Abstract
HSEPI (glucuronyl C5-epimerase) catalyzes the conversion of d-glucuronic acid to l-iduronic acid in heparan sulfate (HS) biosynthesis. Disruption of the Hsepi gene in mice yielded a lethal phenotype with selective organ defects but had remarkably little effect on other organ systems. We have approached the underlying mechanisms by examining the course and effects of FGF2 signaling in a mouse embryonic fibroblast (MEF) cell line derived from the Hsepi−/− mouse. The HS produced by these cells is devoid of l-iduronic acid residues but shows up-regulated N- and 6-O-sulfation compared with wild type (WT) MEF HS. In medium fortified with 10% fetal calf serum, the Hsepi−/− MEFs proliferated and migrated similarly to WT cells. Under starvation conditions, both cell types showed attenuated proliferation and migration that could be restored by the addition of FGF2 to WT cells, whereas Hsepi−/− cells were resistant. Moreover, ERK phosphorylation following FGF2 stimulation was delayed in Hsepi−/− compared with WT cells. Assessment of HS-growth factor interaction by nitrocellulose filter trapping revealed a strikingly aberrant binding property of FGF2 and glia-derived neurotropic factor to Hsepi−/− but not to WT HS. glia-derived neurotropic factor has a key role in kidney development, defective in Hsepi−/− mice. By contrast, Hsepi−/− and WT HS interacted similarly and in conventional mode with FGF10. These findings correlate defective function of growth factors with their mode of HS interaction and may help explain the partly modest organ phenotypes observed after genetic ablation of selected enzymes in HS biosynthesis.
Signaling activities of numerous growth factors and morphogens during development involve cell surface receptor systems consisting of a tyrosine kinase-type receptor along with a heparan sulfate proteoglycan (HSPG)2 co-receptor (1, 2). The complex and heterogeneous HSPG macromolecules occur in the extracellular matrix and on the surfaces of virtually all animal cells (3). HS side chains of HSPGs show great structural variability; hence, they interact with a multitude of proteins and influence a variety of biological processes, including growth factor signaling (4).
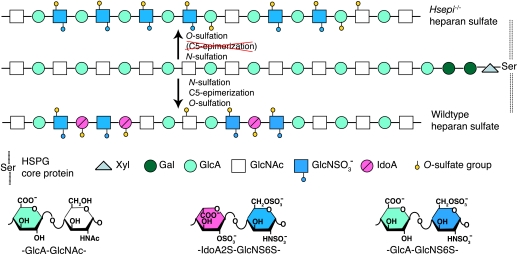
The structural diversity of HS is best envisaged through an account of its biosynthesis (Fig. 1). The process is initiated by glycosylation reactions that generate saccharide sequences composed of alternating GlcA and GlcNAc units covalently bound to a core protein (through a specific “linker” tetrasaccharide sequence). The resulting polymer of (GlcAβ1,4-GlcNAcα1,4-)n disaccharide repeats is modified through a series of reactions, including N-deacetylation/N-sulfation of GlcNAc residues, C5-epimerization of GlcA to l-iduronic acid (IdoA) units, and O-sulfation at various positions of the hexuronic acid and glucosamine residues (4, 5). Modulation of these modification reactions, through as yet poorly understood mechanisms, yields HS chains of strictly regulated saccharide composition, varying with tissue source and age (6, 7). Interactions between protein ligands and HS involve selective ionic binding of peptide sequences containing basic amino acid residues to saccharide domains with clustered sulfate groups and IdoA residues. The latter components are considered to promote ligand apposition through their conformational flexibility (8).
FIGURE 1.
Schematic display of HS biosynthesis. A precursor structure composed of alternating GlcA and GlcNAc residues linked to a core protein is modified through the series of enzymatic reactions indicated and outlined under“Results.” The modifications yield domains of consecutive N-acetylated or N-sulfated disaccharide units, along with mixed sequences. Wild type HS contains both GlcA and IdoA residues, whereas Hsepi−/− HS lacks IdoA units but shows increased N- and 6-O-sulfation. In particular, the mutant HS features extended sequences of -GlcA-GlcNS6S- disaccharide units.
Targeted disruption of genes encoding HS biosynthesis enzymes demonstrated critical roles for HSPGs in developmental processes (9–11). The observed phenotypes vary dramatically in severity, from gastrulation failure to subtle disturbance of organ development. This variability may reflect redundancy due to the occurrence of isoforms for some of the enzymes. Alternatively, developmental events might be critically dependant on HS involvement but not on the fine structure of the polysaccharide chain, such that even structurally deranged HS would fulfill a functional role (12). The glucuronyl C5-epimerase (HSEPI) that catalyzes the conversion of GlcA to IdoA in HS biosynthesis is encoded by a single gene (13). Targeted disruption of this gene in mice resulted in an abnormal HS structure, completely lacking IdoA residues and with severely distorted sulfation pattern. The Hsepi-deficient mice die shortly after birth with multiple developmental defects, such as skeletal malformations and kidney agenesis. Intriguingly, however, other major organ systems, such as the brain and vascular system, known to depend on HS-supported signaling processes, developed seemingly normally (14).
The present study was undertaken to obtain insight into the mechanisms behind the developmental defects in the Hsepi mutant mice, using a model system based on murine embryonic fibroblast (MEF) cell lines. The mutant MEF cells responded poorly to FGF2 in proliferation and migration experiments and showed defective FGF2-dependent intracellular signaling. Moreover, binding studies revealed an aberrant mode of interaction between mutant HS chains and certain growth factors, of potential relevance to growth factor signaling.
EXPERIMENTAL PROCEDURES
MEFs
WT and Hsepi mutant embryos were generated through breeding of heterozygous mice (14). Embryonic fibroblasts were generated from embryonic day 14.5 embryos according to established protocol (15). After dissection of the embryos, head and internal organs were discarded, and remaining parts were finely cut and digested in 0.25% trypsin-EDTA for 20 min at 37 °C. Released cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS and 1% antibiotics (penicillin G/streptomycin). Genotypes of the fibroblasts were ascertained by PCR (14). The cells were immortalized by transformation with recombinant simian virus 40 large T antigen carrying a neo gene (16). A producer cell line, ψ2, containing integrated copies of defective entropic Maloney murine leukemia virus (provided by Dr. Maria Thuvesson, Uppsala University, Sweden), was cultured in DMEM containing 10% FCS and 1% penicillin G/streptomycin. The conditioned medium (0.2 ml) from confluent ψ2 cell culture was used to inoculate MEF cells at ∼60% confluence in a 10-cm dish at 37 °C for 2 h, and then 8 ml of DMEM containing 10% FCS and 1% penicillin G/streptomycin was added. The cells were cultured in the medium supplemented with G-418 antibiotic (1 and 0.2 mg/ml final concentration for mutant and WT, respectively) for 2 weeks. Surviving colonies were selected and propagated. Two clones (supplemental Fig. 1) from each species, WT and Hsepi−/−, were used for the experiments. The null expression of Hsepi in the mutant MEF cells was confirmed by analysis of epimerase activity using a GlcA-C5-3H-labeled, N-sulfoheparosan substrate (14) (data not shown).
Generation of Radiolabeled Polysaccharides
Radiolabeled polysaccharides were generated in two ways, either after incubation of cultured MEF cells with [3H]glucosamine or Na35SO4 or by chemical N-[3H]acetyl labeling of HS isolated from mouse embryos. For metabolic labeling of HS, cells were cultured in DMEM supplemented with 10% FCS to 95% confluence. After adding 100 μCi/ml Na35SO4 (specific activity, 1500 Ci/mmol; PerkinElmer Life Sciences) or 100 μCi/ml d-[6-3H]glucosamine (specific activity, 34 Ci/mmol; GE Healthcare), the cells were maintained in the same medium for 24 h before harvest for HS isolation. Total HS was isolated as described previously (17). Briefly, the cells were lysed in extraction buffer (50 mm Tris-HCl, pH 7.4, 4 m urea, 1% Triton X-100), and the medium fractions were mixed with equal volumes of 2× extraction buffer and incubated at 4 °C for 1 h under gentle shaking. NaOH was added to the mixtures to a final concentration of 0.5 m, and the samples were incubated on ice overnight. After neutralization and centrifugation, the supernatants were diluted ≥20-fold with extraction buffer and applied to 1–2 ml of DEAE-Sephacel columns (GE Healthcare), equilibrated with extraction buffer. Following extensive washing with 50 mm NaAc, pH 4.5, 0.25 m NaCl, bound material was eluted with 50 mm NaAc, pH 4.5, 1.5 m NaCl. Eluates were desalted by dialysis and lyophilized. Products were treated with chondroitinase ABC (25 milliunits; Seikagaku) and Benzonase (60 units; Merck) and reapplied to a DEAE-Sephacel column to eliminate degraded chondroitin sulfate and oligonucleotides. The purity of HS products was confirmed by treatment with HNO2 at pH 1.5 followed by gel chromatography.
Labeled chondroitin sulfate (CS)/dermatan sulfate (DS) was isolated following incubation of MEF cell cultures with 100 μCi/ml Na35SO4 for 24 h. Total polysaccharide was isolated from medium or cell lysates after extraction with 50 mm acetate, pH 5.5, 1% Triton X-100, 6 m urea, and HS was eliminated by nitrous acid degradation, as described (18).
For N-[3H]acetyl labeling, HS was isolated from two WT and two Hsepi mutant embryos (embryonic day 18.5) according to the procedure described previously (17). Purified HS was N-deacetylated by hydrazinolysis followed by re-N-acetylation with N-[3H]acetic anhydride (19). Specific activities of the [3H]acetyl-labeled HS samples were 50 × 103 cpm/μg for both WT and Hsepi mutant samples (polysaccharide amounts based on colorimetry) (20).
Analysis of Polysaccharides
Gel chromatography of 35S-labeled HS chains was done on a Superose-12 column (GE Healthcare). Polyelectrolyte properties were examined by anion exchange chromatography of samples on a DEAE-Sephacel column (1 ml), eluted with a salt gradient (0–1.5 m NaCl in 50 mm NaAc, pH 4.5) connected to an HPLC system.
For analysis of HS domain organization, samples labeled with [3H]glucosamine were subjected to cleavage at N-sulfated glucosamine (GlcNS) residues by treatment with HNO2 at pH 1.5 (21), followed by end reduction with NaBH4. Resultant oligosaccharides were separated by gel chromatography on a column (1 × 200 cm) of Bio-Gel P10 (Bio-Rad) in 0.5 m NaCl at a flow rate of 2 ml/h. The composition of disaccharides generated by deaminative cleavage (HNO2, pH 1.5/NaBH4) of 35S-labeled HS was determined on a Partisil-10 SAX column, as previously described (17).
For analysis of total disaccharide composition, the 35S-labeled CS/DS samples were digested with chondroitinase ABC. The resultant disaccharides were separated on a CarboPac PA-1 HPLC column, followed by liquid scintillation counting (22). The amount and distribution of IdoA along the CS/DS chains were assessed by chondroitinase B and chondroitinase AC-I digestion, respectively, and separation of digests was performed on a Superdex Peptide column (GE Healthcare), as described previously (22).
Cell Proliferation Assay
Cells were seeded on a 24-well plate at a density of 1.5 × 104 cells/well in DMEM supplemented with 10% FCS and cultured for 24 h. After serum starvation (no serum) for 48 h, cells were changed to fresh media (either DMEM supplemented with 10% FCS or DMEM supplemented with FGF2) and were grown for 20 h. Then [3H]thymidine (1 μCi/ml; specific activity, 6.7 Ci/mmol; PerkinElmer Life Sciences) was added to each well and incubated for 4 h. The cells were then trypsinized, washed with cold PBS three times, and incubated in 5% trichloroacetic acid on ice for 30 min. Following centrifugation, the acid-insoluble pellet was dissolved in 2 ml of 0.5 m NaOH, 0.5% SDS solution. Incorporation of [3H]thymidine was determined by scintillation counting of the solubilized cell pellets.
Cell Migration Assay
Cells were seeded on 6-well plates at a density of 5 × 105 cells/well in DMEM supplemented with 10% FCS. At 90% confluence, some cultures were changed to starvation medium (DMEM supplemented with 0.1% serum) and maintained for 24 h. Cell monolayers, cultured in DMEM supplemented with 10% FCS or under starvation conditions (DMEM + 0.1% FCS), were scratched with a fine pipette tip to create a “wound” and washed with PBS to remove the detached cells. Fresh starvation medium was then added with or without FGF2. Wound closure was recorded by image capture at different time points and expressed as the ratio of the average distance between the two wound edges at a given time, compared with the initial distance. The experiment was repeated three times with two different cell clones of each type (WT and Hsepi−/−).
F-actin Staining
Cells were seeded on coverslips in a 24-well plate, at a density of 2 × 104 cells/well, and cultured in DMEM supplemented with 10% FCS until 95% confluence. A wound was made as in the cell migration experiment described above. After washing with PBS twice, the cells were cultured in DMEM containing 10% FCS for 24 h. The medium was then removed, and the coverslips with attached cells were gently washed once with PBS at 37 °C followed by fixation in 4% formaldehyde for 10 min at room temperature. The cells were permeabilized by treatment with 0.5% Triton X-100 in PBS for 5 min. Phalloidin-rhodamine (Cytoskeleton; final concentration 100 nm) in PBS was added to the cells, followed by incubation in the dark for 30 min. After washing three times in PBS, the coverslips were mounted with Glycerol vinyl alcohol aqueous mounting solution (Zymed Laboratories Inc.) containing 4′,6-diamidino-2-phenylindole for counterstaining.
FGF2-induced Cell Signaling and Western Blot Analysis
Cells were seeded on 6-well plates at a density 1 × 105 cells/well in 1 ml of DMEM supplemented with 10% FCS and antibiotics and cultured for 48 h. They were then maintained in starvation medium (no FCS in the DMEM) for 48 h. After changing to fresh starvation medium, FGF2 was added at a 2 ng/ml final concentration and allowed to stimulate for 5, 15, 60, 120, and 360 min. For heparin rescue test, the starved (no serum supplement) cells were stimulated with FGF2 (2 ng/ml) for 10 min in the absence or presence of ∼5 μm heparin.
Following stimulation, medium was removed, and cells were washed with PBS twice before being lysed in 100 μl of sample buffer (0.2 m Tris, pH 8.0, 5 mm EDTA, 0.5 m sucrose, 1% SDS, 0.2 mm Na3VO4, 10 μg/ml pepstatin) and boiled for 4 min. Lysates were then ultrasonicated for 6 s and centrifuged for 15 min at 13,000 rpm; the supernatants were collected, and protein concentration was determined. Samples of 20 μg of total protein were separated by electrophoresis on SDS-PAGE (12%) and electroblotted onto a polyvinylidene difluoride membrane. The membrane was probed using a primary anti-phospho-ERK1/2 antibody kit (phospho-p44/42 mitogen-activated protein kinase (Thr202/Tyr204); Cell Signaling). The signals were developed using an ECL reagent (ECL Plus; GE Healthcare) and exposed to Fuji film. For protein loading control, the membrane was reprobed with anti-glyceraldehyde-3-phosphate dehydrogenase antibody directly (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or with anti-total ERK1/2 antibody (Cell Signaling) after striping with 0.1 m glycine/HCl, pH 2.5. The signals were developed by the same ECL reagent as above. The results were analyzed with ImageJ (National Institutes of Health).
Interactions of Growth Factors with HS
Interactions between growth factors and radiolabeled HS were assessed by nitrocellulose filter trapping, as described previously (23). FGFs were from PeproTech; glia-derived neurotropic factor (GDNF) was kindly provided by Prof. Mart Saarma (Institute of Biotechnology, University of Helsinki, Finland). Briefly, the proteins were incubated with different amounts of radiolabeled HS, isolated from embryos or MEF cells as described above, in 100 or 200 μl of PBS (pH 7.4) containing 0.1 mg/ml bovine serum albumin (Sigma) at room temperature for 2 h. Alternatively, the labeled HS was kept constant and titrated with different amounts of growth factor, as indicated. For calculation of approximate molar concentrations, the HS chains were assigned a molecular mass of 25 kDa (accounting for some depolymerization during the hydrazinolysis step of N-[3H]acetyl labeling). The incubated mixtures were rapidly passed through a PBS-washed nitrocellulose filter (diameter, 25 mm; pore size, 0.45 μm; Sartorius), using a vacuum-assisted filtering apparatus, followed by two washes with 5 ml of PBS. The HS bound to the membrane together with protein was released from the filter with 2 m NaCl, and the radioactivity was measured by scintillation counting. Apparent Kd values and curve fittings were analyzed by GraphPad Prism software (available on the World Wide Web).
RESULTS
The aim of the study was to correlate effects of the HSEPI ablation on HS structure with relevant functional cellular properties. FGF2 stimulation, known to be modulated by HS, was examined regarding effects on cell proliferation and motility as well as intracellular signaling pathway activation. Finally, interactions of Hsepi−/− and WT HS with selected growth factors, including FGF2, were analyzed at the molecular level.
Structural Characterization of HS
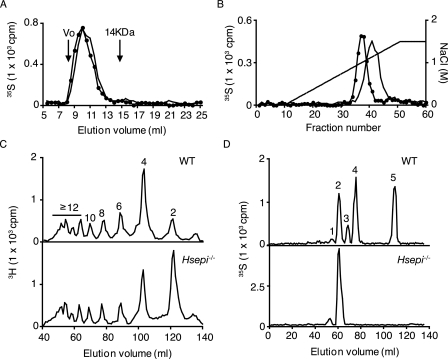
WT and Hsepi−/− MEF cells were cultured in the presence of either [35S]sulfate or [3H]glucosamine for 24 h. Metabolically labeled HS was isolated from cell lysate and assessed for molecular size by gel chromatography (Fig. 2A). Elution profiles were superimposable, indicating similar molecular size of HS from WT and mutant MEFs (estimated molecular mass of 30–60 kDa). However, HS from the mutant cells was more retarded on anion exchange chromatography (Fig. 2B), suggesting a higher net negative charge density in the mutant sample. For analysis of N-sulfation pattern, the [3H]glucosamine-labeled HS samples were cleaved by exhaustive deamination with nitrous acid, under conditions leading to selective cleavage at N-sulfated glucosamine residues. Gel chromatography of the reduced products showed a significantly increased disaccharide fraction in the mutant HS sample compared with the corresponding WT sample, indicating an increased proportion of contiguous N-sulfated disaccharide units (Fig. 2C and supplemental Fig. 2). Determination of peak areas showed an increase in disaccharides, from 11% of total WT deamination products to 33% of total mutant deamination products, along with an overall decrease in larger oligosaccharides (supplemental Fig. 2). These data allowed us to estimate the overall degree of N-sulfation, 39% in WT HS versus 52% in mutant HS. To analyze the disaccharide composition of the N-sulfated domains, 35S-labeled HS was degraded by deamination, and the reduced disaccharides were recovered and further separated by anion exchange HPLC (Partisil-10 SAX). As expected, the mutant HS lacked IdoA residues (Fig. 2D, bottom), since all IdoA-containing disaccharide species present in WT HS (peaks 3, 4, and 5 in Fig. 2D, top) were absent. The N-sulfated domains of mutant HS were extensively 6-O-sulfated and contained also some 2-O-sulfated GlcA residues but were devoid of any trisulfated disaccharide units (Fig. 2D and Table 1). The 6-O-sulfate groups are sterically readily accessible to interaction with the anion exchange matrix, and their abundance thus may explain the retarded elution position on anion exchange chromatography (Fig. 2B). Deduced schematic representations of WT and Hsepi−/− HS are shown in Fig. 1.
FIGURE 2.
Structural analysis of HS from WT and Hsepi−/− MEF cells. HS samples isolated from metabolically 3H- or 35S-labeled WT MEF cells (filled circles) and Hsepi mutant MEF cells (continuous line) were analyzed on a Superose-12 column (A) and a DEAE-Sephacel column connected to an HPLC system and eluted using a salt gradient (0–1.5 m), as described under “Experimental Procedures” (B). Effluent fractions were analyzed for radioactivity by scintillation counting. C, 3H-labeled samples were cleaved at GlcNS residues by deamination with HNO2, and the resultant oligosaccharides were separated on a Bio-Gel P-10 column (1 × 200 cm). Oligomer size is indicated above the peaks. D, for compositional analysis of N-sulfated domains, 35S-labeled samples were similarly degraded with HNO2, and the disaccharide products were recovered and separated further by anion exchange HPLC on a Partisil-10 SAX column. The numbered peaks in D represent -(GlcNS)-GlcA2S-GlcNS- (1), -GlcA-GlcNS6S- (2), -IdoA-GlcNS6S- (3), -IdoA2S-GlcNS- (4), and -IdoA2S-GlcNS6S- (5) sequences in the intact HS chains.
TABLE 1.
Proportion of each disaccharide species within N-sulfated domains
Disaccharides were analyzed as shown inFig. 1D. Results are expressed as the proportion of each disaccharide species. The proportion ofO-sulfate groups at different positions is shown as a percentage of totalO-sulfate residues. ND, none detected.
| HS source |
||||
|---|---|---|---|---|
| MEF |
Embryo |
|||
| WT | Hsepi−/− | WT | Hsepi−/− | |
| % | % | |||
| Disaccharide unit | ||||
| -GlcA2S-GlcNS- | 3.1 | 7.9 | 3.5 | 10.5 |
| -GlcA-GlcNS6S- | 29.6 | 92.1 | 20.0 | 89.5 |
| -IdoA-GlcNS6S- | 9.9 | ND | 15.6 | ND |
| -IdoA2S-GlcNS- | 38.5 | ND | 38.6 | ND |
| -IdoA2S-GlcNS6S- | 18.9 | ND | 22.6 | ND |
| O-Sulfate distribution | ||||
| 2-O- | 50.9 | 7.9 | 52.6 | 10.5 |
| 6-O- | 49.1 | 92.1 | 47.4 | 89.5 |
The N-[3H]acetylated HS samples from whole embryos were not subjected to direct structural analysis. However, previous analysis showed structural differences between whole embryo WT and Hsepi mutant HS samples (Table 1) (14), essentially similar to those observed here for the corresponding MEF cell HS species.
Metabolically labeled galactosaminoglycans (CS/DS) were isolated from WT and Hsepi mutant MEF cells, in addition to HS (see “Experimental Procedures”). Yields of CS/DS from the two types of cells were similar. Analysis of products obtained by exhaustive digestion with chondroitinase ABC showed a predominance of 4-O-monosulfated disaccharides and smaller amounts of additional species, with no significant difference in composition between samples from WT and Hsepi mutant cells (supplemental Table 1). Moreover, size fractionation of products obtained by digestion with chondroitinase AC-I or chondroitinase B failed to reveal any difference in IdoA contents or distribution (supplemental Fig. 3). The Hsepi mutation, as expected, does not seem to affect CS/DS biosynthesis.
FIGURE 3.
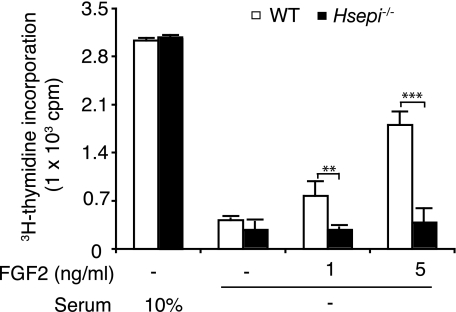
Proliferation of MEF cells. WT and Hsepi−/− cells (1.5 × 104 cells/well) were cultured in 24-well plates in DMEM supplemented with 10% FCS. After 24 h, some cultures were maintained in the same medium, whereas others were changed to starvation medium (no FCS). After further incubation for 48 h, cells were transferred to fresh medium, and FGF2 was added to some of the starved cultures as indicated. Cultures were maintained for an additional 24-h period, with the addition of [3H]thymidine during the last 4 h. Incorporated [3H]thymidine was determined as described under “Experimental Procedures.” Data shown represent triplicate experiments (means ± S.D.). Statistical analysis was done by t test. **, p < 0.01; ***, p < 0.001.
Effect of Hsepi Mutation on FGF2-stimulated Cell Proliferation and Migration
Despite the dramatic alteration in HS structure, the Hsepi-deficient cells did not differ markedly in gross morphology from WT cells (supplemental Fig. 1); nor did they differ in proliferation rate when cultured in medium fortified with 10% FCS, as determined by [3H]thymidine incorporation during exponential growth (Fig. 3). Proliferation of either cell type was essentially abolished in the absence of FCS. Although the addition of FGF2 to the serum-free medium largely restored [3H]thymidine incorporation into WT cells, the growth factor had virtually no effect on the Hsepi−/− cells.
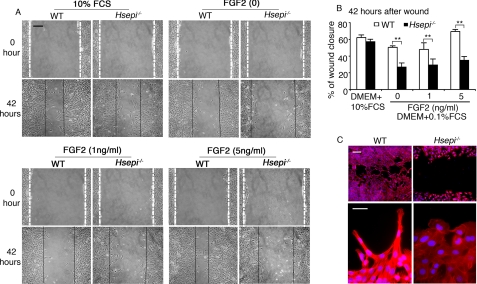
Further effects of the Hsepi mutation were observed in a “cell wound healing” model designed to assess the impact of FGF2 on cell migration. Confluent cultures of Hsepi mutant and WT cells were wounded by scraping with a pipette tip, creating a space free of cells, and migration of cells toward the generated cell-free center was recorded (Fig. 4, A and B). In medium containing 10% FCS, the Hsepi−/− cells migrated almost as well as WT cells. Serum starvation (0.1% FCS in the culture medium) led to moderate inhibition of WT cell migration, which was fully reversed upon the addition of FGF2. By contrast, the Hsepi mutant cells showed marked retardation under serum starvation conditions, which were essentially refractory to FGF2. Although the WT cells thus attained 48 and 70% closure of the gap within 42 h, in response to FGF2 at 1 ng/ml and 5 ng/ml, respectively, the mutant cells managed only 29 and 35% closure under the same conditions. The latter values did not differ significantly from that recorded in the absence of added FGF2. Apparently, FCS contains a factor(s) that stimulates cell migration but shows only marginal dependence on cellular HS containing IdoA. Under conditions of serum starvation, such dependence becomes more marked, as shown by the appreciable difference in wound closure between WT and mutant cells (Fig. 4B). Moreover, the addition of FGF2, which fully restored the migratory ability of WT cells, had limited effect on Hsepi mutants. Similar results were obtained with a different clone of mutant MEF cells (data not shown).
FIGURE 4.
Migration of MEF cells. A, WT and Hsepi−/− MEF cells (5 × 105 cells/well) were seeded on 6-well plates in DMEM supplemented with 10% FCS and cultured until 90% confluence, when a wound was made by a diametral scratch of the cell layers with a pipette tip. Migration of the cells toward the cell-free space was recorded, either in DMEM plus 10% FCS or in starvation medium (DMEM plus 0.1% FCS) fortified with FGF2, as indicated. B, the distance between the wound edges was measured after 42 h as illustrated in A and expressed as percentage of closure of the wound. Results of triplicate experiments are shown. Similar results were obtained with different MEF clones. C, WT and Hsepi−/− MEF cells cultured on coverslips (2 × 104 cells) in DMEM plus 10% FCS after wounding as in A were fixed in 4% formaldehyde and stained with phalloidin-rhodamine and 4′,6-diamidino-2-phenylindole. Scale bars, 25 μm (A), 25 μm (C, upper panels), and 10 μm (C, lower panels). Statistical analysis was done by t test; **, p < 0.01.
Close examination of WT cells at the wound edge, by staining for F-actin with phalloidin-rhodamine, revealed a polarized phenotype, with cell protrusions into the wound and F-actin projecting to the tip of the protrusions. The Hsepi-deficient cells were strikingly different, with relatively uniform flat edges lacking protrusions (Fig. 4C). This finding suggests that the mutant cells lacked the ability to establish proper focal adhesions.
Defective Signaling Response of Hsepi Mutant Cells to FGF2 Stimulation
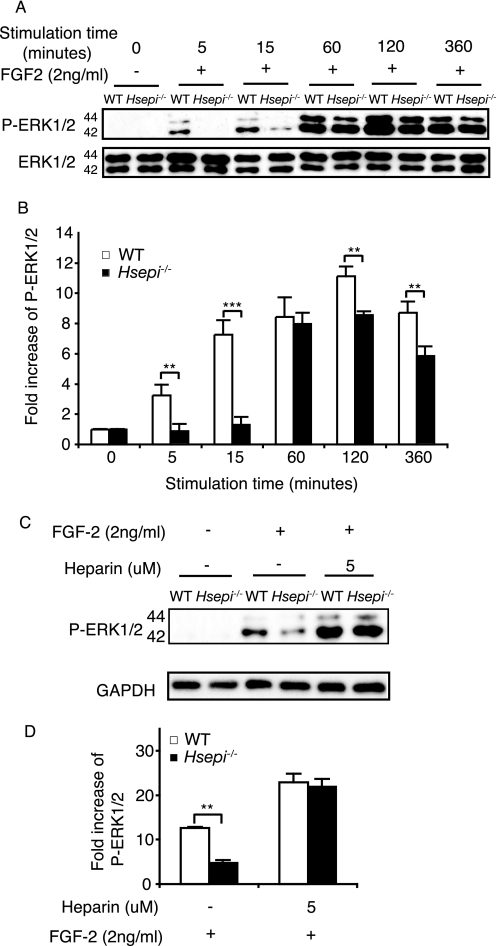
The restricted proliferation and migration of Hsepi mutant cells in response to FGF2 stimulation led us to examine FGF2-induced signaling in mutant and WT MEF cells. We measured phosphorylation of ERK, a downstream transduction component in a major signaling pathway induced by FGF2. Blotting with antibodies against phosphorylated ERK1/2 revealed a significantly weaker signaling response of the mutant compared with WT cells at the initial stage of FGF2 stimulation (Fig. 5, A and B). This difference decreased with increasing stimulation time but reappeared after prolonged FGF2 stimulation (Fig. 5, A and B).
FIGURE 5.
FGF2-induced signaling in WT and Hsepi−/− MEF cells. A, WT and Hsepi−/− MEF cells were cultured in DMEM as described under “Experimental Procedures.” After 48-h starvation, the cells received fresh starvation medium containing 2 ng/ml FGF2. At the indicated time points, cells were collected and lysed. Lysate supernatants were subjected to SDS-PAGE followed by Western blotting using an antibody against phosphorylated forms of ERK1/2 (P-ERK1/2). For loading control, the membranes were reprobed with anti-total ERK1/2 antibody. B, increase of phospho-ERK1/2 signals in FGF2-stimulated compared with nonstimulated cells (intensity quantification of the bands in A). C, cells were cultured as above but with the addition of 5 μm heparin as indicated after the 48-h starvation period. Cultures were maintained for an additional 24-h period before FGF2 (2 ng/ml) stimulation for 10 min. Cell lysates were analyzed by Western blotting. D, increase of phospho-ERK1/2 signals in FGF2-stimulated compared with nonstimulated cells (intensity quantification of the bands in C). Similar results were obtained with different clones. Statistical analysis was done by t test; **, p < 0.01; ***, p < 0.001.
Previous work has shown that defective growth factor signaling due to lack of endogenous HS can be rescued by the addition of exogenous heparin, a fully sulfated HS (24, 25). We therefore examined ERK phosphorylation in the presence of heparin, to confirm that the weak FGF2 signaling of the Hsepi mutant cells was indeed due to aberrant HS structure and not to unspecified signaling deficiency. Adding exogenous heparin to the mutant cells before FGF2 stimulation increased ERK phosphorylation to the level of WT cells (Fig. 5, C and D). Under these conditions, the two types of cells thus showed equal signaling capacity, well above the respective levels attributed to the endogenous HS species.
Aberrant Interaction of IdoA-deficient HS with Growth Factors
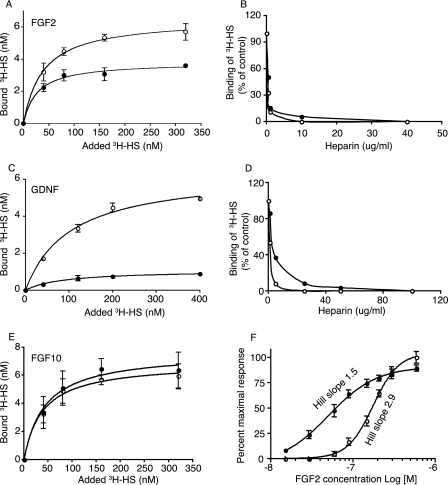
Several growth factors/morphogens utilize cell surface HSPGs as co-receptors, typically in ternary complex formation with cognate primary receptors (17, 26–28). The ability of mutant HS to interact with selected growth factors was tested using nitrocellulose filter trapping of proteins along with chemically [3H]acetylated or metabolically 35S-labeled HS. We focused primarily on the growth factors implicated with the defects observed in the Hsepi mutant mice (14). N-[3H]acetyl-labeled HS samples derived from whole WT and Hsepi−/− embryos both showed specific and saturable binding to FGF2, which was abolished by excess heparin (Fig. 6, A and B). However, whereas affinities seemed similar (Kd ∼30 nm), ∼40% less mutant HS was required to saturate the growth factor compared with WT HS. Similar results were obtained with 35S-labeled HS species isolated from MEF cells (supplemental Fig. 4). An even more pronounced difference in maximal binding between WT and Hsepi−/− HS was observed in corresponding experiments with GDNF (Fig. 6, C and D). Apparently, the growth factor molecules accumulate on a limited fraction of the mutant HS, such that fewer Hsepi−/− compared with WT chains suffice to saturate the proteins. We then reversed the experimental design by titrating a fixed amount of 3H-labeled HS with increasing amounts of FGF2. Indeed, under these conditions, the same amounts of mutant and WT HS were trapped by excess FGF2, indicating that similar proportions of chains (presumably all chains) in the two preparations were capable of interaction (Fig. 6F). However, in comparison with WT HS, the Hsepi−/− HS displayed a clearly sigmoidal saturation curve (Hill slope 2.9) (29), suggestive of cooperative binding of FGF2 molecules along the HS chain. The mode of binding of Hsepi−/− HS to FGF2 (as well as to GDNF) thus differs from that of WT HS; comparison of Kd values under these conditions appears questionable. By contrast, the HS samples from WT and mutant animals did not differ appreciably in their interactions with FGF10 (Fig. 6E).
FIGURE 6.
Interactions of HS with growth factors. Samples of N-[3H]acetyl-labeled HS from WT (empty circles) and Hsepi−/− (filled circles) embryos (same specific radioactivity; see “Experimental Procedures”) were incubated at the concentrations indicated with FGF2 (150 ng, 90 nm final concentration in a 100-μl incubation) (A), GDNF (1 μg, 220 nm final concentration in a 200-μl incubation) (C), and FGF10 (200 ng, final concentration 100 nm in a 100-μl incubation) (E) for 2 h. Bound HS was trapped on a nitrocellulose filter and recovered for radioisotope counting (see “Experimental Procedures”). Data represent triplicate experiments (means ± S.D.). The effects of unlabeled heparin on binding of [3H]HS samples to FGF2 (150 ng, 90 nm; 20 × 103 cpm of HS, 160 nm in a 100-μl incubation) (B) and GDNF (1 μg, 220 nm; 100 × 103 cpm of HS, 800 nm in a 100-μl incubation) (D) were assessed to ascertain specificity of interactions. F, dose-response plot. Samples (1000 cpm, final concentration of 8 nm in a 100-μl incubation) of N-[3H]acetyl-labeled HS from WT (empty circles) and Hsepi−/− (filled circles) embryos were incubated with different amounts of FGF2, as indicated. Bound HS was determined as described above. Data represent triplicate experiments (means ± S.D.).
DISCUSSION
The concept of specificity in HS-protein interactions is a subject of current attention. The large number of distinct interactions implicated with development and homeostasis, along with evidence for strictly regulated HS biosynthesis fostered the notion of distinct or “unique” saccharide sequences generated to bind different protein ligands. However, the highly specific ligand-binding sequences identified to date are few and, furthermore, are distinguished by the occurrence of one or more unusual components, such as 3-O-sulfated or N-unsubstituted glucosamine residues. So far, a protein-binding sequence depending on a unique combination of the common HS constituents (i.e. N-acetylated/N-sulfated glucosamine with or without a 6-O-sulfate group, GlcA, or IdoA with or without a 2-O-sulfate group) remains to be identified (12). Nevertheless, derangement of HS structure in vivo, by ablation of genes encoding selected enzymes required for HS biosynthesis, resulted in phenotype aberrations of varying severity (9, 14, 30). The severe but selective phenotypes of Hsepi−/− and Hs2st−/− mutants are of particular interest, because the associated HS species lack defined sugar and/or sulfate residues yet exhibit essentially unchanged overall negative charge density (14, 31). The present study employed Hsepi−/− MEF cells as a model to evaluate the functional significance of IdoA residues in relation to proliferation, migration, and signaling induced by FGF2, an HS-dependent growth factor.
Structural characterization of HS isolated from WT and Hsepi−/− MEF cells showed that the HS produced by the mutant cells was completely devoid of IdoA, hence of IdoA 2-O-sulfate residues. On the other hand, N- and 6-O-sulfation were up-regulated (Fig. 2), in agreement with previous analysis of HS from whole Hsepi−/− embryos (14). The striking increase in disaccharide deamination products, compared with WT HS, indicated either more numerous or more extended NS domains.
Functional tests, comparing the ability of MEF cells to proliferate (Fig. 3) or migrate (Fig. 4), gave variable results. In the presence of 10% FCS WT and Hsepi−/− cells, both performed well in either assay, without any significant difference between cell types. Switching to starvation conditions dramatically impeded proliferation, again without any marked difference between WT and Hsepi−/− cells. However, whereas proliferation of the WT cells was largely restored by the addition of FGF2, the mutant cells remained refractory to the growth factor (Fig. 3). Similarly, both WT and mutant MEF cells attained efficient wound closure in the culture supplemented with 10% FCS (Fig. 4). Migration of WT cells was only moderately affected by transfer to starvation medium and was completely restored by the addition of FGF2. By contrast, the Hsepi−/− cells were appreciably slowed down in starvation medium and resisted FGF2 treatment. Cell performance in these systems was apparently promoted by factors in FCS that do not strictly depend on cell surface HS of precisely tailored structure. The functional deterioration caused by depletion of these factors was alleviated by the addition of FGF2 to WT cells, whereas Hsepi−/− cells were recalcitrant to treatment. Although we cannot exclude the possibility that these discrepancies may to some extent be influenced by clonal selection of MEF cells, they clearly reflect the difference in composition of WT and mutant HS.
These findings have bearing on the interpretation of phenotypes of mice with genetically manipulated HS biosynthesis. Ablation of either the Hsepi or the Hs2st gene thus resulted in phenotypes with severely perturbed organ systems (e.g. skeletal malformation and kidney agenesis). However, mutant mice also displayed potential targets of HS-dependent signaling that appeared unexpectedly normal, such as the intestinal, central nervous, and vascular systems (9, 14). The seemingly normal functional features of Hsepi−/− MEF cells in serum-containing medium as well as the selective knock-out phenotypes in mice could be explained by various kinds of redundancy. Signaling systems activated by growth factors/morphogens not dependent on HSPGs could be recruited to promote particular responses, such as cell proliferation, migration etc. Alternatively, essential signaling events could be critically reliant on participation of HS chains but less dependent on the detailed structure of these chains. Whereas both mechanisms may well apply, studies of selected organ systems provided clues in favor of the latter alternative. Conditional Ext1 knock-out in mice, selectively eliminating HS biosynthesis, led to severely perturbed gross anatomical features of the brain (32), indicating that processes of fundamental importance to brain development depend on HS participation. Yet brains of Hsepi mutant mice appeared macroscopically normal (14).3 Moreover, deletion of the C-terminal, HS-binding motif of platelet-derived growth factor-BB was found to impair growth factor retention and pericyte recruitment in vascular development in vivo, thereby pointing to an important role for HS in platelet-derived growth factor-BB function. Again, the nearly normal vascular development in Hsepi−/− embryos argues against any critical constraint of HS structure in this context (20). Accordingly, binding to platelet-derived growth factor-BB of HS-related oligosaccharides generated by chemoenzymatic methods was found to depend on overall degree of sulfation, without any apparent requirement for specific sequence.
The phenotype perturbations actually observed following genetic manipulation of HS biosynthesis indicate failure of the resultant abnormal HS to properly interact with critical target proteins. Similarly, functional shortcomings of Hsepi−/− cells could be exposed under starvation conditions by subjecting cells to appropriately selected signaling proteins. Mutant cells thus were unable to mount the functional response (proliferation and migration) to FGF2 shown by WT cells under the same conditions (Figs. 3 and 4). These malfunctions may reflect, in particular, the defective MAPK pathway signaling observed after long term stimulation with FGF2 (Fig. 5) (33). Short term FGF2 signaling of Hs2st−/− MEF cells was similar to that of WT cells (31), suggesting that the signaling defect of Hsepi−/− cells is linked to the lack of IdoA units per se. Whereas both Hsepi−/− HS and Hs2st−/− HS lack IdoA-2-O-sulfate units, the latter HS species contains IdoA residues. Notably, the ability of a particular HS species to promote signaling is not readily predicted from FGF·HS binding data, as shown by the weak interaction between FGF2 and Hs2st−/− HS (31). These findings highlight the need for more detailed information regarding the molecular interactions in binary FGF·HS and ternary FGF·HS·FGF receptor complexes. Data obtained by various techniques, including crystallography, suggest that higher order complexes may be assembled in different ways and have different signaling properties (25, 34, 35). Overall sulfation levels rather than precise sequence appear to be of primary importance in complex formation of saccharides with either FGF1/2 alone (36) or FGFs together with receptors (26), and recent results point to similar requisites in actual (FGF2) signaling.4 The oligosaccharides used in these studies were derived from heparin, hence with IdoA as the predominant hexuronic acid constituent; to our knowledge, there are no previous reports on interaction properties of HS-related saccharides lacking IdoA. The binding experiments based on nitrocellulose filter trapping (Fig. 6) therefore open novel aspects on HS structure-function relations. Whereas FGF2 thus bound WT HS with the expected saturation kinetics, growth factor molecules were preferentially accumulated on a fraction of the Hsepi−/− HS chains, in apparent cooperative mode (Fig. 6, A and F). The relation between this aberrant interaction behavior and the defective signaling and functional responses of Hsepi−/− MEF cells remains unclear. A similar anomaly, although even more marked, was noted in binding of Hsepi−/− HS to GDNF (Fig. 6C), a growth factor implicated in kidney development. Kidney agenesis is a penetrating phenotype in Hsepi−/− (and Hs2st−/−) mice (9, 14). By contrast, Hsepi−/− and WT HS were indistinguishable in binding to FGF10 (Fig. 6E), known to depend primarily on the presence of 6-O-sulfate groups (37). Further work is required to define the mechanism behind the selective, anomalous binding properties of Hsepi−/− HS. An intriguing question is whether this feature is a direct consequence of the lack of IdoA or rather a secondary phenomenon due to perturbed domain organization along the HS chain.
Supplementary Material
Acknowledgments
We acknowledge Prof. Mart Saarma (University of Helsinki, Finland) for providing GDNF protein and Dr. Per Jemth (Department of Medical Biochemistry and Microbiology, University of Uppsala) for valuable discussions in interpretation of the HS-FGF2 interaction.
This work was supported by the Swedish Research Council (32X-15023, 2007-5985; K2009-67X-21128-01-3) and Polysackaridforskning AB (Uppsala, Sweden).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
Y. Yamaguchi, U. Lindahl, and J.-P. Li, unpublished observations.
N. Jastrebova, M. Vanwildemeersch, A. Eriksson, U. Lindahl, and D. Spillmann, unpublished results.
- HSPG
- heparan sulfate proteoglycan
- IdoA
- l-iduronic acid
- MEF
- mouse embryo fibroblast
- WT
- wild type
- DMEM
- Dulbecco's modified Eagle's medium
- FCS
- fetal calf serum
- CS
- chondroitin sulfate
- DS
- dermatan sulfate
- HPLC
- high pressure liquid chromatography
- GlcNS
- N-sulfated glucosamine
- PBS
- phosphate-buffered saline
- FGF
- fibroblast growth factor
- GDNF
- glia-derived neurotropic factor
- ERK
- extracellular signal-regulated kinase
- HS
- heparan sulfate.
REFERENCES
- 1.Lin X. ( 2004) Development 131, 6009– 6021 [DOI] [PubMed] [Google Scholar]
- 2.Patel V. N., Likar K. M., Zisman-Rozen S., Cowherd S. N., Lassiter K. S., Sher I., Gallagher J. T., Yates E. A., Turnbull J. E., Ron D., Hoffman M. P. ( 2008) J. Biol. Chem. 283, 9308– 9317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernfield M., Gotte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. ( 1999) Annu. Rev. Biochem. 68, 729– 777 [DOI] [PubMed] [Google Scholar]
- 4.Esko J. D., Lindahl U. ( 2001) J. Clin. Invest. 108, 169– 173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esko J. D., Selleck S. B. ( 2002) Annu. Rev. Biochem. 71, 435– 471 [DOI] [PubMed] [Google Scholar]
- 6.Feyzi E., Saldeen T., Larsson E., Lindahl U., Salmivirta M. ( 1998) J. Biol. Chem. 273, 13395– 13398 [DOI] [PubMed] [Google Scholar]
- 7.Ledin J., Staatz W., Li J. P., Gotte M., Selleck S., Kjellen L., Spillmann D. ( 2004) J. Biol. Chem. 279, 42732– 42741 [DOI] [PubMed] [Google Scholar]
- 8.Casu B., Lindahl U. ( 2001) Adv. Carbohydr. Chem. Biochem. 57, 159– 206 [DOI] [PubMed] [Google Scholar]
- 9.Bullock S. L., Fletcher J. M., Beddington R. S., Wilson V. A. ( 1998) Genes Dev. 12, 1894– 1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin X., Buff E. M., Perrimon N., Michelson A. M. ( 1999) Development 126, 3715– 3723 [DOI] [PubMed] [Google Scholar]
- 11.Forsberg E., Pejler G., Ringvall M., Lunderius C., Tomasini-Johansson B., Kusche-Gullberg M., Eriksson I., Ledin J., Hellman L., Kjellen L. ( 1999) Nature 400, 773– 776 [DOI] [PubMed] [Google Scholar]
- 12.Kreuger J., Spillmann D., Li J. P., Lindahl U. ( 2006) J. Cell Biol. 174, 323– 327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J. P., Gong F., El Darwish K., Jalkanen M., Lindahl U. ( 2001) J. Biol. Chem. 276, 20069– 20077 [DOI] [PubMed] [Google Scholar]
- 14.Li J. P., Gong F., Hagner-McWhirter A., Forsberg E., Abrink M., Kisilevsky R., Zhang X., Lindahl U. ( 2003) J. Biol. Chem. 278, 28363– 28366 [DOI] [PubMed] [Google Scholar]
- 15.Abbondanzo S. J., Gadi I., Stewart C. L. ( 1993) Methods Enzymol. 225, 803– 823 [DOI] [PubMed] [Google Scholar]
- 16.Jat P. S., Cepko C. L., Mulligan R. C., Sharp P. A. ( 1986) Mol. Cell Biol. 6, 1204– 1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escobar Galvis M. L., Jia J., Zhang X., Jastrebova N., Spillmann D., Gottfridsson E., van Kuppevelt T. H., Zcharia E., Vlodavsky I., Lindahl U., Li J. P. ( 2007) Nat. Chem. Biol. 3, 773– 778 [DOI] [PubMed] [Google Scholar]
- 18.Maccarana M., Olander B., Malmstrom J., Tiedemann K., Aebersold R., Lindahl U., Li J. P., Malmstrom A. ( 2006) J. Biol. Chem. 281, 11560– 11568 [DOI] [PubMed] [Google Scholar]
- 19.Höök G. E., Gilmore L. B. ( 1982) J. Biol. Chem. 257, 9211– 9220 [PubMed] [Google Scholar]
- 20.Abramsson A., Kurup S., Busse M., Yamada S., Lindblom P., Schallmeiner E., Stenzel D., Sauvaget D., Ledin J., Ringvall M., Landegren U., Kjellen L., Bondjers G., Li J. P., Lindahl U., Spillmann D., Betsholtz C., Gerhardt H. ( 2007) Genes Dev. 21, 316– 331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shively J. E., Conrad H. E. ( 1976) Biochemistry 15, 3932– 3942 [DOI] [PubMed] [Google Scholar]
- 22.Lauder R. M., Huckerby T. N., Nieduszynski I. A. ( 2000) Glycobiology 10, 393– 401 [DOI] [PubMed] [Google Scholar]
- 23.Kreuger J., Lindahl U., Jemth P. ( 2003) Methods Enzymol. 363, 327– 339 [DOI] [PubMed] [Google Scholar]
- 24.Guimond S., Maccarana M., Olwin B. B., Lindahl U., Rapraeger A. C. ( 1993) J. Biol. Chem. 268, 23906– 23914 [PubMed] [Google Scholar]
- 25.Goodger S. J., Robinson C. J., Murphy K. J., Gasiunas N., Harmer N. J., Blundell T. L., Pye D. A., Gallagher J. T. ( 2008) J. Biol. Chem. 283, 13001– 13008 [DOI] [PubMed] [Google Scholar]
- 26.Jastrebova N., Vanwildemeersch M., Rapraeger A. C., Gimenez-Gallego G., Lindahl U., Spillmann D. ( 2006) J. Biol. Chem. 281, 26884– 26892 [DOI] [PubMed] [Google Scholar]
- 27.Catlow K. R., Deakin J. A., Wei Z., Delehedde M., Fernig D. G., Gherardi E., Gallagher J. T., Pavao M. S., Lyon M. ( 2008) J. Biol. Chem. 283, 5235– 5248 [DOI] [PubMed] [Google Scholar]
- 28.Sugaya N., Habuchi H., Nagai N., Ashikari-Hada S., Kimata K. ( 2008) J. Biol. Chem. 283, 10366– 10376 [DOI] [PubMed] [Google Scholar]
- 29.Tour E., Hittinger C. T., McGinnis W. ( 2005) Development 132, 5271– 5281 [DOI] [PubMed] [Google Scholar]
- 30.Ringvall M., Ledin J., Holmborn K., van Kuppevelt T., Ellin F., Eriksson I., Olofsson A. M., Kjellen L., Forsberg E. ( 2000) J. Biol. Chem. 275, 25926– 25930 [DOI] [PubMed] [Google Scholar]
- 31.Merry C. L., Bullock S. L., Swan D. C., Backen A. C., Lyon M., Beddington R. S., Wilson V. A., Gallagher J. T. ( 2001) J. Biol. Chem. 276, 35429– 35434 [DOI] [PubMed] [Google Scholar]
- 32.Inatani M., Irie F., Plump A. S., Tessier-Lavigne M., Yamaguchi Y. ( 2003) Science 302, 1044– 1046 [DOI] [PubMed] [Google Scholar]
- 33.Delehedde M., Seve M., Sergeant N., Wartelle I., Lyon M., Rudland P. S., Fernig D. G. ( 2000) J. Biol. Chem. 275, 33905– 33910 [DOI] [PubMed] [Google Scholar]
- 34.Pellegrini L., Burke D. F., von Delft F., Mulloy B., Blundell T. L. ( 2000) Nature 407, 1029– 1034 [DOI] [PubMed] [Google Scholar]
- 35.Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. ( 2000) Mol. Cell 6, 743– 750 [DOI] [PubMed] [Google Scholar]
- 36.Kreuger J., Jemth P., Sanders-Lindberg E., Eliahu L., Ron D., Basilico C., Salmivirta M., Lindahl U. ( 2005) Biochem. J. 389, 145– 150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashikari-Hada S., Habuchi H., Kariya Y., Itoh N., Reddi A. H., Kimata K. ( 2004) J. Biol. Chem. 279, 12346– 12354 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.