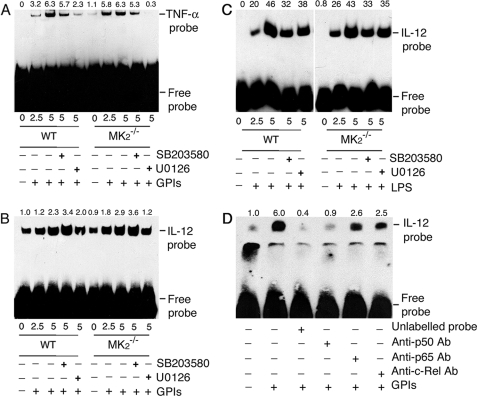
To analyze whether increased NF-κB binding to the gene promoter is responsible for the enhanced mRNA synthesis by MK2−/− macrophages stimulated with GPIs compared with WT cells, we performed EMSA using TNF-α and IL-12p40 promoter-specific DNA probes for NF-κB binding. In both WT and MK2−/− macrophages, GPI stimulation resulted in increased NF-κB binding to the TNF-α and IL-12 DNA probes (Fig. 8, A and B). NF-κB binding to the TNF-α promoter in MK2−/− and WT macrophages was comparable (Fig. 8A). Inhibition of p38 activity and ERK activation weakened the binding in both WT and MK2−/− macrophages, and the results agree with the TNF-α mRNA and protein levels in these cells. The level of NF-κB binding to the IL-12 promoter is slightly higher in MK2−/− macrophages (Fig. 8B). Inhibition of p38 activity caused slightly increased DNA probe binding to NF-κB in both WT and MK2−/− macrophages, whereas inhibition of ERK activation resulted in a noticeable decrease, particularly in MK2−/− macrophages (Fig. 8B). LPS could induce comparable levels of NF-κB binding to the IL-12 promoter probes in both WT and MK2−/− macrophages, and binding was decreased by inhibition of p38 activity or ERK activation (Fig. 8C). The binding of the IL-12 DNA probe to NF-κBs was analyzed by antibody inhibition. Treatment of nuclear extracts from GPI-stimulated macrophages with antibody against p50 prior to the addition of IL-12-specific κB DNA probe resulted in marked inhibition of NF-κB binding to the promoter (Fig. 8D). This is consistent with the requirement of NF-κB p50 homodimer forming a complex with IκBζ (35). The anti-p65 and anti-c-Rel antibodies could also inhibit DNA probe binding to NF-κBs to appreciable extents, demonstrating the binding of these transcription factors to IL-12 gene promoters in response to GPI stimulation. However, the binding of NF-κB c-Rel was higher than that of p65 NF-κB (Fig. 8D).
FIGURE 8.
EMSA analysis of NF-κB binding to TNF-α and IL-12p40 gene promoter in macrophages stimulated with GPIs and LPS. WT and MK2−/− macrophages (∼2 × 106 cells/flask) were stimulated with 200 nm GPIs for the indicated time periods. Both types of macrophages, pretreated with either 10 μm U0126 or 10 μm SB203580 for 1 h, were also stimulated with either 200 nm GPIs or 100 μg/ml LPS. Unstimulated cells were used as controls. The cells were harvested and nuclear extracts prepared (50 μl total volume), and 2 μl of each extract was incubated with biotin-labeled TNF-α κB DNA probe (A) or IL-12p40 κB DNA probe (B, GPI-stimulated cells; C, LPS-stimulated cells) in binding buffer (20 μl) as described under “Experimental Procedure.” The reaction mixtures were electrophoresed on 6% polyacrylamide gels, and protein bands were transferred onto nylon membrane and detected with chemiluminescent nucleic acid detection reagent. D, NF-κB proteins that bound to IL-12p40 κB DNA probe were identified by inhibition with specific antibodies. The nuclear extracts from GPI-stimulated macrophages were incubated with 2 μg each of anti-p50, anti-p65, or anti-c-Rel antibody or 5 pmol (100-fold excess over labeled probe) of unlabeled κB probe prior to the addition of biotin-labeled IL-12p40 κB probe. The samples were analyzed as above. The relative band intensities of specific NF-κB-DNA complex, determined by scanning of x-ray films using Bio-Rad GS-800 densitometer, are shown on the top of each electrophoretogram.
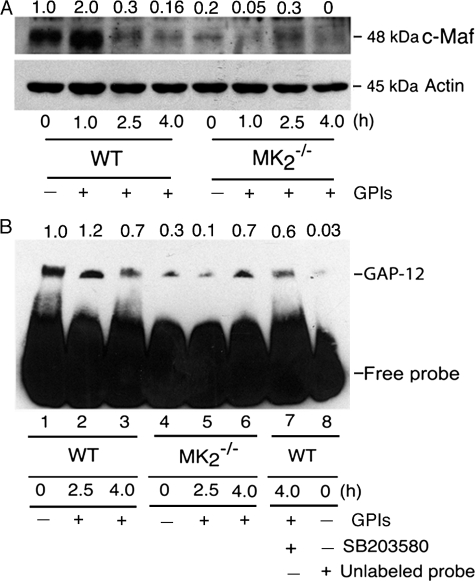
Because NF-κB binding to the IL-12 promoter was only modestly enhanced in MK2−/− macrophages (Fig. 8B), we next analyzed the expression of c-Maf, a repressor factor that regulates IL-12p40 gene transcription, that is constitutively expressed in human monocytes and mouse macrophages (36, 37). Western blot analysis of nuclear proteins showed that c-Maf is present at an appreciable level in the nuclei of unstimulated WT macrophages (Fig. 9A). Upon stimulation with GPIs, the level of c-Maf rapidly increased at 1 h after stimulation by 2-fold, and thereafter drastically decreased. At 4 h after simulation with GPIs, the level of c-Maf was barely detectable in the nucleus of WT cells (Fig. 9A). In contrast, the level of c-Maf was markedly lower in unstimulated and in GPI-stimulated MK2−/− macrophages. Upon stimulation of MK2−/− cells with GPIs, the level of c-Maf in the nucleus was marginally increased with greatly delayed kinetics (maximal at 2.5 h). This is in contrast to WT cells, in which c-Maf level is increased by 2-fold at 1 h with a marked decrease at 2.5 h. Overall, the level of c-Maf was ∼4.5-fold less in MK2−/− cells (Fig. 9A). The markedly low level of c-Maf and its delayed increase in the nucleus of MK2−/− macrophages compared with WT cells correlates with the higher levels of IL-12 mRNA seen in MK2−/− cells.
FIGURE 9.
Analysis of c-Maf expression and GAP-12 binding to IL-12p40 gene promoter in P. falciparum GPI-stimulated macrophages. A, WT and MK2−/− macrophages (5–10 × 106 cells/flask) were stimulated with 200 nm GPIs for 1, 2.5, and 4 h and harvested; nuclear extracts (50 μl total volume) were prepared, and 20-μl aliquots were analyzed by Western blotting using anti-c-Maf antibody (1 μg/ml). The nuclear extracts were also analyzed for β-actin expression as loading control using anti-β-actin antibody. B, WT and MK2−/− macrophages were stimulated with 100 nm GPIs for 2.5 and 4 h, and nuclear extracts were prepared as outlined in Fig. 8. Aliquots of nuclear extracts (2 μl) were incubated with biotin-labeled GAP-12 DNA probe in binding buffer (20 μl). The reaction mixtures were electrophoresed on 6% polyacrylamide gel and transferred onto nylon membranes, and GAP-12 protein bound to DNA probe was detected using HRP-conjugated streptavidin and chemiluminescent detection reagent. The numbers on the top of electrophoretograms indicate the relative band intensities of nuclear C-Maf (A) and GAP-12-DNA complex (B) determined by scanning of x-ray films using Bio-Rad GS-800 densitometer.
GAP-12 has been shown to be another specific repressor of IL-12p40 gene transcription. The binding of nuclear protein, GAP-12, to the GATA sequence (GA-12) in the IL-12p40 promoter has been shown to repress IL-12p40 promoter activity (37, 38). To determine the role of GAP-12 in GPI-induced IL-12 regulation, EMSA was performed using a DNA probe containing the GATA sequence (GA-12) of the IL-12p40 promoter. Unstimulated WT macrophages showed high levels of constitutive GAP-12 binding to the IL-12p40 promoter (Fig. 9B). Upon stimulation with GPIs, the binding of GAP-12 was increased marginally and decreased substantially at 4 h in WT macrophages. Inhibition with unlabeled probe almost completely abolished the binding of GAP-12 (compare lane 8 with lane 1 in Fig. 9B), demonstrating the specificity of GAP-12 binding to the IL-12 promoter. In agreement with the increased level of IL-12 production and IL-12 gene transcription (see Figs. 2 and 3), inhibition of p38 activity by SB203580 prior to stimulation with GPI showed only a marginal decrease in the level of GAP-12 binding (compare lane 7 with lane 3 in Fig. 9B). In MK2−/− macrophages, both the constitutive binding and binding at 2.5 and 4 h after stimulation with GPIs was lower than in WT macrophages; the overall binding of GAP-12 to the IL-12 gene promoter was decreased by 2.6-fold. Interestingly, the binding of GAP-12 occurred with delayed kinetics in MK2−/− cells stimulated with GPIs than in WT macrophages (compare lanes 5 and 6 with lanes 2 and 3 in Fig. 9B). These results also agree with the faster kinetics and higher levels of mRNA in MK2−/− macrophages (see Fig. 3). Thus, the results indicate that increased expression of IL-12 by MK2−/− macrophages is in part due to low level of GAP-12 binding to the IL-12p40 promoter.
DISCUSSION
MK2, a member of downstream MAPK-activated protein kinases, is widely believed to be under the exclusive control of p38 MAPK. Although MK2 is one of several kinases, including MK3, MK5, MNK1, MNK2, MSK1, MSK2 and CK2 that are downstream of p38, MK2 is thought to be the key enzyme involved in the regulation of cytokine biosynthesis (21, 29, 39). MK2-deficient macrophages and spleen cells have been reported to produce markedly decreased levels of proinflammatory cytokines, such as TNF-α, IL-6, and IFN-γ (26, 27, 39–41). In contrast, IL-12 has been reported to produce at increased levels in response to LPS (39). However, the underlying mechanisms have not been studied. In this study, we investigated the role of MK2 in P. falciparum GPI-induced production of TNF-α and IL-12, two of the key proinflammatory cytokines that are believed to be involved in malaria pathogenesis. We also studied the mechanisms by which MK2 up-regulates IL-12 production in macrophages.
The results presented here show that MK2 plays an important role in the GPI-induced production of TNF-α by macrophages. Overall, the level of TNF-α produced by MK2-deficient macrophages in response to GPI stimuli is about 80% lower than that produced by control WT macrophages. A similar marked decrease in TNF-α production was evident with other ligands, Pam3CSK4, LPS, and CpG, which were studied as controls. The decrease in production of TNF-α by MK2−/− macrophages is not because of alterations in the activation of ERK1/2, p38, and JNK MAPK pathways and NF-κB signaling cascades. This conclusion is supported by the observation that all of these signaling pathways are activated in MK2−/− cells stimulated with GPIs and that the level of their activation is comparable with that of WT macrophages (see Fig. 5). The compromised production of TNF-α by MK2-deficient macrophages was also not because of transcriptional defects because MK2−/− macrophages could actually synthesize higher levels of steady state mRNA with faster kinetics than WT cells (see Fig. 3). However, the TNF-α mRNA stability in MK2−/− macrophages stimulated with GPI or another TLR2/TLR1-specific ligand, Pam3CSK4, was significantly lower compared with WT cells. Previously, it has been demonstrated that spleen cells and macrophages deficient in MK2 exhibit markedly decreased levels (80–90%) of TNF-α production in response to LPS stimuli (27, 29, 39, 40). This was found to be due to a substantial decrease in mRNA stability and translation efficiency in MK2−/− cells because of the binding of TTP, a signaling molecule downstream of MK2, to the ARE in the 3′-untranslated region of TNF-α mRNA, thus destabilizing mRNA (21, 26, 27, 42, 43; see below for further details). Consistent with this finding, the TNF-α mRNA level and stability and protein processing and secretion were found to be independent of MK2 when ARE of the TNF-α gene was deleted. Thus, our results when taken together with previously demonstrated mechanisms of TNF-α regulation strongly suggest that MK2 regulates GPI-induced TNF-α production mainly by ARE-dependent mRNA stability and translational control.
We found that inhibition of p38 activity caused only a slight decrease or had no effect on GPI-induced production of TNF-α by WT macrophages, although it resulted in a 2–3-fold increase in TNF-α production by MK2−/− cells (see Fig. 2). This is despite substantial reduction in mRNA levels and decreased NF-κB binding to the gene promoter (see Figs. 3 and 8). The observed increase in TNF-α by GPI-stimulated MK2−/− macrophages agrees with the previously reported marked reduction in TTP expression upon inhibition of p38 activity in LPS-stimulated MK2−/− macrophages (27). TTP is a downstream target of MK2. The p38-MK2-TTP module is involved in post-transcription control of TNF-α mRNA in LPS-stimulated cells (27, 43). In quiescent cells, the TTP is nonphosphorylated and binds to the ARE of TNF-α mRNA, promoting the mRNA degradation. Upon stimulation of macrophages, TTP is phosphorylated by MK2, resulting in decreased affinity for ARE binding and thus its ability to degrade mRNA. Additionally, because TTP induction is p38-dependent, as in the case of LPS, it is likely that TTP expression is markedly decreased in GPI-stimulated MK2−/− macrophages pretreated with p38 inhibitor. The net effect is marked reduction in TTP-ARE-dependent destabilization of TNF-α mRNA. This explains why the treatment of p38 inhibitor led to severalfold increase of TNF-α production by MK2−/−macrophages stimulated with GPIs.
Because the GPI-induced TNF-α production was markedly decreased in MK2−/− macrophages and inhibition of ERK activation by U0126 also caused significant reduction in TNF-α production (see Fig. 2), we predicted that, in addition to p38 MAPK, the ERK pathway regulates TNF-α production through MK2 activation. Although the effect of ERK inhibition on TNF-α production can be explained by the TPL2 pathway regulating TNF-α mRNA nuclear export (44) and TNF-α secretion (45), it is possible that ERK exert its effect partly by activating MK2. We examined this possibility by analyzing the phosphorylation of MK2 and the activity of MK2 in phosphorylating its substrate HSP27. As shown in Figs. 6A and 6C, the phosphorylation of MK2 in WT macrophages stimulated with GPIs was inhibited by pretreatment with either p38 or ERK inhibitors, albeit to a lesser degree with ERK inhibition. Similar results were obtained in Pam3CSK4 and LPS-induced WT macrophages pretreated with either ERK or p38 inhibitors (Fig. 6B and data not shown). Furthermore, pretreatment with inhibitors of both ERK and p38 completely abolished MK2 activity as evident from the complete inhibition of HSP27 phosphorylation (see Fig. 6C). This conclusion agrees with the results of other studies, reporting that both ERK and p38 can activate MK2 (31, 32, 46). Thus, our results suggest that both ERK and p38 pathways regulate the activation P. falciparum GPI-induced activation of MK2. However, further detailed studies using structurally different inhibitors are needed to conclusively establish the role of ERK in activating MK2.
In contrast to TNF-α, the IL-12 production in response to P. falciparum GPIs and other TLR ligands was severalfold higher in MK2−/− macrophages than in WT cells. Unlike TNF-α, IL-12 mRNA lacks typical AREs and hence it does not appear to be subjected to MK2-dependent mRNA stabilization. Accordingly, compared with TNF-α mRNA, the IL-12 mRNA is stable, and the stability is more or less similar in WT and MK2−/− macrophages stimulated with GPI or Pam3CSK4 (see Fig. 4). Therefore, the observed increased production of IL-12 by MK2−/− macrophages in response to GPIs and other TLR ligands compared with WT cells is not related to mRNA stability but dependent at least in part on increased transcription (see Fig. 3).
Previous studies have shown that NF-κB c-Rel is involved in the expression of IL-12 in response to various stimuli (10). Recently, it has been shown that IκBζ, an IκB family nuclear factor that is inducible upon cell stimulation, is critical for the expression of a subset of cytokine genes, including IL-12; deletion of IκBζ caused marked decrease in IL-12 expression (33). However, in our study, there was no noticeable difference in the levels of c-Rel and other NF-κB translocated to the nucleus and/or IκBζ induction in MK2−/− macrophages compared with WT cells (see Fig. 7). Pretreatment with the inhibitor of p38 activity also had no effect on nuclear induction of IκBζ by GPI in WT and MK2−/− macrophages (see Fig. 7B). The nuclear translocation of c-Rel and other NF-κB factors was also normal in WT cells pretreated with p38 inhibitor. Consistent with the increased IL-12 production, pretreatment of WT cells with p38 inhibitor increased the NF-κB binding to the IL-12 promoter (see Fig. 8B). Thus, taken together, our data indicate that increased binding of NF-κB contributes to some extent for enhanced IL-12 gene transcription and IL-12 production.
Two other nuclear factors, c-Maf and GAP-12, have been shown to repress IL-12 gene transcription (36–38). Our data show that these repressors also play important roles in MK2-mediated regulation of microbial ligand-induced IL-12 gene expression. The expression of c-Maf and the binding of GAP-12 in response to GPI stimuli were strongly reduced in MK2−/− macrophages (see Fig. 9, A and B). Furthermore, the residual very low level expression of c-Maf by MK2−/− macrophages stimulated with GPIs occurs with significantly delayed kinetics compared with WT macrophages (see Fig. 9A). The binding of GAP-12 to the IL-12p40 gene promoter was also markedly decreased in MK2−/− macrophages stimulated with GPIs. Moreover, the GAP-12 binding occurs with a substantially delayed kinetics in MK2−/− macrophages; binding is maximal at 2.5 h in WT cells and at 4 h in MK2−/− cells (see Fig. 9B). Thus, increased NF-κB binding to the gene promoter and marked reduction in gene repression collaboratively contribute to the up-regulation of the IL-12 gene transcription by faster kinetics, producing higher levels of mRNA in MK2−/− macrophages and culminating in a higher IL-12 level relative to WT cells.
Our results represent new findings, which support the idea that the expression of TNF-α and IL-12 is differentially regulated by MK2. Although MK2 controls the TNF-α mRNA stabilization and translation, it does not affect IL-12 mRNA stability, but it does regulate binding of transcription factors and repressors to the gene promoter.
Finally, our observation that MK2 differentially regulates TNF-α and IL-12 production in response to several TLR ligands, including malarial GPIs, has broader implications. Proinflammatory cytokines produced in response to microbial ligands are not only involved in pathogen growth control and disease progression, but they also modulate the specificity and effectiveness of adaptive immunity. Thus, regulation of MK2 and other downstream MAPKs using small molecule inhibitors might provide immunomodulatory strategies for controlling pathogen infection and disease pathology in general, and malaria in particular.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AI41139 from NIAID. This work was also supported by the Pennsylvania Department of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- TNF-α
- tumor necrosis factor-α
- GPI
- glycosylphosphatidylinositol
- TLR
- toll-like receptor
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MK2
- MAPK-activated protein kinase 2
- JNK
- c-Jun N-terminal kinase
- NF-κB
- nuclear factor κB
- c-Maf
- c-musculoaponeurotic fibrosarcoma
- GAP-12
- GATA sequence in the IL-12 promoter (GA-12)-binding protein
- EMSA
- electrophoretic mobility shift assay
- FBS
- fetal bovine serum
- RIPA
- radioimmunoprecipitation assay
- PD98059
- 2′-amino-3′-methoxyflavone
- U0126
- 1,4-diamino-2,3-dicyano-1,4-bis(2-aminophenylthio)butadiene
- SB203580
- 4-(4-fluorophenyl)-2-(4-methylsulfinylphenyl)-5-(4-pyridyl)1H-imidazole
- IL
- interleukin
- IFN
- interferon
- DMEM
- Dulbecco's modified Eagle's medium
- PBS
- phosphate-buffered saline
- RT
- reverse transcription
- LPS
- lipopolysaccharide
- WT
- wild type
- HRP
- horseradish peroxidase
- ARE
- AU-rich element
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Sherman I. W. (ed) ( 1998) in Malaria: Parasite Biology, Pathogenesis, and Protection, pp. 1– 575, American Society for Microbiology, Washington, DC [Google Scholar]
- 2.Snow R. W., Craig M., Deichmann U., Marsh K. ( 1999) Bull. WHO 77, 624– 640 [PMC free article] [PubMed] [Google Scholar]
- 3.Sachs J., Malaney P. ( 2002) Nature 415, 680– 685 [DOI] [PubMed] [Google Scholar]
- 4.Miller L. H., Baruch D. I., Marsh K., Doumbo O. K. ( 2002) Nature 415, 673– 679 [DOI] [PubMed] [Google Scholar]
- 5.Malaguarnera L., Musumeci S. ( 2002) Lancet Infect. Dis. 2, 472– 478 [DOI] [PubMed] [Google Scholar]
- 6.Dimopoulos G., Müller H. M., Levashina E. A., Kafatos F. C. ( 2001) Curr. Opin. Immunol. 13, 79– 88 [DOI] [PubMed] [Google Scholar]
- 7.Stevenson M. M., Riley E. M. ( 2004) Nat. Rev. Immunol. 4, 169– 180 [DOI] [PubMed] [Google Scholar]
- 8.Ariyasinghe A., Morshed S. R., Mannoor M. K., Bakir H. Y., Kawamura H., Miyaji C., Nagura T., Kawamura T., Watanabe H., Sekikawa H., Abo T. ( 2006) J. Parasitol. 92, 531– 538 [DOI] [PubMed] [Google Scholar]
- 9.Biron C. A., Gazzinelli R. T. ( 1995) Curr. Opin. Immunol. 7, 485– 496 [DOI] [PubMed] [Google Scholar]
- 10.Sanjabi S., Hoffmann A., Liou H. C., Baltimore D., Smale S. T. ( 2000) Proc. Natl. Acad. Sci. U. S. A. 97, 12705– 12710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su Z., Stevenson M. M. ( 2000) Infect. Immun. 68, 4399– 4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amarante-Paffaro A., Queiroz G. S., Corrêa S. T., Spira B., Bevilacqua E. ( 2004) Reproduction 128, 207– 218 [DOI] [PubMed] [Google Scholar]
- 13.Gasque P. ( 2004) Mol. Immunol. 41, 1089– 1098 [DOI] [PubMed] [Google Scholar]
- 14.Karunaweera N. D., Grau G. E., Gamage P., Carter R., Mendis K. N. ( 1992) Proc. Natl. Acad. Sci. U. S. A. 89, 3200– 3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cot S., Ringwald P., Mulder B., Miailhes P., Yap-Yap J., Nussler A. K., Eling W. M. ( 1994) J. Infect. Dis. 169, 1417– 1418 [DOI] [PubMed] [Google Scholar]
- 16.Artavanis-Tsakonas K., Tongren J. E., Riley E. M. ( 2003) Clin. Exp. Immunol. 133, 145– 152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schofield L., Hackett F. ( 1993) J. Exp. Med. 177, 145– 153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schofield L., Vivas L., Hackett F., Gerold P., Schwarz R. T., Tachado S. ( 1993) Ann. Trop. Med. Parasitol. 87, 617– 626 [DOI] [PubMed] [Google Scholar]
- 19.Krishnegowda G., Hajjar A. M., Zhu J., Douglass E. J., Uematsu S., Akira S., Woods A. S., Gowda D. C. ( 2005) J. Biol. Chem. 280, 8606– 8616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J., Krishnegowda G., Gowda D. C. ( 2005) J. Biol. Chem. 280, 8617– 8627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaestel M. ( 2006) Nat. Rev. Mol. Cell Biol. 7, 120– 130 [DOI] [PubMed] [Google Scholar]
- 22.Yamazaki S., Muta T., Matsuo S., Takeshige K. ( 2005) J. Biol. Chem. 280, 1678– 1687 [DOI] [PubMed] [Google Scholar]
- 23.Naik R. S., Krishnegowda G., Gowda D. C. ( 2003) J. Biol. Chem. 278, 2036– 2042 [DOI] [PubMed] [Google Scholar]
- 24.Naik R. S., Branch O. H., Woods A. S., Vijaykumar M., Perkins D. J., Nahlen B. L., Lal A. A., Cotter R. J., Costello C. E., Ockenhouse C. F., Davidson E. A., Gowda D. C. ( 2000) J. Exp. Med. 192, 1563– 1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos-Payán R., Aguilar-Medina M., Estrada-Parra S., González-Y-Merchand J. A., Favila-Castillo L., Monroy-Ostria A., Estrada-Garcia I. C. ( 2003) Scand. J. Immunol. 57, 439– 445 [DOI] [PubMed] [Google Scholar]
- 26.Neininger A., Kontoyiannis D., Kotlyarov A., Winzen R., Eckert R., Volk H. D., Holtmann H., Kollias G., Gaestel M. ( 2002) J. Biol. Chem. 277, 3065– 3068 [DOI] [PubMed] [Google Scholar]
- 27.Hitti E., Iakovleva T., Brook M., Deppenmeier S., Gruber A. D., Radzioch D., Clark A. R., Blackshear P. J., Kotlyarov A., Gaestel M. ( 2006) Mol. Cell. Biol. 26, 2399– 2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang T., Kruys V., Huez G., Gueydan C. ( 2002) Biochem. Soc. Trans. 30, 952– 958 [DOI] [PubMed] [Google Scholar]
- 29.Kotlyarov A., Yannoni Y., Fritz S., Laass K., Telliez J. B., Pitman D., Lin L. L., Gaestel M. ( 2002) Mol. Cell. Biol. 22, 4827– 4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukas S. M., Kroe R. R., Wildeson J., Peet G. W., Frego L., Davidson W., Ingraham R. H., Pargellis C. A., Labadia M. E., Werneburg B. G. ( 2004) Biochemistry 43, 9950– 9960 [DOI] [PubMed] [Google Scholar]
- 31.Coxon P. Y., Rane M. J., Uriarte S., Powell D. W., Singh S., Butt W., Chen Q., McLeish K. R. ( 2003) Cell. Signal. 15, 993– 1001 [DOI] [PubMed] [Google Scholar]
- 32.Kang Y. J., Chen J., Otsuka M., Mols J., Ren S., Wang Y., Han J. ( 2008) J. Immunol. 180, 5075– 5082 [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto M., Yamazaki S., Uematsu S., Sato S., Hemmi H., Hoshino K., Kaisho T., Kuwata H., Takeuchi O., Takeshige K., Saitoh T., Yamaoka S., Yamamoto N., Yamamoto S., Muta T., Takeda K., Akira S. ( 2004) Nature 430, 218– 222 [DOI] [PubMed] [Google Scholar]
- 34.Muta T. ( 2006) Vitam. Horm. 74, 301– 316 [DOI] [PubMed] [Google Scholar]
- 35.Trinh D. V., Zhu N., Farhang G., Kim B. J., Huxford T. ( 2008) J. Mol. Biol. 379, 122– 135 [DOI] [PubMed] [Google Scholar]
- 36.Cao S., Liu J., Chesi M., Bergsagel P. L., Ho I. C., Donnelly R. P., Ma X. ( 2002) J. Immunol. 169, 5715– 5725 [DOI] [PubMed] [Google Scholar]
- 37.Sugimoto K., Ohata M., Miyoshi J., Ishizaki H., Tsuboi N., Masuda A., Yoshikai Y., Takamoto M., Sugane K., Matsuo S., Shimada Y., Matsuguchi T. ( 2004) J. Clin. Investig. 114, 857– 866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker C., Wirtz S., Ma X., Blessing M., Galle P. R., Neurath M. F. ( 2001) J. Immunol. 167, 2608– 2618 [DOI] [PubMed] [Google Scholar]
- 39.Kotlyarov A., Gaestel M. ( 2002) Biochem. Soc. Trans. 30, 959– 963 [DOI] [PubMed] [Google Scholar]
- 40.Kotlyarov A., Neininger A., Schubert C., Eckert R., Birchmeier C., Volk H. D., Gaestel M. ( 1999) Nat. Cell Biol. 1, 94– 97 [DOI] [PubMed] [Google Scholar]
- 41.Lehner M. D., Schwoebel F., Kotlyarov A., Leist M., Gaestel M., Hartung T. ( 2002) J. Immunol. 168, 4667– 4673 [DOI] [PubMed] [Google Scholar]
- 42.Frevel M. A., Bakheet T., Silva A. M., Hissong J. G., Khabar K. S., Williams B. R. ( 2003) Mol. Cell. Biol. 23, 425– 436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandler H., Stoecklin G. ( 2008) Biochem. Soc. Trans. 36, 491– 496 [DOI] [PubMed] [Google Scholar]
- 44.Dumitru C. D., Ceci J. D., Tsatsanis C., Kontoyiannis D., Stamatakis K., Lin J. H., Patriotis C., Jenkins N. A., Copeland N. G., Kollias G., Tsichlis P. N. ( 2000) Cell 103, 1071– 1083 [DOI] [PubMed] [Google Scholar]
- 45.Rousseau S., Papoutsopoulou M., Symons A., Cook D., Lucocq J. M., Prescott A. R., O'Garra A., Ley S. C., Cohen P. ( 2008) J. Cell Sci. 121, 149– 154 [DOI] [PubMed] [Google Scholar]
- 46.Stokoe D., Campbell D. G., Nakielny S., Hidaka H., Leevers S. J., Marshall C., Cohen P. ( 1992) EMBO J. 11, 3985– 3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.