Abstract
Patients with severe combined immune deficiency (SCID) suffer from defective T-cell Ca2+ signaling. A loss of Ca2+ entry has been linked at the molecular level to single missense mutation R91W in the store-operated Ca2+ channel ORAI1. However, the mechanistic impact of this mutation on ORAI1 function remains unclear. Confocal Förster resonance energy transfer microscopy revealed that dynamic store-operated coupling of STIM1 to ORAI1 R91W was largely sustained similar to wild-type ORAI1. Characterization of various point mutants at position 91 by whole cell patch clamp recordings displayed that neutral or even negatively charged amino acids did not abolish ORAI1 function. However, substitution by hydrophobic leucine, valine, or phenylalanine resulted in non-functional ORAI1 channels, despite preserved STIM1 coupling. Besides conformational constraints at the N terminus/membrane interface predicted for the hydrophobic mutants, additional key factor(s) were suggested to determine ORAI1 functionality. Calculation of the probability for the 1st transmembrane domain and its hydrophobicity revealed a substantial increase for all hydrophobic substitutions that lead to non-functional ORAI1 R91X mutants in contrast to those with hydrophilic residues. Hence, increased hydrophobicity might lead to disrupted permeation/gating, as an ORAI1 channel with increased pore size and R91W mutation failed to recover activity. In conclusion, the increase in hydrophobicity at the N terminus/membrane interface represents the major cause for yielding non-functional ORAI1 channels.
The immune system consists of various cell types such as T- and B-cells that are involved in protecting the body from foreign particles and pathogenic organisms. Defects in T-cell development impair normal immune function and may lead to primary immune deficiency. One subgroup thereof represented by the severe combined immunodeficiency (SCID)4 occurs in 1 of 50,000–100,000 live births, causing an onset of one or more serious infections, such as pneumonia, meningitis, or bloodstream infections, within the first few months of life (1, 2). It is currently known that defective T-cell signaling in SCID patients can arise from mutations in different genes including a point mutation in ORAI1 (3–5). T-cell function and proliferation requires calcium influx mediated by the Ca2+ release-activated Ca2+ channel. It is activated by depletion of intracellular Ca2+ stores induced by the second messenger inositol 1,4,5-trisphosphate (6–9) and this cytosolic Ca2+ entry serves essential functions from secretion to gene expression and cell growth (10).
A combination of RNA interference-based screening and analysis of single nucleotide polymorphism arrays of patients with SCID syndrome has led to the identification of the plasma-membrane protein ORAI1 as a key component of the Ca2+ release-activated Ca2+ channel complex (11, 12). An overexpression of wild-type ORAI1 (4) or a related member ORAI3 (13) in SCID T-cells partially restored store-operated Ca2+ influx. Based on permeability studies of different point mutants in transmembrane regions 1 and 3 of ORAI1, it is suggested to form the pore of the Ca2+ release-activated Ca2+ channel (14–16). ORAI1 acts in concert with the stromal interacting molecule 1 (STIM1) (4, 17–19), a single transmembrane spanning Ca2+ sensor located in the endoplasmic reticulum. We have recently demonstrated that a dynamic coupling of STIM1 to ORAI1 via a putative coiled-coil domain in its C terminus is induced by endoplasmic reticulum store depletion, resulting in Ca2+ influx through the ORAI1 channels (20). The impaired Ca2+ influx into T-cells of SCID patients has been attributed to a single missense mutation R91W in ORAI1, which is conserved among all three ORAI proteins and located at the N terminus/membrane interface (18). Platelets of mice expressing ORAI1 R93W (a homologue to the human ORAI1 R91W) display markedly reduced store-operated Ca2+ entry, reduced integrin expression, as well as impaired degranulation (21). The generation of a series of concatenated tetramers of ORAI1 that include different numbers and arrangements of mutant ORAI1 R91W proteins shows that an increasing number of mutant proteins results in a graded reduction in Ca2+ release-activated Ca2+ channel currents (22). However, the molecular impact that leads to non-functional ORAI1 R91W channels is still unknown.
Here, a set of single point mutations at position 91 of ORAI1 and the adjacent ASSR domain (aa 88–91) were analyzed with confocal Förster resonance energy transfer (FRET) microscopy and the whole cell patch clamp technique. Our functional data together with predictions on the secondary structure suggested that besides conformational constraints within the ASSW domain a substantial increase in hydrophobicity and probability of the first transmembrane segment led to non-functional ORAI channels, yet retained their ability to couple to STIM1 in a store-dependent manner.
MATERIALS AND METHODS
Molecular Cloning and Mutagenesis
Human ORAI1 (ORAI1; accession number NM_032790) was kindly provided by A. Rao, Harvard Medical School. C-terminal-tagged pECFP-N1 and pEYFP-N1/ORAI1 constructs were cloned using the XhoI and BamHI sites of the contemplated vectors. N-terminal-tagged ORAI1 constructs were cloned via SalI and SmaI restriction sites of pECFP-C1 and pEYFP-C1 expression vectors (Clontech). pECFP/pEYFP-C1/ORAI1 and pECFP/pEYFP-N1/ORAI1 served as templates for the generation of the ORAI1 mutants. Suitable primers exchanged the corresponding codon from CGG to TGG (R91W), GGG (R91G), CAC (R91H), GAG (R91E), GTG (R91V), TTT (R91F), CTG (R91L), AGC (R91S), ACG (R91T) and AAT (R91N), from TCC AGC to GGC GGC (S89G/S90G), from TCC AGC to CCC CCC (S89P/S90P), and from GAG to GAT (E106D). pEYFP-C1 ORAI1 R91W was used as template for the generation of ORAI1 R91W/E106D. All mutants were constructed using the QuikChange XL site-directed mutagenesis kit (Stratagene). Human Orai3 (Orai3; accession number NM_152288) was kindly provided by L. Birnbaumer, NIEHS, National Institutes of Health. Orai3 was cloned into the pEYFP-C1 (Clontech) expression vector via its BamHI and XbaI restriction sites for N-terminal labeling. The generation of the YFP-Orai3 S63P/S64P was carried out via the substitution of TCC AGC to CCC CCC using the QuikChange XL site-directed mutagenesis kit (Stratagene). N-terminal ECFP- and EYFP-tagged human STIM1 (STIM1; accession number NM_003156) was kindly provided by T. Meyer, Stanford University. C-terminal-EYFP-tagged STIM1 was purchased from GeneCopoeiaTM (catalog number EX-S0521-M02). All constructs were sequenced and confirmed for their validity.
Cell Culture and Transfection
Transfection of HEK293 cells was performed using either the TransFectin Lipid Reagent (Bio-Rad) or the TransPass Transfection Reagent (New England Biolabs) and following 12–48 h cells were employed for experiments. For reducing cell density, cells were sometimes reseeded >7 h before the experiments started.
Electrophysiology
Electrophysiological recordings comparing characteristics of 2–3 constructs as displayed in the respective figure panels were carried out in paired comparison on the same day. Expression pattern and levels of the various constructs were carefully monitored by confocal fluorescence microscopy and were not significantly changed by the introduced mutations. Electrophysiological experiments were performed at 20–24 °C, using the patch clamp technique in the whole cell recording configuration. For STIM1/ORAI current measurements voltage ramps were usually applied every 5 s from a holding potential of 0 mV, covering a range of −90 to 90 mV over 1 s. The internal pipette solution contained (in mm) 3.5 MgCl2, 145 cesium methane sulfonate, 8 NaCl, 10 HEPES, 10 EGTA, pH 7.2. Extracellular solution consisted of (in mm) 145 NaCl, 5 CsCl, 1 MgCl2, 10 HEPES, 10 glucose, 10 CaCl2, pH 7.4. Cs+ divalent-free solution included 150 CsCl, 10 HEPES, 10 glucose, and 10 EDTA. Currents were leak-corrected by subtracting the leak current obtained in the presence of 10 μm LaCl3.
Confocal FRET Fluorescence Microscopy
Confocal FRET microscopy was performed similarly to Ref. 23. In brief, a QLC100 Real Time Confocal System (VisiTech Int., UK) was used for recording fluorescence images connected to two Photometrics CoolSNAPHQ monochrome cameras (Roper Scientific) and a dual port adapter (dichroic, 505lp; cyan emission filter, 485/30; yellow emission filter, 535/50; Chroma Technology Corp.). This system was attached to an Axiovert 200M microscope (Zeiss, Germany) in conjunction with an argon ion multiwavelength (457, 488, and 514 nm) laser (Spectra Physics). The wavelengths were selected by an Acousto Optical Tuneable Filter (VisiTech Int.). MetaMorph 5.0 software (Universal Imaging Corp.) was used to acquire images and control the confocal system. Illumination times for CFP/FRET and YFP images that were recorded with a minimum delay consecutively were about 900 ms. Prior to calculation the images had to be corrected due to cross-talk as well as cross-excitation. For this, the appropriate cross-talk calibration factors were determined for each of the constructs on the day the FRET experiments were performed. The corrected FRET image (NFRET) was calculated on a pixel to pixel basis after background subtraction and threshold determination using a custom-made software (24) integrated in MatLab 7.0.4 according to the method published by Ref. 25. To minimize an effect of slight variations in expression levels, the corrected FRET was normalized to the square root of CFP × YFP intensities. The local ratio between CFP and YFP might vary due to different localizations of diverse protein constructs, which could lead to the calculation of false FRET values (26). Accordingly, the analysis was limited to pixels with a CFP:YFP ratio between 1:10 and 10:1 (26) to yield reliable results. This occurred rather seldom in our experiments, and ∼90% of ratios were between 1:5 and 5:1.
Secondary Structure Assignments and Bioinformatics
Data were obtained from the Protein Data Bank (27, 28). Redundant entries were removed with the PISCES software (29). Secondary structural assignments were performed with STRIDE (30) and superpositions with the Kabsch algorithm (31, 32), by considering only the Cα atoms. Only four types of secondary structures were considered: helix (STRIDE statuses H, G, or I), extended (statuses E, B, or b), coils (status C), and turn (status T). The numbers of protein structures containing the ASSR, ASSW, ASSL, ASSV, ASSF, ASSG, ASSE, ASSH, ASSS, ASST, ASSN, AGGR, APPR, or AGGW tetrapeptides were 38, 14, 88, 61, 56, 40, 27, 18, 189, 177, 33, 89, 19, and 34, respectively.
Secondary structure prediction was carried out using a Multivariate Linear Regression Combination combining GOR4 (33), SIMPA96 (34), and SOPMA (35) methods. Hydrophobicity profiles (36) were determined with a window size of 9 amino acids. Transmembrane probability prediction based on the Hidden Markov model (37) was calculated.
Statistics
Significance analysis was performed with the two-tailed Mann-Whitney test. The mean ± S.E. values are shown throughout the article.
RESULTS
Lack of Ca2+ Current Activation of ORAI1 R91W Is Not Due to Disrupted Communication with STIM1
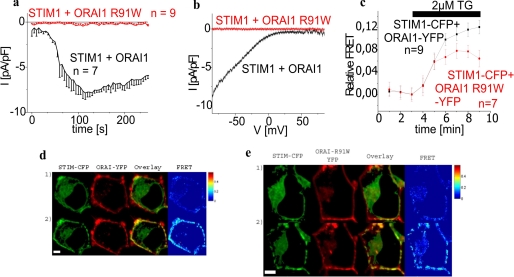
The mechanism of abolished activation of Ca2+ currents resulting from a single point mutation in SCID-related ORAI1 R91W is still unresolved. Co-expression of CFP/YFP-tagged STIM1 and ORAI1 in HEK293 cells yielded robust inward currents following store depletion by intracellular Ca2+ buffering with 10 mm EGTA and a 10 mm Ca2+-containing bath solution (Fig. 1, a and b). In contrast, co-expressed STIM1 and ORAI1 R91W tagged either N or C terminally failed to generate detectable Ca2+ influx in an analogous approach. Moreover, perfusion with a sodium divalent-free bath solution did not reveal significant current activation through ORAI1 R91W (data not shown). In an attempt to analyze whether an altered localization in the cell or a disrupted coupling of ORAI1 R91W to STIM1 is responsible for this complete loss of function, we utilized confocal FRET microscopy of HEK293 cells that expressed C-terminal fluorescent-labeled STIM1 as well as ORAI1 R91W (Fig. 1e). C-terminal in contrast to N-terminal tagging of the latter was preferred in this study to avoid interference with mutations introduced at the N terminus/membrane interface (see below). In support of a previous study (4), localization and expression pattern of the ORAI1 R91W SCID mutant in HEK cells were unaltered compared with wild-type ORAI1 (Fig. 1d). Moreover homomerization of ORAI1 R91W as well as its heteromerization together with ORAI1 wild-type was not impaired (data not shown). The two proteins STIM1 and ORAI1 R91W clearly exhibited clustered co-localization following store depletion (Fig. 1, d and e). This co-clustering occurred along with a substantial increase in FRET (Fig. 1, c, d, and e), yet to a reduced extent probably due to a slight decrease in coupling efficiency when compared with that detected with STIM1 and wild-type ORAI1 or ORAI1 R91W when N-terminal tagged (20) confirming that coupling of STIM1 to ORAI1 R91W was largely sustained (38). Nevertheless, besides intact coupling to STIM1 the molecular process promoting permeation/gating of the channel into the open state might somehow be impaired in ORAI1 R91W causing its non-functionality. In the following we characterized the molecular fingerprint at and around position 91 of ORAI1 by exploring the potential permeation/gating capability of the channel with a set of single point mutations at this prominent position. We specifically focused on the requirement of the positive charge and a certain hydrophobicity (39, 40) at position 91 for permeation/gating together with its impact on predicted conformations and probability as well as hydrophobicity of the first transmembrane segment (41).
FIGURE 1.
ORAI1 R91W SCID mutant displays loss of Ca2+ current activity despite preserved coupling to STIM1. a, time course of whole cell inward currents at −74 mV activated by passive store depletion of HEK293 cells expressing STIM1 with either ORAI1 or ORAI1 R91W. b, respective I/V curves to a from representative cells taken at maximum current density activation. c, time course of relative FRET between STIM1-CFP and ORAI1 R91W-YFP (t = 120 s; p < 0.05) in comparison to STIM1-CFP and ORAI1-YFP. d and e, localization, overlay, and calculated FRET life cell image series of STIM1-CFP and ORAI1-YFP (d) or ORAI1 R91W-YFP (e) (1) before and (2) after store depletion (t = 8 min; p < 0.05).
Loss of a Positive Charge at Position 91 in the ORAI1 R91W SCID Mutant Does Not Account for a Lack of Ca2+ Currents
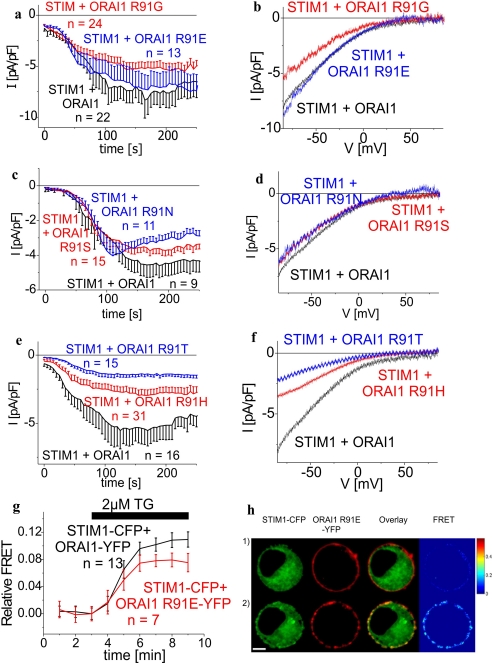
In contrast to the positively charged arginine of wild-type ORAI1 the tryptophan is an uncharged and highly hydrophobic amino acid. To evaluate the impact of the positive charge at position 91 of the arginine for ORAI1-mediated Ca2+ currents, various point mutants, i.e. ORAI1 R91G, ORAI1 R91E, ORAI1 R91H, ORAI1 R91N, ORAI1 R91S, ORAI1 R91T, were generated and examined for their function. Glycine, serine, threonine, asparagines are neutral, glutamate is negatively charged, and histidine is rather neutral at pH 7.2 (42). All these amino acids are clearly more hydrophilic than the tryptophan present in the SCID mutant (see Table 1). ORAI1 R91G, ORAI1 R91E, ORAI1 R91S, ORAI1 R91N, ORAI1 R91H, and ORAI1 R91T exhibited activation of Ca2+ currents following store depletion by intracellular 10 mm EGTA with biophysical (Fig. 2) and pharmacological (2-aminoethyldiphenyl borate (2-APB) sensitivity; data not shown) characteristics similar to wild-type ORAI1. Moreover, ORAI1 R91G (Fig. 2, a and b), ORAI1 R91E (Fig. 2, a and b), ORAI1 R91S (Fig. 2, c and d), and ORAI1 R91T (Fig. 2, c and d) reached maximum current densities slightly below wild-type ORAI1, whereas those of ORAI1 R91H (Fig. 2, e and f) and ORAI1 R91T (Fig. 2, e and f) were significantly reduced. Nevertheless, in line with their functional activation upon store depletion all of these ORAI1 mutants showed co-clustering and an increase in dynamic FRET with STIM1 upon store depletion (exemplarily displayed for ORAI1 R91E in Fig. 2, g and h) further supporting the concept that the positive charge of the arginine at position 91 is not an essential requirement for ORAI1 function. The reason for reduced current density of ORAI1 R91H as well as ORAI1 R91T might be due to increased hydrophobicity when compared with the other substituted amino acids, except that of glycine, which we evaluated in our next approach.
TABLE 1.
Overview on respective currents from STIM1/ORAI1 R91X proteins, their coupling to STIM1, the amino acid hy dro pho bic ity at position 91, and the predicted secondary structure
| Current | STIM1/ORAI1 co-clustering | Hydrophobicitya | Secondary structureb amino acids 73–103 | |
|---|---|---|---|---|
| 91R | 7–9 pA/pF | + | 0.64 | ALSWRKLYLSRAKLKASSRTSALLSGFAMVA |
| hhhhhhhhhhhhhhhccchhhhhhhhhhhhh | ||||
| 91W | 0 pA/pF | + | 0.85 | ALSWRKLYLSRAKLKASSWTSALLSGFAMVA |
| hhhhhhhhhhhhhhhhchhhhhhhhhhhhhh | ||||
| 91G | 6–8 pA/pF | + | 0.72 | ALSWRKLYLSRAKLKASSGTSALLSGFAMVA |
| hhhhhhhhhhhhhhhccccchhhhhhhhhhh | ||||
| 91E | 6–8 pA/pF | + | 0.62 | ALSWRKLYLSRAKLKASSETSALLSGFAMVA |
| hhhhhhhhhhhhhhhccchhhhhhhhhhhhh | ||||
| 91S | 4–5 pA/pF | NDc | 0.66 | ALSWRKLYLSRAKLKASSSTSALLSGFAMVA |
| Hhhhhhhhhhhhhhccccchhhhhhhhhhhh | ||||
| 91N | 4–5 pA/pF | ND | 0.63 | ALSWRKLYLSRAKLKASSNTSALLSGFAMVA |
| hhhhhhhhhhhhhhccccchhhhhhhhhhhh | ||||
| 91H | 3–4 pA/pF | + | 0.78 | ALSWRKLYLSRAKLKASSHTSALLSGFAMVA |
| hhhhhhhhhhhhhhccccchhhhhhhhhhhh | ||||
| 91T | 2 pA/pF | ND | 0.70 | ALSWRKLYLSRAKLKASSTTSALLSGFAMVA |
| hhhhhhhhhhhhhhccccchhhhhhhhhhhh | ||||
| 91L | 0 pA/pF | + | 0.85 | ALSWRKLYLSRAKLKASSLTSALLSGFAMVA |
| hhhhhhhhhhhhhhhhhhhhhhhhhhhhhhh | ||||
| 91V | 0 pA/pF | ND | 0.86 | ALSWRKLYLSRAKLKASSVTSALLSGFAMVA |
| hhhhhhhhhhhhhhhhhhhhhhhhhhhhhhh | ||||
| 91F | 0 pA/pF | ND | 0.88 | ALSWRKLYLSRAKLKASSFTSALLSGFAMVA |
| hhhhhhhhhhhchhchhhhhhhhhhhhhhhh |
a According to Ref. (36).
b Combining three secondary structure prediction programs: SOPMA, GOR4, and SIMPA.
c ND, not determined.
FIGURE 2.
ORAI1 current activation occurs independent of the charge at position 91. a–c and e, time course of whole cell inward currents at −74 mV activated by passive store depletion of HEK293 cells expressing STIM1 with ORAI1 in comparison to (a) ORAI1 R91G (t = 170 s; p > 0.05), ORAI1 R91E (t = 170 s; p > 0.05); (b) ORAI1 R91S (t = 150s, p > 0.05), ORAI1 R91N (t = 150 s, p > 0.05); and (c) ORAI1 R91H (t = 130 s; p < 0.05), ORAI1 R91T (t = 150 s, p < 0.05). b, d, and f, respective I/V curves to a, c, and e from representative cells taken at maximum current density activation. g, time course of relative FRET between STIM1-CFP and ORAI1 R91E-YFP in comparison to STIM1-CFP and ORAI1-YFP. h, localization, overlay, and calculated FRET life cell image series of STIM1-CFP and ORAI1 R91E-YFP before (1) and after (2) store depletion (t = 8 min; p < 0.05).
Hydrophobicity at Position 91 Is an Essential Determinant for ORAI1 Activity
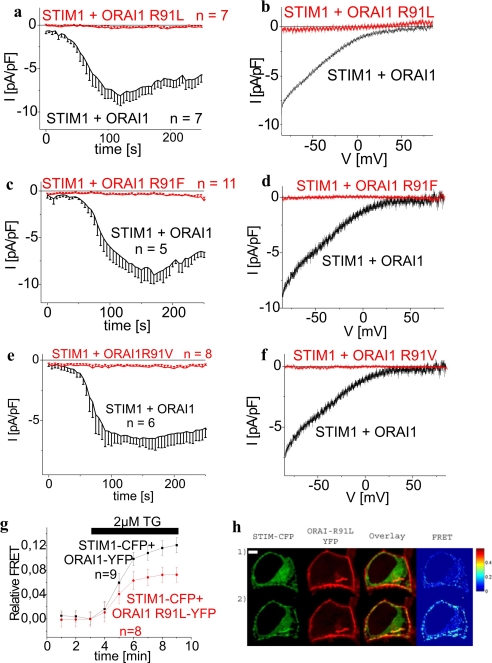
Next we analyzed the impact of hydrophobicity in the context of distinct steric requirements by mutating the hydrophilic arginine of wild-type ORAI1 to the hydrophobic leucine, valine, and phenylalanine. Similar to the SCID mutant, all of these ORAI1 R91L (Fig. 3, a and b), ORAI1 R91F (Fig. 3, c and d), and ORAI1 R91V (Fig. 3, e and f) mutants completely lost their ability to mediate activation of Ca2+ currents. Nevertheless, their expression pattern and particularly their coupling to STIM1 remained largely preserved when compared with wild-type ORAI1 as well as ORAI1 R91W as gauged by the observed co-clustering and FRET with STIM1 following store depletion (exemplarily shown for ORAI1 R91L in Fig. 3, g and h). Thus, the increased hydrophobicity at position 91 drastically affected functionality of the ORAI1 channels in terms of a permeation/gating defect by a mechanism that is still unclear. To get further insight as to how these mutations might have an impact on ORAI1 gating, we carried out bioinformatic analyses focusing both on secondary structure predictions within the region ASSR (aa 88–91) at the N terminus/membrane interface and the probability as well as hydrophobicity of the first transmembrane segment that are highly conserved among all three ORAI proteins.
FIGURE 3.
Hydrophobic amino acids at position 91 result in loss of ORAI1 Ca2+ current activity despite preserved coupling to STIM1. a, c, and e, time course of whole cell inward currents at −74 mV activated by passive store depletion of HEK293 cells expressing STIM1 with either ORAI1 in comparison to ORAI1 R91L (t = 120s; p < 0.05) (a), ORAI1 R91F (t = 150 s; p < 0.01) (c), or ORAI1 R91V (t = 150 s; p < 0.01) (e). b, d, and f, respective I/V curves to a, c, and e from representative cells taken at maximum current density activation. g, time course of relative FRET between STIM1-CFP and ORAI1 R91L-YFP in comparison to STIM1-CFP and ORAI1-YFP. h, localization, overlay, and calculated FRET life cell image series of STIM1-CFP and ORAI1 R91L-YFP before (1) and after (2) store depletion (t = 8 min; p < 0.05).
Lack of Ca2+ Current Activation of the SCID ORAI1 R91W Mutant Might Be Related to an Altered Flexibility in the ASSW Region and an Overall Increased Hydrophobicity of the First Transmembrane Segment
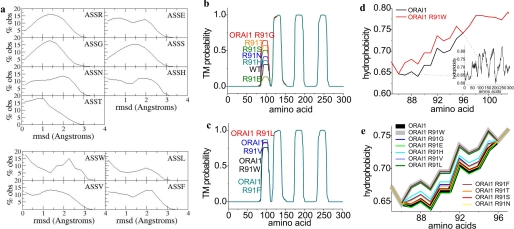
To evaluate structural differences of the ORAI1 R91X (X = Arg, Trp, Gly, Glu, His, Ser, Thr, Asn, Leu, Val, and Phe) point mutants examined above, we initially carried out secondary structure predictions around the conserved region aa 88–91 (ASSX) employing a Multivariate Linear Regression Combination (combining three secondary structure prediction programs: SOPMA, GOR4, and SIMPA; see “Materials and Methods”). It turned out (Table 1) that wild-type ORAI1 as well as all functional hydrophilic ORAI1 mutants contained a short random coil region ranging from position 88 to 91 (aa ASSR) embedded between two predicted helical structures. In contrast, hydrophobic substitution at position 91 led to a predicted transition of random coil to an α helical structure that might be accompanied by a reduction in conformational flexibility (41) and interference with permeation/gating of the ORAI1 channel (Table 1). Apparently, not all secondary structure prediction programs predicted a coil region in the N-terminal/membrane region of ORAI1. Therefore, we extended our bioinformatic analysis by examining the conformational features of the tetrapeptide ASSX (X = Arg, Trp, Leu, Gly, Glu, Ser, Thr, Asn, His, Val, or Phe) moieties when embedded into experimentally determined three-dimensional protein structures that are found in the Protein Data Bank (27, 28). Eight non-redundant sets of structures containing the peptides ASSW, ASSR, ASSL, ASSV, ASSF, ASSG, ASSE, ASSS, ASSN, ASST, and ASSH were assembled. We superposed each pair of tetrapeptides within each set and computed the root mean square deviation values (31, 32) displaying the difference between Cα atom positions between two proteins. For example, given that the set of protein structures with the ASSL moiety contained 88 entries, (88 × 87)/2 = 3828 superpositions were performed and 3828 r.m.s. deviation values were computed. The distributions of the r.m.s. deviation values were then determined, and used to estimate the degree of flexibility of a given polypeptide segment. An r.m.s. deviation distribution with a broad single maximum would suggest a polypeptide string that is conformationally flexible and does not tend to cluster into well defined conformations. On the contrary, if the r.m.s. deviation distribution is bimodal, the polypeptide segment is apparently subject to conformational constraints and tends to assume two or more distinct conformations (43). The r.m.s. deviation distributions of the eight tetrapetide moieties were depicted in Fig. 4. The tetrapeptides ASSR (38 analyzed), ASSG (40 analyzed), ASSS (189 analyzed), ASSN (33 analyzed), and ASST (177 analyzed) were evaluated as conformationally more variable, as their r.m.s. deviation values were well spread and distributed around a single maximum indicating that these moieties could assume many different conformations. On the other hand, conformations of the tetrapeptides ASSW (14 analyzed), ASSV (61 analyzed), ASSF (56 analyzed), and ASSL (88 analyzed) were less variable as gauged from their bimodal distribution of the r.m.s. deviation values indicating the presence of preferred conformational clusters. The other two tetrapeptide ASSE and ASSH moieties revealed an intermediate distribution of r.m.s. deviation values. Thus, secondary structure together with flexibility predictions correlated with our functional data, suggesting that the hydrophobic amino acids tended to reduce flexibility at the ORAI1 N terminus/membrane interface thereby impairing its permeation/gating.
FIGURE 4.
Bioinformatics analyses of ORAI1 constructs. Conformational features for the tetrapeptide moiety ASSX (X = Arg, Gly, Glu, Ser, Asn, Thr, His, Trp, Leu, Phe, and Val) transmembrane (TM) probability as well as hydrophobicity plots from corresponding ORAI1 proteins. a, broad range of conformations for ASSR, ASSG, ASSS, ASSN, ASST, intermediate for ASSE and ASSH, and conformational constraints for ASSW, ASSL, ASSF, and ASSV. b and c, transmembrane probability plots for functional ORAI1 proteins and mutants in comparison to non-functional ORAI mutants. d, hydrophobicity plots for ORAI1 compared with ORAI1 R91W of amino acid sequence 85–103 as shown by the inset displaying the hydrophobicity profile for the whole ORAI1 protein. e, hydrophobicity plots for functional ORAI1 proteins and mutants in comparison to non-functional ORAI mutants.
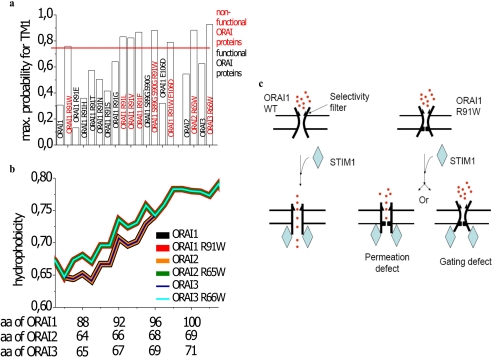
Based on the prominent effect of alteration in hydrophobicity at position 91 on ORAI1 channel function, we further analyzed its impact by changes in the hydrophobicity of the first transmembrane segment. Therefore we carried out transmembrane domain predictions of ORAI proteins and their mutants using TMHMM (Figs. 4, b and c, 7a; see “Materials and Methods”). ORAI1 wild-type displayed a hydrophobic region starting near Arg-91 that predicted to form the first transmembrane segment with a probability of about 0.31 (Figs. 4b and 7a). In contrast, substitution of Arg-91 to Trp yielded a transmembrane probability of about 0.76 assumedly by enhanced hydrophobicity at position 91 (Figs. 4c and 7a). Similarly all other mutants containing a hydrophobic amino acid at position 91 (Leu, Val, and Phe) revealed probabilities higher than 0.76 correlating well with their non-functionality (Figs. 4c and 7a). When employing the same test to the functional ORAI1 mutants that contained more hydrophilic residues, all of them yielded probabilities below that of ORAI1 R91G (Figs. 4b and 7a). The latter, whereas still fully functional, exhibited a probability of 0.64 that was almost 2-fold higher than that of wild-type ORAI1. This value was close to but not as high as that of the non-functional ORAI1 R91W. One reason for the observed functionality of ORAI1 R91G might be found in the fact that glycine by providing the maximum degree of conformational freedom (44) somewhat counterbalances the increase in probability for the first transmembrane segment.
FIGURE 7.
Mechanistic interpretation of loss of ORAI1 R91W function. a, maximum probability is depicted for the first transmembrane (TM) domain of ORAI1, ORAI2, ORAI3, and their respective mutant proteins as determined from TMHMM plots. b, hydrophobicity plots for ORI1, ORAI2, and ORAI3 in comparison to SCID-related mutants ORAI1 R91W, ORAI2 R65W, and ORAI3 R66W. c, scheme depicting probable scenarios for the loss of function of ORAI1 R91W compared with ORAI1 wild-type (WT).
In addition we extended bioinformatic analysis by hydrophobicity profiles particularly along the first transmembrane region employing a prediction program (see “Materials and Methods”) based on the Rose and Lesser hydrophobicity scale. The hydrophobicity profile of ORAI1 (see inset Fig. 4d) displayed four transmembrane domains in correlation with recent studies (45). The mutation of the arginine to the tryptophan at position 91 caused an increase in overall hydrophobicity only at the N terminus/transmembrane interface for the amino acid stretch at 86–96, whereas the rest of the protein was predicted to remain unaffected. All functional ORAI1 mutants in this region displayed an overall hydrophobicity of the first transmembrane domain similar or close to wild-type ORAI1, whereas the hydrophobicity profile of all non-functional mutants perfectly overlapped with that of ORAI1 R91W (Fig. 4e).
In summary, bioinformatics results derived from secondary structure prediction together with analysis of the data extracted from the Protein Data Bank, transmembrane probability plots, and hydrophobicity profiles were in fair correlation with our functional data. Altogether, they suggested a relationship between conformational constraints at the N terminus/membrane interface of ORAI1 protein together with increased hydrophobicity along the first transmembrane region and the capability of the ORAI1 channel to allow permeation/gating.
Hydrophobicity at Position 91 Plays a Major Role in Disrupting Permeation/Gating
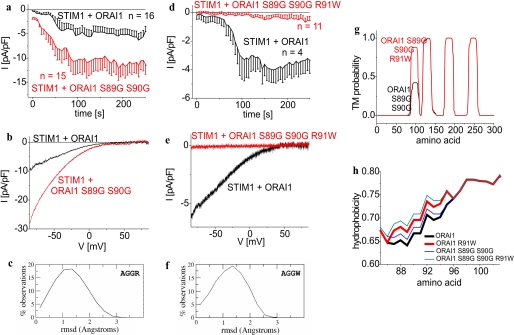
In an attempt to strengthen the above hypothesis, we examined the role of the two flexible (44) serines (Ser-89 and Ser-90) for their impact on ORAI1 function by mutating them to either glycines or prolines, thus promoting or impeding flexibility (44), respectively. According to the r.m.s. deviation distributions it appeared that the AGGR moieties (89 analyzed) comprised a broad range of slightly different conformations (Fig. 5c), resulting in a broad r.m.s. deviation distribution. On the contrary, the APPR (19 analyzed) moieties exhibited a bimodal r.m.s. deviation distribution reflecting conformational clustering and thus less flexibility (data not shown). Functional evaluation of the ORAI1 double glycine mutant (ORAI1 S89G/S90G) revealed Ca2+-current activation to an even higher extent as wild-type ORAI1 (Fig. 5, a and b) in accordance with the bioinformatics predictions (Figs. 5g and 7a). The double proline ORAI1 S89P/S90P mutant failed to generate currents in 10 mm Ca2+ as well as in a sodium divalent-free bath solution (data not shown) in line with the expected substantial conformational constraints by the double proline substitution. Nevertheless, both ORAI1 double mutants exhibited preserved co-clustering and robust FRET increases with STIM1 after store depletion (data not shown).
FIGURE 5.
Function of the SCID ORAI1 R91W mutant cannot be rescued by substituting two glycines for serines at positions 89 and 90 in the ASSW region. a and d, time course of whole cell inward currents at −74 mV activated by passive store depletion of HEK293 cells expressing STIM1 with ORAI1 (a) ORAI1 S89G S90G (t = 120 s; p > 0.05) or (d) ORAI1 S89G/S90G/R91W (t = 150 s; p < 0.01). b and e, respective I/V curves to a and d from representative cells taken at maximum current density activation. c and f, conformational features of the AGGR and AGGW moieties, displaying a broad range of conformations. g, transmembrane probability plots for functional ORAI1 S89G/S90G and non-functional ORAI1 S89G/S90G/R91W mutants. h, hydrophobicity plots for ORAI1 as well as ORAI1 R91W in comparison to functional ORAI1 S89G/S90G and non-functional ORAI1 S89G/S90G/R91W.
In a further approach we tried to restore function of the SCID ORAI1 R91W mutant by substituting two glycines for serines at positions 89 and 90 in the ASSW region. Secondary structure prediction of the AGGW moieties displayed a broad r.m.s. deviation distribution (34 analyzed) (Fig. 5f) suggesting substantial conformational flexibility. Nevertheless, this triple mutant ORAI1 S89G/S90G/R91W remained non-functional following store depletion in both Ca2+ (Fig. 5, d and e) and sodium divalent-free solution (data not shown), suggesting additional key factor(s) that determine functionality. In accordance, transmembrane probability plots and hydrophobicity analysis of ORAI1 S89G/S90G/R91W revealed substantially enhanced probability and overall hydrophobicity for the first transmembrane segment comparable with that of non-functional ORAI1 R91X mutants (Figs. 5g and 7a). Thus, whereas conformational flexibility might have a modulatory impact on ORAI1 activity as evident from predictions on R91X single mutants, the hydrophobicity of the amino acid at position 91 assumedly played a key role in loss of function of the SCID mutant that in the case of the ORAI1 S89G/S90G/R91W mutant was not accounted for in the bioinformatics analysis of tetrapeptide moieties from the Protein Data Bank.
Increased Pore Size Fails to Restore Activation of SCID-related ORAI1 Mutants
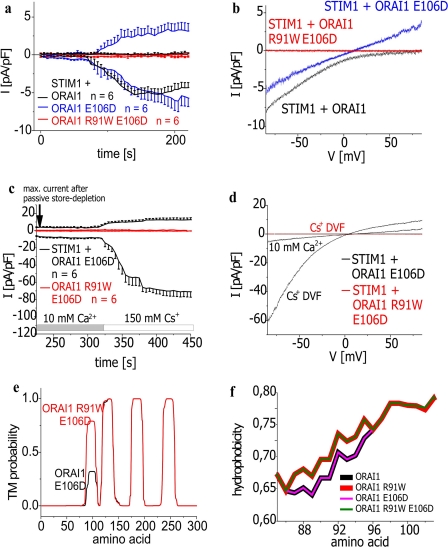
Collapse or constriction of the pore might also contribute to R91W ORAI1 non-functionality. The selectivity filter of the ORAI1 channel is proposed within transmembrane regions I and III (Glu-106, Glu-190) and the I/II loop (Asp-110, Asp-112, Asp-114) based on point mutants with altered permeability properties (14, 46, 47). It has been reported that the single point mutations E106D as well as E190Q increase the pore radius of ORAI1 (14). Consistent with these previous studies we demonstrated that in contrast to the strongly inwardly rectifying STIM1/ORAI1 Ca2+ currents, the mutant ORAI1 E106D displayed a rather linear I/V relationship in the presence of STIM1 after passive store depletion (Fig. 6, a and b). Based on the augmented pore diameter of ORAI1 E106D, Cs+ permeability substantially increased compared with wild-type ORAI1 (Fig. 6, c and d) (14). Introduction of E106D in the ORAI1 SCID mutant would also be expected to increase the radius of the pore thereby possibly antagonizing a potential pore constriction induced by the R91W mutation. However, ORAI1 R91W/E106D containing both mutations clearly failed to restore ORAI1 Ca2+ as well as Cs+ currents (Fig. 6, a, b and c, d) and underscored the dominant effect of the R91W mutation. Consistently, transmembrane probability as well as hydrophobicity predictions underlined our functional data and exhibited comparable values for ORAI1 E106D and wild-type ORAI1 in contrast to the strongly increased ones for both ORAI1 R91W/E106D and ORAI1 R91W (Fig. 6, e and f). The inability of this double mutant to restore neither Ca2+ nor Cs+ permeation despite an increased pore size would be consistent with the view that the R91W mutation more drastically impaired permeation/gating rather than by slight constriction of the pore.
FIGURE 6.
Increase in pore radius fails to restore activity of SCID-related ORAI1 mutant. a, time course of whole cell inward as well as outward currents at −74 mV activated by passive store depletion of HEK293 cells expressing STIM1 with ORAI1, ORAI1 E106D (t = 150s; p > 0.05), or ORAI1 R91W E106D (t = 150 s; p < 0.01) and c, after perfusion of a Cs+-DVF solution. b and d, respective I/V curves to a and c from representative cells taken at maximum current density activation. e, hydropathy plots for functional ORAI1 E106D and non-functional ORAI1 R91W/E106D mutants. f, hydrophobicity plots for ORAI1 as well as ORAI1 R91W in comparison to pore mutants ORAI1 E106D and ORAI1 R91W/E106D.
Similar to E106D point mutation in the ORAI1 selectivity filter, 2-APB (48–51) is able to increase the pore radius of ORAI3 (75 μm) thereby activating currents independent of STIM1 and store depletion, displaying altered permeation properties. We have further shown (50) that ORAI3 R66W, i.e. the analogue to the ORAI1 SCID mutant, also displays loss of store-operated Ca2+ current activity. Similar to the ORAI1 proteins, TMHMM and hydropathy plots of ORAI3 R66W revealed a substantial increase in probability and hydrophobicity for the first transmembrane segment in comparison to wild-type ORAI3 (Fig. 7). Moreover, this ORAI3 R66W mutant also fails to show an activation by 2-APB (50). Analysis of the ORAI3 S64P/S65P mutant generated analogous to ORAI1 S89P/S90P revealed loss of function either activated by store depletion or 2-APB (data not shown). Hence, these findings suggested that these mutations generally prevent permeation/gating of the ORAI1 as well as ORAI3 mutants independent of the mode of activation and a potentially increased pore size.
DISCUSSION
The present study revealed a direct correlation between increased hydrophobicity at the N terminus/transmembrane interface and the loss of function in the ORAI1 channel R91W SCID mutant. This was strengthened by the fact that R91X mutations carrying hydrophilic amino acids did not largely affect channel function of ORAI1, regardless of their charge. However, increased hydrophobicity as obtained with histidine or the neutral threonine or glycine led to more reduced current densities suggesting that hydrophobic rather than neutral amino acid mutations impair ORAI1 R91X function.
Consistently, complete loss of current activation was observed with highly hydrophobic ORAI1 R91X (X = Trp, Leu, Val, and Phe) mutations, yet their functional integrity was similar to wild-type ORAI1 concerning correct targeting to and insertion into the plasma membrane. Additionally, the SCID mutant and the wild-type ORAI1 exhibited similar homomultimerization as judged by FRET microscopy (data not shown), which is key to channel formation. Moreover, all non-functional ORAI1 R91X mutants were able to form heteromeric assemblies with wild-type ORAI1 as monitored by FRET measurements (data not shown). Finally, based on store-operated co-clustering as well as coupling of all tested ORAI1 mutants to STIM1, the intrinsic step initiating ORAI1 activation by its coupling to STIM1 was largely sustained in both functional as well as non-functional ORAI1 R91X mutants. Hence, the single hydrophobic amino acid missense mutation occurring in the SCID ORAI1 mutant induced apparently a defect in ORAI1 gating/permeation properties (Fig. 7c) rather than completely disturbing the overall channel structure.
In contrast to single point mutations within the proposed selectivity filter of the ORAI1 in transmembrane regions I and III (Glu-106, Glu-190) and the I/II loop (Asp-110, Asp-112, Asp-114) (14, 46, 47), mutation of the positive arginine to neutral or particularly negatively charged amino acids at position 91 did not alter Ca2+ selectivity. Hence, we suggest that arginine 91 does not contribute to the selectivity filter. An indirect effect of the R91W substitution potentially leading to substantial pore constriction/collapse seems possible, as attempts to outweigh such an effect by increasing pore size with additional introduction of E106D in the ORAI1 SCID mutant, failed to restore channel activity. Consistently, an increase in pore size that comes along with 2-APB stimulation of ORAI3 similarly failed to reconstitute channel activity in the SCID-related ORAI3 R66W analogue (50), supporting the concept of a more general conformational rearrangement that prevented ion flux. A recent study (22) on the impact of increasing R91W ORAI1 protein monomers within a defined concatamer of four ORAI1 proteins revealed functional channels as long as one R91W monomer is assembled together with three wild-type ORAI1. Such an ORAI1 R91W-ORAI1 1:3 channel complex does not show any change of biophysical characteristics when compared with wild-type ORAI1 concatamers in line with an unaltered selectivity filter. Thus, the single point mutation in the SCID mutant ORAI1 R91W at the N terminus/membrane interface is more likely to affect permeation/gating rather than directly the selectivity filter (Fig. 7c). Another recent study has suggested a defect in permeation rather than channel gating of ORAI1 R91W, based on a similar decline in ORAI1 R91W-ORAI1 R91W FRET after store-operated STIM1 coupling compared with that of ORAI1 homomers (38). As it is not fully clear whether the observed similar change in FRET of ORAI1 R91W and wild-type ORAI1 indeed represents the major gating step, it is safe to conclude that loss of function of SCID mutants results from both a defect in permeation and gating.
Secondary structure predictions revealed that increased hydrophobicity at position 91 probably favored coil to helix transition, which would impose structural constraints in the ASSX region (aa 88–91). Conformational flexibility at the N terminus/membrane interface (ASSX moiety) is assumed to contribute to ORAI1 functionality. A correlation of the degree of flexibility with function has been reported for a variety of proteins, mostly soluble ones (53, 54) but also membrane proteins as for example, the Drosophila Shaker K+ channel (55). Statistical surveys of the Protein Data Bank revealed respective snapshots of the conformational space accessible to the tetrapeptide moieties ASSX (X = Arg, Gly,...). ASSR, ASSG, ASSS, ASST, and ASSN were considerably more flexible than the peptide moieties ASSW, ASSL, ASSV, and ASSF. The tetrapeptides ASSE and ASSH moieties seemed to be characterized by an intermediate degree of flexibility. These predictions were in fair correlation with our electrophysiological data, suggesting a modulatory role of the tetrapeptide conformational flexibility. All of the single point mutations to hydrophobic amino acids Trp, Leu, Val, and Phe displayed complete loss of current activity, despite distinct side chain volumes. This suggests that increased hydrophobicity along the first transmembrane segment in these ORAI1 R91X (X = Trp, Leu, Val, and Phe) as well as ORAI2 R65W and ORAI3 R66W (Fig. 7, a and b) mutants is the major key determinant for their lack of function. Accordingly, the introduction of two flexible glycines at positions 89/90 was not able to outweigh the impact of the tryptophan (ORAI1 S89G/S90G/R91W). Hence it is tempting to speculate that the hydrophobic substitutions by increasing the probability for embedding into the membrane as well as hydrophobicity of the first transmembrane segment generate either a hydrophobic gate (56–61) that imposes a detrimental energetic barrier for gating/permeation and/or favor a lipid-helix interaction (62, 63), which stabilizes the channel in its closed conformation. Interactions of plasma membrane lipids with portions of transmembrane helices (63) are reported to alter the orientation of α-helices (62). For instance, there is evidence that tryptophan- and arginine-flanked membrane peptides, analogues to the mutation in the ORAI1 R91W mutant, display distinct tilt angles (63) and that the hydrophobic length of the α-helix determines the extent of tilt in the plasma membrane (52, 64). Moreover, the positively charged lysine in contrast to tryptophan may reach the interfacial region from a position deeper in the hydrophobic part of the bilayer (snorkeling). This may allow more flexibility in the position of side chains at the interface and hence more freedom in the localization and dynamics of the helices in the bilayer (62). Thus, we suggest that an increase in the probability for the first transmembrane segment above a certain value (0.76) as well as the enhancement of its overall hydrophobicity (Fig. 7, a and b) might lead to a substantial conformational rearrangement thereby impeding gating and/or permeation (Fig. 7c) of the SCID ORAI1 R91W mutant.
In aggregate, our results demonstrate the increase in hydrophobicity along the first transmembrane segment of the SCID ORAI1 R91W mutant as the major cause for a lack of Ca2+ current activation. The initial step in STIM1/ORAI1 communication is largely preserved as judged from their co-clustering and coupling, supporting the concept of a profound defect in gating/permeation of the ORAI1 R91W SCID mutant. To resolve whether this defect transpires via conformational constraints of the first transmembrane segment affecting its tilt and possibly the permeation pathway requires further structural investigations such as x-ray crystallography.
Acknowledgments
We thank S. Buchegger and B. Kenda for excellent technical assistance.
- SCID
- severe combined immunodeficiency
- STIM1
- stromal interacting molecule 1
- 2-APB
- 2-aminoethyldiphenyl borate
- aa
- amino acid(s)
- FRET
- Förster resonance energy transfer
- ECFP
- enhanced cyan fluorescent protein
- EYFP
- enhanced yellow fluorescent protein
- HEK
- human embryonic kidney
- r.m.s.
- root mean square.
REFERENCES
- 1.Buckley R. H. ( 2004) J. Clin. Investig. 114, 1409– 1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaspar H. B., Gilmour K. C., Jones A. M. ( 2001) Arch. Dis. Child 84, 169– 173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll H. P., McNaull B. B., Gadina M. ( 2006) Mol. Interv. 6, 253– 256 [DOI] [PubMed] [Google Scholar]
- 4.Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S. H., Tanasa B., Hogan P. G., Lewis R. S., Daly M., Rao A. ( 2006) Nature 441, 179– 185 [DOI] [PubMed] [Google Scholar]
- 5.Feske S., Prakriya M., Rao A., Lewis R. S. ( 2005) J. Exp. Med. 202, 651– 662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parekh A. B. ( 2006) Nature 441, 163– 165 [DOI] [PubMed] [Google Scholar]
- 7.Parekh A. B., Putney J. W., Jr.( 2005) Physiol. Rev. 85, 757– 810 [DOI] [PubMed] [Google Scholar]
- 8.Dutta D. ( 2000) J. Biosci. 25, 397– 404 [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti R., Chakrabarti R. ( 2006) J. Cell. Biochem. 99, 1503– 1516 [DOI] [PubMed] [Google Scholar]
- 10.Berridge M. J., Bootman M. D., Roderick H. L. ( 2003) Nat. Rev. Mol. Cell Biol. 4, 517– 529 [DOI] [PubMed] [Google Scholar]
- 11.Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. ( 2006) Science 312, 1220– 1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peinelt C., Vig M., Koomoa D. L., Beck A., Nadler M. J., Koblan-Huberson M., Lis A., Fleig A., Penner R., Kinet J. P. ( 2006) Nat. Cell Biol. 8, 771– 773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gwack Y., Srikanth S., Feske S., Cruz-Guilloty F., Oh-hora M., Neems D. S., Hogan P. G., Rao A. ( 2007) J. Biol. Chem. 282, 16232– 16243 [DOI] [PubMed] [Google Scholar]
- 14.Yamashita M., Navarro-Borelly L., McNally B. A., Prakriya M. ( 2007) J. Gen. Physiol. 130, 525– 540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeromin A. V., Zhang S. L., Jiang W., Yu Y., Safrina O., Cahalan M. D. ( 2006) Nature 443, 226– 229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakriya M., Feske S., Gwack Y., Srikanth S., Rao A., Hogan P. G. ( 2006) Nature 443, 230– 233 [DOI] [PubMed] [Google Scholar]
- 17.Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. ( 2005) J. Cell Biol. 169, 435– 445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soboloff J., Spassova M. A., Dziadek M. A., Gill D. L. ( 2006) Biochim. Biophys. Acta 1763, 1161– 1168 [DOI] [PubMed] [Google Scholar]
- 19.Frischauf I., Schindl R., Derler I., Bergsmann J., Fahrner M., Romanin C. ( 2008) Channels 2, 261– 268 [DOI] [PubMed] [Google Scholar]
- 20.Muik M., Frischauf I., Derler I., Fahrner M., Bergsmann J., Eder P., Schindl R., Hesch C., Polzinger B., Fritsch R., Kahr H., Madl J., Gruber H., Groschner K., Romanin C. ( 2008) J. Biol. Chem. 283, 8014– 8022 [DOI] [PubMed] [Google Scholar]
- 21.Bergmeier W., Oh-Hora M., McCarl C. A., Roden R. C., Bray P. F., Feske S. ( 2009) Blood 113, 675– 678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson J. L., Mignen O., Shuttleworth T. J. ( 2009) J. Biol. Chem. 284, 6620– 6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh A., Hamedinger D., Hoda J. C., Gebhart M., Koschak A., Romanin C., Striessnig J. ( 2006) Nat. Neurosci. 9, 1108– 1116 [DOI] [PubMed] [Google Scholar]
- 24.Derler I., Hofbauer M., Kahr H., Fritsch R., Muik M., Kepplinger K., Hack M. E., Moritz S., Schindl R., Groschner K., Romanin C. ( 2006) J. Physiol. 577, 31– 44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia Z., Liu Y. ( 2001) Biophys. J. 81, 2395– 2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berney C., Danuser G. ( 2003) Biophys. J. 84, 3992– 4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. ( 2000) Nucleic Acids Res. 28, 235– 242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr., Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. ( 1977) J. Mol. Biol. 112, 535– 542 [DOI] [PubMed] [Google Scholar]
- 29.Wang G., Dunbrack R. L., Jr.( 2003) Bioinformatics 19, 1589– 1591 [DOI] [PubMed] [Google Scholar]
- 30.Frishman D., Argos P. ( 1995) Proteins 23, 566– 579 [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W. ( 1976) Acta Crystallogr. Sect. A 32, 922– 923 [Google Scholar]
- 32.Kabsch W. ( 1978) Acta Crystallogr. Sect. A 34, 827– 828 [Google Scholar]
- 33.Garnier J., Gibrat J. F., Robson B. ( 1996) Methods Enzymol. 266, 540– 553 [DOI] [PubMed] [Google Scholar]
- 34.Levin J. M. ( 1997) Protein Eng. 10, 771– 776 [DOI] [PubMed] [Google Scholar]
- 35.Geourjon C., Deléage G. ( 1995) Comput. Appl. Biosci. 11, 681– 684 [DOI] [PubMed] [Google Scholar]
- 36.Rose G. D., Geselowitz A. R., Lesser G. J., Lee R. H., Zehfus M. H. ( 1985) Science 229, 834– 838 [DOI] [PubMed] [Google Scholar]
- 37.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. ( 2001) J. Mol. Biol. 305, 567– 580 [DOI] [PubMed] [Google Scholar]
- 38.Navarro-Borelly L., Somasundaram A., Yamashita M., Ren D., Miller R. J., Prakriya M. ( 2008) J. Physiol. 586, 5383– 5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimura K., Batiza A., Kung C. ( 2001) Biophys. J. 80, 2198– 2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raybaud A., Baspinar E. E., Dionne F., Dodier Y., Sauvé R., Parent L. ( 2007) J. Biol. Chem. 282, 27944– 27952 [DOI] [PubMed] [Google Scholar]
- 41.Thunnissen M. M., Franken P. A., de Haas G. H., Drenth J., Kalk K. H., Verheij H. M., Dijkstra B. W. ( 1993) J. Mol. Biol. 232, 839– 855 [DOI] [PubMed] [Google Scholar]
- 42.Kühn F. J., Knop G., Lückhoff A. ( 2007) J. Biol. Chem. 282, 27598– 27609 [DOI] [PubMed] [Google Scholar]
- 43.Micheletti C., Seno F., Maritan A. ( 2000) Proteins 40, 662– 674 [DOI] [PubMed] [Google Scholar]
- 44.Huang F., Nau W. M. ( 2003) Angew. Chem. Int. Ed. Engl. 42, 2269– 2272 [DOI] [PubMed] [Google Scholar]
- 45.Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S. H., Tanasa B., Hogan P. G., Lewis R. S., Daly M., Rao A. ( 2006) Nature 441, 179– 185 [DOI] [PubMed] [Google Scholar]
- 46.Yeromin A. V., Zhang S. L., Jiang W., Yu Y., Safrina O., Cahalan M. D. ( 2006) Nature 443, 226– 229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prakriya M., Feske S., Gwack Y., Srikanth S., Rao A., Hogan P. G. ( 2006) Nature 443, 230– 233 [DOI] [PubMed] [Google Scholar]
- 48.DeHaven W. I., Smyth J. T., Boyles R. R., Bird G. S., Putney J. W., Jr.( 2008) J. Biol. Chem. 283, 19265– 19273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S. L., Kozak J. A., Jiang W., Yeromin A. V., Chen J., Yu Y., Penna A., Shen W., Chi V., Cahalan M. D. ( 2008) J. Biol. Chem. 283, 17662– 17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindl R., Bergsmann J., Frischauf I., Derler I., Fahrner M., Muik M., Fritsch R., Groschner K., Romanin C. ( 2008) J. Biol. Chem. 283, 20261– 20267 [DOI] [PubMed] [Google Scholar]
- 51.Peinelt C., Lis A., Beck A., Fleig A., Penner R. ( 2008) J. Physiol. 583, 3061– 3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Powl A. M., East J. M., Lee A. G. ( 2003) Biochemistry 42, 14306– 14317 [DOI] [PubMed] [Google Scholar]
- 53.Tsong T. Y., Karr T., Harrington W. F. ( 1979) Proc. Natl. Acad. Sci. U. S. A. 76, 1109– 1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wieprecht T., Apostolov O., Beyermann M., Seelig J. ( 1999) J. Mol. Biol. 294, 785– 794 [DOI] [PubMed] [Google Scholar]
- 55.Haris P. I., Ramesh B., Brazier S., Chapman D. ( 1994) FEBS Lett. 349, 371– 374 [DOI] [PubMed] [Google Scholar]
- 56.Beckstein O., Sansom M. S. ( 2006) Phys. Biol. 3, 147– 159 [DOI] [PubMed] [Google Scholar]
- 57.Miyazawa A., Fujiyoshi Y., Unwin N. ( 2003) Nature 423, 949– 955 [DOI] [PubMed] [Google Scholar]
- 58.Payandeh J., Li C., Ramjeesingh M., Poduch E., Bear C. E., Pai E. F. ( 2008) J. Biol. Chem. 283, 11721– 11733 [DOI] [PubMed] [Google Scholar]
- 59.Perozo E. ( 2006) Nat. Rev. Mol. Cell Biol. 7, 109– 119 [DOI] [PubMed] [Google Scholar]
- 60.Long S. B., Tao X., Campbell E. B., MacKinnon R. ( 2007) Nature 450, 376– 382 [DOI] [PubMed] [Google Scholar]
- 61.Kuo A., Domene C., Johnson L. N., Doyle D. A., Vénien-Bryan C. ( 2005) Structure 13, 1463– 1472 [DOI] [PubMed] [Google Scholar]
- 62.Nyholm T. K., Ozdirekcan S., Killian J. A. ( 2007) Biochemistry 46, 1457– 1465 [DOI] [PubMed] [Google Scholar]
- 63.Ozdirekcan S., Rijkers D. T., Liskamp R. M., Killian J. A. ( 2005) Biochemistry 44, 1004– 1012 [DOI] [PubMed] [Google Scholar]
- 64.Powl A. M., East J. M., Lee A. G. ( 2005) Biochemistry 44, 5873– 5883 [DOI] [PubMed] [Google Scholar]