Abstract
Defective membrane repair can contribute to the progression of muscular dystrophy. Although mutations in caveolin-3 (Cav3) and dysferlin are linked to muscular dystrophy in human patients, the molecular mechanism underlying the functional interplay between Cav3 and dysferlin in membrane repair of muscle physiology and disease has not been fully resolved. We recently discovered that mitsugumin 53 (MG53), a muscle-specific TRIM (Tri-partite motif) family protein (TRIM72), contributes to intracellular vesicle trafficking and is an essential component of the membrane repair machinery in striated muscle. Here we show that MG53 interacts with dysferlin and Cav3 to regulate membrane repair in skeletal muscle. MG53 mediates active trafficking of intracellular vesicles to the sarcolemma and is required for movement of dysferlin to sites of cell injury during repair patch formation. Mutations in Cav3 (P104L, R26Q) that cause retention of Cav3 in Golgi apparatus result in aberrant localization of MG53 and dysferlin in a dominant-negative fashion, leading to defective membrane repair. Our data reveal that a molecular complex formed by MG53, dysferlin, and Cav3 is essential for repair of muscle membrane damage and also provide a therapeutic target for treatment of muscular and cardiovascular diseases that are linked to compromised membrane repair.
Membrane recycling and remodeling contribute to multiple cellular functions, including cell fusion events during myogenesis and maintenance of sarcolemma integrity in striated muscle. During the life cycle of striated muscle, membrane repair is a fundamental process in maintaining cellular integrity, as shown by recent studies that link defective membrane repair to the progression of muscular dystrophy (1–3). Repair of the plasma membrane damage requires recruitment of intracellular vesicles to injury sites (4, 5). One protein that has been linked to membrane repair in skeletal muscle is dysferlin (6, 7), which is thought to fuse intracellular vesicles to patch the damaged membrane and restore sarcolemmal integrity following muscle injury. Like dysferlin, caveolin-3 (Cav3)3 is a muscle-specific protein, and many mutations in Cav3, including P104L, R26Q, and C71W, have been linked to muscular dystrophy (8–11). Despite extensive research efforts on Cav3 and dysferlin (12–14), the molecular function of these two proteins in membrane repair in muscle physiology and dystrophy have not been fully defined.
Animal model studies reveal that either loss or gain of Cav3 function both result in dystrophic phenotypes in skeletal muscle (15, 16), suggesting that associated cellular components may be involved in the etiology of Cav3-related dystrophy. Although the discovery of dysferlin highlights the importance of membrane repair in the etiology of muscular dystrophy, dysferlin itself does not appear to participate in recruitment of intracellular vesicles because dysferlin−/− muscle retains accumulation of vesicles near membrane damage sites (7). This indicates that proteins other than dysferlin are required for nucleation of intracellular vesicles at the sites of acute membrane damage. Recently, we discovered that MG53, a muscle-specific TRIM family protein (TRIM72), is an essential component of the acute membrane repair machinery. MG53 acts as a sensor of oxidation to nucleate recruitment of intracellular vesicles to the injury site for membrane patch formation (17). We also found that MG53 can regulate membrane budding and exocytosis in muscle cells, and this membrane-recycling function of MG53 can be modulated through a functional interaction with Cav3 (18).
Here we present evidence that MG53 interacts with dysferlin to facilitate intracellular vesicle trafficking during repair of acute membrane damage. In addition, we show that transgenic overexpression of P104L-Cav3 in striated muscle produces defects in membrane repair that are linked to altered subcellular distribution of MG53 and dysferlin. Our results suggest that altered MG53 localization can be used as a marker for muscular dystrophy involving reduced sarcolemmal membrane repair capacity due to Cav3 mutation, and potentially, in other forms of dystrophy as well.
EXPERIMENTAL PROCEDURES
Cell Culture
C2C12 murine myoblast cell line was purchased from the American Type Culture Collection (Manassas, VA). Cells were grown in a humidified environment at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. C2C12 myoblasts were grown to confluence and switched to Dulbecco's modified Eagle's medium containing 2% horse serum to induce serum withdrawal differentiation. Primary myoblasts were derived from wild-type (WT) or mg53−/− neonatal mouse pups using established techniques. For transient transfections, C2C12 myoblasts were plated at 70% confluence in glass bottom ΔT dishes (Bioptechs, Inc.) and transfected using GeneJammer reagent (Stratagene) per manufacturer's directions. Cells were visualized by live cell confocal imaging at 24–48 h after transfection or at the times indicated for individual experiments.
Plasmid Construction
Cloning and construction of MG53 expression plasmids were previously described (17, 18). Cav3 cDNA were amplified using reverse transcription-PCR from mouse skeletal muscle. Cav3-GFP fusion constructs were generated by inserting Cav3 cDNA into pEGFP-N1 using the XhoI and BamHI restriction enzymatic sites. The various Cav3 mutants, P104L, R26Q, and C71W, were constructed by replacing the appropriate residues in Cav3-GFP using the methods described previously (19). The Cav3-Myc fusion construct was generated by inserting the Cav3 cDNA into the 5′ end of pcDNA3.1A containing the Myc tag (Invitrogen), and the HA-MG53 fusion construct was generated by inserting the mouse MG53 cDNA into the 3′ end of pHM6 plasmid containing the HA tag (Roche Applied Science). GFP-dysferlin plasmid was a generous gift from Dr. Kate Bushby (20).
Western Blot and Immunoprecipitation
Western blotting was performed using standard techniques. In various experiments, C2C12 cells were harvested and lysed with ice-cold modified radioimmune precipitation (RIPA) buffer (150 mm NaCl, 5 mm EDTA, 1% Nonidet P-40, 20 mm Tris-HCl, pH 7.5) supplemented with protease inhibitor mixture (Sigma). For each sample, 10 or 20 μg of total protein was separated on 4–12% SDS-polyacrylamide gradient gels (Invitrogen). A standard protocol was used for co-immunoprecipitation studies of MG53, dysferlin, and Cav3. Briefly, C2C12 cells transfected with Cav3-Myc, HA-MG53, and GFP-dysferlin were lysed in 0.5 ml of modified RIPA buffer. For each sample, 500 μg of total extract was incubated overnight with 5 μg of monoclonal anti-HA (Roche Applied Science), anti-Myc (BD Biosciences), or polyclonal anti-GFP antibody (Molecular Probes). As control, 500 μg of whole cell lysate was incubated with 5 μg of normal mouse IgG. Resulting complexes were collected by protein G-Sepharose beads during a 2-h incubation followed by four washes with RIPA buffer. Co-immunoprecipitation studies with the native skeletal muscle were performed as described in Cai et al. (18).
In Vivo Muscle Transfection
For in vivo transfection of adult skeletal muscle with Cav3-GFP or P104L-GFP, an initial injection of 10 μl of 20 mg/ml hyaluronidase (Sigma) was made into the flexor digitorum brevis (FDB) muscle of wild-type C57Bl/6 male mice (The Jackson Laboratory). After 1 h, 20 μg of plasmid DNA in 0.9% sterile saline was injected into the same muscles. Following 15 min to allow diffusion of DNA, acupuncture needles (Millennia) were inserted longitudinally through the mouse foot to allow application of 20 pulses of a 100 V/cm electrical field at 0.1 Hz (21). Mice were allowed to recover from electroporation for 7 days before experimentation.
Membrane Repair Assay
P104L-Cav3 transgenic mice were generated as described (22), and mg53−/− mice were produced as described elsewhere (17, 23). For isolation of FDB muscle fibers, male mg53−/− mice, P104L-Cav3 transgenic mice, age-matched wild-type control mice, and those electroporated with Cav3-GFP or P104L-GFP were sacrificed by cervical dislocation, and FDB muscles were collected in a Tyrode's solution containing (in mm) 140 NaCl, 5 KCl, 2.5 CaCl2, 2 MgCl2, 10 HEPES (pH 7.2). Muscles were incubated for 120 min at 37 °C in Tyrode's solution supplemented with 2 mg/ml type I collagenase (Sigma). FDB muscles were washed three times in Tyrode's solution, and then fibers were dissociated by several passages through a series of pipette tips with decreasing diameter (24). Membrane repair capacity was determined using previously established techniques (7, 25). Briefly, membrane damage was applied to FDB fibers using a UV laser on a Zeiss-LSM 510 confocal microscope to irradiate a 5 × 5-pixel area at maximum power for 5 s (Enterprise, 80 milliwatts, 351/364 nm). Before cell wounding, 2.5 μm FM1-43 or FM4-64 dye (Molecular Probes) was added to the extracellular solution. X-Y images were captured at 6.6-s intervals, and the mean fluorescence intensity of FM1-43 or FM4-64 at the site of the damage was determined using the Zeiss LSM 510 imaging software (17).
Immunostaining and Confocal Microscopic Imaging
Live cell confocal imaging was used to monitor intracellular trafficking of fluorescent fusion proteins. C2C12 or mg53−/− cells were transfected on ΔT glass bottom dishes (Bioptechs Inc.) and visualized using a Radiance 2100 laser scanning confocal microscope (Bio-Rad) with a ×40 (1.3NA) oil immersion objective at 3.1-s intervals unless otherwise noted. For immunocytochemistry, C2C12 cells were fixed with 100% ethanol at −20 °C for 5 min before anti-MG53, anti-Cav3, or anti-GM130 antibodies were applied. Cells were washed, and fluorescent-coupled secondary antibodies (goat anti-rabbit Alexa Fluor 488 or Alexa Fluor 546) were applied as per the manufacturer's directions (Molecular Probes). For live cell imaging of mechanical membrane damage, C2C12 myoblasts or mg53−/− myotubes were penetrated by the tip of a micropipette attached to a micromanipulator. Fluorescence images were captured using a Radiance 2100 scope as above.
RESULTS
MG53 Ablation and P104L-Cav3 Overexpression Produce Defective Membrane Repair in Skeletal Muscle
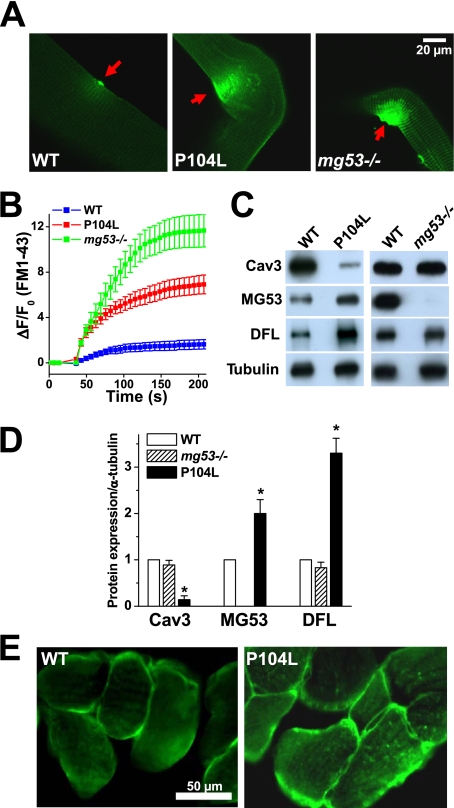
To test whether defective membrane repair may underlie the progression of muscular dystrophy related to mutations in Cav3, we examined the membrane repair capacity of transgenic mice that express P104L-Cav3 (22, 26). Skeletal muscle from the P104L-Cav3 mice displays defective membrane repair following UV laser-induced injury of the sarcolemma (Fig. 1A), similar to those observed with the dysferlin−/− (7) and mg53−/− mice (Fig. 1B). MG53 is a TRIM family protein expressed exclusively in striated muscle, which functions as a sensor of oxidation to nucleate the assembly of the acute membrane repair machinery in skeletal muscle (17). As a result, mg53−/− mice exhibit progressive muscular pathology associated with defective membrane repair capacity, and they share a similarity with the P104L-Cav3 transgenic mice.
FIGURE 1.
Membrane repair defects and altered localization of MG53 in P104L-Cav3 skeletal muscle. A, entry of FM1-43 fluorescent dye into isolated FDB muscle fibers from WT (left), P104L transgenic (middle), or mg53−/− mice (right) following UV laser-induced damage of the sarcolemmal membrane. B, summary time course data of accumulation of FM1-43 dye in UV-damaged FDB muscle fibers derived from WT (n = 30), P104L transgenic (n = 21), or mg53−/− (n = 18) muscle fibers. Data are listed as mean ± S.E. (error bars). C, Western blot shows expression level of Cav3, MG53, and dysferlin (DFL) in WT, P104L transgenic, and mg53−/− gastrocnemius muscle. Consistent with previous observations, expression of Cav3 is reduced in the P104L transgenic muscle (22). 10 μg/lane of total protein was loaded. α-Tubulin was used as a loading control. D, quantitative analysis of protein expression level relative to α-tubulin for Cav3, MG53, and dysferlin. The expression of Cav3, MG53, and dysferlin in wild-type muscle was set as 1, and the level of α-tubulin was used as a loading control. Data are mean ± S.E. (error bars) (n = 3 independent experiments). *, p < 0.01. E, immunostaining of cross sections from WT and P104L transgenic mouse skeletal muscle using polyclonal anti-MG53 antibody shows altered localization of MG53 in P104L muscle.
Western blots show that expression of both MG53 and dysferlin increases in P104L-Cav3 muscle, whereas the expression of Cav3 or dysferlin remains unchanged in mg53−/− muscle (Fig. 1C). Quantitative analysis indicates that the expression of MG53 and dysferlin increases significantly in the P104L-Cav3 muscle when compared with WT muscle (Fig. 1D). Immunohistochemical staining reveals that MG53 displays perisarcolemmal membrane localization in WT muscle that is altered in P104L muscle because MG53 appears more frequently in the center of P104L muscle fibers (Fig. 1E). Thus, although elevated expression of MG53 and dysferlin may represent a compensatory response to the defective membrane repair in P104L-Cav3 muscle, altered MG53 localization may represent a molecular basis for the defective membrane repair in Cav3-related muscular dystrophy.
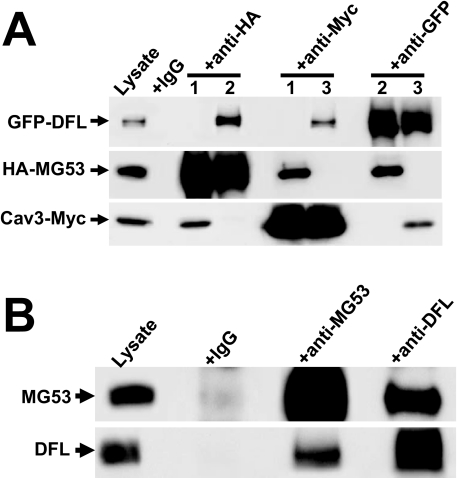
As with Cav3, MG53 and dysferlin are muscle-specific proteins whose expression is associated with myogenesis (7, 18). In particular, C2C12 cells at the myoblast stage do not express any of these proteins, whereas the expression of all three is induced following differentiation of C2C12 myoblasts into myotubes. The absence of endogenous MG53, Cav3, and dysferlin from C2C12 myoblast cells provides a homologous reconstitution system for our functional studies. Using co-immunoprecipitation (Co-IP) assays, we find that MG53, dysferlin, and Cav3 may form a protein complex by physically interacting with one another when co-expressed in C2C12 myoblast cells (Fig. 2A). Such physical interaction between MG53, Cav3, and dysferlin is also observed with endogenous proteins in mouse skeletal muscle (Fig. 2B) (see also Cai et al. (18)).
FIGURE 2.
Co-IP reveals physical interaction between MG53, Cav3, and DFL. A, C2C12 myoblast cells were co-expressed with HA-MG53 and Cav3-Myc (lanes labeled 1), HA-MG53 with GFP-DFL (lanes labeled 2), or Cav3-Myc with GFP-DFL (lanes labeled 3). 24 h after transfection, cell lysates were immunoprecipitated and blotted with antibody to anti-GFP, anti-HA, or anti-Myc. Co-IP of DFL and Cav3 was detected with anti-HA; Co-IP of MG53 and DFL was detected with anti-Myc; and Co-IP of MG53 and Cav3 was detected with anti-GFP. Cell lysates incubated with normal mouse IgG served as control. B, physical interaction between MG53 and DFL is also observed in gastrocnemius muscle from the WT mice. Co-IP of DFL is detected with anti-MG53, and Co-IP of MG53 is detected with anti-DFL. Preimmune IgG was used as a negative control.
Cav3 Regulates MG53 Function during Repair of Acute Membrane Damage
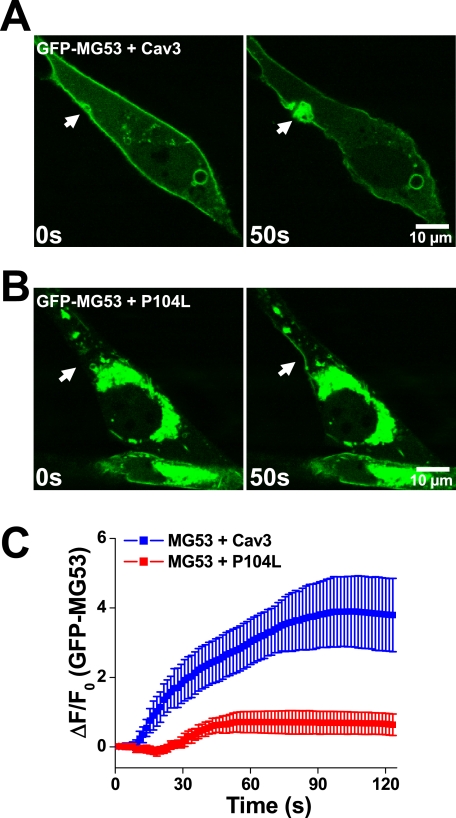
Our previous studies showed that MG53 interacts with phosphatidylserine to associate with intracellular vesicles that display trafficking to and fusion with sarcolemmal membranes, an important process involved in the membrane repair function of MG53 (17). Co-expression studies revealed that the MG53-mediated membrane-recycling activity is tightly controlled via a functional interaction with Cav3 (18). To dissect the impact of Cav3 interaction with MG53 in membrane repair, we performed live cell imaging of GFP-MG53 translocation in C2C12 myoblasts co-expressing either WT Cav3 or P104L-Cav3 following mechanical injury with a microelectrode. Acute injury of the cell leads to rapid translocation of GFP-MG53 toward the injury sites to form a membrane repair patch. Although co-expression of WT Cav3 does not have a major impact on GFP-MG53 translocation (Fig. 3A and supplemental Movie 1), P104L-Cav3 severely attenuates the membrane repair function of MG53, as indicated by mislocalization of GFP-MG53 and subsequent disruption of GFP-MG53 translocation following membrane damage (Fig. 3B and supplemental Movie 2). Although overexpression of WT Cav3 decreases MG53-mediated membrane resealing to some extent (see Fig. 5C), the P104L-Cav3 mutant severely attenuates MG53 translocation to injury sites (Fig. 3C). Thus, compromised MG53-mediated vesicle translocation may provide a mechanistic basis for the reduced membrane repair capacity in P104L-Cav3 muscle.
FIGURE 3.
Defective movement of GFP-MG53-containing vesicles to acute membrane injury sites with co-expression of P104L-Cav3. A, C2C12 myoblast cells transfected with GFP-MG53 and Cav3 were subjected to penetration by a microelectrode. Rapid recruitment of GFP-MG5-containing vesicles toward the injury site (arrow) was observed (see supplemental Movie 1). B, co-transfection of P104L-Cav3 and GFP-MG53 in C2C12 myoblast cells leads to mistargeting of GFP-MG53 in intracellular compartments and compromised GFP-MG53 translocation to injury sites (see supplemental Movie 2). C, summary data for time-dependent accumulation of GFP-MG53 at the injury sites were plotted for C2C12 cells co-transfected with GFP-MG53 and Cav3 or GFP-MG53 and P104L-Cav2. Data are mean ± S.E. (error bars) for n = 18 cells.
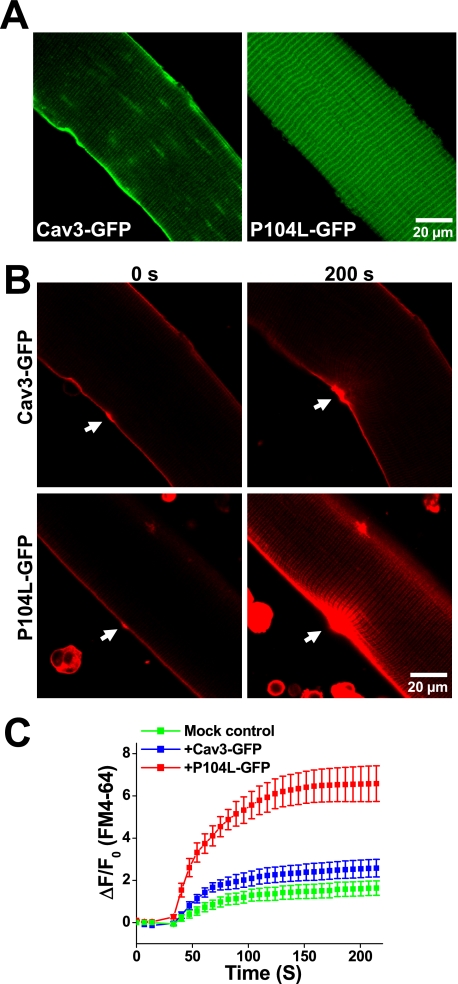
FIGURE 5.
Dominant effect of P104L-Cav3 produces defective membrane repair in native skeletal muscle. A, FDB muscle fibers from WT mice were transfected by in vivo electroporation to allow for transient expression of Cav3-GFP (left) and P104L-GFP (right) that were visualized by confocal microscopy. Confocal images showed that Cav3-GFP mainly targets to the sarcolemmal membrane, whereas P104L-GFP displayed a pattern indicative of intracellular retention of the mutant Cav3 protein. B, measurement of FM4-64 entry revealed severe defects in membrane repair capacity in fibers expressing P104L-GFP (lower), where excessive FM4-64 dye entry is observed following UV laser wounding (arrows) when compared with Cav3-GFP-expressing fibers (upper). C, quantitative assay of FM4-64 dye entry into skeletal muscle transiently expressing Cav3-GFP (blue trace) or P104L-GFP (red trace) or untransfected fibers on the same dish (Mock control, green trace). Data are means ± S.E. (error bars) for n = 18 fibers for each group from 3 independent electroporation with WT mice.
Dominant Effect of P104L-Cav3 Produces Defective Membrane Repair in Skeletal Muscle
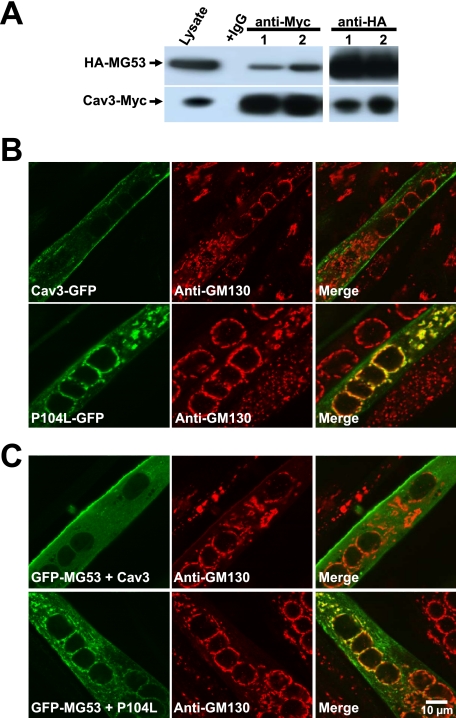
As shown in Fig. 4A, the recombinant MG53 and P104L-Cav3 transiently expressed in C2C12 cells retain their physical interaction based on Co-IP assays. Confocal microscopic imaging shows that Cav3-GFP displays targeting to the plasma membrane in addition to intracellular vesicles (Fig. 4B, top), whereas P104L-GFP is predominantly retained in the Golgi apparatus, as indicated by anti-GM130 immunostaining (9, 20) (Fig. 4B, bottom). Retention of P104L-Cav3 in the Golgi has a significant impact on the subcellular localization of GFP-MG53, for C2C12 myotubes transfected with WT Cav3 display sarcolemma and vesicular localization of GFP-MG53 (Fig. 4C, top), whereas a significant portion of GFP-MG53 appears in the Golgi apparatus of C2C12 myotubes expressing P104L-Cav3 (Fig. 4C, bottom). A similar pattern is observed by confocal imaging in C2C12 myoblast cells with transient co-expression of Cav3-GFP and RFP-MG53 (supplemental Fig. S1). These studies confirm that P104L-Cav3 produces defective MG53 localization to the Golgi that may underlie the defective membrane repair capacity in P104L muscle (Fig. 1B).
FIGURE 4.
P104L-Cav3 causes retention of MG53 at the Golgi network. A, C2C12 cells were co-transfected with HA-MG53 and either Cav3-Myc (lane 1) or P104L-Myc (lane 2). 24 h after transfection, cell lysates were immunoprecipitated and Western blotted with anti-HA or anti-Myc antibody. Cell lysates incubated with normal mouse IgG served as control. B, C2C12 myotubes transfected with Cav3-GFP or P104L-GFP were subjected to immunostaining with anti-GM130, a molecular marker for Golgi apparatus. Confocal images show that Cav3-GFP displays a plasma membrane pattern that does not overlap with GM130 staining (upper). P104L-GFP displays an intracellular localization pattern that co-localizes with GM130 (lower). C, C2C12 myotubes co-transfected with GFP-MG53 and Cav3-Myc or P104L-Myc were subjected to immunostaining with anti-GM130. Confocal images show that co-expression of Cav3 with GFP-MG53 does not impact the plasma membrane-tethering pattern of MG53 (upper), whereas co-expression of P104L with GFP-MG53 results in retention of MG53 at the Golgi apparatus (lower). These are representative images from >30 different cells.
The membrane repair defects observed with the P104L-Cav3 muscle may reflect a direct role of P104L-Cav3 on the subcellular distribution of MG53 or may reflect other adaptive changes in the muscle membrane system due to transgenic expression of the mutant P104L-Cav3. To test the acute effect of P104L-Cav3 on membrane repair in native skeletal muscle, we used electroporation-mediated delivery of plasmid DNA containing Cav3-GFP or P104L-GFP into the FDB of living WT mice. As observed in our previous studies (17), high efficiency delivery of plasmid DNA could be achieved in adult skeletal muscle with this methodology. As shown in Fig. 5A, Cav3-GFP expressed in skeletal muscle primarily localizes to the sarcolemmal membrane (left panel), whereas P104L-GFP displays localization away from the sarcolemmal membrane (right panel), consistent with the intracellular retention pattern observed in Fig. 4A. The impact of P104L expression on membrane repair was assayed by the entry of FM4-64 dye into isolated FDB fibers following UV laser injury. Although overexpression of Cav3-GFP produces some additional dye entry when compared with control fibers (Fig. 5C versus Fig. 1B), severe defects in membrane repair are observed in fibers overexpressing P104L-GFP, as indicated by the excessive entry of FM4-64 dye following injury (Fig. 5C). These results indicate that P104L-Cav3 has a dominant-negative effect on both MG53 translocation and membrane repair in skeletal muscle.
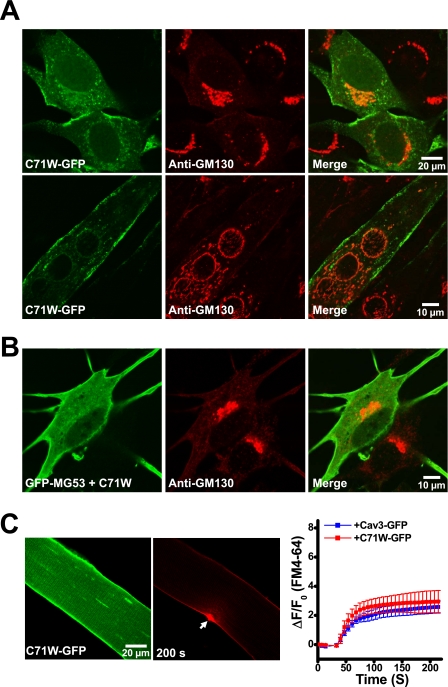
The striking dominant-negative effect of P104L-Cav3 on UV laser-induced injury to the sarcolemmal membrane led us to test two other Cav3 mutations linked to muscular dystrophy. First, another Cav3 mutation (R26Q) that results in aberrant MG53 localization to Golgi (11) also produces severe defects in membrane repair in skeletal muscle (supplemental Fig. S2). Second, a separate Cav3 mutation (C71W) that does not affect MG53 distribution has minimal effect on membrane repair capacity. As shown in Fig. 6A, the subcellular distribution of C71W-GFP expressed in C2C12 myoblasts or myotubes is similar to that observed with the wild-type Cav3-GFP (Fig. 4B, top). Co-expression of GFP-MG53 with the C71W-Cav3 mutant does not cause retention of MG53 at the Golgi apparatus (Fig. 6B), unlike that observed with the P104L-Cav3 mutant (Fig. 4C). Moreover, transient overexpression of C71W-GFP in the WT skeletal muscle does not appear to influence the membrane repair function following UV laser injury because there is essentially no difference observed with the amount of FM4-64 dye entered into the FDB fibers that are transfected with either Cav3-GFP or C71W-GFP (Fig. 6C). Together, our data suggest that altered subcellular localization of MG53 in association with Cav3 mutation may be linked to the defective membrane repair capacity in muscular dystrophy.
FIGURE 6.
C71W-Cav3 does not affect membrane repair capacity or MG53 localization in skeletal muscle. A, C2C12 myoblast or myotube cells transfected with C71W-GFP were subjected to immunostaining with anti-GM130. Confocal images show that C71W-GFP does not co-localize with GM130 in C2C12 cells at either the myoblast stage (upper) or the differentiated myotubes (lower). B, C2C12 myoblast cells co-transfected with GFP-MG53 and C71W-Myc were subjected to immunostaining with anti-GM130. Confocal images show that co-expression of C71W-Cav3 with GFP-MG53 does not cause retention of GFP-MG53 in the Golgi apparatus. C, FDB muscle fibers from WT mice were transfected by in vivo electroporation to allow for transient expression of C71W-GFP (left). Confocal images show that C71W-GFP mainly target to the sarcolemmal membrane. Measurement of FM4-64 entry (middle) shows that transient expression of C71W-GFP in adult WT skeletal muscle produced a similar degree of FM4-64 dye entry following UV laser wounding (arrows) as Cav3-GFP-expressing fibers (right panel, n = 18). Error bars indicate S.E.
MG53 Interacts with Dysferlin to Facilitate Vesicle Trafficking to Sites of Membrane Damage
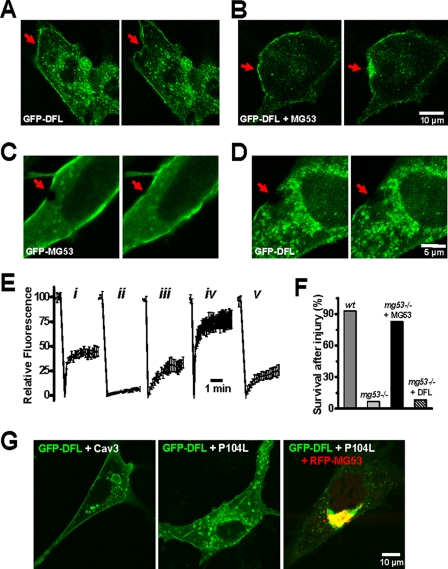
The retention of MG53 in the Golgi by P104L-Cav3 and R26Q-Cav3 observed in this study is similar to a phenomena reported for dysferlin (20). Considering the physical interaction between MG53, Cav3, and dysferlin (Fig. 2), we examined whether MG53 is involved in dysferlin function. Bansal et al. (7) showed that dysferlin is involved in repair of the sarcolemma in skeletal muscle; however, dysferlin itself does not appear to participate in recruitment of intracellular vesicles because dysferlin−/− muscle retains accumulation of vesicles near membrane damage sites. Indeed, GFP-dysferlin expressed in C2C12 myoblasts that do not express endogenous MG53 or dysferlin localizes to intracellular vesicles (Fig. 7A) but appears to be unresponsive to cell injury as there is no recruitment of vesicle toward the injury site created by microelectrode penetration (Fig. 7A and supplemental Movie 3). To test whether MG53 facilitates the membrane repair function of dysferlin, we co-expressed MG53 with GFP-dysferlin in C2C12 cells. By co-expressing untagged MG53, we observed active fusion and trafficking of GFP-dysferlin-containing vesicles toward the plasma membrane in response to acute membrane damage (Fig. 7B and supplemental Movie 4).
FIGURE 7.
MG53 is required for dysferlin function in membrane repair. A, acute microelectrode penetration into the plasma membrane did not cause redistribution of GFP-dysferlin toward the injury site in C2C12 myoblasts (n = 31). B, co-expression of GFP-dysferlin and MG53 can facilitate the recruitment of GFP-dysferlin toward the injury site in C2C12 myoblasts (n = 29). Right panels in A and B are 60 s after acute mechanic injury. For details of the dynamic membrane repair process, see supplemental Movie 3 and supplemental Movie 4. C, laser-induced bleaching of GFP fluorescence at the C2C12 plasma membrane is accompanied with rapid recovery of GFP-MG53 toward the damage site (arrow, n = 20). D, C2C12 myoblasts expressing GFP-dysferlin do not display recovery following UV bleaching (n = 20). Right panels in C and D are 100 s after laser bleaching. E, summary of fluorescence recovery following photo-bleaching in C2C12 myoblasts expressing GFP-MG53 (n = 20) (panel i); C2C12 myoblasts expressing GFP-dysferlin (n = 20) (panel ii); C2C12 myoblasts co-expressing GFP-dysferlin and MG53 (n = 45) (panel iii); C2C12 myotubes expressing GFP-MG53 at 7 days of differentiation (n = 35) (panel iv); and C2C12 myotubes expressing GFP-dysferlin at 7 days of differentiation (n = 35) (panel v). Error bars indicate S.E. F, primary cultured WT or mg53−/− myotubes transfected with either GFP-MG53 or GFP-DFL were subjected to mechanical injury by microelectrode penetration. Survival of mg53−/− myotubes was greatly compromised (n = 3/38) when compared with WT myotubes (n = 32/35) due to the excessive entry of extracellular Ca2+-inducing myotube contraction. Expression of MG53 rescues mg53−/− myotube survival (n = 26/32), whereas DFL does not (n = 3/36). G, mg53−/− myoblast cells were transfected with GFP-DFL and Cav3 (left), GFP-DFL and P104L-Cav3 (middle), or GFP-DFL, RFP-MG53, and P104L-Cav3 (right). These confocal images were representative of n > 20 cells examined.
We next used laser-induced fluorescence bleaching to delineate the functional relationship between MG53 and dysferlin in mediating the dynamic process of acute membrane repair. As shown in Fig. 7C, after laser-induced bleaching, rapid recovery of green fluorescence is observed in C2C12 myoblasts expressing GFP-MG53. However, vesicles in myoblasts expressing only GFP-dysferlin appear to be relatively static as no recovery of fluorescence is observed during the period of the experiment (up to 5 min) (Fig. 7D). Recovery of GFP-dysferlin trafficking toward the injury site is observed with co-expression of MG53 or in C2C12 myotubes that express endogenous MG53 (Fig. 7E). Moreover, MG53-mediated vesicle translocation is essential for membrane resealing and maintenance of cellular integrity because primary cultured mg53−/− myotubes are defective in membrane repair and rarely survive mechanical injury caused by microelectrode penetration (17). The defect in mg53−/− myotube repair is specific to MG53 because MG53 can rescue this defect in mg53−/− myotubes, whereas dysferlin cannot (Fig. 7F), as would be expected if MG53 were required for dysferlin trafficking to injury sites to function in membrane resealing.
A role for MG53 in the defective trafficking of dysferlin associated with Cav3 mutation is tested in primary cultured mg53−/− myoblast cells. GFP-DFL expression in mg53−/− cells displays distinct membrane targeting in addition to intracellular vesicle distribution (Fig. 7G, left), which is consistent with the studies of Bansal et al. (7) and Klinge et al. (6). Co-expression with P104L-Cav3 leads to partial retention of GFP-dysferlin in intracellular vesicles (Fig. 7G, middle), similar to previous studies by Hernández-Deviez et al. (20). Striking differences are observed with co-expression of MG53 together with P104L-Cav3. Under these conditions, a majority of GFP-dysferlin returns to the Golgi apparatus (Fig. 7G, right), suggesting that a functional interaction exists between MG53, Cav3, and dysferlin and that disruption of the function of any of these components can potentially affect the membrane repair capacity of skeletal muscle.
DISCUSSION
In this study, we show that a functional interaction between MG53, Cav3, and dysferlin is an important regulator of membrane repair in skeletal muscle. Our previous studies show that Cav3 can regulate MG53 action by balancing its vesicle trafficking action and prevent the development of filopodia-like structures associated with increased MG53 expression (18). Given the functional interaction between MG53 and Cav3, the effects of Cav3-mutations on dysferlin targeting to the sarcolemma membrane (20, 27) are likely influenced by the action of MG53. We found that MG53 interacts with dysferlin to facilitate membrane repair, with MG53 functioning upstream of dysferlin by providing a nucleation mechanism to recruit intracellular vesicles toward the acute cell injury sites (17). Thus, altered MG53 localization can be used as a marker for muscular dystrophy involving reduced sarcolemmal membrane repair capacity due to Cav3 mutation, and potentially, in other forms of dystrophy as well.
Our results indicate that MG53, Cav3, and dysferlin may form a molecular complex that participates in membrane repair in striated muscles. We find that disruption of the function of one of these components can affect the subcellular localization and membrane repair function of the other components. Specific genetic mutations of either Cav3 or dysferlin can lead to reduced membrane repair capacity and the development of muscular dystrophy in humans (3, 27, 28). It is also possible that molecular interactions between MG53, dysferlin, and caveolin-3 may strengthen the integrity of the sarcolemmal membrane, possibly through networking with the cortical cytoskeleton. Clearly, maintenance of the cytoskeleton and basal lamina structure is important for the integrity and repair capacity of muscle cells. Because our results indicate that the function of these proteins in membrane repair may depend on MG53, it is likely that mutations in the mg53 gene that segregate with muscular dystrophy or cardiac abnormalities will be found in the appropriate patient populations. Identification of such mutants in MG53 will increase our understanding of the functional interplay between MG53, Cav3, and dysferlin in muscle physiology and pathophysiology.
Prior to this study, dysferlin has been well demonstrated to participate in maintenance of muscle membrane integrity (7, 29). It was proposed that dysferlin can function as a fusogen to allow vesicles to form a membrane repair patch. This was based on immunostaining observations that dysferlin concentrates at the injury sites of isolated muscle fibers and the muscular dystrophy that appears in dysferlin knock-out mice (7). However, since the initial study by Bansal et al. (7), there has been no indication that dysferlin itself can facilitate the rapid translocation of vesicles associated with acute membrane damage. Indeed, dysferlin−/− muscle maintains the capacity for vesicle translocation to damage sites on the sarcolemma. This suggests that although dysferlin may participate in the final membrane-resealing process, proteins other than dysferlin are likely required for nucleation of intracellular vesicles toward the acute injury sites. Our data show that MG53 can functionally interact with dysferlin to facilitate repair of acute membrane damage.
Numerous studies have shown that both the loss-of-function and the gain-of function Cav3 mutants result in muscular dystrophy, frequently in an autosomal dominant fashion (8, 30, 31). Our findings of a dominant-negative function for the P104L and R26Q mutations over endogenous Cav3 in membrane repair provide a molecular mechanism for one aspect of the autosomal dominant nature of Cav3 mutations in dystrophy patients. Due to the dominant nature of Cav3 mutations over MG53 function, it is possible that disruption of the physical interaction between MG53 and Cav3 could have functional consequences on improvement of membrane repair capacity in dystrophic patients. Targeting the intermolecular domains that mediate the functional interaction between MG53, Cav3, and dysferlin through either genetic or pharmacological approaches could be an excellent therapeutic intervention for muscular dystrophy and other human diseases where compromised membrane integrity contributes to cellular dysfunction. For example, MG53 and Cav3 are both present in cardiac muscle in addition to skeletal muscle. Because membrane damage contributes to the death of cardiomyocytes during ischemia/reperfusion injury or progression of heart failure (32–34), pursuit of regulators of membrane repair should be an increasing priority for future translational research in the muscle and cardiovascular fields.
Supplementary Material
Acknowledgments
We thank Yi Chu for assistance in data processing and graphic conversions and Dr. Kate Bushby for providing the GFP-dysferlin construct.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-HL069000 and RO1-AG028856 (to J. M. and H. T.) and National Institutes of Health K99 Award K99/R00-AR054793 (to N. W.). This work was also supported by grants from the Ministry of Education, Science, Sports and Culture of Japan (to H. T.) and the American Heart Association (to C. C.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains two supplemental figures and four supplemental movies.
- Cav3
- caveolin-3
- MG53
- mitsugumin 53
- HA
- hyaluronidase
- GFP
- green fluorescent protein
- RFP
- red fluorescent protein
- FDB
- flexor digitorum brevis
- Co-IP
- co-immunoprecipitation
- WT
- wild type
- DFL
- dysferlin
- TRIM
- tri-partite motif.
REFERENCES
- 1.McNeil P. L., Kirchhausen T. ( 2005) Nat. Rev. Mol. Cell Biol. 6, 499– 505 [DOI] [PubMed] [Google Scholar]
- 2.Towler M. C., Kaufman S. J., Brodsky F. M. ( 2004) Traffic 5, 129– 139 [DOI] [PubMed] [Google Scholar]
- 3.Glover L., Brown R. H., Jr.( 2007) Traffic 8, 785– 794 [DOI] [PubMed] [Google Scholar]
- 4.Steinhardt R. A., Bi G., Alderton J. M. ( 1994) Science 263, 390– 393 [DOI] [PubMed] [Google Scholar]
- 5.Miyake K., McNeil P. L. ( 1995) J. Cell Biol. 131, 1737– 1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klinge L., Laval S., Keers S., Haldane F., Straub V., Barresi R., Bushby K. ( 2007) FASEB J. 21, 1768– 1776 [DOI] [PubMed] [Google Scholar]
- 7.Bansal D., Miyake K., Vogel S. S., Groh S., Chen C. C., Williamson R., McNeil P. L., Campbell K. P. ( 2003) Nature 423, 168– 172 [DOI] [PubMed] [Google Scholar]
- 8.Minetti C., Sotgia F., Bruno C., Scartezzini P., Broda P., Bado M., Masetti E., Mazzocco M., Egeo A., Donati M. A., Volonte D., Galbiati F., Cordone G., Bricarelli F. D., Lisanti M. P., Zara F. ( 1998) Nat. Genet. 18, 365– 368 [DOI] [PubMed] [Google Scholar]
- 9.Galbiati F., Volonte D., Minetti C., Chu J. B., Lisanti M. P. ( 1999) J. Biol. Chem. 274, 25632– 25641 [DOI] [PubMed] [Google Scholar]
- 10.Smythe G. M., Eby J. C., Disatnik M. H., Rando T. A. ( 2003) J. Cell Sci. 116, 4739– 4749 [DOI] [PubMed] [Google Scholar]
- 11.Fee D. B., So Y. T., Barraza C., Figueroa K. P., Pulst S. M. ( 2004) Muscle Nerve 30, 375– 378 [DOI] [PubMed] [Google Scholar]
- 12.Han R., Campbell K. P. ( 2007) Curr. Opin. Cell Biol. 19, 409– 416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galbiati F., Razani B., Lisanti M. P. ( 2001) Trends Mol. Med. 7, 435– 441 [DOI] [PubMed] [Google Scholar]
- 14.Han R., Bansal D., Miyake K., Muniz V. P., Weiss R. M., McNeil P. L., Campbell K. P. ( 2007) J. Clin. Investig. 117, 1805– 1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galbiati F., Volonte D., Chu J. B., Li M., Fine S. W., Fu M., Bermudez J., Pedemonte M., Weidenheim K. M., Pestell R. G., Minetti C., Lisanti M. P. ( 2000) Proc. Natl. Acad. Sci. U. S. A. 97, 9689– 9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagiwara Y., Sasaoka T., Araishi K., Imamura M., Yorifuji H., Nonaka I., Ozawa E., Kikuchi T. ( 2000) Hum. Mol. Genet. 9, 3047– 3054 [DOI] [PubMed] [Google Scholar]
- 17.Cai C., Masumiya H., Weisleder N., Matsuda N., Nishi M., Hwang M., Ko J. K., Lin P., Thornton A., Zhao X., Pan Z., Komazaki S., Brotto M., Takeshima H., Ma J. ( 2009) Nat. Cell Biol. 11, 56– 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai C., Masumiya H., Weisleder N., Pan Z., Nishi M., Komazaki S., Takeshima H., Ma J. ( 2009) J. Biol. Chem. 284, 3314– 3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko J. K., Ma J. ( 2005) Am. J. Physiol. Cell Physiol. 288, C1273– 1278 [DOI] [PubMed] [Google Scholar]
- 20.Hernández-Deviez D. J., Martin S., Laval S. H., Lo H. P., Cooper S. T., North K. N., Bushby K., Parton R. G. ( 2006) Hum. Mol. Genet. 15, 129– 142 [DOI] [PubMed] [Google Scholar]
- 21.Pouvreau S., Royer L., Yi J., Brum G., Meissner G., Ríos E., Zhou J. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 5235– 5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sunada Y., Ohi H., Hase A., Ohi H., Hosono T., Arata S., Higuchi S., Matsumura K., Shimizu T. ( 2001) Hum. Mol. Genet. 10, 173– 178 [DOI] [PubMed] [Google Scholar]
- 23.Nishi M., Komazaki S., Kurebayashi N., Ogawa Y., Noda T., Iino M., Takeshima H. ( 1999) J. Cell Biol. 147, 1473– 1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisleder N., Brotto M., Komazaki S., Pan Z., Zhao X., Nosek T., Parness J., Takeshima H., Ma J. ( 2006) J. Cell Biol. 174, 639– 645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNeil A. K., Rescher U., Gerke V., McNeil P. L. ( 2006) J. Biol. Chem. 281, 35202– 35207 [DOI] [PubMed] [Google Scholar]
- 26.Ohsawa Y., Toko H., Katsura M., Morimoto K., Yamada H., Ichikawa Y., Murakami T., Ohkuma S., Komuro I., Sunada Y. ( 2004) Hum. Mol. Genet. 13, 151– 157 [DOI] [PubMed] [Google Scholar]
- 27.Hernández-Deviez D. J., Howes M. T., Laval S. H., Bushby K., Hancock J. F., Parton R. G. ( 2008) J. Biol. Chem. 283, 6476– 6488 [DOI] [PubMed] [Google Scholar]
- 28.Guglieri M., Straub V., Bushby K., Lochmüller H. ( 2008) Curr. Opin. Neurol. 21, 576– 584 [DOI] [PubMed] [Google Scholar]
- 29.Bansal D., Campbell K. P. ( 2004) Trends Cell Biol. 14, 206– 213 [DOI] [PubMed] [Google Scholar]
- 30.McNally E. M., de Sá Moreira E., Duggan D. J., Bönnemann C. G., Lisanti M. P., Lidov H. G., Vainzof M., Passos-Bueno M. R., Hoffman E. P., Zatz M., Kunkel L. M. ( 1998) Hum. Mol. Genet. 7, 871– 877 [DOI] [PubMed] [Google Scholar]
- 31.Galbiati F., Engelman J. A., Volonte D., Zhang X. L., Minetti C., Li M., Hou H., Jr., Kneitz B., Edelmann W., Lisanti M. P. ( 2001) J. Biol. Chem. 276, 21425– 21433 [DOI] [PubMed] [Google Scholar]
- 32.Setsuta K., Seino Y., Ogawa T., Ohtsuka T., Seimiya K., Takano T. ( 2004) Circ. J. 68, 747– 750 [DOI] [PubMed] [Google Scholar]
- 33.Yeh C. H., Chen T. P., Lee C. H., Wu Y. C., Lin Y. M., Lin P. J. ( 2006) Shock 26, 262– 270 [DOI] [PubMed] [Google Scholar]
- 34.Date T., Mochizuki S., Belanger A. J., Yamakawa M., Luo Z., Vincent K. A., Cheng S. H., Gregory R. J., Jiang C. ( 2003) J. Mol. Cell. Cardiol. 35, 811– 821 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.