Abstract
The ExsA protein is a Pseudomonas aeruginosa transcriptional regulator of the AraC/XylS family that is responsible for activating the type III secretion system operons upon host cell contact. Its activity is known to be controlled in vivo through interaction with its negative regulator ExsD. Using a heterologous expression system, we demonstrated that ExsD is sufficient to inhibit the transcriptional activity of ExsA. Gel shift assays with ExsA- and ExsD-containing cytosolic extracts revealed that ExsD does not block DNA target sites but affects the DNA binding activity of the transcriptional activator. The ExsA-ExsD complex was purified after coproduction of the two partners in Escherichia coli. Size exclusion chromatography and ultracentrifugation analysis revealed a homogeneous complex with a 1:1 ratio. When in interaction with ExsD, ExsA is not able to bind to its specific target any longer, as evidenced by gel shift assays. Size exclusion chromatography further showed a partial dissociation of the complex in the presence of a specific DNA sequence. A model of the molecular inhibitory role of ExsD toward ExsA is proposed, in which, under noninducing conditions, the anti-activator ExsD sequesters ExsA and hinders its binding to DNA sites, preventing the transcription of type III secretion genes.
Pseudomonas aeruginosa is an opportunistic pathogen that can cause both acute and chronic infections by exploiting deficiencies in the host defenses. The bacterium turns on/off a battery of different virulence factors depending on the type and the stage of infection (1, 2). The type three secretion system (T3SS)3 is a major virulence determinant of P. aeruginosa that is associated with both early chronic and acute infections (3–5). It allows the bacterium to inject a set of effectors through a syringe-like apparatus directly into the eukaryotic cytoplasm (6, 7). In P. aeruginosa, four translocated effector proteins, namely ExoY, ExoS, ExoT, and ExoU, lead to disruption of the cytoskeletal organization, increased cytosolic cAMP concentration, breaking of eukaryotic cell membranes, and cell death (8, 9).
According to the bacterial metabolism and/or infectious niche, the synthesis of T3SS as well as other virulence factors is finely tuned by general and specific regulatory pathways (10, 11). Molecular links between general pathways and T3SS regulation have generally not been identified yet; however, they all converge toward one transcription factor that is absolutely required for T3SS gene expression, the ExsA protein (10 and references therein and Refs. 12, 13).
ExsA is an activator belonging to the AraC/XylS family of transcription factors, whose common feature is a conserved ∼100-amino acid domain containing two helix-turn-helix (HTH) motifs. Like ExsA, they often possess an additional N-terminal domain that may carry dimerization determinants and/or ligand-binding property capable of modulating their activity (14, 15). Interestingly, although the ligands are usually small molecules (for instance, arabinose for AraC and 3-methyl benzoate for XylS), some AraC/XylS members involved in virulence, and more particularly in T3SS, are regulated positively or negatively through interactions with protein ligands (16). Indeed, InvF of Salmonella typhimurium and MxiE of Shigella flexneri, two transcription factors regulating the expression of T3SS, are activated by T3SS chaperones, respectively, SicA and IpgC (17, 18). In addition, MxiE is also negatively regulated by association with the anti-activator/chaperone OspD1-Spa15 complex (19). The regulation of SPI1 (Salmonella pathogenicity island 1) genes, many of them encoding components of T3SS, involves the HilD factor, another AraC/XylS member negatively modulated by a ligand protein HilE (20).
In P. aeruginosa, ExsA has been reported to be regulated by two inhibitors, ExsD and PtrA (21, 22). The anti-activator ExsD belongs to the dedicated regulatory cascade that links synthesis of T3SS proteins to secretory activity (21). This pathway involves also the anti-anti-activator ExsC (23) and the secreted/translocated protein ExsE (24, 25). Based on in vivo and in vitro studies, a model of regulation has been proposed (10): when the T3SS is induced, after direct contact with the target cell or calcium depletion, the small protein ExsE is translocated through the syringe-like apparatus into the host cell; consequently, ExsC binds ExsD, which releases ExsA that becomes able to activate transcription of the T3SS genes. Indeed, recent biochemical characterization of the ExsE-ExsC and ExsC-ExsD complexes supports this model (26). More recently the second putative ligand called PtrA, unique in P. aeruginosa, has been discovered as an inhibitor of T3SS synthesis under specific environmental conditions, such as copper stress (22). Its direct interaction with ExsA was visualized using a bacterial two-hybrid system, pulldown experiments, and enzyme-linked immunosorbent assays. However, the molecular mechanisms used by these two anti-activators to interact with and prevent ExsA from activating the transcription are still unknown. Notably, no information exists concerning the regulation of any other AraC/XylS members by protein inhibitors, like MxiE in S. flexneri or HilD in Salmonella (19, 20).
This study reports the first biochemical characterization of ExsA in complex with its anti-activator ExsD and the consequence of this interaction on DNA recognition. By combining in vivo (heterologous system) and in vitro approaches (protein purification, analytical ultracentrifugation, size exclusion chromatography, protein/DNA interaction assays), our work shows the following: (i) ExsD is sufficient to inhibit in vivo ExsA transcriptional activity; (ii) ExsD does not bind to DNA and inhibits the binding of ExsA to DNA through direct interaction; (iii) ExsA-ExsD is a 1:1 complex; and (iv) ExsA binds to DNA or ExsD in an exclusive manner. A model for a molecular mechanism by which the protein ligand ExsD inhibits the activity of the AraC/XylS member ExsA is proposed.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The P. aeruginosa strain CHA, a cystic fibrosis clinical isolate (27), was used for gene amplifications. Escherichia coli Top10 strain (Invitrogen) was used for standard cloning experiments and for expression in heterologous systems. The overproduction assays were done using E. coli strain BL21 Star (DE3) (Invitrogen). Cells were grown aerobically in Luria Bertani (LB) medium at 37 °C. Antibiotics were added at the following concentrations (in milligrams/liter): 100 (ampicillin), 34 (chloramphenicol), 25 (kanamycin), 10 (tetracycline).
E. coli Heterologous Expression System
The construction of the reporter gene plasmid, pRK-pC+lacZ, of the exsA expression vector pRSF-pX2ExsA and of the exsD expression vector pIApX2ExsD is detailed in the supplemental material. All three plasmids (pRK-pC+lacZ, control pRSFDuet-1 or pRSF-pX2ExsA, and control pIApX2 or pIApX2ExsD) were transformed into the E. coli Top10 strain by successive transformation. Overnight cultures of the strains grown at 37 °C in LB supplemented with appropriate antibiotics were diluted to an absorbance at 600 nm (A600) of 0.1 and then grown at 37 °C until the A600 reached 1.0. The β-galactosidase activity was measured as described below, and 25 ml of the cells were further sedimented by centrifugation and then resuspended in 0.5 ml of buffer (20 mm Tris/HCl, 500 mm NaCl, 10% glycerol, pH 8). After sonication, the lysates were ultracentrifuged for 30 min at 250,000 × g in a TLA 120.2 rotor (Beckman) at 4 °C to separate the soluble fraction from the insoluble membrane fraction. Then immunoblot analyses on the soluble fractions were performed as described below to assess the amount of ExsA and ExsD produced.
β-Galactosidase Assays
Whole cells (0.5 ml) of E. coli strains at A600 of 1.0 were made permeable by addition of 20 μl of 0.1% SDS and 20 μl of chloroform, followed by vortexing for 1 min. β-Galactosidase activity was then assayed according to Miller (28), with up to 0.1 ml of cells, in 0.9 ml of Z buffer (0.1 m Na2HPO4/NaH2PO4, 10 mm KCl, 1 mm MgSO4, 50 mm 2-mercaptoethanol, pH 7.0) at 28 °C. Reaction was initiated by addition of 0.2 ml of o-nitrophenyl-β-d-galactopyranoside at 4 mg/ml and stopped with 0.5 ml of 1 m Na2CO3. The A420 was then read after sedimentation of cell debris, and the activities were expressed in Miller units ((A420 × 1000)(tmin × Volml × A600)). The reported values for enzyme activities are the average of at least two independent experiments performed in triplicate.
Production and Purification of ExsA, ExsD, and ExsA-ExsD Complex
Construction of overexpression plasmids is detailed in the supplemental material. Production of His6ExsA (HExsA), ExsAHis6 (ExsAH), and ExsD was performed in E. coli BL21 Star (DE3) strains harboring pET15b-HExsA, pET22b-ExsAH, and pACYC-ExsD, respectively, grown in LB at 37 °C. Expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at an A600 of 0.6. Cells were additionally grown overnight at 16 °C and 120 rpm and then harvested by centrifugation. The HExsA and ExsAH proteins were purified and stored as already described for HExsA (29), in 20 mm Tris/HCl buffer, 500 mm NaCl, 1 mm dithiothreitol, 0.5% Tween 20, pH 7.4. For purification of ExsD, the harvested cells were lysed at 4 °C by three passages through a French press cell in lysis buffer (20 mm Tris/HCl, 100 mm NaCl, pH 8.0) supplemented with Protease Inhibitor Mixture (Complete, Roche Applied Science). After centrifugation at 200,000 × g for 45 min at 4 °C, the soluble fraction was loaded onto a 5-ml anion exchange column (HitrapTMQ HP, GE Healthcare). The column was washed with 25 ml of lysis buffer, and the protein was eluted with a 50-ml gradient ranging from 100 mm to 1.5 m NaCl in 20 mm Tris/HCl, pH 8.0. Fractions containing ExsD were pooled, and 6 ml were loaded onto a gel filtration column (Hiload 16/60 SuperdexTM 200, GE Healthcare) previously equilibrated in 20 mm Tris/HCl, 100 mm NaCl, and 2 mm EDTA, pH 8.5. ExsD was eluted with a flow rate of 1 ml/min.
Coproduction of HExsA-ExsD, ExsAH-ExsD, His6ExsA(1–168)-ExsD, or His6ExsA(166–278)-ExsD was performed in E. coli BL21 Star (DE3) strains harboring pACYC-ExsD and pET15b-HExsA, pET22b-ExsAH, pET15b-HNter, or pET15b-HCter, respectively. Culture and induction were performed as described above for individual proteins. Harvested cells were resuspended in 20 mm Tris/HCl buffer, 250 mm NaCl, 20 mm imidazole, pH 8.0, supplemented with Protease Inhibitor Mixture (Complete Roche Applied Science). After cell breakage by French press, the supernatants were cleared by ultracentrifugation at 200,000 × g for 45 min. Then the soluble fractions were loaded onto a nickel affinity column (HisTrapTM chelating, GE Healthcare) and submitted to a step imidazole gradient (20, 50, 100, 200, and 500 mm). Fractions eluted at 100 mm imidazole were pooled, and 6 ml were loaded onto a preparative gel filtration column (Hiload 16/60 SuperdexTM 200, GE Healthcare) at a flow rate of 1 ml/min. Peak protein fractions were concentrated by Vivaspin 20TM (Vivasciences) and analyzed by SDS-PAGE. The same procedure was used for the copurification of the individual N-terminal and C-terminal domains of ExsA with ExsD. Protein concentrations were estimated using BCA Assay (Interchim) and UV measurement.
Analytical Size Exclusion Chromatography (SEC)
500 μl of complex from preparative SEC developed in “binding buffer” (20 mm Tris/HCl, 200 mm KCl, 25 mm NaCl, 1 mm dithiothreitol, 1 mm EDTA, 10% glycerol, pH 8.0) were loaded onto a high resolution gel filtration column (SuperdexTM 200 10/300 GL, GE Healthcare) equilibrated in the same buffer. The complex was eluted at a volume of 14.1 ml with a flow rate of 0.4 ml/min. The column calibration was performed with molecular weight standards of protein, as recommended by the manufacturer (GE Healthcare). For the study of the proteins in the presence of DNA, we used the pC 60-mer DNA probe generated by annealing a complementary pair of oligonucleotides (pC EMSA F and pC EMSA R, see supplemental Table S2). Prior to the chromatography, the HExsA-ExsD complex, HExsA, or ExsD were mixed to the probe in 1:2 molar ratio (protein/DNA) and incubated for 30 min at 25 °C in the binding buffer, in a total volume of 500 μl. Samples containing the proteins, the pC probe, or their mixtures were analyzed on Superdex 200TM 10/300 GL as described (30). Fractions of 250 μl were collected and analyzed by SDS-PAGE or Western blot to monitor the protein elutions.
Antibodies and Immunoblot Analysis
Polyclonal antibodies against the ExsAH protein were raised in rabbits, and antibodies against ExsD were raised in guinea pigs by Eurogentec as described by the manufacturer. The specific antibodies directed against ExsAH were affinity-purified. For the immunoblots, the samples were subjected to SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk before addition of primary antibodies (1:2,000) and a secondary goat anti-rabbit (Sigma) or rabbit anti-guinea pig antibody (Invitrogen) conjugated to peroxidase. Detection was performed by ECL (Amersham Biosciences).
Electrophoretic Mobility Shift Assays (EMSA)
The pC DNA probe was generated by annealing a complementary pair of biotinylated oligonucleotides (5′dR Biot-pC EMSA F and 5′dR Biot-pC EMSA R, see supplemental Table S2). When cytosolic extracts were used, the production of ExsAH, ExsD and PcrV was performed in E. coli BL21 Star (DE3) strains harboring pET22b-ExsAH, pACYC-ExsD, and pET15b-PcrV, respectively. Cultures were grown in LB at 37 °C as described above. 30 ml of cells were resuspended in 2.5 ml of buffer (20 mm Tris-HCl, 500 mm NaCl, 10% glycerol, pH 8.0). After sonication, the lysates were ultracentrifuged, and the concentration of the total soluble proteins was measured (0.7 mg/ml). The protein profiles were analyzed on a 15% Tris/Tricine denaturing gel.
DNA binding assays were performed as follows: 1 μl of each protein preparation (purified proteins or cytosolic extracts) was added to a mixture containing 0.2 nm specific probe, 20 ng/μl poly(dI·dC) (×2,500), 20 mm Tris/HCl, pH 8.0, 200 mm KCl, 10% glycerol, 0.1 mg/ml bovine serum albumin, 2 mm EDTA, in a final reaction volume of 20 μl. The reactions were incubated at 25 °C for 15 min. When indicated, 2 μl of antibodies were added, and the reaction was extended for 5 min. Then the samples were loaded on a native 5% Tris/glycine/EDTA (TGE)-buffered polyacrylamide gel and electrophoresed at 100 V with cold (1×) TGE buffer. After electrotransfer (180 mA, 30 min, in 0.5 × Tris/borate/EDTA buffer) onto a Biodyne® B nylon membrane (Pierce), the probes were cross-linked to the membrane using a Biolinker BLX-E254 UV cross-linker (Vilber Lourmat) and detected using the Lightshift® chemiluminescent EMSA kit (Pierce).
Analytical Ultracentrifugation (AUC)
Experiments were performed using an AN-50 rotor in a Beckman XL-I analytical ultracentrifuge. Samples were freshly prepared by preparative SEC developed in “complex buffer” (20 mm Tris/HCl, 200 mm NaCl, 2 mm EDTA, pH 8.0). Samples of ExsAH/ExsD diluted at 0.95, 0.45, and 0.09 mg/ml and HExsA/ExsD at 0.09 and 0.05 mg/ml were investigated by sedimentation velocity (SV) at 42,000 rpm and 20 °C, at 280 nm and using interference optics, with two-channel centerpieces of 1.2- or 0.3-cm optical path length with sapphire windows. Sedimentation equilibrium (SE) was done at 4 °C for ExsAH/ExsD between 0.14 and 0.57 mg/ml, from SEC followed by a concentration step and dilution series, with two- and six-channel centerpieces of 0.3- and 1.2-cm optical path length, with quartz windows. SE experiments were performed at successively, for more than 24 h at each angular velocity, 8,300, 10,000, and 14,400 rpm, followed by speeding at 42,000 rpm. Radial scans at 280 nm were taken every 2 h. The density (1.0074 g/ml) and viscosity (1.03 millipascal·s) of the buffer as well as the partial specific volume, molar mass, and molar extinction coefficients of the polypeptides and complexes (see supplemental Table S3) were estimated with the program Sednterp. Data analyses were made as described previously (31, 32). We used the refractive index increment ∂n/∂c = 0.186 ml/g to estimate protein concentration from the interference signal and experimental E280,0.1% = 1.44 cm−1 mg−1 ml for determining the complex concentration. SV analyses were done with the program Sedfit in terms of continuous distributions c(s) of sedimentation coefficients (s) (33). As samples were homogeneous, the analysis was made considering one noninteracting particle, allowing the estimates of s, of the protein molar mass (M), and of the Stokes radius (RS). In SE experiments, the equilibrium conditions were checked using Winmatch version 7 software. A global analysis was performed using Sedphat software, assuming one type of ideal species, with mass conservation in each sample, for the determination of M.
RESULTS
ExsD Is Sufficient to Inhibit ExsA Transcriptional Activity
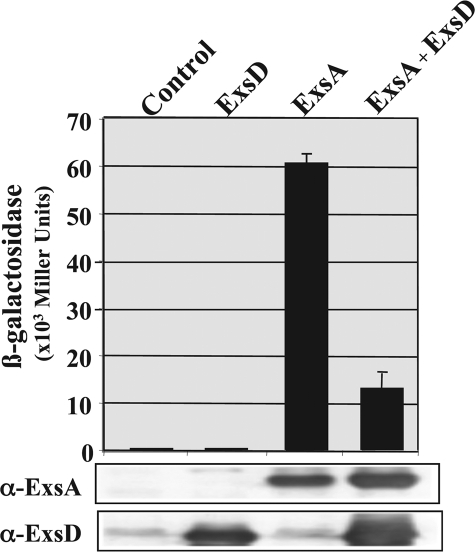
The ExsD protein was shown previously to negatively control ExsA activity in a P. aeruginosa background, and a bacterial two-hybrid system gave evidence for direct interaction between ExsA and ExsD (21). To address the question whether ExsD is sufficient to block ExsA activity, we designed a heterologous transcription assay in E. coli. The ExsA-target promoter of the exsCEBA operon was fused to the lacZ reporter gene (fusion pC+-lacZ) and introduced into E. coli. ExsA and ExsD were produced either alone or in concert, and their effect on pC expression was assessed by measuring the β-galactosidase activity. As shown in Fig. 1, ExsD alone did not affect pC transcription as the β-galactosidase activity was similar to that observed in the control cells. On the contrary, ExsA was able to activate pC expression at a high level (60,500 Miller units). When both ExsA and ExsD were present in the cells, the activity of pC was strongly reduced (4.5-fold) compared with the expression observed with ExsA alone, whereas the amount of ExsA was similar in both cases as visualized by immunoblots (Fig. 1). These data clearly demonstrate that the ExsD protein is sufficient to inhibit in vivo ExsA transcriptional activity.
FIGURE 1.
Direct inhibition of in vivo ExsA activity by the anti-activator ExsD. The histograms present the β-galactosidase activity from E. coli strains containing three plasmids as indicated. The reporter plasmid pRK-pC+lacZ is present in all the strains. The second plasmid is either pRSFDuet-pX2ExsA (ExsA) or pRSFDuet-1 (control). The third plasmid is either pIApX2ExsD (ExsD) or pIApX2 (control). Error bars denote standard deviation. The lower panels represent the anti-ExsA and anti-ExsD immunoblot analyses of the soluble fractions of the corresponding cells.
ExsD Affects ExsA DNA Binding Activity
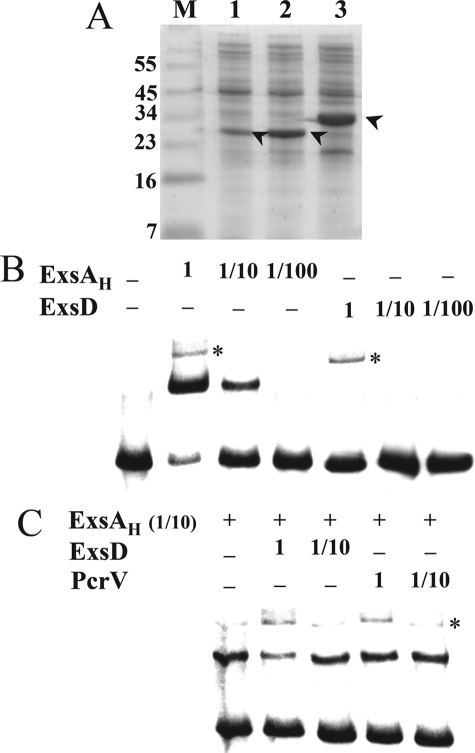
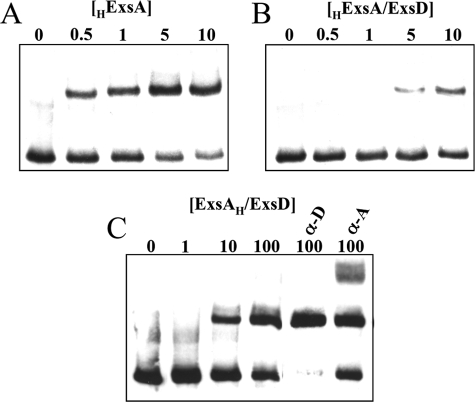
To get insights into the inhibitory effect of ExsD, we assessed the DNA binding activity of both ExsA and ExsD, alone or mixed together. The fusion protein ExsAH (ExsA fused to a His6 tag at its C terminus) and the ExsD protein were overproduced separately in E. coli strains (Fig. 2A). Then increasing dilutions of the cytosolic extracts were used in mobility shift assays using the promoter of the regulatory gene operon (pC) as a probe. As shown Fig. 2B, ExsAH is clearly endowed with DNA binding activity, whereas the anti-activator ExsD does not bind to the probe, not even at the highest concentration. This experiment clearly supports the idea that the anti-activator inhibits ExsA by directly interacting with the transcription factor and not by occluding the DNA targets of ExsA.
FIGURE 2.
ExsD interferes with ExsA binding to DNA. A, cytosolic extracts of cells producing ExsAH (lane 1), ExsD (lane 2), and PcrV (lane 3) were separated by SDS-PAGE and stained with Coomassie Blue. The bands corresponding to the proteins are indicated by an arrowhead. M, protein marker (in kDa). B, EMSA of the pC promoter with ExsAH and ExsD extracts. Biotinylated pC fragment (60-mer, 0.2 nm) was incubated for 15 min at 25 °C in the absence (−) or in the presence of 1 μl of indicated cytosolic fraction, either undiluted (0.7 mg/ml) (lane 1), diluted 10 (lane 1/10), or 100 times (lane 1/100). The samples were then electrophoresed, electrotransferred onto a nylon membrane, and revealed using Lightshift® chemiluminescent EMSA kit. C, 1 μl of ExsAH-cytosolic fraction, 10-fold diluted, was incubated with 1 μl of the indicated fractions (undiluted or diluted 10 times) 15 min prior to addition of pC probe. A nonspecific band (*) is revealed at the highest concentration of all extracts.
Moreover, when the two proteins were mixed together in the binding reaction buffer 15 min prior to the addition of the pC probe, a slight reduction of the amount of shifted probe in presence of a high amount of ExsD could be observed (Fig. 2C). As a negative control, cytosolic extracts of PcrV-overproducing strain did not affect the binding of ExsA to DNA. These data further suggest that it is the DNA binding activity of ExsA that is affected in presence of ExsD.
ExsA and ExsD Form a Complex
To further study the effect of ExsD on the in vitro binding activity of ExsAH, we overproduced and purified the ExsAH and ExsD. Surprisingly, addition of up to 100-fold molar excess of ExsD did not affect the binding of the transcription factor on its target DNA in gel shift experiments (data not shown). Of note, we also never observed by SEC a ExsA-ExsD complex by mixing the two purified proteins in vitro.
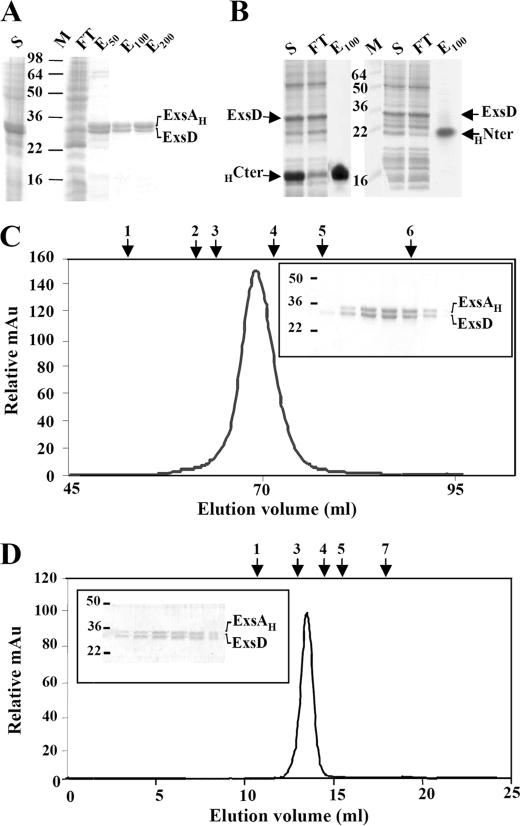
Therefore, to study the DNA-binding properties of ExsA in the presence of ExsD, we attempted to obtain the ExsA-ExsD complex by coproducing the two proteins in BL21 Star (DE3) E. coli and to probe the existence of a complex by copurification. During the Ni2+ affinity chromatography, the untagged ExsD protein coeluted with ExsAH (Fig. 3A), whereas ExsD alone was not retained on the HisTrap column. The fractions containing the two proteins were first injected onto a preparative size exclusion column (Superdex 200 16/60), and the single eluted peak was analyzed by SDS-PAGE (Fig. 3C). This peak should correspond to the ExsA-ExsD complex as, when purified independently in the same conditions, ExsA eluted in the void volume, whereas the ExsD protein eluted at the volume expected for its trimeric form (data not shown and see Ref. 26). The most concentrated fraction was further loaded onto a high resolution gel filtration column (Superdex 200 10/30 GL). One single peak again eluted from this column containing both the ExsAH and ExsD proteins in the same amount, as visualized on SDS-PAGE (Fig. 3D) and confirmed by immunoblots. This second SEC further indicates the existence of a single complex that does not dissociate. The same results were obtained when ExsD was coproduced and copurified with ExsA fused with a His6 tag at its N terminus (HExsA) (data not shown).
FIGURE 3.
Copurification of the ExsAH protein and its anti-activator ExsD. SDS-PAGE analysis of nickel affinity chromatography of coproduced ExsAH-ExsD (A), HCter (HExsA(166–268))-ExsD (B, left panel), and HNter (HExsA(1–168))-ExsD (B, right panel). S, supernatant obtained after ultracentrifugation; M, protein marker (kDa); FT, flow-through; E50, E100, E200, proteins eluted at 50, 100, and 200 mm imidazole, respectively. The gels were stained by Coomassie Blue. C, elution profile of the ExsAH-ExsD complex on size exclusion chromatography (Hiload 16/60 Superdex 200); the complex elutes as a single peak at 69.13 ml. The inset represents the SDS-PAGE monitoring the eluted proteins from 66 to 72 ml. D, elution profile of the ExsAH-ExsD complex on analytical size exclusion chromatography (Superdex 200 10/300 GL); the complex elutes as a single peak at 14.1 ml. The inset represents the SDS-PAGE monitoring the eluted proteins from 13.5 to 15.25 ml. Above the two chromatograms are indicated the elution volumes of the standard proteins with known molecular weight and Stoke radius: arrow 1, ferritin (440 kDa, 61 Å); arrow 2, catalase (232 kDa, 52.2 Å); arrow 3, aldolase (158 kDa, 48.1 Å); arrow 4, bovine serum albumin (67 kDa, 35.5 Å); arrow 5, ovalbumin (43 kDa, 30.5 Å); arrow 6, chymotrypsinogen (25 kDa, 20.9 Å); arrow 7, ribonuclease A (13.7 kDa, 16.4 Å).
ExsA is composed of two distinct domains as follows: the C-terminal domain containing the two HTH motifs and the N-terminal domain supposed to be involved in the regulation of the DNA binding activity. To determine whether any of the two ExsA distinct domains are sufficient to interact with ExsD, ExsD was coproduced with either the first 168 amino acids of ExsA (HExsA(1–168)) or with the C-terminal domain of ExsA (HExsA(166–278)), both domains carrying the hexahistidine tag on N terminus. In both cases, no ExsD could be eluted in the same fractions as ExsA domains during the affinity chromatography. Of note, this also illustrates that untagged ExsD was not retained on the HisTrap column (Fig. 3B). As for the entire ExsA, the recombinant HExsA(1–168) and HExsA(166–278) purified separately were not able to interact with ExsD as assessed by SEC (data not shown). All these data indicated that the ExsA and ExsD proteins do interact and form a complex only when the two partners are coproduced and that the interaction with ExsD requires the entire ExsA protein.
ExsD and ExsA Form a 1:1 Complex
Our results from SEC on the Superdex 200 10/30 GL revealed that the ExsAH-ExsD and HExsA-ExsD complexes eluted at the same volume of 14.1 ml (Fig. 3D, and data not shown), corresponding to a globular macromolecule with a Stokes radius of ∼39.9 Å.
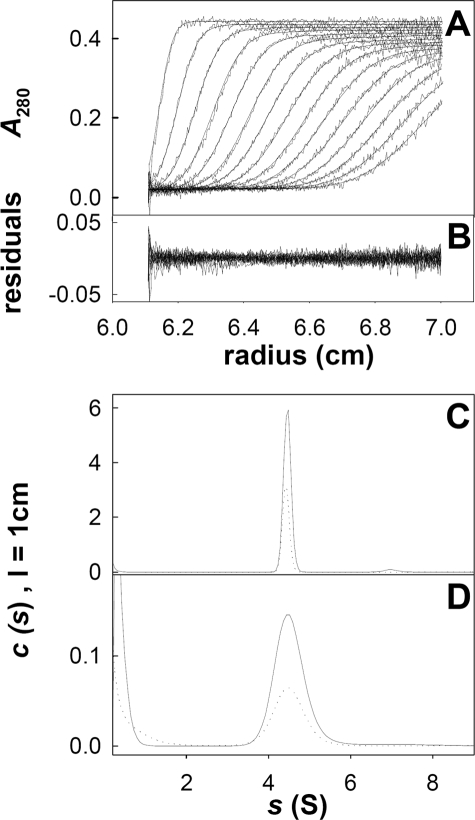
As SEC only allows the estimation of the complex size, we further characterized the stoichiometry of the complex by AUC analysis. SV experiments were performed on fresh samples issued from SEC. For the two complexes ExsAH-ExsD and HExsA-ExsD, investigated in the range 0.09–0.95 and 0.05–0.09 mg/ml, respectively, we observed the sedimentation of a well defined boundary (Fig. 4). The c(s) analysis showed for all investigated samples a species at 4.45 S (s20,w = 4.7 S), and the contributions observed at the lowest s values were most probably artifacts. The sedimentation coefficient did not vary with the His tag position nor with protein concentration suggesting, respectively, that the two complexes present the same stoichiometry and that there is no dissociation at low concentration (Fig. 4, C and D). The s value depends on the molar mass (M) and Stokes radius (RS), of the particle, according to Svedberg Equation 1,
where ρ and η are the solvent density and viscosity; v̄ is the partial specific volume of the particle, and NA is the Avogadro number. RS may be measured as follows: (i) either independently, (ii) from the SV profiles analyzed in the framework of the model of one type of noninteracting species, or (iii) estimated with hypothesis of molar mass and shape. (i) The combination of the s values with RS = 4.0 nm estimated from calibrated SEC gives an estimate for the complex of M = 78 kDa. (ii) The analysis of the SV profiles in the model of one type of noninteracting species gives, from different samples, an estimate of M = 65 kDa and RS = 3.3 nm (±10%). (iii) If considering a frictional ratio of 1.25, which is typical of globular compact shape, the experimental s value corresponds, again, to M = 65 kDa and RS = 3.3 nm values. The molar mass of 65 kDa is the value calculated for a 1:1 complex. Thus, SV supports a compact 1:1 complex, and most probably SEC provides an overestimated value of RS.
FIGURE 4.
Sedimentation velocity experiments of ExsAH/ExsD and HExsA/ExsD. A, superimposition, for ExsAH/ExsD at 0.95 mg/ml, of selected experimental sedimentation profiles obtained at 280 nm during 4 h at 42,000 rpm, at 20 °C, in a 3 mm cell. B, corresponding residuals. C, result of the c(s) analysis for ExsAH/ExsD at 0.95 and 0.45 mg/ml (solid and dashed lines, respectively). D, result of the c(s) analysis for HExsA/ExsD at 0.09 and 0.05 mg/ml (solid and dashed lines, respectively).
SE was done to confirm the association state. The SE profiles were measured for four samples of ExsAH/ExsD in the range 0.14–0.57 mg/ml, at 280 nm and at three angular velocities. A global analysis was first made in the model of one component, with mass conservation constraint for each sample, without fitting any noise and gave M = 70 kDa (see supplemental material and supplemental Fig. S1), close to the value of 65 kDa calculated for a 1:1 complex. When fitting, for each sample, a constant base line or time-independent noise (not shown), slightly larger molar masses of ≈75 kDa were obtained, which remains much below the value of 96–98 kDa calculated for ExsAH(ExsD)2 or (ExsAH)2-ExsD complexes. Taken together, the AUC data demonstrate that both HExsA and ExsAH do form with its anti-activator ExsD a complex of 1:1 stoichiometry.
ExsD Prevents ExsA from Binding to DNA
To precisely characterize the inhibitory effect of ExsD on ExsA within the 1:1 complex, we compared the DNA-binding ability of ExsA alone or in complex with ExsD by performing gel shift assays on the pC promoter. The ExsA protein exhibited a specific and strong binding activity toward the pC probe, as illustrated on Fig. 5A, whereas the anti-activator ExsD by itself was not able to bind the probe in the same experimental conditions and in the same range of concentration (data not shown). These results corroborate the ones observed with cytosolic extracts (Fig. 2).
FIGURE 5.
DNA binding activity of ExsA alone or in complex with ExsD. EMSA of the pC promoter with HExsA (A), HExsA-ExsD complex (B), or ExsAH/ExsD complex (C). Biotinylated pC probe (0.2 nm) was incubated 15 min with indicated concentrations of proteins (in nm). After electrophoresis and electrotransfer onto a nylon membrane, the labeled DNA was revealed using Lightshift® chemiluminescent EMSA kit. C, at the highest concentration of ExsAH-ExsD complex, antibodies directed against ExsD (α-D) or against ExsA (α-A) were added 5 min prior electrophoresis.
When the purified HExsA-ExsD and ExsAH-ExsD complexes, obtained from size exclusion chromatography, were used in EMSA, a shift of the pC promoter was also observed, with an electrophoretic mobility similar to that observed in presence of HExsA alone but that required at least 10 times more protein (Fig. 5, B and C). The same observation was made when another ExsA-target promoter, the promoter of the pcrGVHpopBD operon, was used in EMSA, and addition of unlabeled specific probe strongly reduced the intensity of the shifted band, confirming the specificity of the formed nucleoprotein complex (data not shown). To discriminate between a probe-ExsA-ExsD tripartite complex or a probe-ExsA complex, antibodies directed either against ExsD or against ExsA were added to EMSA reactions. Although the anti-ExsA antibodies (Fig. 5C) or anti-His (data not shown) formed a supercomplex with the probe-ExsA complex, addition of anti-ExsD antibodies led to the almost complete disappearance of the free probe and concomitantly increased the amount of the shifted probe (Fig. 5C).
All these data suggested that the ExsA-ExsD complex cannot bind to DNA and that the shifted probe observed in the presence of the complex was rather due to free ExsA released from the complex in presence of its target DNA. This is supported by the effect of anti-ExsD antibodies that seem to trap ExsD and consequently to increase the amount of free ExsA able to interact with DNA.
ExsA Binds to DNA or ExsD in an Exclusive Manner
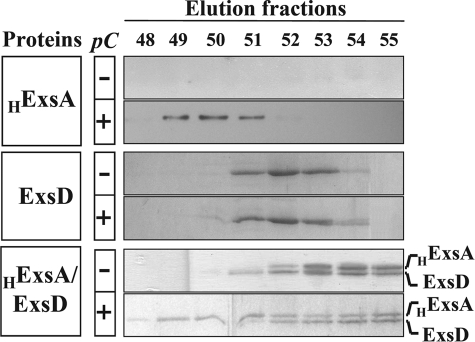
To confirm the partial release of ExsA from the ExsA-ExsD complex in the presence of specific DNA, we analyzed the behavior of ExsA, free or in interaction with ExsD, in the presence of the pC probe. The proteins were incubated either in absence of DNA or in the presence of pC probe at a 1:2 molar ratio (protein/DNA) for 30 min at 25 °C, and the resulting species were separated using analytical SEC (Superdex 200 10/300 GL). The elution profiles were monitored by SDS-PAGE or Western blot (Fig. 6), because the 60-mer probe exhibits high absorbance preventing UV detection of the proteins and nucleoproteins. When HExsA was subjected to SEC analysis, the protein was found in the void volume of the column, probably aggregated (data not shown). However, in the presence of pC, a second peak corresponding to low amounts of HExsA eluted at a volume of 13 ml, as visualized by Western blot (Fig. 6), demonstrating that a part of ExsA is stabilized through the interaction with DNA. ExsD alone, reported to be a trimeric protein (26), was eluted independently of the presence of pC (elution volume of 13.5 ml), confirming its incapability to interact with DNA (Fig. 6). Concerning HExsA-ExsD, the complex was eluted as a single peak at a volume of 14.1 ml as already observed in Fig. 3D. After incubation of the HExsA-ExsD complex with pC, a clear modification of the elution pattern could be observed as follows: a fraction of HExsA eluted earlier than the complex at a volume of 13 ml, which corresponds to the elution of the pC-ExsA nucleoprotein complex described above (Fig. 6). This dissociation was DNA-specific, as an unrelated 60-mer fragment was not able to dissociate ExsA from the complex (data not shown).
FIGURE 6.
Specific DNA partially dissociates the ExsA-ExsD complex. Samples containing HExsA (3 μm), ExsD (4 μm), and the HExsA-ExsD complex (4 μm) were incubated 30 min at 25 °C in binding buffer (see “Experimental Procedures”), either alone or mixed with pC probe in a 1:2 ratio (protein/DNA). The samples were loaded onto an analytical gel filtration column (Superdex 200 10/300 GL); 60 μl of each indicated fraction (from 12.25 to 14.25 ml) were analyzed by SDS-PAGE and Coomassie Blue staining for the complex and ExsD or immunoblot using anti-His6-horseradish peroxidase for HExsA.
These experiments clearly indicate that the inhibitory mechanism used by the anti-activator ExsD to prevent ExsA activity is to sequester the transcription factor and hinder its binding to its target sequences. Indeed the ExsA-ExsD complex was not able to interact with DNA in gel shift assays in our experimental conditions, and the preformed complex partially dissociates in vitro in the presence of target DNA.
DISCUSSION
In this study, we examined the interaction between the key regulator of the T3SS in P. aeruginosa, ExsA, an AraC/XylS member, and one of its two inhibitor proteins, ExsD. The use of a heterologous system indicated that, whereas ExsA can activate by itself the expression of a specific promoter target, the presence of ExsD is sufficient to strongly reduce the ExsA-transcription activation. Furthermore, the ExsD protein of P. aeruginosa (this study) and of Vibrio parahemolyticus (34) is devoid of DNA binding activity on ExsA-DNA targets, and its interaction with ExsA was previously suggested by a two-hybrid system (21). Hence, these data support a model in which ExsD binds to ExsA forming a complex inefficient in transcriptional activation.
The AraC/XylS family of transcriptional regulators of virulence traits can be modulated by ligand proteins in a positive or negative manner, but the molecular mechanisms underlying these interactions are still mostly unknown (16). So far, only activation mechanisms by non-protein ligands have been characterized (35–37). To our knowledge, the inhibition mechanism of AraC/XylS members triggered by ligand binding has not been documented yet. However, based on what is known about non-AraC/XylS transcription factors, several inhibitory molecular mechanisms could be envisioned. One possibility, for instance, would be that the transcription factor may be sequestered by the inhibitor leading to its incapacity to bind DNA and consequently to activate transcription. An example for this is the interaction between the chaperone FliT and FhlDC that prevents FhlDC-dependent expression of flagellar middle genes in Salmonella enterica serovar Typhimurium (38). Another mechanism would be that the activator-inhibitor complex still binds to DNA but is inefficient in transcriptional activation. In P. aeruginosa, this putative mechanism has been illustrated; the anti-activator FleN directly interacts with the enhancer binding protein FleQ without affecting its DNA binding activity (39). Alternatively, the negative regulator TraM in Agrobacterium tumefaciens can bind and inactivate TraR, a quorum-sensing activator of Ti plasmid conjugation, through a two-step process; TraM forms first an unstable tripartite complex with DNA-bound TraR that concomitantly releases the DNA (40).
To decipher the mechanism of ExsA inhibition by its anti-activator ExsD, we undertook the purification of the two proteins. Like most AraC/XylS proteins, ExsA is largely insoluble when overproduced and tends to aggregate when concentrated, both properties which are challenging for biochemical studies. Although a maltose-binding protein-ExsA fusion was used to study the DNA binding activity (41), only recently was the biochemical characterization of His6-ExsA at a low concentration and in presence of Tween 20 reported (29). However, in these conditions separately produced and purified ExsA and ExsD proteins were not able to form any complex in vitro, and our attempts to see an effect of ExsD on the in vitro binding activity of purified ExsA repeatedly failed (data not shown). Therefore, to study the DNA-binding properties of ExsA in the presence of ExsD, we undertook the purification of the expected ExsA-ExsD complex after coproduction of the two partners in E. coli. This approach was successfully used for two other AraC/XylS members in interaction with their positive ligand proteins as follows: InvF-His6 with the chaperone SicA (17) and the regulator maltose-binding protein-MxiE associated to the chaperone IpgC (42). We could obtain the ExsAH-ExsD and HExsA-ExsD complexes by a two-step purification, including metal ion affinity chromatography followed by a size exclusion chromatography. Of note, the protein/protein interaction with ExsD stabilizes ExsA in solution.
AUC analysis revealed that ExsD is engaged with ExsA in a 1:1 complex. On the other hand, free ExsD was reported to be a trimer and to form a 2:2 complex with its own inhibitor, the anti-anti-activator ExsC (26). Free ExsC is a homodimeric chaperone (26), and ExsA was recently shown to be a monomer (29) (Fig. 7). Therefore, it is possible that ExsD has important and surprising plasticity allowing it to be a trimer, dimer, or monomer depending on its interacting partner (ExsD, ExsC, and ExsA, respectively), and that this modulating oligomeric state is important in the regulatory cascade. Accordingly, Zheng et al. (26). observed that subunit assembly within trimeric ExsD is dynamic, with the subunits associating and dissociating. However, it is also probable that, in P. aeruginosa, the protein is always engaged in a complex, either as a monomer in interaction with ExsA or two monomers interacting with the dimeric ExsC protein. Indeed, whereas the ExsD protein is able to self-associate, this property is not absolutely required for its regulatory activity as observed by the monohybrid system and complementation experiment using several mutants of the protein (43).
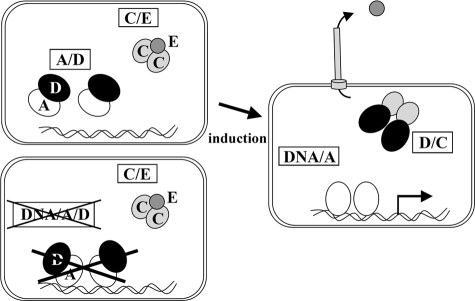
FIGURE 7.
Model of ExsD inhibitory role on ExsA. Under noninducing T3SS conditions, ExsD captures ExsA within a 1:1 complex, making the transcription activator unable to bind target promoters. The second putative mechanism of inhibition, implying the DNA-binding of an ExsA-ExsD complex inefficient in transcriptional activation, was ruled out. The two other components of the regulatory cascade coupling T3SS secretion and synthesis, ExsC and ExsE, can form a 2:1 complex (11, 26). Upon induction of the T3SS system, ExsE is translocated into host cell, and the free ExsC protein interacts with ExsD. Released ExsA then binds to DNA and activate T3SS operons.
The effect of ExsD on ExsA DNA binding activity was then characterized by gel shift experiments. In the presence of the complex, we observed a shifted band with a mobility identical to that of the ExsA-dependent shift but that required a protein amount at least 10-fold higher for the complex compared with ExsA alone. One could have expected either no binding to the probe or a shifted probe with a lower mobility than DNA/ExsA, which could reflect a tripartite DNA-ExsA-ExsD complex, as observed for probe-FleQ-FleN or TraM-TraR-DNA (39, 40). Notably, the presence of anti-ExsD antibodies did not provoke any supershift of the probe, but rather it increased the amount of the shifted probe. These data suggested a release of a fraction of ExsA from the complex in the presence of specific DNA. This release was further confirmed by analyzing the novel protein species resulting from the mixture of DNA with the complex by SEC. Therefore, ExsA was shown to be able to bind to ExsD and to DNA in an exclusive manner.
In the AraC/XylS family, non-protein ligand binding is mediated by the N-terminal domain of the transcription factor that is associated to the C-terminal domain containing the two HTH motifs. We probed if the N-terminal domain of ExsA has a regulatory role and possesses this capacity of ligand binding. However, no complex was observed between the isolated domain and ExsD, after coproduction of the putative partners or in vitro after mixing the purified proteins. Similarly, when the independently purified C-terminal domain was assayed, no complex could be formed, suggesting that both domains of ExsA are required for the interaction with ExsD. Of note, the overproduction of the N-terminal domain in P. aeruginosa does not lead to the up-regulation of the T3SS by ExsD titration, as an unexpected dominant negative effect was observed.4 Therefore, the role of the N-terminal domain remains puzzling. The mutagenesis of ExsA is underway to identify the determinants important for the ExsD interaction, and the three-dimensional structure of the complex would be another way to define the molecular basis of the inhibitory interaction.
Taken together, this work provides strong evidence for a negative effect of ExsD on the DNA binding activity of ExsA through protein/protein interaction. As illustrated Fig. 7, we propose a model of the molecular inhibitory role of ExsD that sequesters the transcriptional activator ExsA and abolishes the binding to DNA in absence of a signal inducing T3SS. Upon induction by host cell contact or calcium depletion, rearrangements of the different protein complexes lead to the release of ExsA that can bind to its DNA recognition sites. Consequently, ExsA activates gene transcription, and the synthesis of the P. aeruginosa T3SS is switched on.
Supplementary Material
Acknowledgments
We thank Aline Appourchaux from the Analytical Ultracentrifugation Platform of the Institut de Biologie Structurale (Grenoble, France) for technical assistance. We also thank Tammy Bohn-Chang and François Cretin for comments regarding the manuscript, Caroline Gébus for technical assistance, and the entire team for helpful discussions.
This work was supported in part by institutional grants from the CNRS, Commissariat à l'Energie Atomique, and Université Joseph Fourier and a grant from Région Rhône-Alpes (Cluster 10 Infectiologie).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Tables S1–S3, and Fig. S1.
J. Thibault and S. Elsen, unpublished results.
- T3SS
- type III secretion system
- HTH
- helix-turn-helix
- SEC
- size exclusion chromatography
- EMSA
- electrophoretic mobility shift assays
- AUC
- analytical ultracentrifugation
- SV
- sedimentation velocity
- SE
- sedimentation equilibrium
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Furukawa S., Kuchma S. L., O'Toole G. A. ( 2006) J. Bacteriol. 188, 1211– 1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman A. L., Kulasekara B., Rietsch A., Boyd D., Smith R. S., Lory S. ( 2004) Dev. Cell 7, 745– 754 [DOI] [PubMed] [Google Scholar]
- 3.Banwart B., Splaingard M. L., Farrell P. M., Rock M. J., Havens P. L., Moss J., Ehrmantraut M. E., Frank D. W., Barbieri J. T. ( 2002) J. Infect. Dis. 185, 269– 270 [DOI] [PubMed] [Google Scholar]
- 4.Bodey G. P., Bolivar R., Fainstein V., Jadeja L. ( 1983) Rev. Infect. Dis. 5, 279– 313 [DOI] [PubMed] [Google Scholar]
- 5.Lyczak J. B., Cannon C. L., Pier G. B. ( 2002) Clin. Microbiol. Rev. 15, 194– 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornelis G. R. ( 2006) Nat. Rev. Microbiol. 4, 811– 825 [DOI] [PubMed] [Google Scholar]
- 7.Galán J. E., Wolf-Watz H. ( 2006) Nature 444, 567– 573 [DOI] [PubMed] [Google Scholar]
- 8.Kipnis E., Sawa T., Wiener-Kronish J. ( 2006) Med. Mal. Infect. 36, 78– 91 [DOI] [PubMed] [Google Scholar]
- 9.Engel J., Balachandran P. ( 2009) Curr. Opin. Microbiol. 12, 61– 66 [DOI] [PubMed] [Google Scholar]
- 10.Yahr T. L., Wolfgang M. C. ( 2006) Mol. Microbiol. 62, 631– 640 [DOI] [PubMed] [Google Scholar]
- 11.Brutinel E. D., Yahr T. L. ( 2008) Curr. Opin. Microbiol. 11, 128– 133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dacheux D., Attree I., Toussaint B. ( 2001) Infect. Immun. 69, 538– 542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yahr T. L., Hovey A. K., Kulich S. M., Frank D. W. ( 1995) J. Bacteriol. 177, 1169– 1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin R. G., Rosner J. L. ( 2001) Curr. Opin. Microbiol. 4, 132– 137 [DOI] [PubMed] [Google Scholar]
- 15.Egan S. M. ( 2002) J. Bacteriol. 184, 5529– 5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plano G. V. ( 2004) Mol. Microbiol. 54, 287– 290 [DOI] [PubMed] [Google Scholar]
- 17.Darwin K. H., Miller V. L. ( 2001) EMBO J. 20, 1850– 1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mavris M., Page A. L., Tournebize R., Demers B., Sansonetti P., Parsot C. ( 2002) Mol. Microbiol. 43, 1543– 1553 [DOI] [PubMed] [Google Scholar]
- 19.Parsot C., Ageron E., Penno C., Mavris M., Jamoussi K., d'Hauteville H., Sansonetti P., Demers B. ( 2005) Mol. Microbiol. 56, 1627– 1635 [DOI] [PubMed] [Google Scholar]
- 20.Baxter M. A., Fahlen T. F., Wilson R. L., Jones B. D. ( 2003) Infect. Immun. 71, 1295– 1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCaw M. L., Lykken G. L., Singh P. K., Yahr T. L. ( 2002) Mol. Microbiol. 46, 1123– 1133 [DOI] [PubMed] [Google Scholar]
- 22.Ha U. H., Kim J., Badrane H., Jia J., Baker H. V., Wu D., Jin S. ( 2004) Mol. Microbiol. 54, 307– 320 [DOI] [PubMed] [Google Scholar]
- 23.Dasgupta N., Lykken G. L., Wolfgang M. C., Yahr T. L. ( 2004) Mol. Microbiol. 53, 297– 308 [DOI] [PubMed] [Google Scholar]
- 24.Urbanowski M. L., Lykken G. L., Yahr T. L. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 9930– 9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rietsch A., Vallet-Gely I., Dove S. L., Mekalanos J. J. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 8006– 8011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Z., Chen G., Joshi S., Brutinel E. D., Yahr T. L., Chen L. ( 2007) J. Biol. Chem. 282, 6136– 6142 [DOI] [PubMed] [Google Scholar]
- 27.Toussaint B., Delic-Attree I., Vignais P. M. ( 1993) Biochem. Biophys. Res. Commun. 196, 416– 421 [DOI] [PubMed] [Google Scholar]
- 28.Miller J. H. ( 1972) Experiments in Molecular Genetics, pp. 352– 355, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29.Brutinel E. D., Vakulskas C. A., Brady K. M., Yahr T. L. ( 2008) Mol. Microbiol. 68, 657– 671 [DOI] [PubMed] [Google Scholar]
- 30.Chen G., Malenkos J. W., Cha M. R., Fuqua C., Chen L. ( 2004) Mol. Microbiol. 52, 1641– 1651 [DOI] [PubMed] [Google Scholar]
- 31.Lebowitz J., Lewis M. S., Schuck P. ( 2002) Protein Sci. 11, 2067– 2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebel C. ( 2007) in Protein Structures: Methods in Protein Structure and Stability Analysis ( Uverski V., Permyakov E. A. eds) pp. 229– 260, Nova Science Publishers, New York [Google Scholar]
- 33.Schuck P. ( 2000) Biophys. J. 78, 1606– 1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X., Shah D. H., Konkel M. E., Call D. R. ( 2008) Mol. Microbiol. 69, 747– 764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schleif R. ( 2003) BioEssays 25, 274– 282 [DOI] [PubMed] [Google Scholar]
- 36.Domínguez-Cuevas P., Marín P., Busby S., Ramos J. L., Marqués S. ( 2008) J. Bacteriol. 190, 3118– 3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J., Hart E., Tauschek M., Price G. D., Hartland E. L., Strugnell R. A., Robins-Browne R. M. ( 2008) Mol. Microbiol. 68, 314– 327 [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto S., Kutsukake K. ( 2006) J. Bacteriol. 188, 6703– 6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dasgupta N., Ramphal R. ( 2001) J. Bacteriol. 183, 6636– 6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin Y., Su S., Farrand S. K. ( 2007) J. Biol. Chem. 282, 19979– 19991 [DOI] [PubMed] [Google Scholar]
- 41.Hovey A. K., Frank D. W. ( 1995) J. Bacteriol. 177, 4427– 4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilonieta M. C., Munson G. P. ( 2008) J. Bacteriol. 190, 2249– 2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lykken G. L., Chen G., Brutinel E. D., Chen L., Yahr T. L. ( 2006) J. Bacteriol. 188, 6832– 6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.