Abstract
Alzheimer disease (AD) is characterized by senile plaques, which are mainly composed of β amyloid (Aβ) peptides. Aβ is cleaved off from amyloid precursor protein (APP) with consecutive proteolytic processing: β-secretase, followed by γ-secretase. Here, we show that BRI3, a member of the BRI gene family that includes the familial British and Danish dementia gene BRI2, interacts with APP and serves as an endogenous negative regulator of Aβ production. BRI3 colocalizes with APP along neuritis in differentiated N2a cells; endogenous BRI3-APP complexes are readily detectable in mouse brain extract; reducing endogenous BRI3 levels by RNA interference results in increased Aβ secretion. BRI3 resembles BRI2, because BRI3 overexpression reduces both α- and β-APP cleavage. We propose that BRI3 inhibits the various processing of APP by blocking the access of α- and β-secretases to APP. However, unlike BRI2, the binding of BRI3 to the β-secretase cleaved APP C-terminal fragment is negligible and BRI3 does not cause the massive accumulation of this APP fragment, suggesting that, unlike BRI2, BRI3 is a poor γ-cleavage inhibitor. Competitive inhibition of APP processing by BRI3 may provide a new approach to AD therapy and prevention.
About 1% of humans aged 60–64 years have AD,2 increasing steadily to as many as 35–40% after age 85 (1). AD progressively leads to a severely impaired state and complete social dependence. At autopsy, cerebral atrophy, neurofibrillary tangles, and amyloid plaques are observed in the hippocampus, entorhinal cortex, amygdala, and other areas. Tangles consist of intraneuronal masses of helically wound filaments of the hyperphosphorylated protein, Tau. Plaques are extracellular deposits of Aβ, a peptide derived from cleavage of APP, surrounded by dystrophic neurites. Most AD cases present Aβ deposits in cortical and/or meningeal microvessels. In a minority of cases, this vascular cerebral amyloid angiopathy is rather severe (2).
APP is a type I transmembrane protein that undergoes a series of proteolytic cleavages (3). β-Secretase cleaves APP into a soluble ectodomain (sAPPβ) and a membrane-bound C-terminal fragment of 99 amino acids (C99). C99 is cleaved by the γ-secretase, which consists of a multicomponent complex composed of presenilins (PS1 and PS2), Nicastrin, PEN2, and APH1 (4). The γ-cleavage releases two peptides: Aβ peptide, consisting of 2 major species of 40 and 42 amino acids (Aβ40 and Aβ42, respectively) and an intracellular product APP intracellular domain (AID)/AICD. Several findings point to the short AID/AICD as a biologically active intracellular peptide, which may modulate cell death, Notch signaling, gene transcription, and Ca2+ homeostasis (5–19). In an alternative pathway, APP is processed by α-secretase within the Aβ sequence, leading to the production of the soluble sAPPα ectodomain and a membrane-bound C-terminal fragment of 83 amino acids (C83). For aged patients, a family history of dementia is a major risk factor for AD, and 10–15% of all AD subjects have a family history consistent with an autosomal dominant trait. These familial cases are due to mutations in APP and in presenilins, because they alter the rate of APP processing and Aβ42 generation.
Given the role of APP processing by secretases to AD pathology and APP-mediated functions, identifying the molecules that regulate APP cleavage is physiologically relevant and of therapeutic interest. A genetic screen aimed to the identification of regulators of APP processing led to the identification of BRI2 as an APP ligand that inhibits Aβ formation both in vitro (20) and in vivo (21). BRI2 is a type II membrane protein of 266 amino acids of unknown function that is mutated in AD-like familial British dementia (22) and familial Danish dementia (23).
In the same screening (20), we found BRI3 as an APP-interacting protein as well. The physiological functions of BRI3 remained largely unclear. Because BRI2, the homolog of BRI3, interacts with APP and inhibits APP processing (20, 21, 24) we investigated the role of BRI3 in regulating the processing of APP.
MATERIALS AND METHODS
Split-ubiquitin Screening
The construction of APP as bait and screening method were described elsewhere (20).
Cell Culture, Transfection, Plasmids, and Antibodies
Cell lines, transfection methods, mammalian expression constructs of APP, FLAG-BRI2, APP-Ncas, GFP-AID, APP-GFP, and BACE were described before (20, 25). BRI3 from the two-hybrid screening and BRI1 cDNA purchased from Open Biosystems (GenBankTM accession number BC040437) were cloned into pcDNA3.1 vector (Invitrogen) with an N-terminal FLAG tag. London and Swedish mutations were introduced into APP by QuikChange XL (Stratagene). FLAG-BRI3-Myc has an Myc tag and a glycine insertion (GEQKLISEEDL) just before the stop codon of FLAG-BRI3. Human APLP2 were cloned in pcDNA3.1/Hygro. A mammalian expression construct of Fas ligand was kindly provided by Dr. Lenardo. To make shRNA vectors, oligonucleotides pairs were annealed and cloned into a modified pSUPER (26), which is driven by an H1 promoter. Oligonucleotide pairs used were: sh12 (5′-gatccccgtctacatctacagatactttcaagagaagtatctgtagatgtagacttttt-3′ and 5′-agctaaaaagtctacatctacagatacttctcttgaaagtatctgtagatgtagacggg-3′) and m1 (5′-gatccccgcacatgattattactgatttcaagagaatcagtaataatcatgtgcttttt-3′ and 5′-agctaaaaagcacatgattattactgattctcttgaaatcagtaataatcatgtgcggg-3′). pcDNA6-Myc-BRI1 and pcDNA6-Myc-BRI3 used for stable transfectants were created by cloning BRI1 and BRI3 fragments from FLAG-BRI1 and FLAG-BRI3 into pcDNA6/Myc His C (Invitrogen) with an Myc tag linker, which inserts MGEQKLISEEDLVPGSA at the N termini of both genes. GFP-BRI3 was created by cloning BamHI/XhoI fragment from FLAG-BRI3 into BglII/SalI of pEGFP-C1, which adds GFP to the N terminus of BRI3.
The following antibodies and antibody beads were used: αFLAG (M2, Sigma F1804); α Myc (Cell Signaling, 2276); αAPP (22C11, Chemicon MAB348); αsAPPα (IBL 11088); αsAPPβ (IBL 18957); αAPP C-terminal fragments (CTF) (αAPPct, Invitrogen/Zymed Laboratories Inc. 36-6900); αAPLP2 (EMD/Calbiochem 171616); αsAPLP2 (R&D Systems MAB4945); α Fas ligand (BD Pharmingen 556387); FLAG M2 beads (Sigma A2220). Rabbit αBRI2 was described before (21). Rabbit polyclonal antibody against BRI3 (αBRI3) was raised against the peptide LTPAREERPPRHRS (corresponding to amino acids 36–49) and purified using the peptide. Rabbit polyclonal antibody and horseradish peroxidase-conjugated secondary antibodies were from Southern Biotechnology.
Immunoprecipitation from HeLa Cells
HeLa cells were transfected with indicated combinations of plasmids. The transfected cells were lysed in the Hepes-Triton buffer (20 mm Hepes/NaOH, pH 7.4, 1 mm EDTA, 150 mm NaCl, 0.5% Triton-X 100) on ice. The lysates were cleared by spinning at 20 kg for 10 min. To precipitate the immune complex, the cleared lysates were mixed with FLAG beads or with indicated antibodies and protein A beads (Pierce). A rabbit polyclonal antibody was used as a negative control of αAPPct precipitation. After being washed three times with the same buffer, the precipitated beads were boiled in 2× SDS Buffer and analyzed by Western blot.
Staining of Differentiated N2A
N2A cells were maintained in Dulbecco's modified Eagle's medium supplemented with penicillin, streptomycin, and 10% fetal bovine serum in a 5% CO2 incubator. N2A cells were plated on coverslips coated with poly l-lysine (Sigma) and were transfected with indicated plasmids. The differentiation of N2A cells was induced by 0.1% fetal bovine serum and 2 μm retinoic acid for 24–36 h (27). The cells on coverslips were fixed, permeabilized with 0.2% Triton X-100, stained as described (28), and analyzed by confocal microscopy (Bio-Rad Radiance 2000). The images shown are representative of the phenotypes observed in at least three independent experiments.
Immunoprecipitation from Mouse Brain Extract
A whole mouse brain was homogenized in 2 ml of the Hepes-sucrose buffer (20 mm Hepes/NaOH, pH 7.4, 1 mm EDTA, 1 mm EGTA, 0.25 m sucrose) supplemented with complete protease inhibitor without EDTA (Roche Applied Science). Unbroken tissues and nuclei were cleared by spinning at 1000 × g for 10 min. The pellet was homogenized in the same buffer and cleared again. This procedure was repeated three times in total. All the supernatants were combined, and the brain membrane was collected by spinning at 100 kg for 45 min. The membrane pellets were extracted with Hepes-Triton buffer at 2 mg/ml and were cleared by spinning at 100 kg for 45 min. The cleared extracts were then pre-cleared by mixing with protein A beads. The protein A beads were removed, and the extracts were mixed with αBRI3 or a control antibody and fresh protein A beads. After the incubation, beads were washed three times with the Hepes-Triton buffer, boiled in 2× SDS buffer, and subjected to Western blot.
Measurement of APP Fragments of HEK293APP Cells
HEK293APP cells were transfected with pcDNA3 or with FLAG-BRI3. One day after the transfection, the cells were conditioned for 24 h. The conditioned media were collected and cleared by spinning at 20 kg for 10 min. Aβ in the media was measured using the ELISA kit for Aβ40 and Aβ42 (IBL 17713 and 17711, respectively). Cells were lysed in the Hepes-Triton buffer, and protein concentration was determined by Bradford method using bovine serum albumin as a standard. Equal amounts of total lysates and the cleared media were analyzed by Western blot.
Analysis of shRNA-transfected HEK293APP Cells
The cloned shRNA vectors were transfected into HEK293APP cells. Two days after the transfection, total mRNA was prepared from the transfected cells and untransfected cells by using an RNeasy mini kit (Qiagen). The RNA was reverse transcribed with Superscript III (Invitrogen) with random hexamers, and quantitative PCR analysis were performed with pairs of primers (β-actin, 5′-aaggccaaccgcgagaagat-3′ and 5′-tatccctgtacgcctctgg-3′; BRI3, 5′-gcatggtcgtgctgctcat-3′ and 5′-gcacaccacagcggaagaa-3′) using a SYBR Green PCR Master Mix (Applied Biosystems) and ABI PRISM 7900HT Sequence Detection System (Applied Biosystems).
Two days after the transfection, the cells were conditioned for 24 h, and Aβ in the media or the cells were processed as above. A comparable amount was loaded and analyzed by Western blot.
FACS Analysis of shRNA-transfected HEK293APP Cells
HEK293APP cells were transfected with the indicated combinations of dsRed (Clontech), GFP-BRI3, m1, and sh12. Two days after the transfection, the cells were collected in phosphate-buffered saline containing 2% fetal bovine serum and analyzed with FACSCalibur (BD Biosciences). Live cells were gated and dsRed and GFP-BRI3 were detected in FL2 and FL1 channels with appropriate compensation to separate singly or dually transfected cells. The geometric average of dsRed-positive cells was calculated with the cells gated as dsRed-positive and cotransfected.
Metabolic Labeling of HEK293APP Cells and HeLa Cells
HEK293APP cells were transfected with pcDNA3 or FLAG-BRI3. One day after the transfection, the cells were starved for 90 min by incubating in Dulbecco's modified Eagle's medium without cysteine and methionine. The cells were labeled at 500 μCi/ml of [35S]cysteine and methionine in the same media and chased in the complete Dulbecco's modified Eagle's medium for the indicated times. After the chase, the cells were lysed and immunoprecipitated with αAPPct as in HeLa cells. The precipitants were analyzed by SDS-PAGE and autoradiography. The metabolic labeling of HeLa cells was performed similarly except the cells were transfected also with APP, and a longer chase time was used.
AID Production Measurement
Gal4-driven luciferase assay was described elsewhere (20). Briefly, HEK293T cells were transfected with or without BRI3, together with APP fused to Gal4 (APP-Gal4), pG5E1b-luc, and β-galactosidase. When AID is produced from the APP-Gal4 fusion protein, Gal4 is released from membrane-bound APP and increases the transcription under the Gal4-dependent G5E1b promoter. Luciferase activity was normalized with β-galactosidase activity.
Stably Transfected γ30 Cells
The γ30 cells (generously provided by Dr. Dennis Selkoe) are Chinese hamster ovary cells stably expressing APP, PS1, AphI, and Pen2 (29). The γ30 cells were transfected with pcDNA6-Myc-BRI1 or pcDNA6- Myc-BRI3, and the transfectants were selected in complete Dulbecco's modified Eagle's medium containing 200 μg/ml G418, 2.5 μg/ml puromycin, 250 μg/ml Zeocin, and 250 μg/ml hygromycin. After limiting dilution, each clone was analyzed for the expression of BRI proteins by α-Myc Western blot.
The immunoprecipitation was performed as in transfected HeLa cells. APP fragments were analyzed as in HEK293APP cells except that conditioning started 1 day after plating without further transfection.
Computer Analysis
The sequence alignment was done using CLUSTAL 2.0.10 multiple sequence alignment (available on the web). The BRICHOS and the transmembrane domains were determined using SMART (available on the web).
RESULTS
BRI3 and BRI2 Bind APP but BRI1 Does Not
To identify APP ligands, we screened human brain cDNA library with split-ubiquitin system using APP as bait (20). One of the clones isolated contained the entire BRI3 cDNA. BRI3 was originally cloned as a type II transmembrane protein of 267 amino acids (30, 31). Similar to BRI2, BRI3 is also cleaved by furin or furin-like convertases, and the C-terminal peptide is secreted (32).
The BRI3 cDNA obtained in the two-hybrid screening was cloned into a FLAG-tagged mammalian expression vector, and cotransfected into HeLa cells together with APP. Immunoprecipitation with FLAG beads indicated that BRI3 indeed binds APP (Fig. 1, B and C). This binding of BRI3 to APP is similar to that of BRI2 (20), a dementia gene mutated in familial British and familial Danish dementia (22, 23). Just like BRI2, BRI3 does not bind C83 (the 83-amino acid APP CTF produced by the α-secretase cleavage). However, in contrast to BRI2, BRI3 shows minimal or no binding to C99 (the 99-amino acid APP CTF) (Fig. 1B). To confirm the interaction, FLAG-BRI3 was cotransfected with APP into HeLa cells, and the lysates were immunoprecipitated with the APP CTF antibody (αAPPct) or a control rabbit polyclonal antibody (RP). The APP antibody specifically precipitated BRI3, confirming that BRI3 binds APP (Fig. 1C).
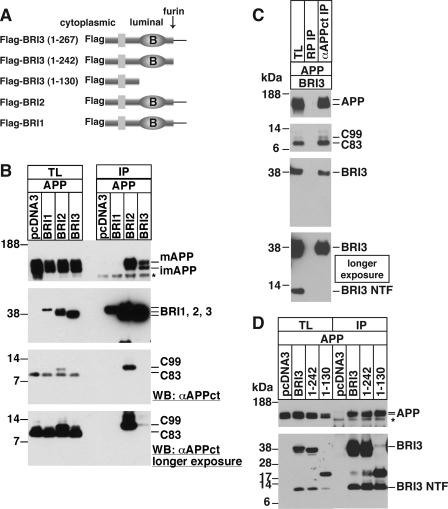
FIGURE 1.
APP bind BRI2 and BRI3 but not BRI1. A, a schematic of FLAG-BRI3, two deletion constructs, a FLAG-BRI2, and a FLAG-BRI1 construct. The locations of the FLAG tag, the site of furin cleavage, the cytoplasmic and luminal domains, the BRICHOS domain (B), and the number of amino acids coded by the constructs are indicated. B, the members of the BRI gene family (BRI1, BRI2, and BRI3) or pcDNA3 were cotransfected with APP into HeLa cells, and BRIs were precipitated. Total lysates (TL) and FLAG immunoprecipitants (IP) were analyzed for full-length APP, APP CTFs, and BRI3 by Western blot with 22C11, αAPPct, or αFLAG antibodies, respectively. Mature and immature APP, the APP CTFs of 99 or 83 amino acids (C99 or C83) are indicated. The band marked with an asterisk was attributed to the FLAG antibody used in the immunoprecipitation. C, FLAG-BRI3 (BRI3) was transfected into HeLa cells together with APP. Total lysates (TL), samples immunoprecipitated with either rabbit polyclonal antibody (RP) or with αAPPct were probed for APP and BRI3 as in B. The N-terminal fragment of BRI3 (BRI3 NTF) is indicated. D, amino acids 1–130 of BRI3 are sufficient for the BRI3-APP interaction. Two deletion constructs (BRI3-(1–242) or BRI3-(1–130)) and indicated plasmids were cotransfected with APP into HeLa cells, and the FLAG immunoprecipitates were isolated and analyzed as in A.
The results that two members of BRI family bind APP prompted us to test whether this is also true for the other member of the family, BRI1. BRI1 is also a type II transmembrane protein of 264 amino acids, which is primarily expressed in chondrocytes (31). FLAG-BRI1 shares overall topology with FLAG-BRI3 as depicted in Fig. 1A. Each member of the BRI family was transfected into HeLa cells together with APP, and BRIs were precipitated from the prepared lysates with FLAG beads. As shown in Fig. 1B, BRI1 did not bind APP, whereas BRI2 and BRI3 did. As described above, BRI2 and BRI3 share similar binding characteristics, because they both bind to APP but not to C83 (Fig. 1B). However, there two clear distinctions: the increase of C99 in total lysates was undetectable in BRI3-transfected cells, and C99 bound to BRI3 was minimal when compared with that bound to BRI2.
To determine the domain responsible for the BRI3-APP interaction, a BRI3 construct lacking the C-terminal peptide released by furin cleavage (FLAG-BRI3-(1–242)), another lacking the entire BRICHOS domain (33) (FLAG-BRI3-(1–130)), and another BRI3 tagged with both FLAG and Myc were tested for their interaction. As shown in Fig. 1D, all tested BRI3 constructs bound similarly to APP. Expression of BRI3 results in the appearance of a shorter N-terminal fragment (BRI3-NTF, Fig. 1C). This fragment, which may perhaps derive by ADAMs cleavage of BRI3 in the ectodomain like for BRI2 (34), does not interact with APP (Fig. 1C). These data indicate that the BRICHOS domain and the secreted peptide are dispensable for this interaction. These data, together with the evidence that BRI3-NTF does not bind APP (Fig. 1C), suggest that the APP-interacting domain of BRI3 is NH2-terminal to amino acid 130 and in the extracellular/lumen region of BRI3 juxtaposed to the membrane. However, it is also possible that BRI3-NTF is generated in different subcellular compartments where it cannot access APP.
BRI3 Specifically Interacts with APP via the Juxtamembrane Region
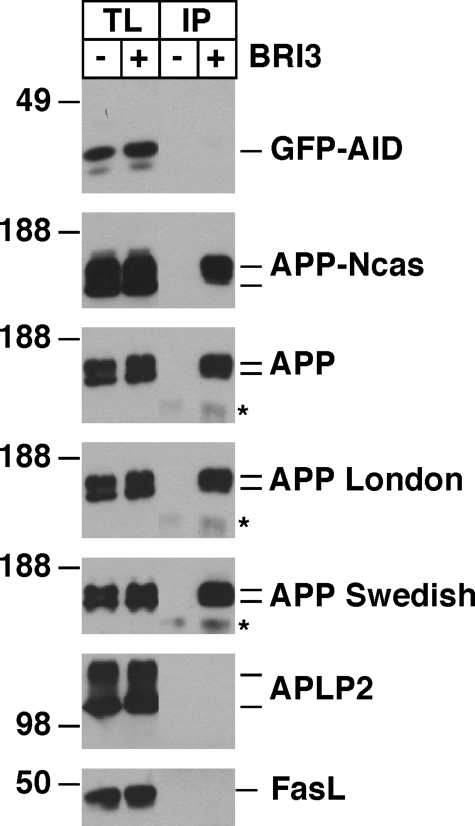
To investigate the specificity of BRI3-APP binding, a GFP fusion construct of APP intracellular domain (GFP-AID), an APP construct lacking intracellular C-terminal 31 amino acids (APP-Ncas), two familial AD mutants of APP (APP London and APP Swedish), an APP homolog (APLP2), or unrelated type II transmembrane protein (Fas ligand) was cotransfected to HeLa cells with or without FLAG-BRI3. Isolated FLAG immunocomplex showed that BRI3 binds APP, APP-Ncas, and London and Swedish mutants of APP but does not bind GFP-AID (Fig. 2A). This indicates that deleting 31 amino acids of APP-CTF or the two Alzheimer mutations around the Aβ region does not affect BRI3-APP binding, and that the intracellular domain of APP is not sufficient for the binding. Although we cannot exclude that C83 and BRI3 do not interact, because they are segregated in distinct subcellular compartments, these data suggest that the domain of APP required for the interaction should be at least N-terminal to α-cleavage site, and probably N-terminal to β-cleavage site. The lack of interaction with APLP2 or Fas ligand further attests the specificity of the BRI3-APP interaction.
FIGURE 2.
BRI3 binds the extracellular region of APP. GFP-AID, APP-Ncas, APP, APP London mutant, APP Swedish mutant, APLP2, or Fas ligand (FasL) was tested for their interaction with BRI3 as in Fig. 1A. The total lysates and FLAG immunoprecipitants were analyzed by Western blot with FLAG (BRI3), 22C11 (APP, APP-Ncas, APP London, APP Swedish), αAPPct (GFP-AID), or antibodies against APLP2 or Fas ligand.
BRI3 and APP Colocalize in Differentiated N2A Cells
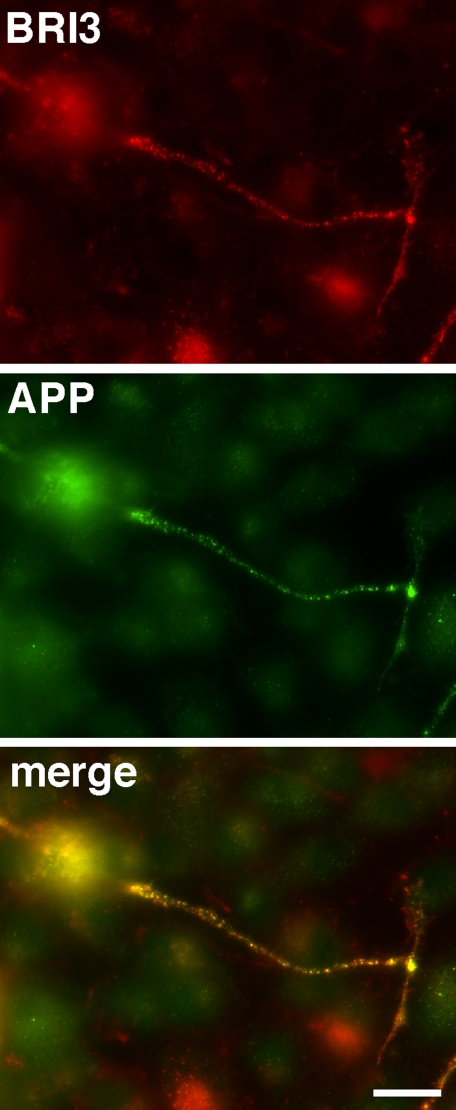
FLAG-tagged BRI3 and GFP-APP were cotransfected into N2A cells. The transfected cells were differentiated with retinoic acid, and the location of BRI3 and APP were visualized with αFLAG antibody and GFP fluorescence, respectively. As shown in Fig. 3, BRI3 and APP colocalize nicely as a dotted structure along the neurites, further supporting the idea that BRI3 and APP form a complex in vivo.
FIGURE 3.
BRI3 shows clear colocalization with APP along neurites of differentiated N2A cells. N2A cells were transfected with FLAG-BRI3 and GFP-APP. BRI staining (red), GFP-APP fluorescence (green), and merged photographs are displayed. Bar = 10 μm.
BRI3 Overexpression Reduces Aβ and APP Intracellular Domain Production
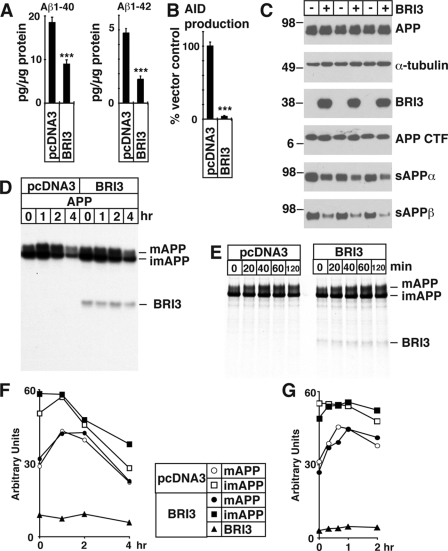
Next, we asked if BRI3 changes the production of Aβ. HEK293APP cells, which stably express APP, were transfected with pcDNA3 or with FLAG-BRI3. The transfectants were conditioned, and the secreted Aβ40 and Aβ42 were measured with Aβ ELISA. As shown in Fig. 4A, exogenous expression of BRI3 decreased both the secreted Aβ40 and Aβ42.
FIGURE 4.
A, BRI3 suppresses Aβ secretion. HEK293APP cells were transfected with pcDNA3 or FLAG-BRI3, and the secreted Aβ in the media was measured. The averages and the standard deviations of the triplicates are normalized by the protein concentration of the total lysates of the transfected cells. The probability of Student's t test was <0.001 (***). B, BRI3 suppresses AID production. HEK293T cells were transfected with or without BRI3, together with APP-Gal4, pG5E1b-luc, and β-galactosidase. AID production was measured as luciferase activity induced by AID-Gal4 fusion, which was cleaved from APP-Gal4. The average and standard deviation values of the luciferase activity of the triplicate samples were normalized by β-galactosidase activity measured simultaneously. The p value of Student's t test was <0.001 (***). C, BRI3 decreases sAPPα and sAPPβ. HeLa cells were transfected with pcDNA3 (−) or FLAG-BRI3 (+). sAPPα or sAPPβ in the media, and APP, α-tubulin, FLAG-BRI3, and APP CTF in the lysates were analyzed by Western blot. D, there is no clear difference of APP maturation upon BRI3 transfection. HeLa cells were transfected with the combinations of APP, pcDNA3, or FLAG-BRI3, as indicated. The cells were pulsed for 30 min with [35S]methionine-cysteine and chased for indicated times. APP was precipitated with αAPPct and visualized by autoradiography. Coprecipitated BRI3 is indicated. E, HEK293APP cells were transfected with pcDNA3 or FLAG-BRI3. Pulse chase was performed as in D, except the cells were chased for indicated minutes. F, quantification of D. Changes of mAPP, imAPP, and BRI3 are indicated. G, quantification of E. Legends are the same as in F.
We next investigated whether BRI3 is involved in AID production. The rationales of the luciferase assay used here are 1) that APP fused to entire Gal4 releases AID fused to Gal4 upon cleavage by γ-secretase and 2) then the released Gal4 fusion translocates to the nucleus and activates the transcription of luciferase (20). HEK293T cells were transfected with pcDNA3 or FLAG-BRI3, together with APP-Gal4, pG5E1b-luc, and β-galactosidase. One day after the transfection, the cells were lysed and the luciferase activity was measured. As shown in Fig. 4B, overexpression of BRI3 dramatically reduced AID release.
BRI3 Overexpression Reduces sAPPα and sAPPβ
Next, we tested how other APP metabolites were affected by the overexpression of BRI3. HeLa cells were transfected with APP together with pcDNA3 or FLAG-BRI3, and 1 day later, the cells were conditioned for 24 h. APP fragments in the media and in the cell were analyzed by Western blot. As shown in Fig. 4C, BRI3 overexpression decreased the secretion of both sAPPα and sAPPβ. Full-length APP is slightly increased, and there is no noticeable change in APP CTFs. Intracellular transport and localization of APP are critical components of Aβ production. In fact, α-secretase cleaves mAPP en route to or on the plasma membrane. β-Secretase predominantly cleaves mAPP in early endosomes (35, 36), whereas C99 and C83 are processed by the γ-secretase in endocytic compartments (36). Notably, pulse-chase experiments on metabolically labeled BRI3 and control transfected cells shows that BRI3 neither altered the imAPP/mAPP ratio nor changed the rate of maturation of APP (Fig. 4, D–G), indicating that BRI3 does not alter APP maturation and trafficking.
Effect of BRI3 on APP Processing Is Specific
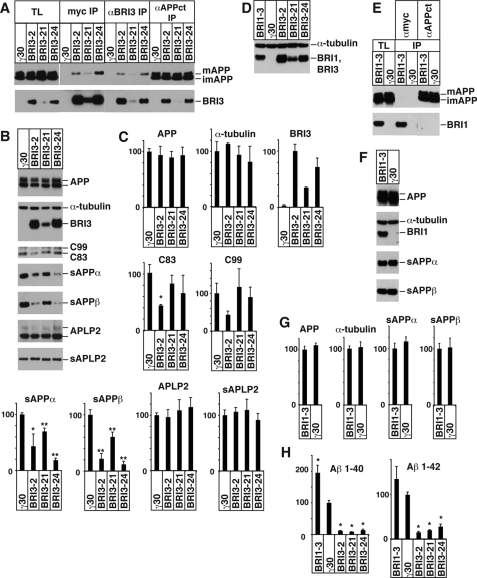
Because overexpression artifacts inherent in transient transfection system could cause potential problems for coimmunoprecipitation experiments and for determining functional effects on APP processing, we have established γ30 cell lines (29) stably expressing BRI3. Three representative clones, expressing distinct levels of Myc-tagged BRI3, are shown in Fig. 5A. Immunoprecipitation with αMyc and αBRI3 antibodies shows that BRI3 specifically interacts with mature APP, just like BRI2 (20, 21, 24). The reverse immunoprecipitation shows that an αAPPct antibody copurifies BRI3 (Fig. 5A). Analysis of APP processing in these clones shows reduction in sAPPα and sAPPβ (Fig. 5, B and C), and Aβ40 and Aβ42 (Fig. 5H) in all three BRI3-expressing clones as compared with the parental γ30 cell line. Notably, the levels of APP are similar in all four cell types studied (Fig. 5, B and C).
FIGURE 5.
A, BRI3 binds only mature APP in BRI3 stable cells. Total lysates were prepared from BRI3 stable clones and their parental cell line (γ30), and immunoprecipitated with αMyc, αBRI3, or αAPPct antibodies. APP and BRI3 in the total lysates and precipitants were detected with 22C11 and αMyc Western blot. Precipitants correspond to 8 volumes of total lysates. B, BRI3 stables secrete less sAPPs. The total lysates and the conditioned media of indicated stables were analyzed as in Fig. 4C. APLP2 and sAPLP2 were detected with corresponding antibodies. C, the average and standard deviation of the densitometry analysis of B. The average of γ30 cells was set to 100, except the amount of BRI3, in which case BRI3–2 was set to 100. The p values of Student's t test against γ30 cells were <0.05 (*) or 0.01 (**). D, Western blot with the αMyc antibody shows that the levels of transgenic BRI1 expressed by the BRI1 stable clone (BRI1–3) are comparable to the levels of BRI3 expressed by the BRI3 stable clones BRI3–21 and BRI3–24. α-Tubulin was used as a loading control. E, BRI1 does not interact with APP in the stable clone. The total lysates and immunoprecipitants of αMyc or αAPPct antibodies were probed with αMyc and 22C11 as in A. F, a representative Western blot shows that BRI1 has no effect on secretion of sAPPs as compared with parental γ30 cells. G, densitometry analysis of the experiment in F was displayed as in C. There is no significant difference of APP fragments between the two clones. H, BRI3 stables produce less Aβ. The Aβ 1–40 and 1–42 in the conditioned media of BRI1 and BRI3 stables were measured by ELISA, as in Fig. 4A. Aβ in the media of γ30 cells was set to 100. The asterisks are as in C. Error bars represent S.D.
Next, we tested whether BRI3 affects activity of secretases by studying processing of another substrates of secretases. We selected APLP2, because it is structurally similar to APP and is processed by γ-, α-, and β-secretases in the same way (37, 38). Analysis of fragments derived from either α- or β-secretase cleavage of ALPL2 (soluble APLP2s (sAPLP2s)) shows that BRI3 does not alter APLP2 processing, because the levels of sAPLP2s in the conditioned media of BRI3 stable clones are unchanged (Fig. 5, B and C). It must be noted that these are the same cell culture supernatants, which show reduced sAPPα and sAPPβ levels. Thus, like BRI2 (21), BRI3 does not affect the activity of secretases but only cleavage of APP by secretases.
As a further specificity control and to rule out a possible nonspecific effect of overexpressed BRI3, we analyzed cells transfected with a cDNA encoding another type II membrane protein. We choose BRI1, because it belongs to the same gene family as BRI3 but does not bind APP (Fig. 1B). These features make BRI1 the most stringent control we could use. We studied APP processing in the BRI1 stable γ30 clone that expressed the highest levels of Myc-BRI1 protein (BRI1–3, data not shown). These levels are comprised between the levels of Myc-BRI3 expressed by clones BRI3–21 and BRI3–24 (Fig. 5D). The stable clone confirms that BRI1 does not bind APP (Fig. 5E) and does not affect APP processing by either α- or β-secretase, because the levels of sAPPα and sAPPβ were unchanged as compared with the parental γ30 cell (Fig. 5, F and G). As shown in Fig. 5H, unlike BRI3, Aβ40 was increased in the conditioned media of BRI1–3 cells. The increase in Aβ42 was not statistically significant. In conclusion, the decrease of Aβ40 and -42 in BRI3 transfectants was not a nonspecific phenomenon caused by exogenous expression of membrane proteins.
Thus, BRI3 specifically interferes with APP processing. This effect is not dependent on inhibition of secretase activity but, rather, it correlates with binding to mature APP.
Reducing Endogenous BRI3 Increases the Secretion of Aβ
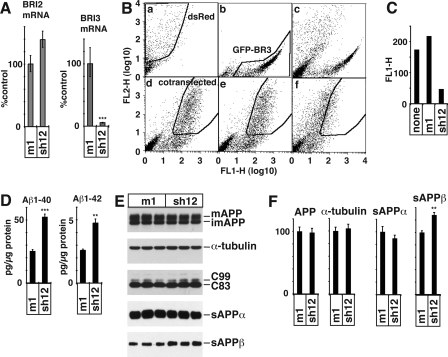
To determine whether inhibition of APP processing by BRI3 overexpression represented an artifact of overexpression rather than reflecting the physiological function of BRI3, we knocked down endogenous BRI3 by RNA interference using a shRNAs corresponding to human BRI3 (sh12) or a point mutant of sh12 (m1). HEK293APP cells were transfected with BRI3 shRNA construct (sh12) or control (m1). Quantitative PCR showed BRI2 mRNA are comparable in both shRNA-transfected cells, but BRI3 mRNA level was reduced to ∼5% of the control, indicating that the reduction was specific (Fig. 6A). To determine whether the levels of BRI3 proteins were also reduced, we transfected cells with a construct coding for a BRI3-GFP fusion protein, because we do not posses an antibody that can detect endogenous BRI3 in Western blot analysis. As shown in Fig. 6 (B and C), sh12 efficiently reduced BRI3-GFP levels 48 h post-transfection. Thus, from these two experiments we infer that the levels of endogenous BRI3 protein are reduced by the shRNA transfection. Next, we determined whether lowering BRI3 expression has any impact on APP processing; we measured the levels of Aβ peptides, sAPPα and sAPPβ, in the conditioned media of HEK293APP cells and found that BRI3 down-regulation significantly augmented Aβ40 and Aβ42 (Fig. 6D) as well as sAPPβ levels (Fig. 6, E and F). Notably sAPPα did not significantly change upon BRI3 down-regulation, suggesting perhaps that amyloidogenic APP processing is more sensitive to endogenous BRI3 levels. Thus, although BRI3 overexpression reduces Aβ, suppressing endogenous BRI3 levels has the opposite effect and increases levels of Aβ peptides. These data suggest that BRI3 is a physiological inhibitor of Aβ production.
FIGURE 6.
A, BRI3 shRNA reduces BRI3 mRNA. HEK293APP cells were transfected with BRI3 shRNA (sh12) or with control (m1), and mRNA was measured by quantitative PCR. BRI3 mRNA was specifically reduced by the transfection of sh12. B, FACS analysis shows sh12 reduces GFP-BRI3 protein expression. FACS analysis of HEK293APP cells transfected with dsRed alone (a), GFP-BRI3 alone (b), the mixture of the two (c), or the cells cotransfected with dsRed and GFP-BRI3 (d), dsRed, GFP-BRI3, and m1 (e), and dsRed, GFP-BRI3, and sh12 (f). The area representing cells singly transfected with dsRed or GFP-BRI3, or those cotransfected with both are indicated. C, the geometric average of FL1 channel signals of all dsRed-positive cells of B (d–f). D, knockdown of BRI3 increases the secretion of both Aβ40 and Aβ42. Aβ secreted from shRNA-transfected cells was measured by ELISA. The p values of Student's t test are <0.001 (***) or <0.01 (**). E, BRI3 knockdown increases sAPPβ. The Western blots of the total lysates and the conditioned media of m1 or sh12 transfected HEK293APP. The samples were analyzed as in Fig. 5B. F, the densitometry analysis of E. **, p value < 0.01. Error bars represent S.D.
BRI3 Binds Immature BACE, but Not Mature BACE
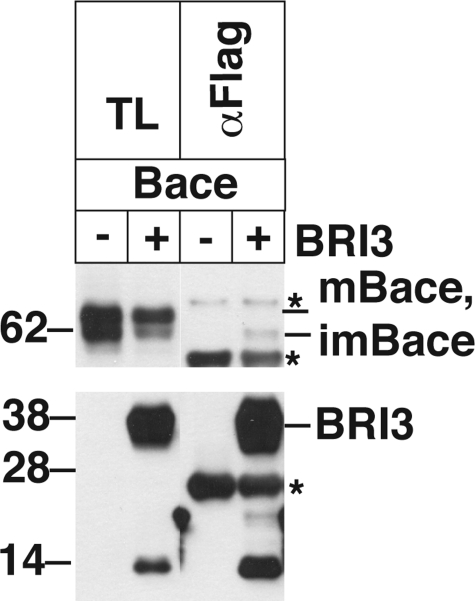
The β-secretase has been identified as BACE (39), and it has been reported that BRI3 and BACE interact (32). Thus the inhibitory effect of BRI3 on β-cleavage of APP could be explained by the BRI3-BACE interaction rather than the BRI3-APP interaction. As shown in Fig. 7, HeLa cells were transfected with BACE, pcDNA3, or FLAG-BRI3, and the total lysates were prepared from the transfected cells. FLAG immunoprecipitation showed that traces of immature BACE were precipitated by BRI3. However, the mature form of BACE was not detected in the precipitants. We could not detect endogenous BACE-BRI3 complexes in mouse brain lysate (data not shown). Considering that BACE is supposed to work on APP on the raft-mediated endocytotic pathway (36), these suggest that BRI3 does not form a complex with BACE where BACE acts on APP.
FIGURE 7.
BRI3 does not bind mature BACE. HeLa cells were transfected with Myc-tagged BACE and FLAG-BRI3. The total lysates (TL) were precipitated by FLAG antibody (αFlag) and probed for the presence of BACE (αmyc) and BRI3 (αFlag). Bands marked with an asterisk are attributed to FLAG antibody used in the precipitation. Mature (mBACE) and immature BACE (imBACE) are also indicated.
Endogenous BRI3 Binds Endogenous APP in Mouse Brain Lysate
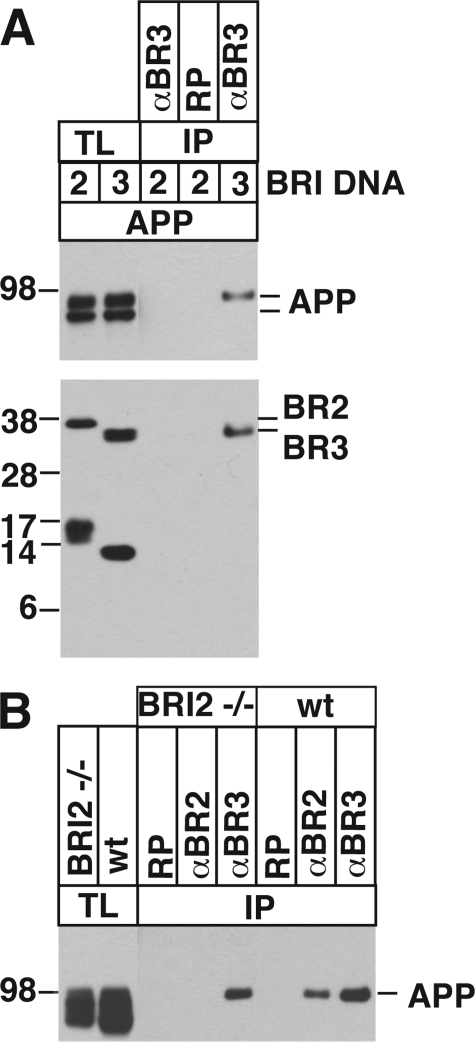
This interaction is independent of BRI2. We raised a rabbit antibody against BRI3 and tested whether the antibody can specifically immunoprecipitate BRI3. HeLa cells were transfected with APP and FLAG-BRI2 or FLAG-BRI3 as indicated in Fig. 8A. The transfected cells were lysed 1 day later, and the lysates were precipitated with a rabbit control antibody, or with the BRI3 antibody. The BRI3 antibody precipitated the BRI3-APP complex, but did not precipitate the BRI2-APP complex, indicating that the antibody can specifically precipitate BRI3. Unfortunately, the antibody was not strong enough to detect endogenous BRI3 in mouse brain extract by Western blot (data not shown).
FIGURE 8.
A, the rabbit polyclonal antibody raised against BRI3 can precipitate BRI3. HeLa cells were transfected with FLAG-BRI2, FLAG-BRI3, or APP as indicated. Total lysates (TL) were immunoprecipitated (IP) with the BRI3 antibody (αBRI3) or the control antibody (RP). The total lysates and immunoprecipitants were probed for the presence of APP and BRIs by Western blot using 22C11 and αFlag, respectively. B, endogenous BRI3 binds APP regardless of the presence or absence of BRI2. Membrane extract was prepared from Bri2−/− or wild-type (wt) mouse brain. The extracts were precipitated with a control antibody (RP), the BRI2 (αBRI2), or BRI3 (αBRI3) antibodies. Precipitated APP was detected as in A.
Next, we tested whether the endogenous BRI3-APP interaction is detectable, and, if so, whether it depends on BRI2-APP interaction. Membrane extracts were prepared from Bri2−/− (21) and wild-type mouse brains, and the extracts were precipitated with a control rabbit polyclonal antibody, and with antibodies against BRI2 or BRI3 as shown in Fig. 6B. The BRI3 antibody precipitated APP from both Bri2−/− and wild-type brain extracts, indicating that the BRI3-APP interaction exists at an endogenous level, and it is independent of BRI2-APP interaction (Fig. 8B). Of relevance, only mature APP binds BRI3 endogenously. This resembles what we observed in BRI3-stable transfected clones (Fig. 5A) and what we have previously described for BRI2 (20, 21, 29). As expected, the BRI2 antibody precipitated APP from wild-type mouse brain extract but did not precipitate APP from Bri2−/− brain extracts. The control antibody did not precipitate APP from any extracts. This finding, together with the evidence that down-regulation of endogenous BRI3 expression increases Aβ, further stresses the biological relevance and the physiological consequences of the BRI3-APP interaction.
DISCUSSION
We isolated BRI3, together with the family member BRI2, during a genetic screening for membrane-bound APP ligands (20). Mouse BRI3 was originally cloned as E25 by cDNA subtraction (31) but was not further characterized. Human BRI3 was cloned and mapped to chromosome 2q37 (30), and there is no known genetic disease caused by BRI3.
We had previously shown that the dementia gene BRI2 is an inhibitor of APP processing (20, 21). Here, we have established that BRI3, a member of the same gene family, binds APP and inhibits the production of Aβ. Like the other two members of the BRI gene family, which comprises also BRI1, BRI3 is type II transmembrane proteins and contains a BRICHOS domain (Figs. 1A and 7) whose functions are unknown. BRI3 can immunoprecipitate APP (Fig. 1A). Reciprocal immunoprecipitation shows APP can precipitate BRI3 (Fig. 1C). The N-terminal 130 amino acids of BRI3 are enough for the interaction (Fig. 1D). On the APP side, the smallest region of APP required for the BRI3-APP interaction seems to be equal to or larger than C99, because the BRI3-C99 interaction is much weaker than the BRI2-C99 interaction (Fig. 1A). C83 and AID do not interact with BRI3, and the last 31 amino acids of APP intracellular region, which consists of 47 amino acids, are dispensable for the interaction (Fig. 2). The regions required for the BRI3-APP interaction are largely similar to those mediating the BRI2-APP interaction: 1) the juxtamembrane region of BRIs is required, but the C-terminal fragment, including the entire BRICHOS, is dispensable; 2) the short segment of extracellular domain of APP is required.
It is noteworthy that the highest amino acid sequence homology among the BRIs is found in the BRICHOS domain and the COOH-terminal peptide region, but not the region responsible for the binding (Fig. 9).
FIGURE 9.
Amino acid sequence comparison of human BRI family proteins. Gaps (-), identical amino acids (*), conserved substitutions (:), and semi-conserved substitutions (.) are indicated. The predicted transmembrane sequence is in bold. The BRICHOS domain is boxed. The end of BRI3-(1–130) is indicated by an arrow. Amino acid numbers are indicated at the right side.
BRI3 colocalizes with APP in dotted structure in the neurites of differentiated N2A cells. APP in neurons is transported by fast forward axonal flow (40). Our observation that BRI3 colocalizes with APP in the vesicular structures along the neurites suggests the possibility that BRI3 can regulate APP processing during APP transport through the neurites.
Numerous lines of evidence shown here indicate that BRI3 plays a physiological role in APP processing. BRI3 physically associates with APP, and this interaction serves to suppress APP processing by secretases thereby reducing sAPPα, sAPPβ, Aβ, and AID production. Increasing BRI3 levels by overexpression reduces APP cleavage by α- and β-secretase (Figs. 4 and 5). On the contrary, reducing BRI3 levels in cell lines by shRNA increases APP processing. In fact, sAPPβ, Aβ40, and Aβ42 levels are increased in BRI3 knockdown cells (Fig. 6). Moreover, endogenous BRI3-APP complexes are found in vivo in the mouse brain. Altogether, these data demonstrate that BRI3 is an important physiological regulator of α- and β-cleavage of APP and hint to the possibility that factors interfering with the regulation of APP processing by BRI3 may contribute to AD pathogenesis. To this end, it is noteworthy that BRI2 is ubiquitous, but BRI3 is predominantly neuronal (30).
Is the anti-amyloidogenic mechanism of BRI3 similar to that of BRI2? We have shown that BRI2 masks the cleavage sites of β- and α-secretase on APP and the γ-secretase docking site on C99 (21). A previous report showed that BRI3 binds BACE in an overexpression context (32), thus is it possible that BRI3 reduces sAPPβ and Aβ by inhibiting β-secretase? We think this is unlikely because of the following reasons. First, BRI3 does not bind the mature form of BACE (Fig. 5) and BACE cleaves APP mainly inside raft clusters and endocytic vesicles (35). Moreover, BACE-mediated inhibition does not explain why BRI3 reduces the secretion of sAPPα. Second, in γ30 cells stably expressing BRI3, sAPLP2 was not changed while both sAPPα and sAPPβ were reduced.
Thus we propose that, like BRI2, BRI3 is directly working on APP, not on secretases, but in different processes: the BRI3-APP interaction masks the access of α- and β-secretases to APP, thereby inhibiting the processing of APP by the secretase; the BRI3-C99 interaction is minimal making BRI3 a poor inhibitor of γ-processing of C99. This hypothesis can explain all aspects of our observations: 1) BRI3 decreases sAPPα, sAPPβ, Aβ, and AID, because BRI3 binds full-length APP and limits the access of α- and β-secretase to APP; 2) BRI3 does not cause the accumulation of C99 or C83, because BRI3 binds C99 poorly, and it does not bind C83; therefore, it has no effect on the processing of C99 and C83.
BRI2 may influence both Aβ formation and deposition. Recent findings indicate that the Bri2–23 peptide released from the BRI2 protein by furin/furin-like protease cleavage can inhibit Aβ aggregation in vitro (41). Whether the furin-derived BRI3-soluble peptide has a similar function remains to be determined.
The mechanism of inhibition of amyloid formation by BRI3 and BRI2 suggests that is should be possible to inhibit access of secretases to APP rather than secretases activity. This method would represent an alternative therapeutic approach to AD, which would avoid toxic effects due to inhibiting processing of other substrates of secretases (42) or even a direct pathogenic effect of γ-secretase inhibitors, as postulated by others (43, 44). Evidence that the transgenic expression of BRI2 reduces APP processing (21) and amyloid pathology in mouse models of AD (21, 41) supports this notion.
Our results, that BRI3 can bind APP in the absence of BRI2 (Fig. 6B), indicate that, if the therapeutic intervention exploiting this substrate-oriented inhibition is possible, in addition to BRI2, BRI3 provides an independent route to modulate APP processing.
Acknowledgments
We thank Robert Tamayev for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 AG22024, RO1 AG21588, and R21 AG027139 from the NIA. This work was also supported by Alzheimer's Association Grant IIRG-05-14511 (to L. D.).
- AD
- Alzheimer disease
- APP
- amyloid precursor protein
- sAPP
- soluble APP
- C99
- C-terminal fragment of 99 amino acids
- C83
- C-terminal fragment of 83 amino acids
- shRNA
- short hairpin RNA
- GFP
- green fluorescent protein
- CTF
- C-terminal fragment
- ELISA
- enzyme-linked immunosorbent assay
- FACS
- fluorescence-activated cell sorting
- RP
- rabbit polyclonal antibody
- AID
- APP intracellular domain
- Aβ
- β amyloid peptide.
REFERENCES
- 1.Breteler M. M., Claus J. J., van Duijn C. M., Launer L. J., Hofman A. ( 1992) Epidemiol. Rev. 14, 59– 82 [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D. J., Podlisny M. B. ( 2002) Annu. Rev. Genomics Hum. Genet. 3, 67– 99 [DOI] [PubMed] [Google Scholar]
- 3.Selkoe D., Kopan R. ( 2003) Annu. Rev. Neurosci. 26, 565– 597 [DOI] [PubMed] [Google Scholar]
- 4.Sisodia S. S., St George-Hyslop P. H. ( 2002) Nat. Rev. Neurosci. 3, 281– 290 [DOI] [PubMed] [Google Scholar]
- 5.Hamid R., Kilger E., Willem M., Vassallo N., Kostka M., Bornhovd C., Reichert A. S., Kretzschmar H. A., Haass C., Herms J. ( 2007) J. Neurochem. 102, 1264– 1275 [DOI] [PubMed] [Google Scholar]
- 6.Madeira A., Pommet J. M., Prochiantz A., Allinquant B. ( 2005) FASEB J. 19, 1905– 1907 [DOI] [PubMed] [Google Scholar]
- 7.Passer B., Pellegrini L., Russo C., Siegel R. M., Lenardo M. J., Schettini G., Bachmann M., Tabaton M., D'Adamio L. ( 2000) J. Alzheimers Dis. 2, 289– 301 [DOI] [PubMed] [Google Scholar]
- 8.Cao X., Sudhof T. C. ( 2001) Science 293, 115– 120 [DOI] [PubMed] [Google Scholar]
- 9.Cupers P., Orlans I., Craessaerts K., Annaert W., De Strooper B. ( 2001) J. Neurochem. 78, 1168– 1178 [DOI] [PubMed] [Google Scholar]
- 10.Pardossi-Piquard R., Petit A., Kawarai T., Sunyach C., Alves da Costa C., Vincent B., Ring S., D'Adamio L., Shen J., Muller U., St George Hyslop P., Checler F. ( 2005) Neuron 46, 541– 554 [DOI] [PubMed] [Google Scholar]
- 11.Liu Q., Zerbinatti C. V., Zhang J., Hoe H. S., Wang B., Cole S. L., Herz J., Muglia L., Bu G. ( 2007) Neuron 56, 66– 78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Rotz R. C., Kohli B. M., Bosset J., Meier M., Suzuki T., Nitsch R. M., Konietzko U. ( 2004) J. Cell Sci. 117, 4435– 4448 [DOI] [PubMed] [Google Scholar]
- 13.Kim H. S., Kim E. M., Lee J. P., Park C. H., Kim S., Seo J. H., Chang K. A., Yu E., Jeong S. J., Chong Y. H., Suh Y. H. ( 2003) FASEB J. 17, 1951– 1953 [DOI] [PubMed] [Google Scholar]
- 14.Baek S. H., Ohgi K. A., Rose D. W., Koo E. H., Glass C. K., Rosenfeld M. G. ( 2002) Cell 110, 55– 67 [DOI] [PubMed] [Google Scholar]
- 15.Checler F., Sunyach C., Pardossi-Piquard R., Sevalle J., Vincent B., Kawarai T., Girardot N., St George-Hyslop P., da Costa C. A. ( 2007) Curr. Alzheimer Res. 4, 423– 426 [DOI] [PubMed] [Google Scholar]
- 16.Leissring M. A., Murphy M. P., Mead T. R., Akbari Y., Sugarman M. C., Jannatipour M., Anliker B., Muller U., Saftig P., De Strooper B., Wolfe M. S., Golde T. E., LaFerla F. M. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 4697– 4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giliberto L., Zhou D., Weldon R., Tamagno E., De Luca P., Tabaton M., D'Adamio L. ( 2008) Mol. Neurodegener. 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheinfeld M. H., Matsuda S., D'Adamio L. ( 2003) Proc. Natl. Acad. Sci. U. S. A. 100, 1729– 1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roncarati R., Sestan N., Scheinfeld M. H., Berechid B. E., Lopez P. A., Meucci O., McGlade J. C., Rakic P., D'Adamio L. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 7102– 7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuda S., Giliberto L., Matsuda Y., Davies P., McGowan E., Pickford F., Ghiso J., Frangione B., D'Adamio L. ( 2005) J. Biol. Chem. 280, 28912– 28916 [DOI] [PubMed] [Google Scholar]
- 21.Matsuda S., Giliberto L., Matsuda Y., McGowan E. M., D'Adamio L. ( 2008) J. Neurosci. 28, 8668– 8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vidal R., Frangione B., Rostagno A., Mead S., Revesz T., Plant G., Ghiso J. ( 1999) Nature 399, 776– 781 [DOI] [PubMed] [Google Scholar]
- 23.Vidal R., Revesz T., Rostagno A., Kim E., Holton J. L., Bek T., Bojsen-Moller M., Braendgaard H., Plant G., Ghiso J., Frangione B. ( 2000) Proc. Natl. Acad. Sci. U. S. A. 97, 4920– 4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fotinopoulou A., Tsachaki M., Vlavaki M., Poulopoulos A., Rostagno A., Frangione B., Ghiso J., Efthimiopoulos S. ( 2005) J. Biol. Chem. 280, 30768– 30772 [DOI] [PubMed] [Google Scholar]
- 25.Scheinfeld M. H., Roncarati R., Vito P., Lopez P. A., Abdallah M., D'Adamio L. ( 2002) J. Biol. Chem. 277, 3767– 3775 [DOI] [PubMed] [Google Scholar]
- 26.Zazzeroni F., Papa S., Algeciras-Schimnich A., Alvarez K., Melis T., Bubici C., Majewski N., Hay N., De Smaele E., Peter M. E., Franzoso G. ( 2003) Blood 102, 3270– 3279 [DOI] [PubMed] [Google Scholar]
- 27.Stamer K., Vogel R., Thies E., Mandelkow E., Mandelkow E. M. ( 2002) J. Cell Biol. 156, 1051– 1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda S., Yasukawa T., Homma Y., Ito Y., Niikura T., Hiraki T., Hirai S., Ohno S., Kita Y., Kawasumi M., Kouyama K., Yamamoto T., Kyriakis J. M., Nishimoto I. ( 2001) J. Neurosci. 21, 6597– 6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chyung J. H., Raper D. M., Selkoe D. J. ( 2005) J. Biol. Chem. 280, 4383– 4392 [DOI] [PubMed] [Google Scholar]
- 30.Vidal R., Calero M., Revesz T., Plant G., Ghiso J., Frangione B. ( 2001) Gene 266, 95– 102 [DOI] [PubMed] [Google Scholar]
- 31.Deleersnijder W., Hong G., Cortvrindt R., Poirier C., Tylzanowski P., Pittois K., Van Marck E., Merregaert J. ( 1996) J. Biol. Chem. 271, 19475– 19482 [DOI] [PubMed] [Google Scholar]
- 32.Wickham L., Benjannet S., Marcinkiewicz E., Chretien M., Seidah N. G. ( 2005) J. Neurochem. 92, 93– 102 [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Pulido L., Devos D., Valencia A. ( 2002) Trends Biochem. Sci. 27, 329– 332 [DOI] [PubMed] [Google Scholar]
- 34.Martin L., Fluhrer R., Reiss K., Kremmer E., Saftig P., Haass C. ( 2008) J. Biol. Chem. 283, 1644– 1652 [DOI] [PubMed] [Google Scholar]
- 35.Kaether C., Schmitt S., Willem M., Haass C. ( 2006) Traffic 7, 408– 415 [DOI] [PubMed] [Google Scholar]
- 36.Ehehalt R., Keller P., Haass C., Thiele C., Simons K. ( 2003) J. Cell Biol. 160, 113– 123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q., Sudhof T. C. ( 2004) J. Biol. Chem. 279, 10542– 10550 [DOI] [PubMed] [Google Scholar]
- 38.Scheinfeld M. H., Ghersi E., Laky K., Fowlkes B. J., D'Adamio L. ( 2002) J. Biol. Chem. 277, 44195– 44201 [DOI] [PubMed] [Google Scholar]
- 39.Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. ( 1999) Science 286, 735– 741 [DOI] [PubMed] [Google Scholar]
- 40.Koo E. H., Sisodia S. S., Archer D. R., Martin L. J., Weidemann A., Beyreuther K., Fischer P., Masters C. L., Price D. L. ( 1990) Proc. Natl. Acad. Sci. U. S. A. 87, 1561– 1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J., Miller V. M., Levites Y., West K. J., Zwizinski C. W., Moore B. D., Troendle F. J., Bann M., Verbeeck C., Price R. W., Smithson L., Sonoda L., Wagg K., Rangachari V., Zou F., Younkin S. G., Graff-Radford N., Dickson D., Rosenberry T., Golde T. E. ( 2008) J. Neurosci. 28, 6030– 6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evin G., Sernee M. F., Masters C. L. ( 2006) CNS Drugs 20, 351– 372 [DOI] [PubMed] [Google Scholar]
- 43.Saura C. A., Choi S. Y., Beglopoulos V., Malkani S., Zhang D., Shankaranarayana Rao B. S., Chattarji S., Kelleher R. J., 3rd, Kandel E. R., Duff K., Kirkwood A., Shen J. ( 2004) Neuron 42, 23– 36 [DOI] [PubMed] [Google Scholar]
- 44.Shen J., Kelleher R. J., 3rd. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 403– 409 [DOI] [PMC free article] [PubMed] [Google Scholar]