Abstract
Mutations in transforming growth factor-β (TGF-β) receptor superfamily members underlie conditions characterized by vascular dysplasia. Mutations in endoglin and activin-like kinase receptor 1 (ALK1) cause hereditary hemorrhagic telangiectasia, whereas bone morphogenetic protein type II receptor (BMPR-II) mutations underlie familial pulmonary arterial hypertension. To understand the functional roles of these receptors, we examined their relative contributions to BMP signaling in human pulmonary artery endothelial cells (HPAECs). BMP9 potently and selectively induced Smad1/5 phosphorylation and Id gene expression in HPAECs. Contrary to expectations, BMP9 also stimulated Smad2 activation. Furthermore, BMP9 induced the expression of interleukin 8 and E-selectin. Using small interfering RNA, we demonstrate that the type I receptor, ALK1, is essential for these responses. However, small interfering RNA and inhibitor studies showed no involvement of ALK5 or endoglin. We further demonstrate that, of the candidate type II receptors, BMPR-II predominantly mediated IL-8 and E-selectin induction and mitogenic inhibition by BMP9. Conversely, activin receptor type II (ActR-II) contributed more to BMP9-mediated Smad2 activation. Only abolition of both type II receptors significantly reduced the Smad1/5 and Id responses. Both ALK1 and BMPR-II contributed to growth inhibition of HPAECs, whereas ActR-II was not involved. Taken together, our findings demonstrate the critical role of type II receptors in balancing BMP9 signaling via ALK1 and emphasize the essential role for BMPR-II in a subset of BMP9 responses (interleukin 8, E-selectin, and proliferation). This differential signaling may contribute to the contrasting pathologies of hereditary hemorrhagic telangiectasia and pulmonary arterial hypertension.
Pulmonary arterial hypertension (PAH)3 and hereditary hemorrhagic telangectasia (HHT) are diseases characterized by dysregulated smooth muscle and endothelial cell proliferation in the pulmonary circulation. In PAH, progressive muscularization of arterial vessels decreases the luminal area and elasticity of these vessels (1). Mutations in the gene (BMPR2) encoding the bone morphogenetic type II receptor (BMPR-II), a member of the transforming growth factor (TGF) receptor superfamily, underlie most familial PAH cases and a quarter of idiopathic PAH cases (2–4). HHT involves a progressive loss of capillaries and the emergence of directly linked dilated arterioles and venules, termed arteriovenous malformations (5). Although this mainly occurs in nasal and dermal blood vessels, pulmonary arteriovenous malformations occur in more than 20% of HHT patients (5, 6). The two main forms of HHT arise due to mutations in two other TGF receptor superfamily members, endoglin (HHT1) and ALK1 (HHT2) (7, 8). Although the pathologies of PAH and HHT differ, mutations in ALK1 are associated with severe PAH in some HHT2 families, suggesting overlapping characteristics (9–11). HHT1 is not associated with PAH, but pulmonary arteriovenous malformations are more prevalent than in HHT2 patients (5).
TGFβ superfamily receptors form heteromeric receptor complexes of type I and type II receptors in combinations that dictate the ligand selectivity of the complex (12). There are 7 known type I receptors (ALK1–7) and 5 type II receptors (ActR-II, ActR-IIB, BMPR-II, TGFβR-II, and AMHR-II). Activated BMP receptors mediate signaling via C-terminal phosphorylation of the receptor Smad (R-Smad) proteins, Smad1, Smad5, and Smad8 (12–15). In contrast, TGFβ receptors typically phosphorylate and activate Smad2 and Smad3. Upon phosphorylation, both the BMP and TGFβ R-Smads associate with the co-Smad, Smad4, and the R-Smad·Smad4 complex translocates and associates with other specific transcription factors to modulate the expression of specific genes (16–19).
ALK1, in complex with ALK5 and TGFβR-II in endothelial cells, is reported to mediate TGFβ receptor signaling via the Smad1/5/8 pathway rather than the Smad2/3 pathway (20, 21). More recently, BMP9 and BMP10 have been identified as ALK1 ligands, stimulating the Smad1/5/8 pathway and transcription of the Id genes (22–24). BMP9/10 signaling may underlie the potent induction of genes such as IL8 by a constitutively active ALK1 receptor, but not by TGFβ1, in endothelial cells (25). These new insights into ALK1 function in the endothelium have led to a more BMP-centered hypothesis of vascular dysfunction in HHT (26). Moreover, the potential role of BMPR-II in BMP9/ALK1 signaling implies that BMPR-II mutations in PAH may also alter endothelial cell responses to BMP9 (22, 23). Given that BMP9 is reported to act as a circulating vascular quiescence factor, mutations in ALK1 and BMPR-II may result in vascular instability, a feature of both PAH and HHT (24).
We hypothesized that the ligand selectivities of ALK1 and BMPR-II may explain the differences and overlapping consequences of particular mutations. Therefore, we explored the functional responses of HPAECs to a range of BMPs and TGFβ1, and established the receptors involved. We show that BMP9 is the major ALK1 ligand mediating Smad phosphorylation and transcriptional induction in HPAECs. Contrary to previous reports, we observed the novel response that BMP9 stimulated the phosphorylation of Smad1/5 and Smad2 via the same type I receptor, ALK1, in HPAECs. These Smad responses were independent of ALK5 or endoglin, but were abolished by co-transfection of siRNAs for ActR-II and BMPR-II. In addition, cotransfection of siRNAs for ActR-II and BMPR-II was required to abrogate the BMP9-induced Id1 and Id2 transcription and Smad1/5 phosphorylation. BMPR-II preferentially mediates BMP9-mediated IL-8 and E-selectin induction and HPAEC growth inhibition. Conversely, ActR-II mediates a greater proportion of the Smad2 response to BMP9. Taken together, our data imply that ALK1 mutations in HHT2 will impact upon a wider spectrum of BMP9 responses than BMPR-II mutations in HPAECs. These data support a role for dysfunctional BMP9 signaling in both HHT and PAH, and highlight the relative impact and functional consequences of BMPR-II versus ALK1 deficiency in these conditions.
EXPERIMENTAL PROCEDURES
Cell Culture
HPAECs and human aortic endothelial cells (HAECs) were purchased from Lonza Wokingham. Cells were propagated according to the instructions supplied. The human microvascular endothelial cell line HMEC-1 was obtained from the Center for Disease Control (CDC, Atlanta, GA). Human pulmonary artery smooth muscle cells (HPASMCs) were isolated in our laboratory by explant cultures as previously described (27).
RNA Interference
ECs were seeded in 6-well plates (2 × 105 cells/well) for RNA studies or 6-cm dishes (4.38 × 105 cells/dish) for protein extraction and grown for 2 days in EGM-2 (Lonza). Prior to transfection, ECs were incubated in Opti-MEM I for 3 h. ECs were transfected with 10 or 15 nm siRNA (DharmaconTM BMPR-II siGenomeTM Smartpool® (siBMPR-II), Dharmacon On-TARGETplus siRNAs for ActR-II (siActR-II), ALK1 (siALK1), ALK5 (siALK5), Endoglin (siEng), Smad2 (siS2), Smad3 (siS3), Smad4 (siS4), or siControl Non-targeting Pool (siCP) (Perbio Science UK Ltd.)) in complex with DharmaFECT1TM (4 μl/well for 6-well plate or 8.75 μl/dish for 6-cm dish) diluted in Opti-MEM I. Cells were incubated with the complexes for 4 h at 37 °C, followed by replacement with EGM-2 for 24 h. For treatments, cells were serum-restricted in M199, 0.1% FBS (0.1%) for 16 h and treated with the relevant ligands in 0.1% for the times described in the figure legends. For Western blot siRNA experiments, parallel wells were transfected for RNA extraction. Specific reduction of the relevant RNA was quantified using qPCR and specific reduction of the relevant protein was also confirmed by Western blotting wherever possible.
Western Blotting
Cells were grown to confluence in 6-cm dishes and serum-restricted in M199, 0.1% FBS for 16 h. Cells were then treated with recombinant human BMP2, BMP4, BMP6, BMP9, or TGFβ1 (R&D Systems) diluted in 0.1% for 1 h. For ALK5 inhibition experiments, cells were preincubated for 30 min with SD208 (TGFβ Receptor kinase inhibitor V, Calbiochem) or dimethyl sulfoxide vehicle control in 0.1%, prior to treatment with ligands. Cells were snap-frozen and lysed in 150 μl of ice-cold lysis buffer (125 mm Tris-HCl, pH 7.4, 10% (v/v) glycerol, 2% (w/v) SDS containing an EDTA-free protease inhibitor mixture (Roche Diagnostics Ltd., Lewes, East Sussex, UK)). Lysates were sonicated and frozen at −20 °C until protein assay and Western blot analysis. Cell lysates (15–40 μg of total protein) were separated on SDS-PAGE gels and proteins transferred to polyvinylidene fluoride membranes by semi-dry blotting. Blots were then blocked and probed with the relevant antibodies (rabbit monoclonals to C-terminal phospho-Smad1/5, Smad1, C-terminal phospho-Smad2, Smad2, Smad3, or rabbit polyclonal to Smad4 (Cell Signaling Technology), C-terminal phospho-Smad3 rabbit polyclonal (R&D Systems), Id1 rabbit monoclonal (Biocheck Inc., Foster City, CA), Id2 rabbit polyclonal, myc mouse monoclonal, ACTR-IIA goat polyclonal (Santa Cruz Biotechnology Ltd., Santa Cruz, CA), BMPR-II, or endoglin mouse monoclonals (BD Transduction Laboratories)). The ALK1 rabbit polyclonal was kindly provided by Professor D. A. Marchuk. All blots were reprobed with anti-human β-actin monoclonal antibody (Sigma).
Quantitative Reverse Transcriptase-PCR
DNase-digested total RNA (300–700 ng) was reverse transcribed using a high capacity cDNA reverse transcription kit (Applied Biosystems) as described in the manufacturers instructions. qPCR reactions were prepared with 45 ng of cDNA using the SYBR® Green JumpstartTM Taq ReadymixTM (Sigma) containing 200 nm of the relevant sense and antisense primers and 10 nm fluorescein (Invitrogen). The following primer sequences were used for BMPR-II (sense, 5′-CAAATCTGTGAGCCCAACAGTCAA-3′; antisense, 5′-GAGGAAGAATAATCTGGATAAGGACCAAT-3′), Id2 (sense, 5′-GACCCGATGAGCCTGCTATAC-3′; antisense, 5′-GGTGCTGCAGGATTTCCATCT-3′), and β-actin (sense, 5′-GCACCACACCTTCTACAATGA-3′; antisense, 5′-GTCATCTTCTCGCGGTTGGC-3′). Reactions were amplified on an iCycler (Bio-Rad) using the primers above or Quantitect Primers for Id1, E-selectin, L-selectin, P-selectin, fibroblast growth factor 2, IL-6, IL-8, ALK1, ALK5, Endoglin, and ActR-II (Qiagen). The efficiency of each primer set was confirmed to be 90–110%. The relative expression of target mRNAs was normalized to β-actin using the ΔΔCT method (28) and expressed as the fold-change relative to the control (0.1% or Dharmafect1).
IL-8 Enzyme-linked Immunosorbent Assay
HPAECs were serum depleted in M199, 0.1% FBS for 3 h. This was removed and replaced with M199, 0.1% FBS in the presence or absence of exogenous BMP9 (1 ng/ml). After 24 h, supernatants were collected, centrifuged at 1500 × g for 10 min at 4 °C, and assayed using a custom enzyme-linked immunosorbent assay, as previously reported (29). The cells were trypsinized and counted to allow normalization of IL-8 values to cell number.
[3H]Thymidine Incorporation
The thymidine incorporation assay has been described previously (30). Briefly, HPAECs were seeded in 24-well plates at 5 × 104 cells/well and left to adhere overnight. HPAECs were transfected as described above and incubated in EGM-2 overnight. The medium was changed to 0.1% for 8 h prior to treatment with M199, 5% FBS or M199, 5% FBS containing 1 ng/ml BMP9 at 28 h post-transfection. After a further 18 h, 0.25 μCi of [methyl-3H]thymidine (25 μCi/mmol, GE Healthcare) was added to each well and cells washed and lysed 6 h later (52 h post-transfection). The lysates were quantified by liquid scintillation counting. Parallel plates in M199, 5% FBS alone were lysed at 28 and 52 h for RNA extraction to determine receptor expression.
Statistical Analysis
Statistical analysis was performed using a one-way analysis of variance for all mRNA expression data or an unpaired Student's t test to analyze the [3H]thymidine data (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
BMP9 Selectively Induces mRNA Expression in HPAECs
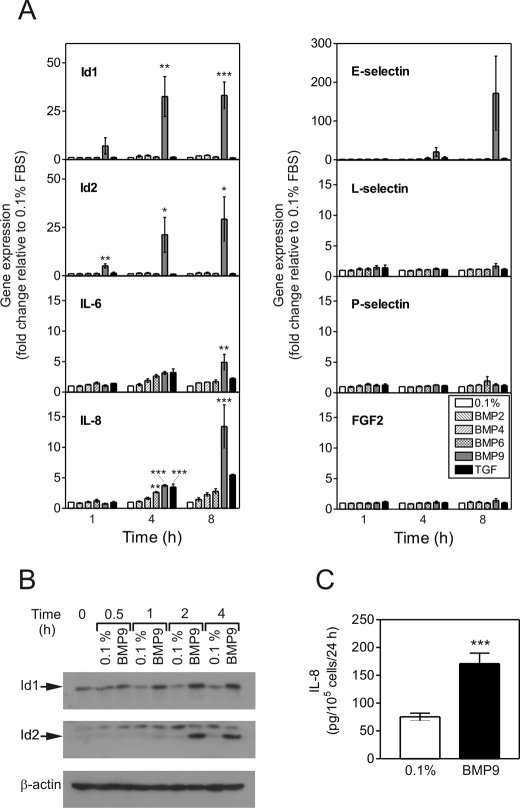
We initially sought to determine the potential roles of different BMP ligands and TGFβ1 on the transcription of genes with potential involvement in angiogenic processes (Fig. 1A). BMP9 potently and selectively induced the transcription of Id1, Id2, E-Selectin, IL-6, and IL-8. Compared with cells treated with 0.1%, Id1 and Id2 transcription were significantly increased by BMP9 at 4 h (Id1, 32.6 ± 17.9-fold, p < 0.01; Id2, 21.2 ± 15.6-fold, p < 0.05) and 8 h (Id1, 33.2 ± 12.1-fold, p < 0.001; Id2, 29.3 ± 20.0-fold). At 8 h, BMP9 induced expression of E-selectin (171.6 ± 166.2-fold), IL-6 (4.9 ± 2.2-fold), and IL-8 (13.4 ± 6.1-fold, p < 0.001). Of the other ligands examined, TGFβ1 also induced the expression of IL-8 at 8 h (5.5 ± 0.2-fold), albeit to a lesser extent than BMP9. In comparison to BMP9, weak mRNA responses were stimulated by BMP2, BMP4, BMP6, and TGFβ1 in HPAECs. The activity of the BMP2 and BMP4 used in these studies was confirmed by their abilities to induce Id1 and Id2 mRNA expression in HPASMCs treated in parallel (data not shown).
FIGURE 1.
BMP9 selectively activates mRNA transcription in HPAECs. A, confluent serum-restricted HPAECs were treated with BMP2, BMP4, BMP6, BMP9 (10 ng/ml), or TGFβ1 (2 ng/ml) in M199, 0.1% FBS (0.1%) for 1, 4, or 8 h. Total RNA was extracted and cDNA prepared. The expression of Id1, Id2, IL-6, IL-8, E-selectin, L-selectin, P-selectin, and fibroblast growth factor 2 (FGF2) were determined by qPCR, with expression being normalized to β-actin and expressed as the fold-change relative to 0.1% at that time point. Data are presented as the mean ± S.E. of three experiments. *, p < 0.05; **, p < 0.01; or ***, p < 0.001 compared with 0.1% FBS (0.1%). B, serum-restricted HPAECs were treated with BMP9 (1 ng/ml) in M199, 0.1% FBS (0.1%) for 0.5, 1, 2, and 4 h. Immunoblotting was performed with antibodies against Id1 and Id2. All blots were reprobed for β-actin to ensure equal loading. C, HPAECs were treated with BMP9 (1 ng/ml) in M199, 0.1% FBS (0.1%) for 24 h. Conditioned medium was assayed for IL-8 using a specific enzyme-linked immunosorbent assay and data are expressed as picograms of IL-8/105 cells. Data are mean ± S.E. (n = 6) from a representative experiment from 3 repeats.
We also confirmed the induction of Id1 and Id2 protein expression by Western blotting (Fig. 1B), demonstrating induction of Id1 protein from 1 to 4 h, and Id2 protein from 2 to 4 h. We also confirmed that IL-8 release is increased from HPAECs treated with BMP9 compared with M199, 0.1% FBS (Fig. 1). Furthermore, BMP9 treatment increased cell surface E-selectin expression in HPAECs (supplemental Fig. S1).
BMP9 Stimulates Phosphorylation of Smad1/5 and Smad2, but not Smad3 in Endothelial Cells
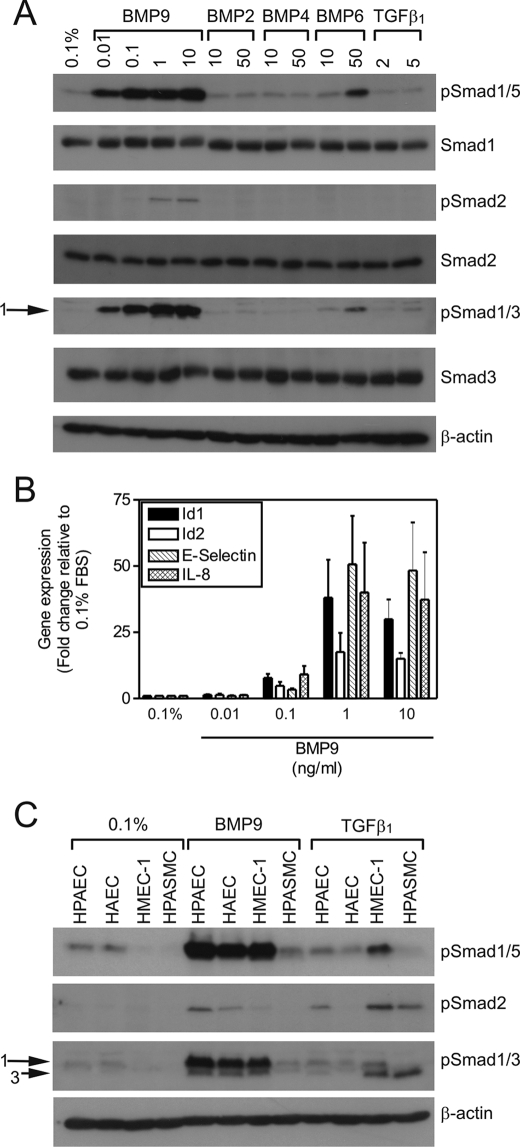
To establish whether our observed pattern of mRNA responses correlated with Smad activation, we examined Smad phosphorylation in response to BMPs and TGFβ1 (Fig. 2A). BMP9 did not alter the total levels of Smad1, Smad2, or Smad3 in HPAECs. BMP9 stimulated Smad1/5 C-terminal phosphorylation at concentrations of 0.01–10 ng/ml, with maximal stimulation at 0.1 ng/ml. In addition to phospho-Smad1/5, our novel observation was the induction of Smad2 phosphorylation at concentrations of 1 and 10 ng/ml BMP9. Although the Smad3 antibody often detected two bands in response to BMP9, Smad3 is not activated by BMP9. The band of greater molecular mass is phospho-Smad1, as it mimics the phospho-Smad1/5 responses and migrates with a higher mass than Smad3 (supplemental Fig. S2, A and B). The band of slightly lower mass does not migrate in exactly the same position as the phospho-Smad3 band in response to TGFβ1 (supplemental Fig. S3, A and B). Furthermore, our studies of Smad3 siRNA (supplemental Fig. S2C) in HMEC-1 cells show this band remains in BMP9-treated cells, whereas the Smad3 band in response to TGFβ1 is reduced. In accordance with the mRNA expression data in Fig. 1, BMP2 and BMP4 did not induce robust Smad1/5 phosphorylation. BMP6 was able to induce Smad1/5 phosphorylation, but did not alter Smad2 or Smad3 phosphorylation. In comparison to BMP9, TGFβ1 was a weaker agonist of Smad phosphorylation in HPAECs.
FIGURE 2.
BMP9 induction of Smad phosphorylation and mRNA transcription in HPAECs is concentration-dependent. A, serum-restricted HPAECs were treated with BMP9 (0.01–10 ng/ml), BMP2 (10 or 50 ng/ml), BMP4 (10 or 50 ng/ml), BMP6 (10 or 50 ng/ml), or TGFβ1 (2 or 5 ng/ml) in M199, 0.1% FBS (0.1%) for 1 h. Immunoblotting was performed with antibodies against phospho-Smad1/5, Smad1, phospho-Smad2, Smad2, phospho-Smad1/3, or Smad3. All blots were reprobed for β-actin to ensure equal loading. B, serum-restricted HPAECs were treated with BMP9 (0.01–10 ng/ml) in 0.1% for 8 h. Total RNA was extracted and cDNA prepared. The expression of Id1, Id2, IL-8, and E-selectin were determined by qPCR, with expression being normalized to β-actin and expressed as the fold-change relative to 0.1%. Data are presented as the mean ± S.E. of three experiments. C, serum-restricted HPAECs, HAECs, HMEC-1, and HPASMCs were treated with BMP9 (1 ng/ml) or TGFβ1 (5 ng/ml) in M199,.
We also assessed the concentration dependence of BMP9-mediated mRNA induction (Fig. 2B). BMP9 stimulated the induction of Id1, Id2, E-selectin, and IL-8 transcription, with a maximal response at 1 ng/ml BMP9. Therefore, we opted to use 1 ng/ml BMP9 in all subsequent experiments.
We questioned whether these Smad responses to BMP9 were peculiar to HPAECs. We compared BMP9- and TGFβ1-mediated Smad activation in HPAECs, HAECs, and HMEC-1 cells in parallel to HPASMCs. HPASMCs should express ALK5 and exhibit robust Smad responses to TGFβ1 (31). All three endothelial cell types exhibited Smad1 and Smad2 phosphorylation in response to BMP9, whereas HPASMCs did not (Fig. 2C). As we would expect from the high ALK5 and low ALK1 expression in smooth muscle cells (31), TGFβ1 stimulated Smad2 and Smad3 phosphorylation in HPASMCs, whereas no Smad1/5 response was evident. The endothelial cell responses to TGFβ1 were more variable. TGFβ1 stimulated Smad2 phosphorylation in HPAECs, but to a lesser extent than BMP9. In contrast, TGFβ1 stimulated Smad2 and Smad3 phosphorylation to a greater extent than BMP9 in HMEC-1 cells, whereas HAECs were unresponsive to TGFβ1. As the BMP9-mediated Smad1/2/5 responses were common to endothelial cells, we sought to examine these responses further in HPAECs.
BMP9 Requires ALK1 for Signaling, but Does Not Require ALK5 or Endoglin
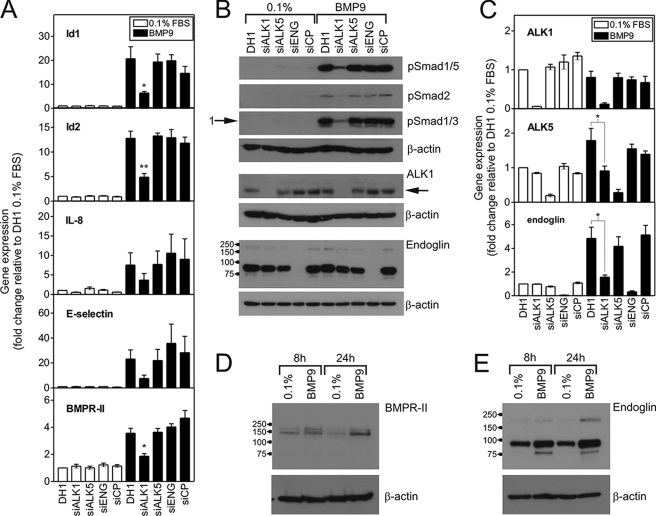
We examined the potential roles of relevant type I receptors and endoglin in BMP9 responses in HPAECs. Reduction of endogenous ALK1 expression significantly attenuated BMP9-mediated phosphorylation of Smad1/5 and Smad2 (Fig. 3A). Furthermore, transfection of HPAECs with ALK1 siRNA (Fig. 3B) attenuated the BMP9-mediated transcription of Id1 (20.66 ± 5.04 versus 6.39 ± 0.65-fold, DH1 and siALK1), Id2 (12.79 ± 1.44 versus 4.89 ± 0.74-fold, DH1 and siALK1), E-selectin (23.16 ± 7.30 versus 4.89 ± 0.74-fold, DH1 and siALK1), IL-8 (7.52 ± 3.14 versus 3.67 ± 1.67-fold, DH1 and siALK1), and BMPR-II (3.56 ± 0.36 versus 1.86 ± 0.19-fold, DH1 and siALK1). In contrast, we did not observe any significant alteration of BMP9-mediated Smad phosphorylation or mRNA induction in cells transfected with siRNA for ALK5 or Endoglin (Fig. 3, A and B). We also confirmed this for ALK5 in HMEC-1 cells, which exhibit robust Smad responses to BMP9 and TGFβ1 consistent with high expression of ALK1 and ALK5 (supplemental Fig. S3A). In these cells, reduction of ALK5 expression by 62.1 ± 3.2% consistently attenuated the Smad responses to TGFβ1, but not BMP9. As a further approach, we examined the effect of the selective ALK5 kinase inhibitor, SD208 (32). This inhibitor did not alter BMP9-mediated phosphorylation of Smad1/5 or Smad2 in HMEC-1 or HPAECs (supplemental Fig. 3, B and C). In contrast, 500 nm SD208 reduced the Smad1/5 and Smad1/3 responses to TGFβ1. Of interest, SD208 had less effect on the Smad2 response.
FIGURE 3.
siALK1 selectively inhibits BMP9-mediated Smad phosphorylation and mRNA induction in HPAECs. A, HPAECs were transfected with siALK1, siALK5, siEng, or siCP (all 10 nm) using Dharmafect1 (DH1). After 28 h, cells were serum-restricted for 16 h, followed by treatment with BMP9 (1 ng/ml) in 0.1% for 8 h. Total RNA was extracted and cDNA prepared. The expression of Id1, Id2, IL-8, and E-selectin were determined by qPCR, with expression being normalized to β-actin and expressed as the fold-change relative to 0.1%. Data are presented as the mean ± S.E. of four experiments. B, HPAECs were transfected as described above and treated with BMP9 (1 ng/ml) in 0.1% for 1 h. Immunoblotting was performed with antibodies against phospho-Smad1/5, phospho-Smad2, or phospho-Smad1/3. Immunoblotting was also performed to confirm the specific loss of ALK1 (65 kDa) and endoglin with their respective siRNAs. The migration positions of the molecular mass markers (kDa values) are shown on the endoglin blot. C, the expression of ALK1, ALK5, and endoglin were determined in the samples for panel B, confirming each siRNA was selective. Data are presented as the mean ± S.E. of four experiments. D and E, untransfected serum-restricted HPAECs were treated with BMP9 (1 ng/ml) in 0.1% for 8 or 24 h. Immunoblotting was performed with antibodies against (D) BMPR-II or (E) endoglin to confirm the increase in these proteins due to BMP9 treatment. Blots are representative of three separate experiments. *, p < 0.05 or **, p < 0.01 compared with DH1 plus BMP9. All blots were reprobed for β-actin to ensure equal loading.
We confirmed selective loss of ALK1 or endoglin protein by siALK1 or siEng transfection, respectively, by Western blotting (Fig. 3A). ALK1 protein migrated with a mass of 65 kDa. Endoglin migrated as a major band with a mass of 90 kDa and two minor bands with masses of 170 and 74 kDa. Fig. 3C shows confirmation of the specific reduction of mRNA for ALK1 (94.3 ± 0.3% reduction), ALK5 (80.7 ± 4.0% reduction), and endoglin (94.9 ± 1.1% reduction) by each siRNA. In addition, siALK1 led to a failure of BMP9 to induce the transcription of ALK5 (1.78 ± 0.35 versus 0.90 ± 0.14-fold, DH1 and siALK1) and endoglin (4.84 ± 0.95 versus 1.57 ± 0.18-fold, DH1 and siALK1).
As BMP9 induced BMPR-II and Endoglin mRNA expression at 8 h (Fig. 3, A and C), we examined whether this was reflected by increased protein expression. Treatment of HPAECs with BMP9 for 8 or 24 h increased the protein expression of BMPR-II (Fig. 3D) and endoglin (Fig. 3E), corresponding to increases in RNA expression of BMPR-II (8 h, 6.84 ± 0.50-fold; 24 h, 2.77 ± 0.54-fold) and endoglin (8 h, 5.52 ± 0.56-fold; 24 h, 3.70 ± 0.83-fold).
BMPR-II and ActR-II Differentially Contribute to BMP9-mediated Responses
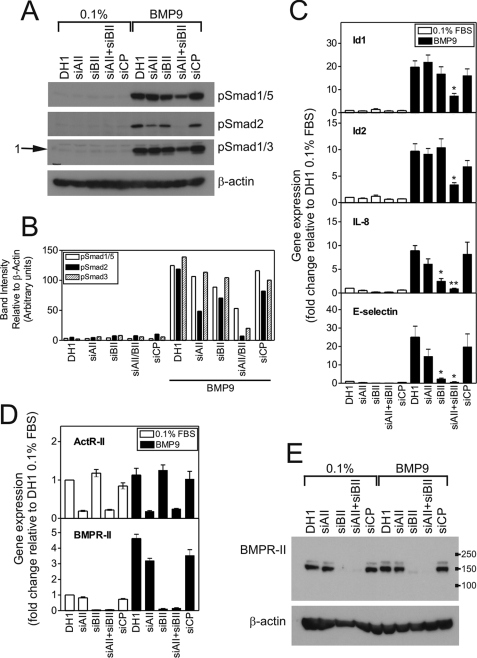
Having demonstrated a role for ALK1 in BMP9 responses, we assessed the contributions of the type II receptors, BMPR-II and ActR-II, to BMP9-mediated Smad signaling and transcriptional regulation in HPAECs. Although siBMPR-II had a small impact (20–25% reduction) on BMP9-mediated Smad1/5 phosphorylation in HPAECs, cotransfection of siActR-II and siBMPR-II was required to significantly attenuate the Smad1/5 response (Fig. 4, A and B). In contrast, siActR-II reduced BMP9-mediated Smad2 phosphorylation to a greater extent than siBMPR-II. When siActR-II and siBMPR-II were co-transfected, the reduction in BMP9-mediated phosphorylation of Smad1/5 or Smad2 was greater than for either siRNA alone.
FIGURE 4.
siActR-II and siBMPR-II differentially alter BMP9-mediated Smad phosphorylation and mRNA induction in HPAECs. A, HPAECs were transfected with siActR-II (siAII), siBMPR-II (siBII), siActR-II + siBMPR-II or siCP (10 nm each siRNA) using Dharmafect1 (DH1). After 28 h, cells were serum-restricted for 16 h, followed by treatment with BMP9 (1 ng/ml) in 0.1% for 1 h. Immunoblotting was performed with antibodies against phospho-Smad1/5, phospho-Smad2, or phospho-Smad1/3. All blots were reprobed for β-actin to ensure equal loading. The blots are representative of three separate experiments. B, graph showing densitometry for the blots in panel A. Band intensities were quantified using Image J and normalized to the β-actin blots corresponding to each Smad blot. C, HPAECs were treated as described above for 8 h. Total RNA was extracted and cDNA prepared. The expression of Id1, Id2, IL-8, and E-selectin were determined by qPCR, with expression being normalized to β-actin and expressed as the fold-change relative to 0.1%. Data are presented as the mean ± S.E. of four experiments. D, the expression of ActR-II and BMPR-II were determined in the samples for panel C, confirming each siRNA was selective. Data are presented as the mean ± S.E. of four experiments. E, cell lysates were immunoblotted for BMPR-II to confirm the specific loss of receptor expression. The migration positions of the molecular mass markers (kDa) are shown. *, p < 0.05 or **, p < 0.01 compared with DH1 plus BMP9.
We also examined the contributions of these type II receptors to BMP9-mediated mRNA induction (Fig. 4C). Only co-transfection of siActR-II and siBMPR-II significantly abrogated BMP9-mediated transcription of Id1 (19.68 ± 2.73 versus 7.26 ± 1.07-fold, DH1 and siActR-II + siBMPR-II) and Id2 (9.70 ± 1.40 versus 3.37 ± 1.15-fold, DH1 and siActR-II + siBMPR-II). Transfection of siBMPR-II or siActR- II alone abrogated the induction of IL-8 and E-selectin, with siBMPR-II having the greater effect. Co-transfection of siActR-II and siBMPR-II caused the greatest abrogation of BMP9-mediated transcription of IL-8 (8.90 ± 1.10 versus 6.12 ± 1.08 versus 2.49 ± 0.56 versus 0.90 ± 0.09-fold; DH1, siActR-II, siBMPR-II, and siActR-II + siBMPR-II, respectively) and E-selectin (25.06 ± 5.97 versus 14.66 ± 3.86 versus 2.35 ± 0.43 versus 0.69 ± 0.15-fold, DH1, siActR-II, siBMPR-II, and siActR- II + siBMPR-II, respectively).
By qPCR (Fig. 4D), we confirmed the specific reduction of mRNA for ActR-II (80.9 ± 1.8% reduction) and BMPR-II (95.5 ± 1.2% reduction) by the individual siRNAs and similar reductions in these receptors were observed with co-transfection of both siRNAs (ActR-II, 78.3 ± 1.4% reduction; BMPR-II, 94.7 ± 1.1% reduction). Western blotting for the BMPR-II protein confirmed the specificity of the siRNA (Fig. 4E), the major band migrating with a mass of ∼150 kDa, as previously shown (33). We tested a range of ActR-IIA antibodies in endothelial cells, but the protein expression was too low to obtain a clear signal, consistent with our previous report of low mRNA expression of this receptor (33). To confirm that siActR-II was capable of reducing the receptor protein expression, we cotransfected HeLa cells with a plasmid encoding myc-tagged ActR-II and siActR-II. The siRNA reduced endogenous mRNA expression by 80% and the overexpressed receptor mRNA by 65% compared with the transfection reagent alone. We further confirmed by Western blotting for ActR-II and the myc tag that siActR-II reduced the overexpressed protein (supplemental Fig. 4). ActR-IIA-myc migrated with a mass of ∼80–90 kDa.
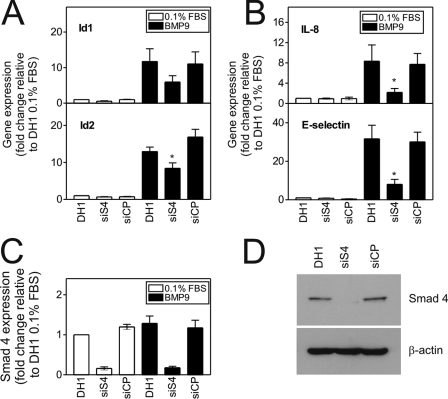
BMP9-mediated mRNA Induction Is Smad4-dependent
To address whether the observed transcriptional activity of BMP9 was Smad-dependent, we examined the effect of siSmad4 on mRNA induction. Notably, reduction of Smad4 by 80% or greater was required to have an effect (data not shown). Under these conditions (Fig. 5, A and B), siSmad4 transfection abrogated BMP9-mediated transcription of Id1 (17.66 ± 4.10 versus 7.88 ± 1.15-fold, DH1 and siSmad4, respectively, 49.9 ± 6.6% reduction), Id2 (12.95 ± 1.21 versus 8.39 ± 1.50-fold, DH1 and siSmad4, respectively, 33.7 ± 11.9% reduction, p < 0.05), IL-8 (8.33 ± 3.21 versus 2.18 ± 0.77-fold, DH1 and siSmad4, respectively, 69.2 ± 5.5% reduction, p < 0.05), and E-selectin (31.66 ± 7.08 versus 8.04 ± 2.57-fold, DH1 and siSmad4, respectively, 71.5 ± 7.7% reduction, p < 0.05). Interestingly, siSmad4 transfection also abrogated BMP9-mediated transcription of BMPR-II (6.02 ± 1.08 versus 2.10 ± 0.28-fold, DH1 and siSmad4, respectively, 63.7 ± 4.0% reduction, p < 0.05) and endoglin (9.78 ± 1.89 versus 4.89 ± 0.62-fold, 49.9 ± 6.6% reduction, DH1 and siSmad4, p < 0.05) (supplemental Fig. 5A). We confirmed the reduction of Smad4 protein by Western blotting (Fig. 5C) and mRNA (84.1 ± 3.9% reduction) by qPCR (Fig. 5D).
FIGURE 5.
BMP9 induction of mRNA transcription is Smad4-dependent. A and B, HPAECs were transfected with siSmad4 or siCP (15 nm each) using Dharmafect1 (DH1). After 28 h, cells were serum-restricted for 16 h, followed by treatment with BMP9 (1 ng/ml) in 0.1% for 8 h. Total RNA was extracted and cDNA prepared. The expression of Id1 and Id2 (A) and IL-8 and E-selectin (B) were determined by qPCR, with expression being normalized to β-actin and expressed as the fold-change relative to 0.1%. Data are presented as the mean ± S.E. of five experiments. C, the expression of Smad4 was determined using qPCR, confirming selectivity. Data are presented as the mean ± S.E. of five experiments. D, cell lysates were immunoblotted for Smad4 to confirm the specific loss of receptor expression. *, p < 0.05 compared with DH1 plus BMP9.
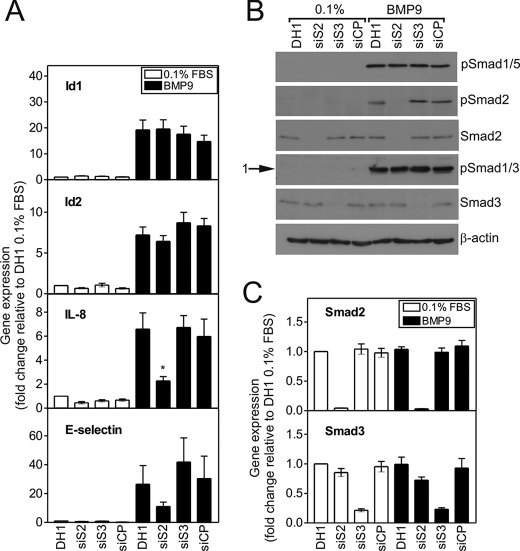
Smad2 Regulates BMP9-mediated IL-8 and E-selectin mRNA Induction
Having shown that BMP9 regulates the activation of Smad1/5 and Smad2 signaling, we questioned whether Smad2 regulates the BMP9-mediated transcriptional responses studied, and also assessed the involvement of Smad3. Neither siSmad2 nor siSmad3 altered the BMP9-mediated induction of Id1 or Id2 (Fig. 6A), or BMPR-II or endoglin (supplemental Fig. S5B). However, Smad2 siRNA transfection led to an abrogation of the IL-8 induction by BMP9 (6.58 ± 1.36 versus 2.27 ± 0.36-fold, DH1 and siSmad2, respectively, 62.1 ± 6.0% reduction, p < 0.05). In addition, siSmad2 inhibited E-selectin induction by BMP9, whereas siSmad3 slightly augmented E-selectin induction (26.44 ± 12.97 versus 11.05 ± 2.99 versus 41.85 ± 16.69-fold, DH1, siSmad2, and siSmad3, respectively). We confirmed the selective reduction of Smad2 or Smad3 protein and the identities of the protein bands by Western blotting (Fig. 6B). We also confirmed selective reduction of Smad2 (95.6 ± 0.3% reduction) and Smad3 (78.6 ± 2.5% reduction) by qPCR (Fig. 6C).
FIGURE 6.
The BMP9-mediated induction of IL-8 and E-selectin is regulated by Smad2. A, HPAECs were transfected with siSmad2, siSmad3, or siCP (10 nm each) using Dharmafect1 (DH1). After 28 h, cells were serum-restricted in M199, 0.1% FBS (0.1%) for a further 16 h, followed by treatment with BMP9 (1 ng/ml) in 0.1% for 8 h. Total RNA was extracted and cDNA prepared. The expression of Id1, Id2, IL-8, and E-selectin were determined using qPCR, with expression being normalized to β-actin and expressed as the fold-change relative to 0.1%. Data are presented as the mean ± S.E. of five experiments. B, HPAECs were treated as described above for 1 h. Immunoblotting was performed with antibodies against phospho-Smad1/5, phospho-Smad2, Smad2, phospho-Smad1/3, or Smad3. All blots were reprobed for β-actin to ensure equal loading. The blots are representative of three separate experiments. C, the expression of Smad2 and Smad3 were determined using qPCR, confirming selectivity. Data are presented as the mean ± S.E. of five experiments. *, p < 0.05 compared with DH1 plus BMP9.
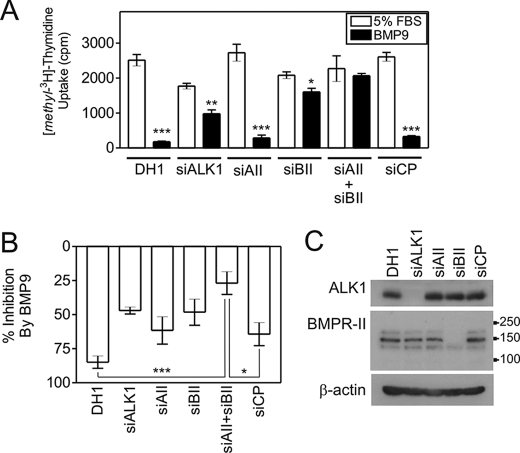
BMP9 Inhibits DNA Synthesis via ALK1 and BMPR-II in HPAECs
BMP9 potently inhibited DNA synthesis, as determined by [methyl-3H]thymidine uptake, in HPAECs. Fig. 7A shows a representative experiment. When data were normalized for four separate experiments (Fig. 7B), BMP9 inhibited DNA synthesis in DH1-treated cells (84.8 ± 4.5%) and cells transfected with siCP (64.3 ± 8.4%). Furthermore, siActR-II transfection did not significantly alter the effect of BMP9 (61.6 ± 10.1% inhibition). However, the inhibitory effect of BMP9 was abrogated by transfection of siALK1 (47.0 ± 2.5% inhibition) or siBMPR-II (48.2 ± 9.6% inhibition). The inhibitory effect of BMP9 was reversed to the greatest extent after cotransfection of siActR-II and siBMPR-II (26.9 ± 8.4% inhibition). We confirmed the specific reduction of mRNA at 28 and 52 h for ALK1 (28 h, 93.4 ± 3.0% reduction; 52 h, 91.0 ± 1.5% reduction), ActR-II (28 h, 82.1 ± 0.4% reduction; 52 h, 74.4 ± 2.5% reduction), BMPR-II (28 h, 96.7 ± 0.2% reduction; 52 h, 96.4 ± 0.5% reduction), and ActR-II plus BMPR-II (28 h, ActR-II, 71.5 ± 1.7% reduction, BMPR-II, 96.5 ± 0.6% reduction; 52 h, ActR-II, 61.4 ± 4.5% reduction, BMPR-II, 95.4 ± 0.8% reduction). We also demonstrate reduction of the ALK1 and BMPR-II proteins (Fig. 7C).
FIGURE 7.
BMP9 inhibits DNA synthesis via ALK1 and BMPR-II. A, HPAECs were transfected with siALK1, siActR-II (siAII), siBMPR-II (siBII), siActR-II + siBMPR-II, or siCP (10 nm each siRNA) using Dharmafect1 (DH1). Cells were incubated in EGM-2 overnight and the medium changed to M199, 0.1% FBS for 8 h. At 28 h post-transfection, cells were incubated with M199, 5% FBS (5% FBS) or M199, 5% FBS containing 1 ng/ml BMP9 (BMP9). After a further 18 h, 0.25 μCi of [methyl-3H]thymidine was added to each well for 6 h. The lysates were quantified by liquid scintillation counting. Quantitative PCR analysis confirmed the reduction in mRNA at 28 and 52 h. The graph is representative of five experiments (n = 4 wells/treatment). *, p < 0.05; **, p < 0.01; or ***, p < 0.001 compared with 5% FBS with Student's unpaired t test. B, mean normalized data from four experiments showing the percentage inhibition of [methyl-3H]thymidine uptake by BMP9 in HPAECs transfected with siRNAs as described above. *, p < 0.05; or ***, p < 0.001 compared with DH1 with Student's unpaired t test. C, protein lysates were immunoblotted with antibodies against ALK1 or BMPR-II to confirm selective reduction of these proteins. The migration positions of the molecular mass markers are shown on the BMPR-II blot.
DISCUSSION
Mutations in the genes encoding ALK1, BMPR-II, and endoglin underlie HHT2 (ALK1), HHT2 with PAH (ALK1), PAH (BMPR-II), and HHT1 (endoglin), implying these proteins are critical regulators of pulmonary vascular structure (2–4, 7, 8, 10, 11, 34). Although these receptors exhibit promiscuous ligand selectivities, we found that BMP9 selectively induces potent Smad responses and the induction of Id1, Id2, E-selectin, and IL-8 transcription in HPAECs. We present novel data demonstrating that BMP9 activates both the Smad1/5 and Smad2 pathways via ALK1. We confirm that ActR-II and BMPR-II compensate for each other with respect to Smad1/5 phosphorylation and Id mRNA induction, but show that ActR-II has a greater role in mediating Smad2 phosphorylation. Conversely, BMPR-II contributes more to the E-selectin and IL-8 responses than ActR-II. We also show that BMP9-mediated mRNA transcription, including induction of BMPR-II and endoglin, is Smad4-dependent. Our data suggest that the Smad1/5/8 and Smad2 pathways mediate induction of IL-8. Our findings imply that BMPR-II mutations affect a subset of BMP9-responsive pathways and may alter the balance of Smad signaling toward Smad2 in endothelial cells in PAH. In contrast, ALK1 mutations will have a wider impact on BMP9-mediated functions in HHT2, also affecting the regulation of the Id, endoglin and BMPR-II.
As mutations in TGFβ and BMP receptors are associated with diseases characterized by pulmonary vascular instability, we asked whether BMPs and TGFβ1 regulate the expression of HPAEC genes associated with vascular structure, leukocyte recruitment, and differentiation. BMP9 inhibits endothelial cell migration and proliferation via activation of ALK1 (22, 23). In addition, studies with a constitutively active ALK1 (caALK1) receptor suggest this receptor mediates the maturation phase of angiogenesis via inhibition of endothelial cell proliferation and migration, in a similar manner to TGFβ1 (35, 36). Here, we show that BMP9 elicited potent transcriptional responses in HPAECs, whereas BMP2, BMP4, BMP6, and TGFβ1 weakly induced mRNA induction. BMP9 potently induced Id1 and Id2 in HPAECs, consistent with previous studies with BMP9 and caALK1 in endothelial cells (22, 25). However, the functional roles of the Id proteins are not entirely clear, as Id1 is reported to be essential for BMP6-mediated migration in bovine aortic and murine embryonic endothelial cells (37). These contrasting roles suggest the Id proteins may function to promote or inhibit angiogenesis in a context- and ligand-specific manner through integration with different signaling pathways.
We also show the first evidence for BMP9 inducing E-selectin expression via ALK1 in endothelial cells. The selectins are endothelial adhesion molecules that mediate cell-cell contact and leukocyte recruitment (38–40). Depending on the cellular context, E-selectin may promote or inhibit angiogenesis (39, 41). We also show that BMP9 induces IL-8 expression in HPAECs more potently than TGFβ1, consistent with a previous study showing that caALK1 potently induces IL-8 in HMEC-1 cells, whereas TGFβ1 had little effect (25). IL-8 is an angiogenic cytokine that can act as a chemoattractant and mitogen for vascular smooth muscle cells (42), but may also promote leukocyte adhesion (38). Furthermore, IL-6, also induced by BMP9, is involved in lung angiogenesis and inflammation (43). Further studies are required to establish the contributions of E-selectin, IL-8, and IL-6 to BMP9 functions in the pulmonary vasculature.
We also assessed the ligand responsiveness of HPAECs by examining Smad phosphorylation. BMP9 potently induced Smad1/5 phosphorylation in HPAECs, consistent with recent studies (22, 23). BMP6 exerted a weaker Smad1/5 phosphorylation and BMP2/4 had little effect, consistent with the low ALK3 and ALK6 expression in HPAECs (33). BMP9 did not stimulate Smad3 phosphorylation, consistent with previous reports that caALK1 does not induce Smad2/3 phosphorylation (35) and BMP9 does not induce the Smad3-responsive CAGA-luciferase reporter in human dermal microvascular endothelial cells (22). Contrary to our expectations, BMP9 stimulated Smad2 phosphorylation in HPAECs and other endothelial cell types. The previously reported lack of Smad2 activation by caALK1 is consistent with the observation that the Q201D substitution in caALK1 does not phosphorylate Smad2 (44). Therefore, our observed Smad2 response may be mediated via an alternative ALK1 domain or require the presence of Type II receptors.
We explored the contributions of ALK1, ALK5, and endoglin to our observed BMP9 responses using siRNA. We confirm that ALK1 mediates BMP9-induced Smad phosphorylation and mRNA induction in HPAECs (22, 23). Previous studies of endothelial cells have shown that ALK1 requires ALK5 to mediate TGFβ1 signaling via Smad1/5 (20). Furthermore, overexpression of endoglin in murine fibroblasts was reported to enhance the BMP9-mediated activity of the BRE-luciferase reporter (22). Here, we see no effect of endoglin siRNA on BMP9 responses in HPAECs, although we cannot exclude a concentration-response effect. We observed greater TGFβ1-mediated Smad2/3 activation in HMEC-1 cells than HPAECs, suggesting that endothelial ALK5 expression may vary with cell type and may be low in HPAECs. Here, we see no effect of siALK5 or ALK5 inhibition upon BMP9 signaling in HPAECs or HMEC-1 cells, whereas TGFβ1 signaling was attenuated in HMEC-1 cells. Our data suggests ALK5 is not required for BMP9 signaling via ALK1, whereas TGFβ1 responses require ALK5.
Having confirmed the central role of ALK1 in BMP9 signaling, we sought to establish the contributions of BMPR-II and ActR-II to these responses. We show, for the first time, that ActR-II and BMPR-II are differentially involved in Smad signaling and transcriptional responses in HPAECs. It was reported that cotransfection of siActR-II and siBMPR-II was required to abrogate BMP9-driven BRE-luciferase responses in murine fibroblasts (22). Our study confirms that cotransfection of siActR-II and siBMPR-II is required to diminish BMP9-mediated Smad1/5 phosphorylation and Id mRNA transcription in HPAECs. However, we also present novel data demonstrating that BMPR-II preferentially contributes to BMP9-mediated E-selectin induction, IL-8 induction, and mitogenic inhibition compared with ActR-II. Conversely, ActR-II contributes more to the Smad2 response. We also show, using Smad4 siRNA, the Smad dependence of transcription of Id1, Id2, IL-8, E-selectin, BMPR-II, and endoglin. This is of particular relevance to the association of Smad4 mutations with HHT (45), and juvenile polyposis syndrome with associated HHT (46).
The IL-8 and E-selectin transcriptional responses were shown to be Smad2-dependent, a novel finding. Conversely, siSmad3 resulted in enhanced E-selectin expression in response to BMP9, suggesting that Smad3 may limit this particular response. The weak induction of IL-8 expression by TGFβ1 may reflect the weak Smad2 response in HPAECs (25). However, we cannot exclude a role for Smad1/5/8 in the induction of IL-8 and E-selectin, given that Smad1/5-linked siBMPR-II also abrogates IL-8 and E-selectin induction. These observations are of particular interest in the context of PAH, where BMPR-II dysfunction may cause altered IL-8 and E-selectin regulation and a reduction of Smad1/5 signaling pushing the signaling balance in favor of Smad2. In contrast, less of a role is suggested for the Id genes, which are relatively unaffected due to compensation by ActR-II.
Our data supports dysregulated BMP9 signaling having direct consequences in HHT2 (ALK1), but not HHT1 (endoglin). However, ALK1 mutations may cause disease by attenuating the functions of endoglin or BMPR-II. BMP9 and caALK1 induce the transcription of BMPR-II and endoglin in endothelial cells (22, 25). Here, we also show that BMP9 induces BMPR-II and endoglin protein expression and that reduced ALK1 expression inhibits BMP9-mediated transcription of endoglin and BMPR-II. The circulating concentration of BMP9 is 2–15 ng/ml (24), so BMP9 may maintain the expression of BMPR-II and endoglin in the pulmonary vasculature. It is plausible that dysfunctional ALK1 signaling in HHT2 could lead to reduced endoglin signaling, which may underlie both forms of HHT. Indeed, blood outgrowth endothelial cells from patients with HHT1 or HHT2 have reduced endoglin protein (47). Furthermore, endoglin-null endothelial cells exhibit altered TGFβ1 responses (48, 49), so we cannot exclude the effect of dysfunctional TGFβ responses due to dysregulated endoglin in the pathogenesis of HHT. In patients with HHT2 and PAH, the PAH component may represent dysfunctional BMPR-II signaling rather than ALK1 signaling. In patients with HHT2 and PAH, most of the mutations lead to intracellular retention of ALK1 (34). We recently reported that cysteine-substituted BMPR-II mutants are retained in the cytoplasm with wild-type ALK3 (50). Therefore, if the retained ALK1 mutants cause BMPR-II protein retention coupled with impaired induction of BMPR-II mRNA expression, this may drive a PAH phenotype.
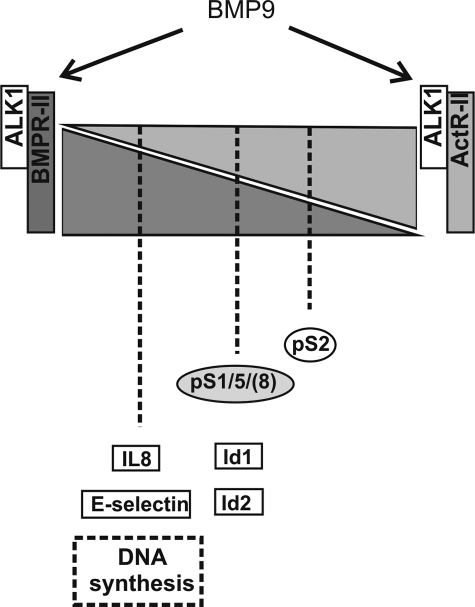
In conclusion, we propose that BMPR-II and ActR-II differentially regulate Smad and mRNA transcription in response to activation of ALK1 by BMP9 (Fig. 8). These observations provide an insight regarding how the differing functional consequences of mutations in ALK1 and BMPR-II may lead to the different pathologies HHT and PAH.
FIGURE 8.
Summary of the relative contributions of BMPR-II and ActR-II to BMP9 signaling through ALK1. Summary figure representing the relative contributions of ALK1, ActR-II, and BMPR-II to BMP9-mediated Smad signaling and transcriptional regulation in HPAECs. The shaded triangles represent the relative contribution and compensation by BMPR-II and ActR-II. The Smad, gene, and functional responses are positioned according to the type II receptor involvement/compensation, such that both receptors compensate for loss of the other with regard to Smad1/5 and Id responses. In contrast, loss of BMPR-II has a dramatic effect upon IL-8 and E-selectin expression, and inhibition of DNA synthesis by BMP9. Conversely, reduction of ActR-II has a greater impact on Smad2 phosphorylation than BMPR-II reduction, but loss of both receptors abolishes the Smad2 response.
Supplementary Material
Acknowledgments
We thank Professor D. A. Marchuk (Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, NC) for kindly providing the ALK1 antibody. We also thank Dr. Y. Sidis for kindly providing the ACTR-IIA-myc plasmid construct and Dr. P. B. Yu for advice regarding this manuscript (Massachusetts General Hospital, Boston, MA). P. U. thanks J. Deighton for technical assistance with the IL-8 enzyme-linked immunosorbent assay (Department of Medicine, University of Cambridge, Cambridge, UK).
This work was supported by British Heart Foundation Programme Grant RG/03/005 (to N. W. M. and R. C. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S2–S5.
- PAH
- pulmonary arterial hypertension
- EC
- endothelial cell
- HHT
- hereditary hemorrhagic telangectasia
- BMPR-II
- bone morphogenetic protein type II receptor
- TGF
- transforming growth factor
- ALK1
- activin-like kinase receptor 1
- HPAEC
- human pulmonary artery endothelial cell
- caALK
- constitutively active ALK
- siRNA
- small interfering RNA
- HPASMC
- human pulmonary artery smooth muscle cell
- FBS
- fetal bovine serum
- qPCR
- quantitative PCR
- TGFβ1
- transforming growth factor β1
- IL
- interleukin.
REFERENCES
- 1.Tuder R. M., Marecki J. C., Richter A., Fijalkowska I., Flores S. ( 2007) Clin. Chest Med. 28, 23– 42, vii [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane K. B., Machado R. D., Pauciulo M. W., Thomson J. R., Phillips J. A., 3rd, Loyd J. E., Nichols W. C., Trembath R. C. ( 2000) Nat. Genet. 26, 81– 84 [DOI] [PubMed] [Google Scholar]
- 3.Deng Z., Morse J. H., Slager S. L., Cuervo N., Moore K. J., Venetos G., Kalachikov S., Cayanis E., Fischer S. G., Barst R. J., Hodge S. E., Knowles J. A. ( 2000) Am. J. Hum. Genet. 67, 737– 744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machado R. D., Aldred M. A., James V., Harrison R. E., Patel B., Schwalbe E. C., Gruenig E., Janssen B., Koehler R., Seeger W., Eickelberg O., Olschewski H., Elliott C. G., Glissmeyer E., Carlquist J., Kim M., Torbicki A., Fijalkowska A., Szewczyk G., Parma J., Abramowicz M. J., Galie N., Morisaki H., Kyotani S., Nakanishi N., Morisaki T., Humbert M., Simonneau G., Sitbon O., Soubrier F., Coulet F., Morrell N. W., Trembath R. C. ( 2006) Hum. Mutat. 27, 121– 132 [DOI] [PubMed] [Google Scholar]
- 5.Sadick H., Sadick M., Götte K., Naim R., Riedel F., Bran G., Hörmann K. ( 2006) Wien. Klin. Wochenschr. 118, 72– 80 [DOI] [PubMed] [Google Scholar]
- 6.Cottin V., Plauchu H., Bayle J. Y., Barthelet M., Revel D., Cordier J. F. ( 2004) Am. J. Respir. Crit. Care Med. 169, 994– 1000 [DOI] [PubMed] [Google Scholar]
- 7.McAllister K. A., Grogg K. M., Johnson D. W., Gallione C. J., Baldwin M. A., Jackson C. E., Helmbold E. A., Markel D. S., McKinnon W. C., Murrell J. ( 1994) Nat. Genet. 8, 345– 351 [DOI] [PubMed] [Google Scholar]
- 8.Johnson D. W., Berg J. N., Baldwin M. A., Gallione C. J., Marondel I., Yoon S. J., Stenzel T. T., Speer M., Pericak-Vance M. A., Diamond A., Guttmacher A. E., Jackson C. E., Attisano L., Kucherlapati R., Porteous M. E., Marchuk D. A. ( 1996) Nat. Genet. 13, 189– 195 [DOI] [PubMed] [Google Scholar]
- 9.Trembath R. C. ( 2001) J. Heart Lung Transplant. 20, 175. [DOI] [PubMed] [Google Scholar]
- 10.Abdalla S. A., Gallione C. J., Barst R. J., Horn E. M., Knowles J. A., Marchuk D. A., Letarte M., Morse J. H. ( 2004) Eur. Respir. J. 23, 373– 377 [DOI] [PubMed] [Google Scholar]
- 11.Trembath R. C., Thomson J. R., Machado R. D., Morgan N. V., Atkinson C., Winship I., Simonneau G., Galie N., Loyd J. E., Humbert M., Nichols W. C., Morrell N. W., Berg J., Manes A., McGaughran J., Pauciulo M., Wheeler L. ( 2001) N. Engl. J. Med. 345, 325– 334 [DOI] [PubMed] [Google Scholar]
- 12.van den Driesche S., Mummery C. L., Westermann C. J. ( 2003) Cardiovasc. Res. 58, 20– 31 [DOI] [PubMed] [Google Scholar]
- 13.Ebisawa T., Tada K., Kitajima I., Tojo K., Sampath T. K., Kawabata M., Miyazono K., Imamura T. ( 1999) J. Cell Sci. 112, 3519– 3527 [DOI] [PubMed] [Google Scholar]
- 14.Liu F., Hata A., Baker J. C., Doody J., Cárcamo J., Harland R. M., Massagué J. ( 1996) Nature 381, 620– 623 [DOI] [PubMed] [Google Scholar]
- 15.Hoodless P. A., Haerry T., Abdollah S., Stapleton M., O'Connor M. B., Attisano L., Wrana J. L. ( 1996) Cell 85, 489– 500 [DOI] [PubMed] [Google Scholar]
- 16.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. ( 1998) EMBO J. 17, 3091– 3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verrecchia F., Vindevoghel L., Lechleider R. J., Uitto J., Roberts A. B., Mauviel A. ( 2001) Oncogene 20, 3332– 3340 [DOI] [PubMed] [Google Scholar]
- 18.Chen X., Weisberg E., Fridmacher V., Watanabe M., Naco G., Whitman M. ( 1997) Nature 389, 85– 89 [DOI] [PubMed] [Google Scholar]
- 19.Hoodless P. A., Tsukazaki T., Nishimatsu S., Attisano L., Wrana J. L., Thomsen G. H. ( 1999) Dev. Biol. 207, 364– 379 [DOI] [PubMed] [Google Scholar]
- 20.Goumans M. J., Valdimarsdottir G., Itoh S., Lebrin F., Larsson J., Mummery C., Karlsson S., ten Dijke P. ( 2003) Mol. Cell 12, 817– 828 [DOI] [PubMed] [Google Scholar]
- 21.Goumans M. J., Valdimarsdottir G., Itoh S., Rosendahl A., Sideras P., ten Dijke P. ( 2002) EMBO J. 21, 1743– 1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.David L., Mallet C., Mazerbourg S., Feige J. J., Bailly S. ( 2007) Blood 109, 1953– 1961 [DOI] [PubMed] [Google Scholar]
- 23.Scharpfenecker M., van Dinther M., Liu Z., van Bezooijen R. L., Zhao Q., Pukac L., Löwik C. W., ten Dijke P. ( 2007) J. Cell Sci. 120, 964– 972 [DOI] [PubMed] [Google Scholar]
- 24.David L., Mallet C., Keramidas M., Lamandé N., Gasc J. M., Dupuis-Girod S., Plauchu H., Feige J. J., Bailly S. ( 2008) Circ. Res. 102, 914– 922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lux A., Salway F., Dressman H. K., Kröner-Lux G., Hafner M., Day P. J., Marchuk D. A., Garland J. ( 2006) BMC Cardiovasc. Disord. 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park S. O., Lee Y. J., Seki T., Hong K. H., Fliess N., Jiang Z., Park A., Wu X., Kaartinen V., Roman B. L., Oh S. P. ( 2008) Blood 111, 633– 642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrell N. W., Yang X., Upton P. D., Jourdan K. B., Morgan N., Sheares K. K., Trembath R. C. ( 2001) Circulation 104, 790– 795 [DOI] [PubMed] [Google Scholar]
- 28.Livak K. J., Schmittgen T. D. ( 2001) Methods 25, 402– 408 [DOI] [PubMed] [Google Scholar]
- 29.Cowburn A. S., Deighton J., Walmsley S. R., Chilvers E. R. ( 2004) Eur. J. Immunol. 34, 1733– 1743 [DOI] [PubMed] [Google Scholar]
- 30.Morrell N. W., Upton P. D., Kotecha S., Huntley A., Yacoub M. H., Polak J. M., Wharton J. ( 1999) Am. J. Physiol. 277, L440– 448 [DOI] [PubMed] [Google Scholar]
- 31.Seki T., Hong K. H., Oh S. P. ( 2006) Lab. Investig. 86, 116– 129 [DOI] [PubMed] [Google Scholar]
- 32.Uhl M., Aulwurm S., Wischhusen J., Weiler M., Ma J. Y., Almirez R., Mangadu R., Liu Y. W., Platten M., Herrlinger U., Murphy A., Wong D. H., Wick W., Higgins L. S., Weller M. ( 2004) Cancer Res. 64, 7954– 7961 [DOI] [PubMed] [Google Scholar]
- 33.Upton P. D., Long L., Trembath R. C., Morrell N. W. ( 2008) Mol. Pharmacol. 73, 539– 552 [DOI] [PubMed] [Google Scholar]
- 34.Harrison R. E., Flanagan J. A., Sankelo M., Abdalla S. A., Rowell J., Machado R. D., Elliott C. G., Robbins I. M., Olschewski H., McLaughlin V., Gruenig E., Kermeen F., Halme M., Räisänen-Sokolowski A., Laitinen T., Morrell N. W., Trembath R. C. ( 2003) J. Med. Genet. 40, 865– 871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamouille S., Mallet C., Feige J. J., Bailly S. ( 2002) Blood 100, 4495– 4501 [DOI] [PubMed] [Google Scholar]
- 36.Pepper M. S. ( 1997) Cytokine Growth Factor Rev. 8, 21– 43 [DOI] [PubMed] [Google Scholar]
- 37.Valdimarsdottir G., Goumans M. J., Rosendahl A., Brugman M., Itoh S., Lebrin F., Sideras P., ten Dijke P. ( 2002) Circulation 106, 2263– 2270 [DOI] [PubMed] [Google Scholar]
- 38.Szekanecz Z., Koch A. E. ( 2000) Arthritis Res. 2, 368– 373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen M., Strubel N. A., Bischoff J. ( 1993) Nature 365, 267– 269 [DOI] [PubMed] [Google Scholar]
- 40.Bullard D. C., Kunkel E. J., Kubo H., Hicks M. J., Lorenzo I., Doyle N. A., Doerschuk C. M., Ley K., Beaudet A. L. ( 1996) J. Exp. Med. 183, 2329– 2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu Y., Moulton K. S., Khan M. K., Vineberg S., Boye E., Davis V. M., O'Donnell P. E., Bischoff J., Milstone D. S. ( 2004) Proc. Natl. Acad. Sci. U. S. A. 101, 8005– 8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue T. L., Wang X., Sung C. P., Olson B., McKenna P. J., Gu J. L., Feuerstein G. Z. ( 1994) Circ. Res. 75, 1– 7 [DOI] [PubMed] [Google Scholar]
- 43.McClintock J. Y., Wagner E. M. ( 2005) J. Appl. Physiol. 99, 861– 866 [DOI] [PubMed] [Google Scholar]
- 44.Chen Y. G., Massagué J. ( 1999) J. Biol. Chem. 274, 3672– 3677 [DOI] [PubMed] [Google Scholar]
- 45.Gallione C. J., Richards J. A., Letteboer T. G., Rushlow D., Prigoda N. L., Leedom T. P., Ganguly A., Castells A., Ploos van Amstel J. K., Westermann C. J., Pyeritz R. E., Marchuk D. A. ( 2006) J. Med. Genet. 43, 793– 797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallione C. J., Repetto G. M., Legius E., Rustgi A. K., Schelley S. L., Tejpar S., Mitchell G., Drouin E., Westermann C. J., Marchuk D. A. ( 2004) Lancet 363, 852– 859 [DOI] [PubMed] [Google Scholar]
- 47.Fernandez-L. A., Sanz-Rodriguez F., Zarrabeitia R., Pérez-Molino A., Hebbel R. P., Nguyen J., Bernabéu C., Botella L. M. ( 2005) Cardiovasc. Res. 68, 235– 248 [DOI] [PubMed] [Google Scholar]
- 48.Pece-Barbara N., Vera S., Kathirkamathamby K., Liebner S., Di Guglielmo G. M., Dejana E., Wrana J. L., Letarte M. ( 2005) J. Biol. Chem. 280, 27800– 27808 [DOI] [PubMed] [Google Scholar]
- 49.Lee N. Y., Ray B., How T., Blobe G. C. ( 2008) J. Biol. Chem. 283, 32527– 32533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sobolewski A., Rudarakanchana N., Upton P. D., Yang J., Crilley T. K., Trembath R. C., Morrell N. W. ( 2008) Hum. Mol. Genet. 17, 3180– 3190 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.