Abstract
Vascular smooth muscle cells (VSMC) exhibit phenotypic plasticity and change from a quiescent contractile phenotype to a proliferative synthetic phenotype during physiological arteriogenesis and pathological conditions such as atherosclerosis and restenosis. Platelet-derived growth factor (PDGF)-BB is a potent inducer of the VSMC synthetic phenotype; however, much less is known about the role of fibroblast growth factor-2 (FGF2) in this process. Here, we show using signal transduction mutants of FGF receptor 1 (FGFR1) expressed in rat VSMC that the adaptor protein FRS2 is essential for FGFR1-mediated phenotypic modulation and down-regulation of VSMC smooth muscle α-actin (SMA) gene expression. In addition, we show that PDGF-BB and FGF2 act synergistically to induce cell proliferation and down-regulate SMA and SM22α in VSMC. Furthermore, we show that PDGF-BB induces tyrosine phosphorylation of FGFR1 and that this phosphorylation is mediated by PDGF receptor-β (PDGFRβ), but not c-Src. We demonstrate that FRS2 co-immunoprecipitates with PDGFRβ in a complex that requires FGFR1 and that both the extracellular and the intracellular domains of FGFR1 are required for association with PDGFRβ, whereas the cytoplasmic domain of FGFR1 is required for FRS2 association with the FGFR1-PDGFRβ complex. Knockdown of FRS2 in VSMC by RNA interference inhibited PDGF-BB-mediated down-regulation of SMA and SM22α without affecting PDGF-BB mediated cell proliferation or ERK activation. Together, these data support the notion that PDGFRβ down-regulates SMA and SM22α through formation of a complex that requires FGFR1 and FRS2 and prove novel insight into VSMC phenotypic plasticity.
Phenotypic modulation of vascular smooth muscle cells (VSMC)3 is an important step in the development of several pathophysiological processes including atherosclerosis, restenosis, and vascular remodeling (1, 2). During these processes VSMC change from a contractile phenotype to a synthetic phenotype characterized by increased proliferation, migration, increased extracellular matrix production, and decreased expression of contractile proteins, including smooth muscle α-actin (SMA), SM22α, calponin, and myosin heavy chain. Several growth factors including platelet-derived growth factor-BB (PDGF-BB), fibroblast growth factor 2 (FGF2), and thrombin have been implicated in the induction of the synthetic phenotype (3). These growth factors bind cell surface receptors and activate intracellular signaling pathways that result in changes in gene expression and cellular phenotype. Understanding the interactions between these pathways may provide insights into mechanisms of phenotypic modulation of VSMC and provide new targets for therapeutic intervention in vascular disease.
Experimental evidence using various in vitro and in vivo models points to a role for FGF-FGFR in the phenotypic modulation of VSMC. FGFs and FGFRs are expressed in VSMC and are up-regulated during vascular injury and in atherosclerotic plaque formation (4–6). Balloon injury of rat arteries led to an increase in FGFR expression in VSMC. The up-regulation of FGF and FGFR suggests that they contribute to the pathogenesis of vascular disease. In support of this hypothesis, administration of anti-FGF2 antibodies and FGFR tyrosine kinase inhibitors results in decreased VSMC proliferation, migration, and attenuated neointimal thickening (7).
PDGF-BB binds to PDGFRβ and activates several intracellular signaling pathways including ERK, phosphatidylinositol 3-kinase/Akt, and mammalian target of rapamycin (mTOR) (8). Studies have indicated that PDGF-BB induces the release of FGF2 and activation FGFR1, resulting in sustained ERK activation and proliferation of human VSMC (9). When FGFR1 expression was inhibited by RNA interference, PDGF-BB induced transient but not sustained ERK activation.
Binding of FGF2 to FGFR1 activates the ERK and phosphatidylinositol 3-kinase/Akt pathways via the adaptor protein FRS2 (10, 11). Upon FGF2 binding, FGFR1 phosphorylates FRS2 on six tyrosine residues that function as docking sites for the SH2 domain-containing proteins Grb2 and SHP2 (12, 13). Grb2 binds Gab1 leading to activation of phosphatidylinositol 3-kinase/Akt, whereas SHP2 activates the Ras-Raf-ERK pathway. FRS2 binds to FGFR1 via a Val-Thr dipeptide in the juxtamembrane region of FGFR1 (14, 15). Deletion of these two amino acids abrogates binding of FRS2 to FGFR1. To determine the role of FRS2 in FGFR1-mediated VSMC phenotypic modulation and to determine the interaction of PDGFRβ with the FGFR1 signaling pathway, we developed a set of FGFR1 signaling pathway deficient mutants and stably expressed them in rat VSMC. In this study we report that PDGFRβ, FGFR1, and FRS2 form a multi-protein complex that is essential for VSMC phenotypic modulation and that stable knockdown of FRS2 inhibits PDGF-BB-mediated down-regulation of VSMC marker gene expression but not PDGF-BB-mediated VSMC proliferation.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
293T cells (human embryonic kidney cells, ATCC CRL-11268), PAC1 cells, A7r5 cells (16), primary bovine aortic vascular smooth muscle cells (BVSMC) cells, and primary rat aortic vascular smooth muscle cells (RASMC) cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (FBS) and penicillin-streptomycin. Recombinant human FGF-2 (100–18B) and PDGF-BB (100–14B) were purchased from PeproTech. Dulbecco's modified Eagle's medium and FBS were obtained from HyClone Laboratories Inc. SM α-actin and SM22α promoter luciferase plasmids were provided by Dr. Gary K. Owens (University of Virginia) (17). PDGFRβ, PDGFRβ D850N, and PDGFRβ K634A plasmids were provided by Dr. Carl-Henrik Heldin (Uppsala University) (18). GST-FRS2αPTB construct was provided by Dr. Patrik Ernfors (Karolinska Institute) (19). FRS2αPTB construct was provided by Dr. John Heath (University of Birmingham) (14). fms/PDGFRβ chimera construct was provided by Dr. Jonathan Cooper (Fred Hutchinson Cancer Research Center) (20).
Generation of FGFR1 Pathway-deficient Mutant Constructs (14, 21, 22)
Plasmids containing single or multiple mutations were generated using the QuikChange site-directed mutagenesis kit (Stratagene) employing the Xenopus constitutively active FGFR1 K562E construct (23) as template according to the manufacturer's recommendation. To study FGFR1-mediated FRS2 signaling pathway, we created the FGFR1 K562E: −Crk/−PLCγ construct in which Crk and PLCγ binding sites were mutated (Y463F and Y766F) (see Fig. 1A, construct 7). For the FGFR1-mediated Crk signaling pathway, we created the FGFR1 K562E: −FRS2/−PLCγ mutant (construct 6) in which the FRS2-binding site was deleted and the PLCγ-binding site was mutated (Y766F). To study the FGFR1-mediated PLCγ signaling pathway, we created FGFR1 K562E: −FRS2/−Crk (construct 5) in which the FRS2-binding site was deleted and the Crk-binding site was mutated (Y463F). To examine the cross-regulation between Crk and PLCγ pathways, we created the FGFR1 K562E: −FRS2 construct (construct 2). To study the cross-regulation between FRS2 and PLCγ pathways, we created the FGFR1 K562E: −Crk construct (construct 3). To study the cross-regulation between the FRS2 and Crk pathways, we created the FGFR1 K562E: −PLCγ construct (construct 4). Finally, we created a triple mutant construct (FGFR1 K562E: −FRS2/−Crk/−PLCγ) (construct 8) to examine whether other tyrosine kinase residues within the FGFR1 intracellular domain have the potential to activate downstream signals. The following primers were used for mutagenesis (the mutated sequences are in bold): The FRS2-binding site deletion mutation was introduced using 5′-GCA TCC CCG TGC GCA GAC AGG TTT CAG GGG ACT CCA GCT C-3′ and 5′-GAG CTG GAG TCC CCT GAA ACC TGT CTG CGC ACG GGG ATG C-3′; the Crk Y463F mutation was introduced using 5′-GTT GTC TGG ACT ATC GGA ATT TGA GCT TCC AGA AGA TCC AC-3′ and 5′-GTG GAT CTT CTG GAA GCT CAA ATT CCG ATA GTC CAG ACA AC-3′; the PLCγ Y766F mutation was introduced using 5′-CTG AGT TCC AAT CAG GAA TTT CTT GAT CTC TCC ATG CCA GTG-3′ and 5′- CAC TGG CAT GGA GAG ATC AAG AAA TTC CTG ATT GGA ACT CAG-3′. Each construct was verified by sequencing on an ABI Prism 310 sequencer at the Maine Medical Center Research Institute Core Facility.
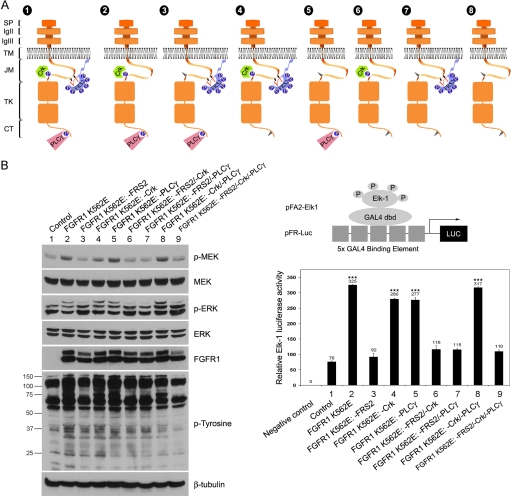
FIGURE 1.
Diagrams of FGFR1 mutants and biochemical characterization. A, schematic representation of FGFR1 K562E mutant constructs. Construct 1, FGFR1 K562E; construct 2, FGFR1 K562E: −FRS2; construct 3, FGFR1 K562E: −Crk; construct 4, FGFR1 K562E: −PLCγ; construct 5, FGFR1 K562E: −FRS2/−Crk; construct 6, FGFR1 K562E: −FRS2/−PLCγ; construct 7, FGFR1 K562E: −Crk/−PLCγ; construct 8, FGFR1 K562E: −FRS2/−Crk/−PLCγ. SP, signal peptide; Ig, immunoglobulin-like domain; TM, transmembrane domain; JM, juxtamembrane domain; TK, tyrosine kinase; CT, C-terminal regulatory tail. B, effects of FGFR1 K562E mutants on ERK signaling in PAC1 VSMC. Left panel, PAC1 stable cell lines were cultured in 10% FBS medium for 24 h and then switched to 0.5% FBS medium overnight. The cells were lysed, and the cell lysates were subjected to Western blot analysis and probed with indicated antibodies. β-Tubulin served as a loading control. The molecular masses are indicated on the left in kilodaltons. Right panel, upper panel, Diagram of the PathDetect transreporting system used in this study (pFA/pFR-Luc; Stratagene). Lower panel, PAC1 cells were transfected with pFA2-CMV and pFR-Luc. The pFA2-dbd plasmid was used as a negative control. After 24 h, the cells were lysed, and luciferase activities were measured. Graphed are the means ± S.D. of triplicate samples. Statistical analysis was performed by using one-way ANOVA test. ***, p < 0.001, compared with the control. All of the results are representative of three separate experiments.
Retrovirus Production and Creation of PAC1 Stable Cell Lines
BamHI-cleaved cDNAs fragments encoding Xenopus constitutively active FGFR1 K562E, and its derivative mutants were inserted into the retroviral amphotropic vector pWZL. The resulting retroviral vectors were transfected into an amphotropic retrovirus packaging cell line, BOSC 23 cells. Forty-eight h after transfection, viral supernatants were filtered (0.45 μm), supplemented with polybrene (4 μg/ml, Sigma), and incubated with PAC1 cells. The cells expressing FGFR1 K562E mutants were selected by culturing them in medium containing hygromycin (300 μg/ml) and were pooled for analysis.
Cell Lysis, Immunoprecipitation, and Western Blot Analysis
To obtain cell lysates, the cells were washed with cold phosphate-buffered saline (PBS) and lysed for 10 min on ice in a hypotonic HNTG lysis buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 1.5 mm MgCl2, 1.0 mm EGTA) containing Complete Protease inhibitor mixture (Roche Applied Science), 1 mm sodium orthovanadate (Sigma), and 1 mm sodium fluoride (Sigma) as phosphatase inhibitors. The lysates were centrifuged at 13,000 rpm at 4 °C for 15 min. For immunoprecipitations, equal amounts of cell lysate were incubated with the indicated antibodies for 16–18 h at 4 °C with rotation. Protein A/G-agarose beads (30 μl; Santa Cruz) were then added to the lysates to capture the immune complex for 1 h. The immunoprecipitates were collected by centrifugation in a microcentrifuge at 2,500 rpm for 30 s. The supernatant was discarded, whereupon the pellet was washed four times with HNTG buffer and one time with PBS, and then the immune complexes were eluted from the beads by boiling in 2× Laemmli sample buffer (125 mm Tris-HCl, 10% 2-mercaptoethanol, 4% SDS, 20% glycerol, 0.01% bromphenol blue). For immunoblot analysis, proteins were loaded into 6% or 10% polyacrylamide gels, separated by SDS-PAGE, and transferred onto nitrocellulose membranes (Bio-Rad) at 20 V for 70 min in transfer buffer (25 mm Tris, 192 mm glycine, and 20% methanol). The uniform transfer of proteins to the nitrocellulose membrane was monitored by transiently staining the membranes with Ponceau S stain (Fisher) before application of the blocking procedure. Subsequently, the membranes were blocked in 5% nonfat milk or 5% bovine serum albumin for 1 h at room temperature and probed for specific primary antibodies at 4 °C overnight. After washing with TBST (10 mm Tris, pH 7.4, 150 mm NaCl, 0.05% Tween 20) three times 10 min each, the membranes were further incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000) for 1 h at room temperature. The membranes were washed for three times 10 min each with TBST and detected by Supersignal® West Pico chemiluminescent substrate (Pierce) according to the manufacturer's instructions. The membranes were then stripped with stripping buffer (62.5 mm Tris-HCl, pH 6.8, 100 mm 2-mercaptoethanol, 2% SDS) at 50 °C for 30 min and redetected, as described above, using different primary and secondary antibodies. The apparent molecular masses of proteins were estimated based on migration of prestained molecular mass standards (Bio-Rad). Antibody against SM22α (ab14106, 1:2000) was purchased from abcam. Antibodies against HA tag (2367, 1:2000; for immunoblotting), p44/p42 MAPK (9102, 1:1000), phospho-FRS2 (Tyr196, 1:1000), phospho-FRS2 (Tyr436, 1:1000), PDGFRα (3164, 1:1000), PDGFRβ (3175, 1:50; for immunoprecipitation), PDGFRβ (3169, 1:1000; for immunoblotting) were purchased from Cell Signaling. Antibody against FGFR1 (Tyr653/654) (44-1140G, 1:1000) was purchased from Invitrogen. Antibody against HA tag (600-401-384, 1:100; for immunoprecipitation) was purchased from Rockland. Antibodies against β-tubulin (T7816, 1:10,000) and diphosphorylated ERK1/2 (M9692, 1:10,000) were purchased from Sigma. Antibodies against FGFR1 (H-76, 1:1000), FGFR2 (C-17, 1:1000), FGFR3 (C-15, 1:1000), FRS2 (H-91, 1:1000), c-Fms/CSF-1R (H-300, 1:1000), c-Src (Src 2, 1:1000), and cyclin D1 (DCS-6, 1:1000) were purchased from Santa Cruz. Antibody against phosphotyrosine clone 4G10 (05–321, 1:2000) was purchased from Upstate. Mouse monoclonal antibody against Xenopus transmembrane region of FGFR1 (5G11, 1:300) and rabbit polyclonal antibody against C-terminal region of FGFR1 (FB817, 1:1000) have been described previously (24).
Quantification of Western Blots
Images of blot signals on blue film (DENVILLE) were scanned on a transmission scanner (Canon) using Adobe PhotoShop® 7.0 software. The images were then viewed in ImageQuant software (Molecular Dynamics) for data analysis. Signal intensities of individual bands were determined using volume analysis with object average background correction applied followed the ImageQuant user's guide. The data were exported to MicrosoftTM Excel to generate the plot. To obtain the mean, standard deviation, and test for significant differences between samples, we averaged the relative band intensities from three independent experiments. The data are presented as fold change in protein expression for the experimental groups compared with the control group after normalized to loading controls (β-tubulin or total phospho-protein). For presentation, the data are plotted as fold change versus the samples. The error bars show the calculated standard deviation. The statistical significance was calculated by Student's t test (comparing two groups) or one-way ANOVA and Tukey's post-hoc test (comparing multiple groups). p values of < 0.05 were considered significant and are indicated with asterisks.
Promoter Analysis and Transient Transfection in VSMC
The cells were plated at a density of 1 × 105 cells/well in 12-well plates and transiently transfected with 0.3 μg of the SM α-actin or SM22α promoter luciferase plasmid and together with 50 ng of pRL-TK Renilla plasmid. Twenty-four h after transfection, the cells were changed to 0.5% FBS medium overnight. Reporter gene activity was measured by using dual luciferase reporter assay (Promega) according to the manufacturer's technical manual.
RNA Isolation and Real Time PCR
Total RNA was extracted from PAC1 stable cell lines by using RNeasy mini kit (Qiagen) and following the manufacturer's instructions. First strand cDNA was synthesized by random priming using the NEB first stand synthesis system (New England Biolabs) following the manufacturer's instructions. Real time quantitative PCR analysis was performed using SYBR green (Bio-Rad). For each reaction, 25 μl of 2× SYBR Master mix and 0.1 μm of forward and reverse primers (Rat Acta2 and Rat Hprt; BASciences) along with 1 μl each of appropriate transcripts were mixed using a Bio-Rad iCycler. Cycling parameters were as follows: 95 °C for 15 min and 40 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. Relative abundance of mRNAs was calculated using the comparative threshold cycle method and normalized with Hprt as an internal control. The data are the means of three individual experiments.
Immunohistochemical Staining of FRS2
Mouse aortas were fixed, embedded, and sectioned as for immunohistochemistry. The slides were dewaxed in a series of three xylene washes for 5 min each, then rehydrated in a graded ethanol series (100, 90, and 70%), and washed in PBS. After rehydration, the slides were pretreated in 3% H2O2 in PBS. Antigen retrieval was performed by microwaving slides in Tris-EDTA buffer (10 mm Tris-base, 1 mm EDTA, 0.05% Tween 20, pH 9.0). The sections were incubated in blocking buffer (5% bovine serum albumin, 0.1% Tween 20, and 0.1% Triton X-100) for 1 h at room temperature. Anti-FRS2 primary antibody (H-91; Santa Cruz, 1:50) was then added to the blocking buffer, and the sections were incubated at 4 °C overnight. The next day, the sections were washed five times in PBS containing 0.5% Triton X-100, following by incubation with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. After extensive washes, the sections were mounted with mounting solution (Vector Laboratories) and sealed for analysis. For negative control staining, the sections were incubated without primary antibody.
Proliferation Assays
For MTT assay, the cells were incubated with 50 mg/ml of MTT (Sigma) for 1 h. Next, 0.5 ml of solubilized solution (composition: 0.1 n HCl in isopropanol) was added to resolved the water-insoluble formazan salt, and the 550 nm absorbance of the medium was measured with a Beckman DU-640 Spectrophotometer. For bromodeoxyuridine (BrdUrd) labeling, a 5-bromo-2′-deoxy-uridine labeling and detection kit (Roche Applied Science) was used for quantification of cell DNA synthesis and cellular proliferation according to the manufacturer's instructions. The cells grown on coverslips were pulse-labeled with 10 μm BrdUrd and then fixed with ethanol for 30 min at −20 °C. Primary antibody to BrdUrd was diluted 1:10 with incubation buffer supplied with the kit for 30 min at 37 °C. Secondary antibody to anti-mouse IgG conjugated with Alexa Fluor® 546 was employed in a dilution of 1:1000 for 30 min at 37 °C. Finally, the coverslips were washed in PBS and mounted in Hard set mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories). The proliferation rate was calculated as the number of BrdUrd-positive nuclei divided by the total cell number found in individual microscopic fields at 200× magnification, multiplied by 100, using an Olympus fluorescence microscope. At least 200 cells were counted per sample.
Transient Transfection
Transient transfection of 293T cell lines was optimized using GeneJuice transfection reagent (Invitrogen) according to the manufacturer's instructions. Briefly, 50–60% confluent cells were incubated with the GeneJuice/DNA mix. To ensure optimal protein expression levels, experiments with transiently transfected cells are generally performed between 24 and 48 h following transfection. The transfection efficiency was greater than 90% monitored by green fluorescent protein expression.
GST Pulldown Assays
All of the GST-fused proteins were purified from Escherichia coli as described previously (25). For pulldown assays, the cells were rinsed once with PBS and lysed with the HNTG lysis buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 1.5 mm MgCl2, 1.0 mm EGTA, 1 mm sodium orthovanadate, 1 mm sodium fluoride, and EDTA-free protease inhibitor mixture). The cell lysates were then centrifuged at 13,000 rpm at 4 °C for 15 min. The cell lysates were precleared with glutathione-Sepharose and then were incubation with GST fusion proteins conjugated with glutathione-Sepharose beads at 4 °C for 2 h. After the beads were washed with ice-cold HNTG lysis buffer, the bound proteins were eluted in 2× Laemmli sample buffer and subjected to SDS-PAGE and immunoblot analysis.
FRS2 Knockdown by RNA Interference
To stably suppress FRS2 expression, PAC1 VSMC were transfected with 5 μg of rat FRS2 retrovirus shRNA constructs (KR47278P; BASciences) in 10-cm plates, 24 h after plating at 30% density, using FuGENE 6 reagent (Roche Applied Science) following the manufacturer's instructions. We also used a pSilencer plasmid encoding a hairpin RNA whose sequence is not found in the mouse, human, or rat genome data based supplied with the kit as nontargeting control. After 24 h of transfection, the cells were changed to selection medium containing 0.75 μg/ml puromycin.
Statistical Analysis
All of the experiments were independently repeated at least three times. Comparisons between control and treatment groups were analyzed by one-way ANOVA and Tukey's post-hoc test or Student's t test. A probability value of p < 0.05 was considered statistically significant.
RESULTS
Signaling by FGFR1 Influences Vascular Smooth Muscle Marker Gene Expression
The regulation of VSMC phenotype and marker gene expression by PDGF-BB has been extensively studied (1, 2). In addition, PDGF-BB has been shown to stimulate VSMC proliferation but not migration in part via an FGFR1-dependent mechanism (3, 9). We used PAC1 cells, an immortalized cell line established from the pulmonary artery of adult Sprague-Dawley rats, for these studies (16). These cells have been used extensively as a model system for studying transcriptional control of VSMC differentiation and growth. To investigate whether FGFR1 contributes to the loss of VSMC contractile protein expression, we stably transfected PAC1 VSMC with a constitutively activated mutant FGFR1 or FGFR1 mutants deficient in specific downstream signaling pathways (14, 21, 22) (Fig. 1A). Analysis using FGFR1 signaling pathway mutants revealed that FRS2 is required for linking FGFR1 to the MAPK pathway including MAPK/ERK kinase (MEK), ERK, and the ERK substrate Elk-1 in PAC1 cells (Fig. 1B, lanes 2, 4, 5, and 8).
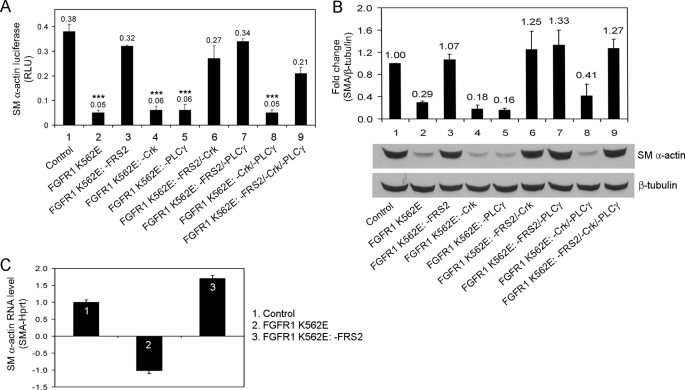
To investigate the involvement of FGF signaling in smooth muscle contractile protein expression, we analyzed SMA. We used a previously characterized rat SMA promoter luciferase reporter construct (26). FGFR1 K562E mutants with an intact FRS2 signaling pathway expression in PAC1 cells decreased SMA reporter activity ∼6-fold compared with control cells (Fig. 2A, lanes 1, 2, 4, 5, and 8). PAC1 cells that expressed FGFR1 K562E mutant receptors deficient in the FRS2-binding site exhibited SMA reporter activity similar to control cells (Fig. 2A, lanes 1, 3, 6, 7, and 9). Consistent with the SMA reporter activity, SMA protein expression was reduced in PAC1 cells expressing FGFR1 K562E mutants with an intact FRS2-binding site compared with control cells (Fig. 2B, lanes 1, 2, 4, 5, and 8). SMA protein levels in PAC1 cells expressing FGFR1 K562E mutants lacking the FRS2-binding site were similar to controls (Fig. 2B, lanes 1, 3, 6, 7, and 9). Real time quantitative PCR shows that SMA mRNA levels were deceased in FGFR1 K562E expressing PAC1 cells compared with control cells (Fig. 2C). In contrast, the FRS2-binding site deletion mutant FGFR1 K562E: −FRS2 showed a small increase in SMA mRNA expression relative to control.
FIGURE 2.
Effects of FGFR1 FRS2 deletion mutants on SM α-actin gene expression. A, PAC1 stable cell lines expressing FGFR1 mutants were transiently transfected with SM α-actin luciferase reporter and pRL-TK Renilla luciferase. The cells were serum-starved overnight and analyzed for luciferase activity. The results are displayed as the means ± S.D. ***, p < 0.001, as compared with the control (ANOVA test; n = 3). RLU, relative luciferase units. B, PAC1 stable cell lines were analyzed for SM α-actin expression by immunoblotting. Quantification of SM α-actin and normalization were described under “Experimental Procedures.” The values were reported as the means ± S.D. and compared with the control (ANOVA test; n = 3). C, PAC1 stable cell lines were analyzed for SM α-actin mRNA expression by real time quantitative PCR. The data were normalized to Hprt and were expressed as the fold difference from the vector control cells. The results are representative of three separate experiments.
We took advantage of several FGFR1 mutants that have graded levels of tyrosine kinase activity to investigate the role of intensity of tyrosine kinase activity on SMA expression (23). FGFR1 with different mutations exhibited graded inhibition of SMA reporter activity (supplemental Fig. S1). These effects were also observed in the rat VSMC line A7r5. FGFR1 K562E down-regulated both SMA and SM22α promoter activity in a manner similar to PAC1 cells (supplemental Fig. S1). Immunoblot analysis showed that SMA protein levels were decreased in FGFR1 K562E-expressing A7r5 cells compared with control A7r5 cells (supplemental Fig. S1). Thus, these data confirm a role for FGFR1 in regulating VSMC marker gene expression.
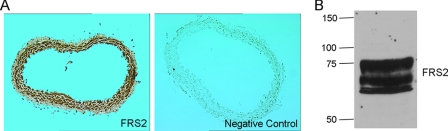
Because FRS2 plays an important role in the regulation of FGFR1-mediated down-regulation of SM marker gene expression, we next looked at FRS2 expression pattern within blood vessel wall in vivo. Immunohistochemistry (Fig. 3A) and immunoblotting of mouse aorta (Fig. 3B) reveal that FRS2 is abundant in the vessel wall and likely a mediator of normal and pathological signaling via FGFR1.
FIGURE 3.
FRS2 expression in vessels. A, immunohistochemical staining of FRS2 in mouse aorta sections. B, expression of FRS2 in mouse aorta extracts. Three aortas from 2-month-old FVB mice were isolated. Tissues were lysed and subjected to immunoblot analysis. The molecular masses are indicated on the left in kilodaltons. The results are representative of three separate experiments.
FGF2 and PDGF-BB Have Synergistic Effects on Smooth Muscle Marker Gene Expression
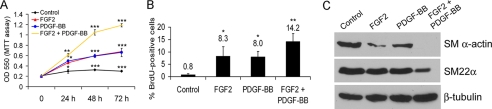
The loss of smooth muscle contractile protein expression is a prerequisite for acquisition of a proliferative and migratory phenotype, and PDGF-BB plays a role in this dedifferentiation (1, 27). To investigate whether FGF2 and PDGF-BB signaling pathways interact to regulate smooth muscle contractile protein expression, we performed a time course study that revealed that FGF2 or PDGF-BB stimulation increased cell proliferation (Fig. 4, A and B) and decreased SMA protein expression in primary BVSMC (Fig. 4C). Together, FGF2 and PDGF-BB stimulation have a synergistic effect on both increased cell proliferation and down-regulation of VSMC marker gene expression (Fig. 4).
FIGURE 4.
Effects of FGF2 and PDGF-BB on primary BVSMC cell proliferation and VSMC marker gene expression. A, BVSMC were serum-starved overnight and treated with growth factor(s) (20 ng/ml FGF2, 50 ng/ml PDGF-BB) or left untreated for the indicated time points. Cell proliferation was determined using the MTT assay. Statistical analysis was performed by Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001, compared with each time point of the control. B, BVSMC were serum-starved overnight and treated with growth factor(s) (20 ng/ml FGF2, 50 ng/ml PDGF-BB) or left untreated for 24 h. The cells were pulse-labeled with 10 μm BrdUrd for 1 h, and the BrdUrd-positive cells were quantified. Statistical analysis was performed by Student's t test. *, p < 0.05; **, p < 0.01, compared with the control. C, BVSMC were serum-starved overnight, stimulated with growth factor(s) (20 ng/ml FGF2, 50 ng/ml PDGF-BB) or left untreated for 72 h, and analyzed for smooth muscle marker gene expression by immunoblotting. β-Tubulin served as a loading control. All of the results are representative of three separate experiments.
PDGF-BB Induces FGFR1 Tyrosine Phosphorylation
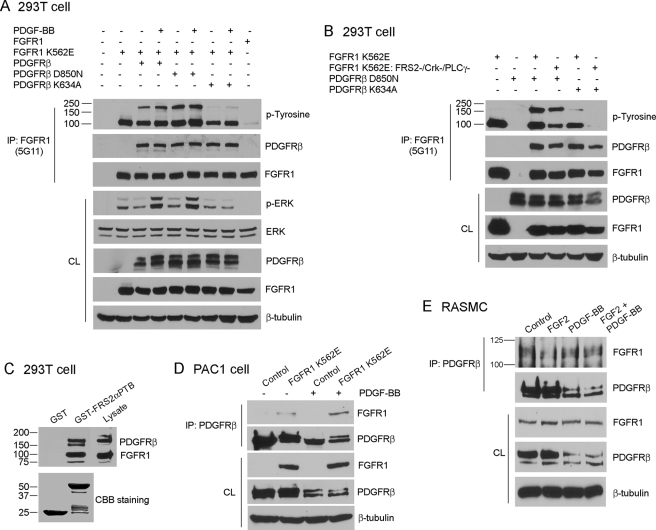
To gain insight into the synergistic effect of PDGF-BB and FGF2 on VSMC marker gene expression and to determine the potential for cross-talk of PDGFR and FGFR1 in this regard, we stimulated BVSMC with either FGF2 or PDGF-BB. Immunoblot analysis reveals that stimulation of BVSMC with FGF2 results in tyrosine phosphorylation of FGFR1 and FRS2 as expected (Fig. 5A). In addition, stimulation with PDGF-BB resulted in FGFR1 and FRS2 (Tyr196) tyrosine phosphorylation to a similar extent as FGF2. Because PDGFRβ is the predominant form of PDGFR expressed in VSMC (Fig. 5B) and because Src is involved in PDGFR cross-talk with other receptors (28, 29), we wanted to determine whether Src-mediated transactivation of FGFR1 by PDGFRβ. We transfected 293T cells with a constitutively active form of PDGFRβ (PDGFRβ D850N) together with a kinase deficient FGFR1 (FGFR1 K562E: −FRS2/−Crk/−PLCγ) and either a constitutively activated form of Src (CA-Src) or a dominant negative form of Src (DN-Src). The results show that PDGFRβ D850N induced phosphorylation of FGFR1 independent of Src because DN-Src had no effect on PDGFRβ-mediated tyrosine phosphorylation of FGFR1, and CA-Src alone did not result in phosphorylation of FGFR1 (Fig. 5C).
FIGURE 5.
PDGF-BB transactivates FGFR1. A, BVSMC were serum-starved overnight, stimulated with growth factors (20 ng/ml FGF2, 50 ng/ml PDGF-BB) or left untreated for 24 h, and analyzed for FGFR1 and FRS2 phosphorylation by immunoblotting. β-Tubulin served as a loading control. B, VSMC were analyzed for expression of PDGFRs and FGFRs by immunoblotting. C, 293T cells were transiently transfected with different constructs as indicated. After serum starvation overnight, FGFR1 was immunoprecipitated (IP) and subjected to immunoblot analysis. The amount of transfected proteins in the cell lysate (CL) were also analyzed by immunoblotting. β-Tubulin served as a loading control. All of the results are representative of three separate experiments.
FGFR1 and PDGFRβ Form a Multiprotein Complex
To determine whether PDGFRβ mediates tyrosine phosphorylation of FGFR1 by forming a multiprotein complex, we transfected 293T cells with FGFR1 and PDGFRβ mutants, followed by stimulation with PDGF-BB and immunoprecipitation with FGFR1 antibodies. The results show that PDGFRβ co-immunoprecipitates with FGFR1 and that PDGFRβ kinase activity is dispensable for this association (Fig. 6A). In addition, FGFR1 kinase activity was dispensable for the association with PDGFRβ (Fig. 6B). However, whereas an activated FGFR1 was unable to induce tyrosine phosphorylation of a kinase-deficient PDGFRβ (Fig. 6A), an activated PDGFRβ was able to induce tyrosine phosphorylation of a tyrosine kinase-deficient FGFR1 (FGFR1 K562E: −FRS2/−Crk/−PLCγ) (Fig. 6B). These data suggest that PDGFRβ may transactivate FGFR1 through formation of a multiprotein complex.
FIGURE 6.
FGFR1 and PDGFRβ form a complex. A and B, 293T cells were transiently transfected with different constructs as indicated. After serum starvation overnight, FGFR1 was immunoprecipitated (IP) and subjected to immunoblot analysis. The amount of transfected proteins in the cell lysate (CL) were also analyzed by immunoblotting. β-Tubulin served as a loading control in all experiments. C, 293T cells were transiently transfected with FGFR1 and PDGFRβ constructs. After serum starvation overnight, the cells were lysed, and the cell lysates were precleared with glutathione-Sepharose alone before incubation with GST or GST-FRS2αPTB fusion proteins bound to glutathione-Sepharose. The precipitates were subjected to immunoblot analysis. D, PAC1 stable cell lines were stimulated with 50 ng/ml PDGF-BB or left untreated for 24 h after 0.5% FBS starvation overnight. PDGFRβ was immunoprecipitated and subjected to immunoblot analysis. β-Tubulin served as a loading control. E, RASMC were stimulated with growth factor(s) (20 ng/ml FGF2, 50 ng/ml PDGF-BB) or left untreated for 24 h after 0.5% FBS starvation overnight. PDGFRβ was immunoprecipitated and subjected to immunoblot analysis. β-Tubulin served as a loading control. The molecular masses are indicated on the left in kilodaltons. All of the results are representative of three separate experiments.
To gain additional insight into PDGFRβ-FGFR1 complex formation, we co-transfected 293T cells with wild-type FGFR1 and PDGFRβ followed by pulldown assays using either GST or GST-FRS2αPTB. GST-FRS2αPTB was able to pulldown both PDGFRβ and FGFR1, further supporting that these two receptors form a multiprotein complex (Fig. 6C). To determine whether FGFR1 and PDGFRβ form a complex in VSMC, FGFR1 K562E-expressing PAC1 cells or primary RASMC were treated with vehicle or growth factor(s) for 24 h, followed by immunoprecipitation with antibodies to PDGFRβ. Results show that FGFR1 and PDGFRβ form a complex in both PAC1 cells and RASMC (Fig. 6, D and E).
The Extracellular Domain and Intracellular Domain of FGFR1 and PDGFRβ Interact
To determine the domains of FGFR1 and PDGFRβ responsible for complex formation, we used two truncated FGFR1 mutant receptors (DN-FGFR1 and FGFR1 IC) and PDGFRβ chimera receptor (fms/PDGFRβ) (supplemental Fig. S2). Our results showed that DN-FGFR1 or FGFR1 IC co-immunoprecipitates with PDGFRβ, and the fms/PDGFRβ chimera co-immunoprecipitate with FGFR1 (supplemental Fig. S2). These results indicate that FGFR1 and PDGFRβ complex formation is mediated through their extracellular and intracellular domains.
FRS2 Potentiates PDGFRβ-mediated ERK Signaling through FGFR1
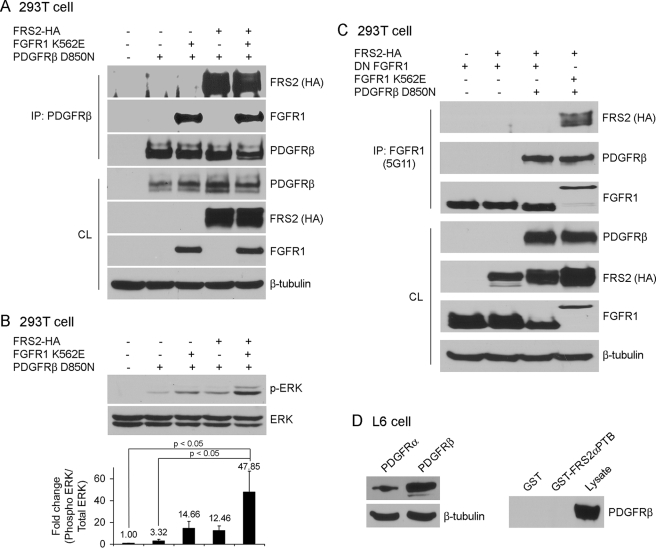
Because FRS2 binds to FGFR1 and FRS2 was tyrosine-phosphorylated in response to PDGF-BB in BVSMC, we examined whether FRS2 participates in PDGFRβ signaling via complex formation with FGFR1. We transfected 293T cells with FGFR1 K562E, PDGFRβ D850N, and HA-tagged FRS2 as indicated and then immunoprecipitated with PDGFRβ antibodies (Fig. 7A). FRS2-HA was immunoprecipitated with PDGFRβ antibodies efficiently in the absence or presence of transfected FGFR1 K562E. Furthermore, PDGFRβ D850N only weakly induced ERK phosphorylation, and this was increased in the presence of FGFR1 K562E (Fig. 7B). However, the greatest induction of ERK phosphorylation was seen when FRS2 was co-transfected with PDGFRβ D850N and FGFR1 K562E, suggesting that this tripartite complex functions synergistically to activate ERK signaling. The association of FRS2 with PDGFRβ D850N suggests that either FRS2 is able to associate with PDGFRβ directly or that FRS2-HA is associated with endogenous FGFR1 in 293T cells, which then associates with PDGFRβ D850N. To distinguish between these two possibilities, we transfected 293T with FRS2-HA, PDGFRβ D850N, and dominant negative FGFR1 (DN-FGFR1) in various combinations. Immunoprecipitation with FGFR1 antibodies shows that PDGFRβ D850N associated with DN-FGFR1; however, FRS2-HA was not present in the complex (Fig. 7C). FRS2 does not associate with DN-FGFR1; therefore these data indicate that FRS2 association with PDGFRβ is likely through its association with FGFR1. To verify these results, we performed GST pulldown assays on L6 myoblasts that express PDGFRβ but lack expression of FGFRs (30, 31). In this FGFR-deficient context, GST-FRS2αPTB failed to associate with PDGFRβ (Fig. 7D), further supporting the idea that the association of FRS2 with PDGFRβ is via a complex with FGFR1.
FIGURE 7.
FRS2 participates in PDGFRβ signaling. A–C, 293T cells were transiently transfected with different constructs as indicated. After serum starvation overnight, PDGFRβ (A) or FGFR1 (C) was immunoprecipitated (IP) and subjected to immunoblot analysis. The amount of transfected proteins in the cell lysates (CL) were also analyzed by immunoblotting. B, quantification of ERK phosphorylation and normalization were described under “Experimental Procedures.” The values were reported as the means ± S.D., and compared with the control (ANOVA test; n = 3). D, left panel, L6 myoblasts were analyzed for PDGFRα and PDGFRβ expression by immunoblotting. Right panel, L6 myoblasts were lysed, and the cell lysates were precleaned with glutathione-Sepharose alone before incubation with GST or GST-FRS2αPTB fusion proteins bound to glutathione-Sepharose. The precipitates were subjected to immunoblot analysis. The input of GST and GST-FRS2αPTB construct levels were the same as shown in Fig. 6C. All of the results are representative of three separate experiments.
RNA Interference-mediated Knockdown of FRS2 Reverses PDGF-BB-mediated Down-regulation of VSMC Marker Gene Expression
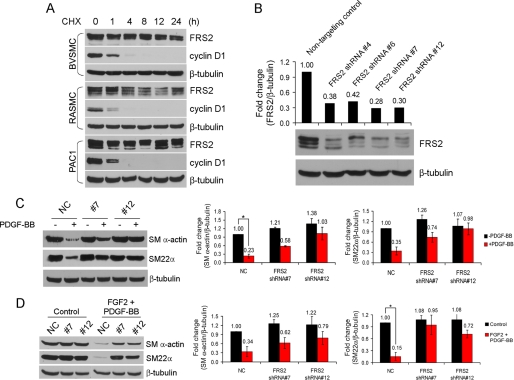
To investigate the role of FRS2 in PDGF-BB-mediated inhibition of VSMC marker gene expression, shRNA was used to knockdown FRS2 expression. Because FRS2 has a long half-life (Fig. 8A), we used retroviral vectors to stably knockdown FRS2 in PAC1 cells. Four different shRNAs targeting different regions of the rat FRS2 coding region were tested. Individually, these shRNAs achieved ∼50% knockdown (data not shown); therefore PAC1 cells were transfected with two sets of shRNAs and individual colonies selected after antibiotic selection. This approach resulted in two clones of PAC1 cells with knockdown of FRS2 of ∼70% (Fig. 8B).
FIGURE 8.
Effects of FRS2 knockdown on growth factor-mediated down-regulation of smooth muscle marker gene expression. A, VSMC were treated with 25 μg/ml cycloheximide (CHX) for the indicated time points and analyzed for FRS2 and cyclin D1 expression by immunoblotting. β-Tubulin served as a loading control. B, PAC1 FRS2 knockdown and nontargeting control cells were analyzed for FRS2 expression by immunoblotting. The amount of FRS2 protein was quantified by ImageQuant software and normalized to β-tubulin. C and D, PAC1 cells were serum-starved overnight, treated with growth factors (20 ng/ml FGF2, 50 ng/ml PDGF-BB) or left untreated for 72 h, and analyzed for smooth muscle marker gene expression by immunoblotting. NC, nontargeting control. Quantification of SM α-actin and SM22α and normalization were described under “Experimental Procedures.” The values are reported as the means ± S.D. and compared with the control (ANOVA test; n = 3). *, p < 0.05.
Control and FRS2 knockdown PAC1 cells were treated with vehicle or growth factor(s) for 72 h. Immunoblotting of lysates from control cells shows that the SMA and SM22α were down-regulated in response to FGF2 and PDGF-BB stimulation of PAC1 cells expressing a nontargeting control vector (Fig. 8, C and D). In contrast, SMA and SM22α expression was essentially unaffected in PAC1 cells where FRS2 expression was reduced by shRNA. Together, these data indicate that FRS2 is a critical mediator of PDGF-BB- and FGF2-mediated down-regulation of VSMC marker gene expression and that complex formation between PDGFR, FGFR1, and FRS2 is likely important to this regulation.
FRS2 Knockdown Does Not Affect PDGF-BB-mediated VSMC Growth
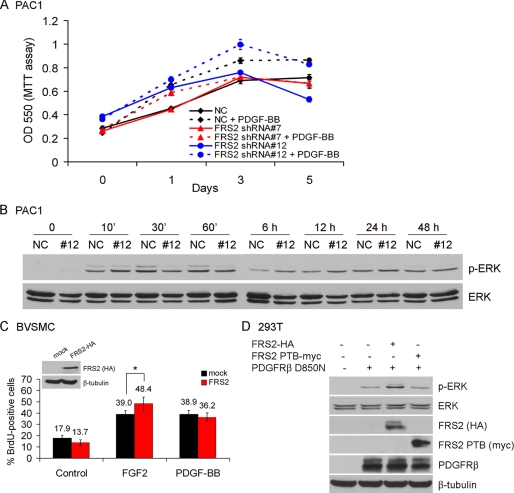
PDGF-BB transactivates FGFR to induce proliferation of VSMC by inducing the release of endogenous FGF2 (9). Because we have clearly demonstrated that PDGFRβ transactivates FGFR1 and FRS2 and that FRS2 plays an essential role in the down-regulation of VSMC marker gene expression, we wanted to determine whether knockdown of FRS2 diminished the proliferative response of VSMC to PDGF-BB. Proliferation assays show that there is little difference in the proliferation of PAC1 cells in response to PDGF-BB with FRS2 stably knocked down and PAC1 cells expressing a nontargeting control (Fig. 9A). In support of this observation, there was little difference in the sustained activation of ERK in response to PDGF-BB between PAC1 cells where FRS2 was knocked down and control PAC1 cells (Fig. 9B). The results presented in Fig. 7B indicate that PDGFRβ, FGFR1, and FRS2 synergize to induce ERK activation. However, the role of FRS2 in PDGF-BB-mediated cell proliferation was not addressed. Therefore, we expressed HA-tagged FRS2 in BVSMC, stimulated with FGF2 or PDGF-BB for 24 h, and quantified the cell proliferation event by BrdUrd labeling (Fig. 9C). We found that FGF2 but not PDGF-BB increased S phase cells in the presence of FRS2 compared with the control. In addition, the dominant negative form of FRS2 (FRS2αPTB) has no effect on ERK activation by PDGFRβ-D850N (Fig. 9D). Taken together, these results indicate that FRS2 is not important in PDGFRβ-mediated ERK phosphorylation and cell proliferation. This is in contrast to the requirement for FRS2 binding to FGFR1 in PDGF-BB-mediated down-regulation of smooth muscle marker gene expression.
FIGURE 9.
Effects of FRS2 knockdown on PDGF-BB-mediated VSMC proliferation. A, PAC1 cells were serum-starved overnight and treated with 100 ng/ml PDGF-BB or left untreated for the indicated time points. The cell proliferation rate was determined using MTT assay. B, PAC1 cells were serum-starved overnight, stimulated with 100 ng/ml PDGF-BB for the indicated time points, and analyzed for ERK activation by immunoblot. The level of total ERK served as a loading control. C, BVSMC were transiently transfected with FRS2-HA construct. After serum starvation overnight, the cells were stimulated with growth factor (20 ng/ml FGF2, 50 ng/ml PDGF-BB) or left untreated for 24 h, following a 60-min pulse of BrdUrd labeling. BrdUrd was visualized by immunofluorescence staining and quantified by cell counting. *, p < 0.05, as compared with the nontransfected control cells treated with FGF2 (Student's t test). D, 293T cells were transiently transfected with different constructs as indicated. After serum starvation overnight, the cell lysates were prepared and subjected to Western blot analysis. The amount of transfected proteins in the cell lysates were also analyzed by immunoblotting. β-Tubulin served as a loading control. All of the results are representative of three separate experiments.
DISCUSSION
The present study has identified a new mechanism by which PDGF-BB acts synergistically with FGF-2 in phenotypic modulation of vascular smooth muscle cells from a contractile toward a synthetic phenotype characterized by a decrease in the expression of the smooth muscle marker genes SMA and SM22α. Evidence for this synergistic interaction includes: (i) FGF2 and PDGF-BB together result in a greater decrease in SMA and SM22α expression in VSMC than either one alone, and this correlates with a synergistic increase in VSMC proliferation; (ii) PDGF-BB induces tyrosine phosphorylation of FGFR1 and FRS2 to a similar extent as FGF2; (iii) PDGFRβ and FGFR1 form a tripartite complex with FRS2, which is required for maximal ERK activation; and (iv) shRNA-mediated knockdown of FRS2 results in abrogation of PDGF-BB-mediated down-regulation of VSMC marker gene expression, while leaving the proliferative response to PDGF-BB intact.
The regulation of proliferation and smooth muscle marker gene expression in VSMC by PDGF-BB has been studied extensively; however, much less is known about the role of FGF2 in these processes. To study the role of FGFR1 signaling in VSMC and to avoid the confounding effects of heparan sulfate proteoglycans on FGF receptor signaling, we developed a constitutively activated FGFR1 with mutations that abolish signaling through specific pathways. These mutants eliminate the need for FGF2 stimulation, which will abrogate FGF-mediated signaling through syndecans (32, 33) and allows dissection of signaling via FGFR1. These constitutively active FGFR1 mutants reveal that the majority of ERK signaling occurs through FGFR1 interaction with FRS2, although low level ERK signaling occurred with FGFR1 K562E: −FRS2. There was strong concordance between FGFR1 mutants that had an intact FRS2-binding site and the down-regulation of smooth muscle marker gene expression. FRS2 also mediates the activation of phosphatidylinositol 3-kinase through the formation of a FRS2-Gab1-PI3 kinase complex (12). Phosphatidylinositol 3-kinase activation leads to activation of a signaling cascade of Akt, mTOR, and S6 kinase, resulting in suppression VSMC marker gene expression (1, 2, 34). Interestingly, in PAC1 cells expressing the mutant FGFR1 K562E, there was a small decrease in Akt phosphorylation that differed little from FGFR1 K562E: −FRS2-expressing PAC1 cells (36). In addition, SMA luciferase reporter assays indicate that a dominant negative Akt was unable to reverse the down-regulation of smooth muscle marker gene expression in FGFR1 K562E PAC1 cells. Together, these data indicate that the major pathway mediating down-regulation of VSMC marker gene expression in FGFR1 K562E expressing PAC1 cells is via the ERK pathway. The ERK pathway also leads S6 kinase activation independent of mTOR (2), likely represents an additional pathway by which smooth muscle marker genes are down-regulated, and will require additional study.
Once we established that FGFR1 signaling via FRS2 results in down-regulation of smooth muscle marker gene expression, we queried whether FGFR1 and PDGFRβ signaling pathways act synergistically to down-regulate VSMC marker gene expression. Treatment of VSMC with either FGF2 or PDGF-BB results in down-regulation of SMA and increased VSMC proliferation. In addition, when added together, FGF2 and PDGF-BB had a synergistic effect on both processes. Furthermore, PDGF-BB stimulation of BVSMC resulted in tyrosine phosphorylation of FGFR1 and FRS2 to the same extent as that of FGF2. Together, these data suggest cross-talk between these two pathways at the level of receptor-receptor interactions. Indeed, FGFR1 and PDGFRβ co-immunoprecipitate, and PDGFRβ is able to induce tyrosine phosphorylation of a FGFR1 kinase-deficient mutant. This tyrosine phosphorylation was neither inhibited by DN-Src nor enhanced by a CA-Src, suggesting that the transactivation of FGFR1 by PDGFRβ is likely direct. This is in contrast to the transactivation of the PDGFRβ in VSMC by G-protein-coupled receptors, which is Src-dependent (29).
We next questioned whether FRS2 was an important component of FGFR1-PDGFRβ cross-talk. In 293T cells PDGFRβ D850N stimulated ERK phosphorylation and was significantly enhanced by the presence FRS2. The presence of FRS2 in PDGFRβ immunoprecipitates could be completely abrogated by co-transfection with a dominant negative FGFR1. Furthermore, in L6 myoblasts that are devoid of detectable FGFR but express PDGFRα and PDGFRβ, GST-FRS2αPTB did not pull down either PDGFR; however, in 293T cells expressing PDGFRβ the GST-FRS2αPTB efficiently pulled down a complex composed of FGFR1 and PDGFRβ, indicating that the association of PDGFRβ with FRS2 is indirect, through association with FGFR1. These data suggest a unique mechanism whereby FRS2 and FGFR1 contribute to the signaling output of PDGF-BB in the down-regulation of VSMC marker gene expression.
Because PDGF-BB and FGF2 down-regulate smooth muscle marker gene expression in a synergistic manner and because FGFR1, PDGFRβ, and FRS2 form a multiprotein complex, we performed shRNA mediated knockdown of FRS2 to determine its contribution to PDGF-BB-mediated down-regulation of SMA and SM22α. We chose to target FRS2 for knockdown because it will likely abrogate signaling through FGFR1, FGFR2, and FGFR3 and eliminate the need to knockdown all FGFRs expressed in VSMC. Stable knockdown of FRS2 in two independent clones of PAC1 cells abrogated PDGF-BB-mediated down-regulation of SMA and SM22α. These data suggest that FGFR1 and FRS2 are major components of PDGF-BB-mediated signaling in the down-regulation of SMA and SM22α. Although we cannot rule out the direct activation of the Akt-mTOR-S6 kinase pathway by PDGFRβ in PAC1 VSMC, our data suggest that FRS2 via FGFR1 is a major regulator of VSMC phenotype and that ERK activation is a component of this regulation.
VSMC proliferation is mediated in part via PDGF-BB-mediated activation of FGFR1 via the release of FGF2 (9). Sustained ERK activation is crucial to VSMC proliferation, and the early phase of ERK activation is mediated by PDGF-BB and is independent of FGF2, whereas the later phase of ERK activation is FGF2-dependent and required for entry into the S phase. Here we show that PDGF-BB-mediated VSMC growth is independent of FRS2 because PDGF-BB-mediated ERK activation and cell growth are unaffected by knockdown of FRS2 or increase in FRS2 expression. This is in contrast to the significant effect knockdown of FRS2 on PDGF-BB-mediated down-regulation of SMA and SM22α. There are some possible explanations for this apparent paradox. First, our knockdown of FRS2 is ∼70%, and the remaining FRS2 protein in PAC1 cells may be sufficient to mediate ERK activation and cell growth by PDGF-BB activation of the PDGFRβ-FGFR1-FRS2 complex but not down-regulation of VSMC marker gene expression. However, this is likely not to be the case in this study. We found that there was no increase in PDGF-BB-mediated cell proliferation in BVSMC transfected with FRS2 compared with PDGF-BB-stimulated nontransfected cells. The second possibility is that PDGF-BB may activate ERK via a secondary mechanism, perhaps through the adaptor protein Shc, which binds Grb2, and activates the Ras-Raf-ERK pathway. Shc signaling to ERK can be activated by PDGF-BB and FGF2, making this a plausible explanation for these differences (35).
In summary, the present study demonstrates that in VSMC, PDGF-BB through a multiprotein complex consisting of PDGFRβ, FGFR1, and FRS2 synergize to down-regulate VSMC marker gene expression and increase VSMC growth. Knockdown of FRS2 abrogated PDGF-BB-mediated down-regulation of SMA and SM22α but had little effect on PDGF-BB-mediated cell growth, suggesting that FRS2 has a unique function in the synergistic interaction between the PDGFRβ and FGFR1. Because PDGF and FGF2 signaling plays a role in vascular diseases such as atherosclerosis and restenosis, this study indicates that FRS2 may be a new target for therapeutic intervention in vascular disease.
Supplementary Material
Acknowledgments
We thank Charles P. Emerson, Jr., for advice and helpful discussions. We thank Carl-Henrik Heldin for reagents. We also thank Norma Albrecht for expert editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-DK073871 (to R. F.), R01-HL53793 (to M. S.), and P20 RR1555 (to R. F.). This work was also supported by American Heart Association Founders Affiliate Predoctoral Fellowship 0715788T (to P.-Y. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- VSMC
- vascular smooth muscle cell(s)
- SMA
- smooth muscle α-actin
- FGF
- fibroblast growth factor
- FGFR
- FGF receptor
- FRS2
- fibroblast growth factor receptor substrate 2
- PDGF
- platelet-derived growth factor
- ERK
- extracellular signal-regulated kinase
- mTOR
- mammalian target of rapamycin
- BVSMC
- bovine aortic VSMC
- RASMC
- rat aortic VSMC
- FBS
- fetal bovine serum
- GST
- glutathione S-transferase
- PLC
- phospholipase C
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin
- MAPK
- mitogen-activated protein kinase
- ANOVA
- analysis of variance
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- BrdUrd
- bromodeoxyuridine
- shRNA
- small hairpin RNA.
REFERENCES
- 1.Owens G. K. ( 2007) Novartis Found. Symp. 283, 174– 193 [DOI] [PubMed] [Google Scholar]
- 2.Rzucidlo E. M., Martin K. A., Powell R. J. ( 2007) J. Vasc. Surg. 45, ( Suppl. A) 25– 32 [DOI] [PubMed] [Google Scholar]
- 3.Millette E., Rauch B. H., Kenagy R. D., Daum G., Clowes A. W. ( 2006) Trends Cardiovasc. Med. 16, 25– 28 [DOI] [PubMed] [Google Scholar]
- 4.Brogi E., Winkles J. A., Underwood R., Clinton S. K., Alberts G. F., Libby P. ( 1993) J. Clin. Investig. 92, 2408– 2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindner V. ( 1995) Z. Kardiol. 84, ( Suppl. 4) 137– 144 [PubMed] [Google Scholar]
- 6.Lindner V., Reidy M. A. ( 1993) Circ. Res. 73, 589– 595 [DOI] [PubMed] [Google Scholar]
- 7.Lindner V., Reidy M. A. ( 1991) Proc. Natl. Acad. Sci. U. S. A. 88, 3739– 3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrae J., Gallini R., Betsholtz C. ( 2008) Genes Dev. 22, 1276– 1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millette E., Rauch B. H., Defawe O., Kenagy R. D., Daum G., Clowes A. W. ( 2005) Circ. Res. 96, 172– 179 [DOI] [PubMed] [Google Scholar]
- 10.Thisse B., Thisse C. ( 2005) Dev. Biol. 287, 390– 402 [DOI] [PubMed] [Google Scholar]
- 11.Ornitz D. M. ( 2000) Bioessays 22, 108– 112 [DOI] [PubMed] [Google Scholar]
- 12.Ong S. H., Hadari Y. R., Gotoh N., Guy G. R., Schlessinger J., Lax I. ( 2001) Proc. Natl. Acad. Sci. U. S. A. 98, 6074– 6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ong S. H., Guy G. R., Hadari Y. R., Laks S., Gotoh N., Schlessinger J., Lax I. ( 2000) Mol. Cell. Biol. 20, 979– 989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgar H. R., Burns H. D., Elsden J. L., Lalioti M. D., Heath J. K. ( 2002) J. Biol. Chem. 277, 4018– 4023 [DOI] [PubMed] [Google Scholar]
- 15.Xu H., Lee K. W., Goldfarb M. ( 1998) J. Biol. Chem. 273, 17987– 17990 [DOI] [PubMed] [Google Scholar]
- 16.Firulli A. B., Han D., Kelly-Roloff L., Koteliansky V. E., Schwartz S. M., Olson E. N., Miano J. M. ( 1998) In Vitro Cell Dev. Biol. Anim. 34, 217– 226 [DOI] [PubMed] [Google Scholar]
- 17.Yoshida T., Sinha S., Dandre F., Wamhoff B. R., Hoofnagle M. H., Kremer B. E., Wang D. Z., Olson E. N., Owens G. K. ( 2003) Circ. Res. 92, 856– 864 [DOI] [PubMed] [Google Scholar]
- 18.Chiara F., Goumans M. J., Forsberg H., Ahgren A., Rasola A., Aspenstrom P., Wernstedt C., Hellberg C., Heldin C. H., Heuchel R. ( 2004) J. Biol. Chem. 279, 42516– 42527 [DOI] [PubMed] [Google Scholar]
- 19.Lundgren T. K., Scott R. P., Smith M., Pawson T., Ernfors P. ( 2006) J. Biol. Chem. 281, 29886– 29896 [DOI] [PubMed] [Google Scholar]
- 20.Sachsenmaier C., Sadowski H. B., Cooper J. A. ( 1999) Oncogene 18, 3583– 3592 [DOI] [PubMed] [Google Scholar]
- 21.Larsson H., Klint P., Landgren E., Claesson-Welsh L. ( 1999) J. Biol. Chem. 274, 25726– 25734 [DOI] [PubMed] [Google Scholar]
- 22.Mohammadi M., Dionne C. A., Li W., Li N., Spivak T., Honegger A. M., Jaye M., Schlessinger J. ( 1992) Nature 358, 681– 684 [DOI] [PubMed] [Google Scholar]
- 23.Neilson K. M., Friesel R. ( 1996) J. Biol. Chem. 271, 25049– 25057 [DOI] [PubMed] [Google Scholar]
- 24.Mood K., Friesel R., Daar I. O. ( 2002) J. Biol. Chem. 277, 33196– 33204 [DOI] [PubMed] [Google Scholar]
- 25.Yang X., Webster J. B., Kovalenko D., Nadeau R. J., Zubanova O., Chen P. Y., Friesel R. ( 2006) Calcif. Tissue Int. 78, 233– 240 [DOI] [PubMed] [Google Scholar]
- 26.Mack C. P., Owens G. K. ( 1999) Circ. Res. 84, 852– 861 [DOI] [PubMed] [Google Scholar]
- 27.Holycross B. J., Blank R. S., Thompson M. M., Peach M. J., Owens G. K. ( 1992) Circ. Res. 71, 1525– 1532 [DOI] [PubMed] [Google Scholar]
- 28.Saito Y., Haendeler J., Hojo Y., Yamamoto K., Berk B. C. ( 2001) Mol. Cell. Biol. 21, 6387– 6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanimoto T., Lungu A. O., Berk B. C. ( 2004) Circ. Res. 94, 1050– 1058 [DOI] [PubMed] [Google Scholar]
- 30.Vainikka S., Joukov V., Wennstrom S., Bergman M., Pelicci P. G., Alitalo K. ( 1994) J. Biol. Chem. 269, 18320– 18326 [PubMed] [Google Scholar]
- 31.Wang J. K., Gao G., Goldfarb M. ( 1994) Mol. Cell. Biol. 14, 181– 188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen B. L., Filla M. S., Rapraeger A. C. ( 2001) J. Cell Biol. 155, 845– 858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rauch B. H., Millette E., Kenagy R. D., Daum G., Fischer J. W., Clowes A. W. ( 2005) J. Biol. Chem. 280, 17507– 17511 [DOI] [PubMed] [Google Scholar]
- 34.Muto A., Fitzgerald T. N., Pimiento J. M., Maloney S. P., Teso D., Paszkowiak J. J., Westvik T. S., Kudo F. A., Nishibe T., Dardik A. ( 2007) J. Vasc. Surg. 45, ( Suppl. A) 15– 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamin C. W., Jones D. A. ( 1994) J. Biol. Chem. 269, 30911– 30916 [PubMed] [Google Scholar]
- 36.Chen P. Y., Friesel R. ( 2009) Biochem. Biophys. Res. Commun. 382, 424– 429 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.