Abstract
Discrete chromatin domains (ChrD), containing an average of ~1Mbp DNA, represent the basic structural units for the regulation of DNA organization and replication in situ. In this study a bio-computational approach is employed to simultaneously measure the translational motion of large populations of ChrD in the cell nucleus of living cells. Both movement and configurational changes are strikingly higher in early S- phase replicating ChrD compared to those that replicate in mid and late S-phase. The chromatin dynamics was not sensitive to transcription inhibition by α-amanitin, but was significantly reduced by actinomycin D treatment. Since a majority of active genes replicate in early S-phase, our results suggest a correlation between levels of chromatin dynamics and the transcriptionally active state of chromatin. Analysis of ChrD co-localization with transcription sites and cDNA with ChrD and transcription sites further supports this proposal.
Keywords: Nuclear Organization, Chromatin Dynamics, Chromatin Domains, DNA Replication, cDNA
Introduction
Microscopy studies of living cells have resulted in a remarkable breakthrough in our understanding of nuclear organization. Earlier views of chromatin as a static structure are being replaced with dynamic models (Fraser and Bickmore 2007; Kumaran, et al. 2008; Lanctot, et al. 2007). The vast majority of investigations that have led to this new dynamic view of chromatin are based on the exogenous introduction and labeling of DNA sequences at a limited number of chromatin sites in the cell nucleus using the lac operator/repressor method (Chubb, et al. 2002; Heun, et al. 2001; Li, et al. 1998; Tsukamoto, et al. 2000; Tumbar and Belmont 2001; Verschure, et al. 2005). In contrast, studies of the dynamic properties of the overall native chromatin in the cell nucleus have lagged behind (Abney, et al. 1997).
Replication labeling of DNA enables visualization of large regions of native chromatin in the cell nucleus with the distinctive morphological patterns characteristic of early, mid or late S- phase (Ma, et al. 1998; Nakayasu and Berezney 1989). The newly replicating DNA occurs at numerous chromatin domains (ChrD) containing an average of ~1Mbp of DNA (Jackson and Pombo 1998; Koberna, et al. 2005; Ma, et al. 1998). Upon completion of replication, the pattern of labeled ChrD is maintained as the cells progress through the cell cycle and into subsequent generations (Jackson and Pombo 1998; Ma, et al. 1998). Furthermore, the entire population of ChrD replicates during S-phase of subsequent cell cycles at the same time as the replication sites from which they were derived (Dimitrova and Gilbert 1999; Ma, et al. 1998; Sadoni, et al. 1999), which supports the view that ChrD are basic structural units for DNA organization in situ (Jackson and Pombo 1998). The ChrD are further spatially compartmentalized into higher order domains of precisely regulated replication timing (Berezney 2002; Wei, et al. 1998).
In a pioneering study, replicating DNA was labeled in living cells by microinjection of fluorochrome-coupled nucleotides into the cell nucleus (Zink, et al. 1998). Following several cell divisions, a subset of labeled chromosome territory regions were observed in accordance with segregation of the labeled chromosomes by semi-conservative replication (Zink, et al. 1998). Within each chromosome territory region, numerous sub-chromosomal foci were detected that corresponded to the ~1Mbp ChrD identified in fixed cells (Ma, et al. 1998). Time lapse observations further revealed constrained positional transformations in the labeled chromosome territory regions (Zink, et al. 1998). Surprisingly, only limited studies involving relatively long time lapse intervals (e.g., 20 min or longer) have been performed on the mobility properties of the ChrD visualized with this in vivo approach (Bornfleth, et al. 1999; Edelmann, et al. 2001; Zink, et al. 1998) and the relationship between native chromatin dynamics and its function has yet to be established.
With this in mind, we investigated the dynamics properties of chromatin associated with active genes that replicate in early S-phase compared to predominantly inactive chromatin that replicates in mid and late S-phase using time-lapse intervals as short as 3 min for collection of 3-D image sets. We report that early S- labeled chromatin is significantly more dynamic than chromatin that replicates in mid or late S-phase. This higher level dynamics of early S-replicated chromatin is not inhibited by α-amanitin treatment but is significantly reduced with actinomycin D. We, therefore, propose that chromatin motion is not directly dependent on ongoing transcription. Instead, a high level of dynamics may be characteristic of the transcriptionally competent state of native chromatin and a crucial prerequisite for transcriptional regulation in vivo.
Results
Visualizing Early, Mid and Late S-phase Replicated Chromatin in Live Cells
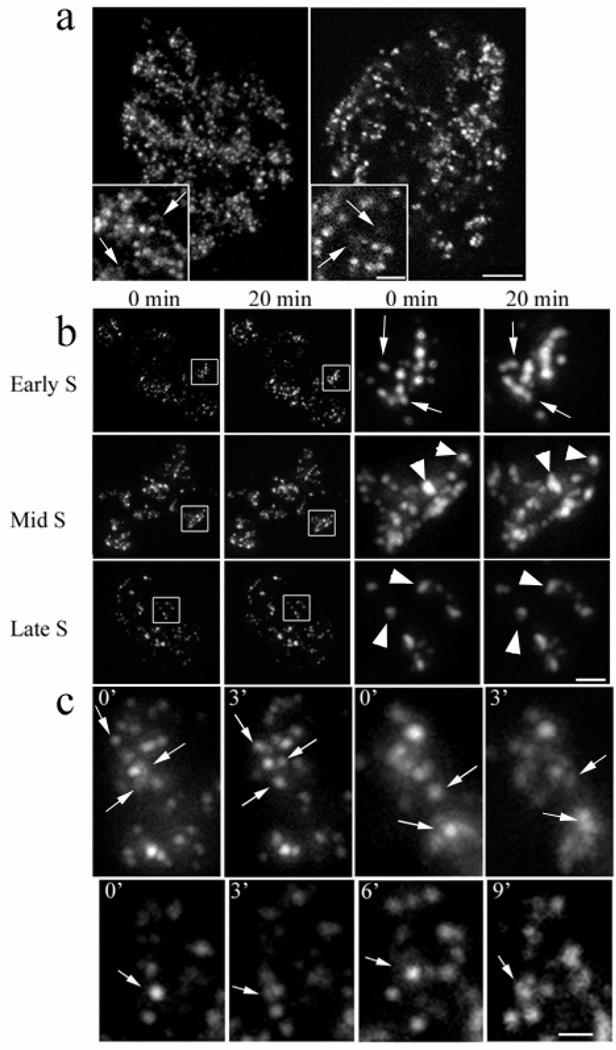
Exponentially growing HeLa cells, synchronized with a single aphidicolin block or non-synchronized in control cells, were microinjected with fluorochrome coupled nucleotides: Cy3- or Alexa 488-dUTP. Incorporation of the microinjected nucleotides into numerous replication sites was observed along with the characteristic replication patterns of early- mid- and late-S-phase (Nakayasu and Berezney 1989). As predicted by semiconservative replication, several days after microinjection, the fluorescent signal was confined to a few segregated areas, largely corresponding to individual chromosome territories (Fig. 1a) (Edelmann, et al. 2001; Zink, et al. 1998). Discrete ChrD of 0.3–0.5 µm in diameter were often surrounded by a more diffuse and less intense signal (Fig. 1a). This was especially clear in early S-phase labeled chromatin. Identical looking foci surrounded by diffusely labeled chromatin were also present in fixed cells labeled in vivo with BrdU (not shown). Time lapse microscopy at 20 min intervals revealed translational motion and configurational changes in the individual ChrD, which were far more pronounced in early S-phase labeled cells than in cells labeled in mid to late S-phase (Fig. 1b).
Fig. 1.
(a) Early S-phase labeled chromatin in living HeLa cells 3 days following microinjection of Cy3- dUTP Left: 2D maximal intensity projection of a deconvolved z-stack of optical sections acquired with an inverted fluorescence microscope. Right: A single confocal section. Labeled chromosome territory regions begin to segregate at this stage (notice unlabeled areas in nuclei). As it is seen on enlarged insets, both images show discrete and more diffuse (arrows) labeled chromatin regions. Scale bars are 5 µm and 2 µm for inset.
(b) Comparison of the dynamics of early, mid and late S-phase replicating chromatin domains (ChrD). HeLa cells were microinjected with fluorochrome-coupled nucleotide during either early, mid or late S-phase. Approximately three generations later the cells were observed at 20 min intervals. The two columns to the left show segregation of the label to individual chromosome territories; white-outlined representative chromosome territories were enlarged and shown in two right columns. Note the striking positional and configuration changes in the early S labeled ChrD (arrows). In contrast, chromatin labeled in mid and late S-phase is virtually static (arrowheads). Scale bars are 5 µm and 1 µm for enlarged regions.
(c) Chromatin domains (ChrD) fluctuate between discrete and diffuse configurations in live cells. HeLa cell labeled in early S-phase was acquired in time-lapse mode and a representative area was cropped. Arrows point to ChrD undergoing transitions between discrete and diffuse organization. In mid- and late S-phase labeled cells these changes are not observed. Scale bar is 1µm.
The dynamics of the ChrD derived from early S was then studied at shorter 3 min intervals where many shifts in position were observed (Fig. 1c). Moreover, while the configuration of chromatin labeled in mid to late S-phase appeared to be predominantly fixed (Fig 1c, bottom panels), early S-phase labeled chromatin exhibited extensive changes in shape (Fig. 1c, top panels). These shape transformations typically involve the reorganization of individual ChrD from discrete to more diffuse domains or the reverse. We have documented numerous cases where fluorescent foci change from discrete to diffuse states and then back in a several min interval (Fig. 1c, bottom panels). Detailed examination revealed that most ChrD derived from early S replicating DNA exhibit at least some degree of shape change. Since projections images of the entire 3-D stack of z-sections were examined (see Materials and Methods), the characteristic changes from discrete to diffuse sites observed in this study cannot be a result of the translational motion of the ChrD in the z-axis plane. We, therefore, conclude that the bulk of early S-phase labeled ChrD are not permanent structural entities, but fluctuate between condensed and more relaxed configurations.
A Bio-computational Approach for Measuring Chromatin Dynamics
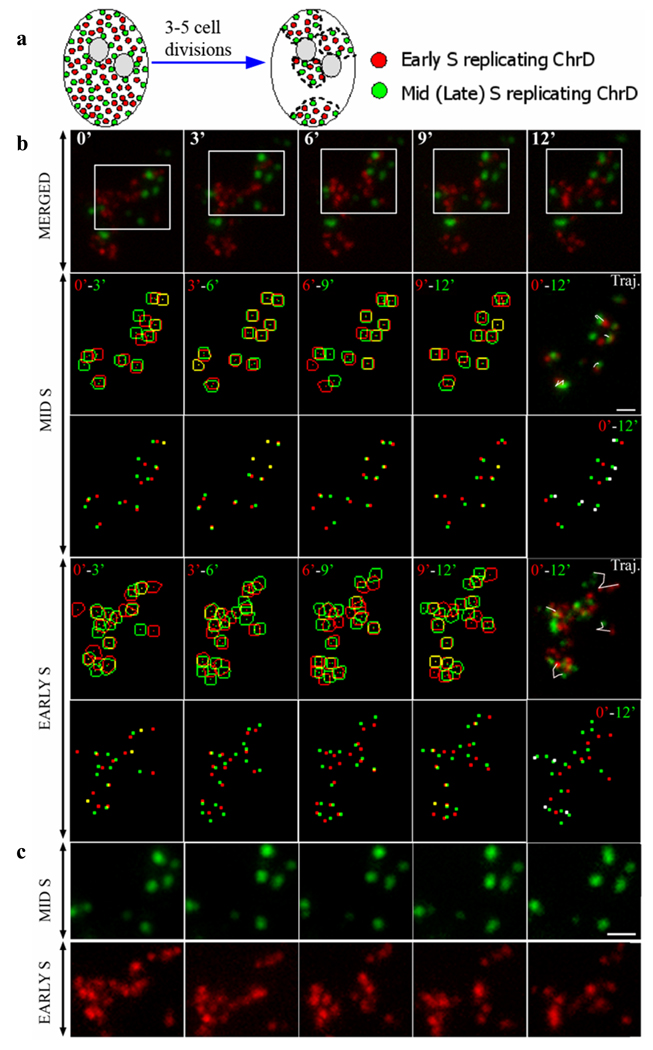
Previous experiments utilizing the lac-operator/ repressor system, showed that the position and configuration of chromatin is altered by transcription activation or through the onset of DNA replication (Li, et al. 1998; Mahy, et al. 2002; Osborne, et al. 2004; Tumbar and Belmont 2001; Volpi, et al. 2000) and that chromatin motion correlates with the intranuclear location of chromatin loci (Chubb, et al. 2002). To offset potential variability in overall chromatin dynamics, we developed a bio-computational approach to measure the linear motion of the discrete ChrD with different replication timing within the same chromosome territory. Double microinjection experiments were performed on cells synchronized at the G1/S border to label replicating chromatin in early S and, following appropriate chase times, mid- or late S-phase (Fig. 2 a). Following cell proliferation for 3–5 days, the fluorescent label is segregated and limited to a few chromosomal territories (Fig. 2a). Sets of z-axis optical sections were acquired every 3 min, with acquisition times not exceeding 10 seconds per stack, deconvolved and transformed into maximum intensity pixel projections. Segmentation of the individual ChrD was performed with spot-based segmentation software developed in our laboratory (Malyavantham, et al. 2008a; Samarabandu, et al. 1995) and corrected for translational motion of the cell and nucleus with image registration software (Jahne 2002). Contoured segments of ChrD at sequential time points were then overlaid individually for each probe (Fig. 2 b). The distance between centers of gravity (centroids) at corresponding time points was measured to quantify positional changes of individual sites. This enabled us to measure the centroid-to-centroid distances in pixels at sub-diffraction resolutions (1 pixel ≈ 0.07 µm) and to directly compare the translational motion of ChrD with different replication timing (i.e., derived from early versus mid- or late-S replicating chromatin) within chromosome territories of the same nucleus (Fig. 2 b, c).
Fig. 2. Chromosome territory analyzed several generations after dual labeling for early and mid S replicating chromatin domains (ChrD).
(a) Schematic illustrating the segregation of replication labeled chromatin. Cell is labeled in two channels for early- (red) and mid- or late- (green) S-phase replicating chromatin. Several generations following labeling, segregation of the label allows visualization of individual chromosome territories.
(b) Chromosome territory exhibiting signal for both early and mid S replicating ChrD (red and green channels, respectively) acquired with 3 min intervals (upper row). Second and fourth row: mid and early S replicating ChrD, respectively were segmented, corrected for cell movement and merged. Earlier time point contours (red), subsequent time point (green). Trajectories are shown for representative early (row 4, right panel) and mid replicating ChrD (row 2, right panel). Third and fifth rows: enlarged centers of gravity (centroids) of contoured early and mid S replicating ChrD at consecutive 3 min intervals were merged as described above. Centroids of early (row 5, right panel) and mid S (row 3, right panel) replicating ChrD at 0 min and 12 min were merged. Corresponding centroids that shifted 4 pixels (~0.3 µm) or less were colored white. Note that over 50% of the mid-S centroids were shifted in white versus < 20% for the early S centroids.
(c) White outlined area on the upper row was enlarged for each channel individually to show pronounced differences in the degree of configuration and position changes of early versus mid S labeled ChrD. Scale bars are 1µm.
Chromatin Dynamics is Correlated with Replication Timing
Significant variability was detected in the average translational motion of ChrD in different cells as well as in different chromosome territories of the same cell (Fig. 3 a, b). Despite these variations, the averaged motion of early S phase labeled ChrD was in every instance significantly higher than their mid- and late S-phase replicated counterparts from the same chromosome territory (Fig. 3 a, b) . Many cases were observed where striking motion changes in early S-labeled ChrD were in close spatial apposition to relatively immobile mid- or late-S-labeled ChrD (Fig. 2 c). Significant differences in the mobility of ChrD based on their replication timing were further emphasized by representative trajectory plots over 3 min intervals (Fig. 2 b, rows two and four, far right) and by visualization of the centroids for the populations of ChrD at each time interval (Fig. 2b). For example, in the mid-S ChrD centroid image merged between 0 and 12 min time points (Fig. 2 b; third row, far right), >50% of the centroids were among the least mobile (within 4 pixels or < 0.3 µm) as indicated by their white color, while this proportion was reduced to < 20% in the corresponding early-S ChrD centroids (Fig 2b, fifth row, far right)
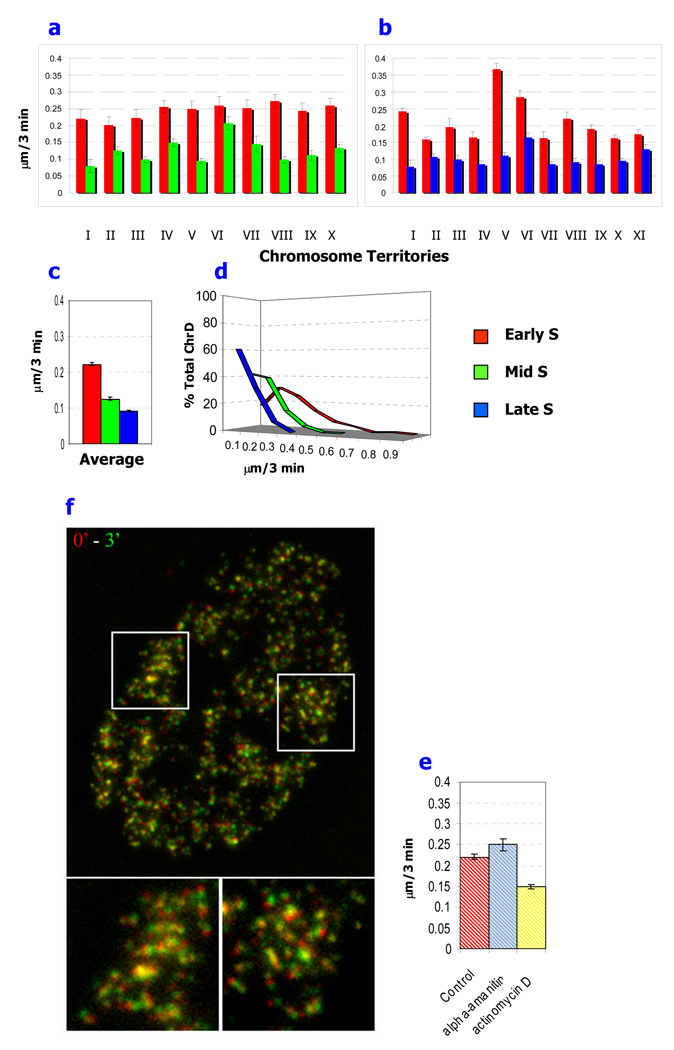
Fig.3. Comparison of the average translation motion of early-, mid- and late S-phase replicating chromatin domains (ChrD).
ChrD were double labeled in (a) early- and mid-S phase or (b) early- and late- S phase. Linear displacements of individual ChrD were measured on five sequential time points and presented as the mean.
(c) The average rate of motion is displayed for the total population of measured early-, mid- and late- S labeled ChrD, respectively. (d) The distribution of translational motion for the total population of measured early-, mid-, and late-S ChrD. Note the much larger population of virtually immobilized ChrD in late S compared to early S.
(e, f) Effect of transcription inhibitors on chromatin dynamics. Cells with ChrD labeled in early S phase were treated with actinomycin D or α-amanitin. Images were acquired with 3 min intervals in 3D. (e) Translational motion of the early S replicated ChrD is significantly reduced by actinomycin D treatment with no significant reduction by α-amanitin. (f) Time lapse analysis for motion following actinomycin D treatment. Images of maximum intensity projections were pseudocolored into red and green channels for the first and subsequent time point, respectively and overlaid. Under these conditions many ChrD are essentially static (yellow color). Two representative areas (white outlines) are enlarged at the bottom panel. Scale bars are 5µm and 1µm. Data are presented as the mean ± SEM (error bars) for a, b, c and e. *- Differences are significant by Anova test; p< 0.05
Linear displacement of labeled ChrD, averaged among individual chromosome territories, ranged from 0.16–0.37 µm/ 3 min, 0.08–0.21 µm/ 3 min and 0.08–0.16 µm/ 3 min for early-, mid- and late S-phase, respectively (Fig 3. a, b). The overall average linear displacements based on 1420 measurements were 0.23 µm/ 3 min, 0.13 µm/ 3 min and 0.08 µm/ 3 min for ChrD labeled in early, mid and late S-phase, respectively (Fig. 3. c). These differences in mobility were statistically significant. Distribution analysis demonstrated that the majority of ChrD labeled in late S-phase were virtually immobile with 60% <0.1 µm/ 3 min, while <1% exceeded 0.3 µm/ 3 min (Fig. 3 d). The mid S-phase labeled ChrD were also relatively immobile with 42% <0.1 µm/ 3 min and <3% exceeding 0.3 µm/ 3 min (Fig. 3 d). In contrast, only 17% of the early S-phase labeled ChrD were immobile (<0.1 µm/ 3 min), and almost one-third had positional changes exceeding 0.3 µm/ 3 min (Fig. 3d). These double labeling results also reinforce our conclusions from single labeling experiments (Fig. 1b, c) that ChrD labeled in early S-phase undergo significantly greater configurational changes compared to their counterparts from mid- or late S-phase and extend them to the level of individual chromosome territories observed in the same cell nucleus (Fig. 2).
Since the majority of actively transcribed genes replicate in early S-phase (Goldman, et al. 1984; Schubeler, et al. 2002), a correlation between active gene expression and chromatin dynamics is possible. As a step toward elucidating the interrelation of gene expression and chromatin dynamics, we incubated replication labeled cells with two transcription inhibitors (α-amanitin and actinomycin D) having different mechanisms of action. Inhibition of transcription by α-amanitin, which interacts with the transcription machinery, did not decrease the dynamics of chromatin in replication labeled cells (Fig. 3 e). Consistent with these results, DRB, a transcription inhibitor which interferes with pre-mRNA elongation, but not directly with chromatin, showed no significant effect on the motility of genomic loci (Chubb, et al. 2002; Thomson, et al. 2004). Similarly, a recent study visualizing chromatin via GFP fused to histone H4 did not indicate any significant influence of this transcription inhibitor on the chromatin dynamics (Wiesmeijer, et al. 2008). Preliminary results in our system have confirmed the lack of effect of DRB on chromatin motion (data not shown).These findings suggest that the dynamic state of chromatin is not a result of active transcription per se. Instead, it may be an epigenetic property characteristic of chromatin poised for active transcription.
Consistent with this view, actinomycin D, which inhibits transcription by intercalation into DNA, resulted in a significant decrease in the motion of the early S-derived ChrD and a dramatic reduction in configuration changes (Fig. 3 e, f). A unique mechanism of action has been proposed for actinomycin D involving the targeting of transcription competent pre-melted DNA conformation (Sobell 1985). We, therefore, propose that structural alterations in the chromatin induced by actinomycin D binding results in a more static chromatin structure (a “frozen state”) that is transcriptionally inactive (see Discussion).
Detecting Gene Expression Outside of Discrete ChrD
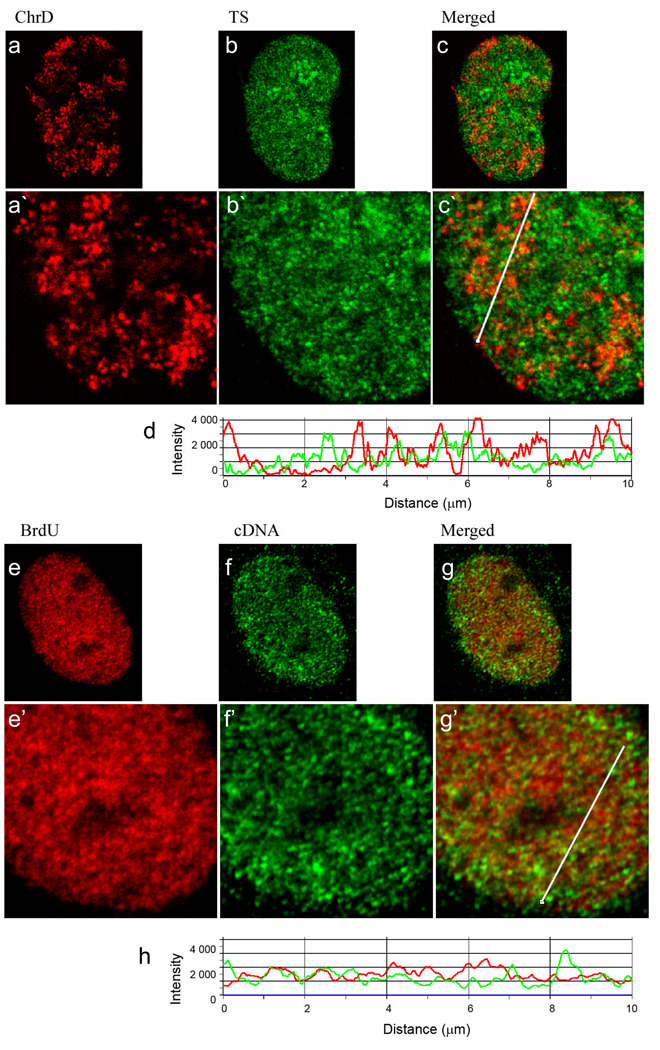
As an initial step in studying the functional significance of chromatin motion and reconfiguration, we examined the localization of ChrD with either the transcription signal or gene-rich chromatin visualized via cDNA probe. Transcription sites (TS) were labeled in vivo by F-U incorporation into nascent RNA. Numerous sites of RNA synthesis were observed. in the nucleoplasm and, under milder fixation conditions, in the nucleolus (see Fig S1 and Materials and Methods). Although TS were very abundant only a very limited overlap of TS with the discrete early S-phase replicated ChrD was detected by direct observation of images as well as by line scan profiles (Fig. 4a–d; Fig. S1). Instead, TS were significantly enriched in more diffuse chromatin regions surrounding the discrete sites (Fig. 4a–d, Fig 1).
Fig. 4. Visualization of the transcription sites, cDNA and early S-labeled chromatin indicates association of transcription with relaxed chromatin surrounding the discrete chromatin domains (ChrD).
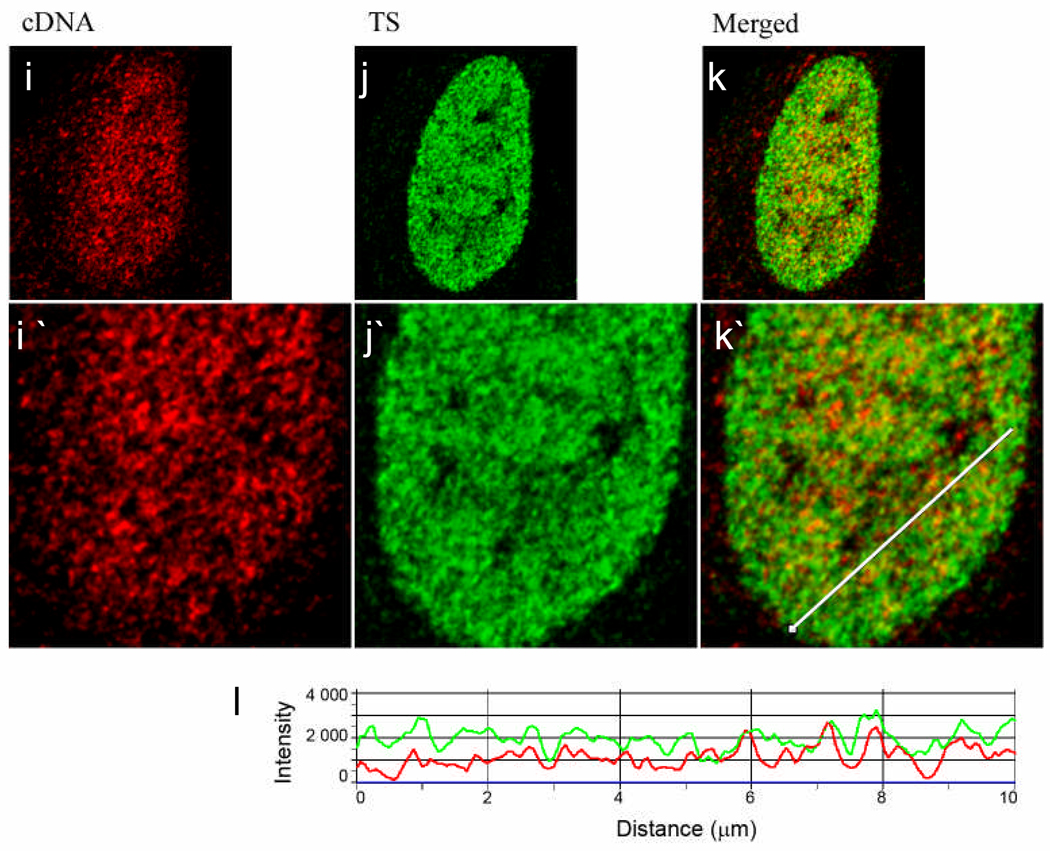
Single confocal optical sections (0.5 µm) are presented. (a) Cells were labeled for early S replicated DNA by microinjection and chased for 3 days (red, left panel) followed by in vivo labeling of transcription sites (Tsukamoto, et al.) with fluorouridine (green, middle panel) (b). (c) Displays the merged images (a) and (b). (d) A line profile through (c) with ChrD in red and TS in green. Red arrowheads point to ChrD that do not overlap with TS. Intensity peaks corresponding to ChrD foci which co-localize with TS are marked by yellow arrowheads. As is demonstrated by the virtual absence of yellow signal in the merged image and the line profile, little colocalization was seen of the TS with the discrete ChrD. Instead, the TS were enriched within the lower intensity, relaxed chromatin found between the discrete ChrD (see also Fig 1). (e,f) Cells were double labeled for ChrD (red, left panel) and chromatin enriched in transcribed sequences with a cDNA FISH probe, (green, middle panel). In this case the ChrD was labeled by in vivo pulsing with BrdU (20 min) followed by a 150 min chase to ensure that all of the labeled ChrD have finished replication (see Ma et al. 1998). (g) Displays the merged images (e) and (f); (h) shows a line profile through (g) with ChrD in red and TS in green. Similar to the results with TS, the majority of ChrD foci do not co-localize with the cDNA. (i, j) Cell was double labeled for cDNA (red, left panel) and TS (green, middle panel). (k) Displays the merged images (i) and (j). (l) A line profile through (k) demonstrates a high degree of association of cDNA intensity peaks with TS peaks. Scale bar is 5 µm. Average Pearson Coefficients calculated for experiments demonstrated in Figure 4 (c, g, kj, i) are~ 0.30, 0.24 and 0.62, respectively; n=15 (see also Fig S1). All lines used for intensity profiles (Fig 4 c, g, k) were artificially thickened to enhance their visibility.
As an approach to label chromatin enriched in genes that are actively transcribed in the overall cell population, a total cDNA probe was prepared from exponentially growing HeLa cells. Numerous studies have shown that cDNA probes effectively label individual genes on chromosomes (Galli-Stauber, et al. 1998; Kamei, et al. 1998; Ohbayashi, et al. 1999; Thomsen, et al. 1998; Wang, et al. 1996; Yamashita, et al. 2000) and a study of cDNA probe generated to individual human chromosomes revealed specific staining across only the chromosome of interest (Rouquier, et al. 1995). We reasoned that cDNA generated to the total poly A+ mRNA would enable labeling of a significant portion of the genes that were either actively transcribing or poised for transcription in the interphase nucleus especially those that produce the more abundant mRNAs. The cDNA fluorescent signal was organized into multiple foci, scattered throughout the nucleoplasm and excluded from the nucleolus (Fig 4). Although ChrD and cDNA foci were present in close proximity, only a small proportion of ChrD co-localized with the cDNA probe, as detected both visually and by line scan profiles (Fig. 4e–h). In contrast, we detected a much higher level of overlap of cDNA with labeled TS (Fig. 4i–l). The degree of co-localization was then quantitated by determination of the Pearson correlation coefficients for each double labeled image set. This value ranges from −1 for completely separated to +1 for perfectly co-localized channels. Consistent with the line profile analysis, the average Pearson coefficient was 0.62 for TS-cDNA images compared to 0.30 and 0.24 for ChrD-TS and ChrD-cDNA images, respectively (n=15 cells for each group; Fig S1).
Taken together these findings support the view that active genes replicated during early S phase unfold for active transcription into relaxed chromatin regions adjacent to the discrete ChrD. The constant configurational transitions that we observe in early S labeled ChrD, may, therefore, reflect a corresponding dynamics of gene expression wherein genes are relocated in or out of discrete ChrD accordingly to their transcriptional status.
Discussion
In this study we have used the nucleotide microinjection approach (Zink, et al. 1998) to investigate the dynamic properties of the ~ 1 mbp chromatin domains (ChrD) (Ma, et al. 1998) in the cell nucleus of live HeLa cells. Following synchronization at the G1/S border and a double injection approach (Fig 1), we have been able to simultaneously measure translational motion and configurational changes in ChrD replicated in early S versus mid- or late-S phase in the chromosome territories of the same nuclei. Our results demonstrate a significantly higher level of translational motion for ChrD which were replicated several generations before in early S-phase compared to mid- and late- S phase counterparts. (Fig 1–Fig 3). In addition, high levels of configurational changes were detected in the early S phase replicated ChrD which were virtually devoid in the mid- and late S- phase replicated ChrD.
Since chromatin replicated in early S-phase is highly enriched in actively transcribed genes (Goldman, et al. 1984; Schubeler, et al. 2002), we further investigated the possible relationship of this dynamic behavior to the transcriptional state of the chromatin. We report that actinomycin D decreases the mobility of the early S-phase replicated ChrD and completely prevents the configurational changes (Fig. 3). Co-localization studies in fixed cells of labeled ChrD versus transcription sites or cDNA labeling revealed that the sites of active chromatin transcription were not localized within the confines of individual ChrD, but rather along the periphery in more diffuse regions of chromatin, where the cDNA probe showed high levels of association with sites of transcription (Fig. 4 and Fig. S1).
A profound difference between the structure organization of gene-rich and gene-poor genome regions has been reported, which correlates with either open, de-condensed or compact chromatin fibers (Gilbert, et al. 2004). This feature of genomic organization is consistent with our findings of striking configurational transformations of the early S replicated ChrD in contrast to mid and late S-phase chromatin, and the localization of the actively transcribed genes in more diffuse regions of the chromatin. Furthermore, the linear motion and configurational transformations of early S replicated ChrD could provide the basis for the long range spatial interactions of genes which have been described in a variety of experimental systems (Brown, et al. 2008; Osborne, et al. 2004; Simonis, et al. 2006; Volpi, et al. 2000) as well as enabling the interaction of the gene sequences with regulatory subnuclear domains (Lawrence and Clemson 2008).
In addition, the dynamic properties of the early S replicated ChrD may be reflecting a constant modulation of gene expression in the cell nucleus. Predisposition to constant motion and configuration changes of the gene-rich ChrD would likely enhance molecular interactions that are productive for transcription regulation. Whether or not chromatin motion is a stochastic or an energy consuming and directional process, a dynamic assembly and disassembly of genes in ChrD and in close proximity to transcription regulatory subnuclear domains (Brown, et al. 2008; Lawrence and Clemson 2008; Osborne, et al. 2004; Thompson, et al. 2003; Xu and Cook 2008) can act as an additional layer of control, modulating the access of chromatin to regulatory binding proteins that affect gene expression (Malyavantham, et al. 2008b; Zaidi, et al. 2007).
Furthermore, the observed configurational changes of the ChrD may correspond to the unfolding or refolding of chromatin loops as they associate or disassociate from transcription factories or other regulatory elements of nuclear architecture (Malyavantham, et al. 2008a,b). Under this scenario, the observed movement of ChrD may enable distal genes from different ChrD to reach a common site where their expression is regulated. We, therefore, propose that chromatin dynamics is a critical factor in epigenetic transcription regulation of the thousands of genes that are active in the cell nucleus of a mammalian cell. In this way, chromatin motion can be a prerequisite requirement for the complex interactions among the multiple components of the gene regulatory machinery.
Materials and Methods
Cell Culture and Drug Treatment
HeLa cells were obtained from ATTC (line CCL2) and were cultured in DMEM (Sigma) supplemented with 5% fetal bovine serum (Gibco), 1% penicillin/streptomycin and 3.7 g/liter NaHCO3 at 37°C in a humidified atmosphere containing 5% CO2. Before microinjection cells were placed on thin glass bottom dishes (World Precision Instruments) or on 12 mm circular cover slips for immunolabeling and FISH experiments. Partial synchronization of the cells (~ 70%) was achieved at the G1/S border by a single aphidicolin block: the cells were incubated in DMEM containing 10 µg/ml of aphidicolin (14 h), washed with PBS and returned to regular media. As indicated, cells were treated with α-amanitin (50 µg/ ml for 5 h) or actinomycin D (0.7µg/ml for 10 min) prior to image acquisition. During the imaging these transcription inhibitors were present in the medium.
Preparation of the total cDNA probe
RNA was isolated from HeLa cells using RNAqueous-4PCR (Ambion, Austin, TX) according to manufacturer’s instructions. Total cDNA was generated by reverse transcription with M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA) using an anchored oligo-dT primer. Degenerate-oligonucleotide-primed PCR was performed with the following conditions 94°C for 3 min, 8 cycles of (94°C for 1 min, 30°C for 1min, 30–72°C for 3 min, 72°C for 2 min), 35 cycles of (94°C for 1 min, 56°C for 1 min, 72°C for 2 min) with the last extension at 72°C for 5 min. DOP-PCR products were purified using a PCR purification column (Qiagen, Valencia, CA). To control product length and incorporate label, a second round of PCR amplification was performed with the addition of 30% biotin14-dCTP and 70% dCTP and an elongation time of 30 sec. Products were verified by gel electrophoresis to range between 200–700 bp.
Microinjection, Immunolabeling and FISH
Microinjection was performed using the Eppendorf FemtoJet microinjector and Eppendorf InjectMan micromanipulator. Observations of the cells were performed on an Olympus IX-70 epifluorescence inverted microscope equipped with a z-motorized stage and a Sensicam CCD camera (Cooke Corporation), controlled by ImagePro Plus (Media Cybernetics). During microinjection and microscopic observation, cells were kept at 37°C in a heat controlled cell cultivation chamber (Olympus). For the labeling of early, mid and late S-phase replicating chromatin, 100 µM Cy3-dUTP (Amersham) and/or Alexa 488-dUTP (Molecular Probes) in PBS were microinjected into each nucleus at 1, 7 and 11 h , respectively after the block removal. In the majority of replicating cells, the observed labeling pattern (early, mid or late S-phase) was in agreement with the synchronization protocol (Nakayasu and Berezney 1989). Cells showing replication patterns different from that predicted by the synchronization protocol were excluded from observation. In control experiments, no differences were observed in labeling patterns between synchronized and non-synchronized cells. Cells were observed three to five days after the injection and no significant deviations were observed in the growth rate of microinjection-labeled cells.
Replication and transcription sites were labeled as previously described with BrdU and FU (Ma, et al. 1998; Malyavantham, et al. 2008a) For cDNA visualization, cells were fixed in 4% PFA/PBS for 15 min, permeabilized for 10 min in 0.5% Triton X-100 and denatured in 70 % formamide at 75°C for 8 min. Hybridization with cDNA probe (20 ng/cover slip) was performed overnight at 37°C. BrdU and FU labeling were visualized with rat anti-BrdU antibody (Harlan Sera Lab) following by anti-rat IgG coupled to Alexa 488 (Invitrogen). Hybridized cDNA was detected with rabbit anti- biotin antibody (Enzo) followed by anti- rabbit secondary antibody coupled either to Alexa 568 (Invitrogen) or Cy3 (Jackson ImmunoResearch). Under these harsher fixation conditions to optimize cDNA labeling, we observed only very limited staining of TS within the nucleoli. Control experiments involving more typical milder fixation with 2% PFA for 10 min revealed typical strong decoration of nucleoli with TS (Fig. S1)
Microscopy and Image Processing
All images were collected as z-stacks in separate channels; with a step of 0.5 µm, deconvolved using a nearest neighbor algorithm incorporated into Slidebook (Intelligent Imaging Innovations) and 2D images were generated using maximum intensity pixel projection. Average acquisition time for each stack was ~ 10 s. The stacks in each time lapse sequence were acquired with constant intervals. Alternatively, 0.5 µm thick optical sections were acquired on a Bio-Rad MRC-1024 laser scanning confocal microscope imaging system equipped with a Nikon Optiphot 2 microscope, a Nikon 60x, 1.4 NA objective, and a krypton/argon laser (l = 488/565 nm). For single labeling experiments, sixty cells labeled in either early, mid or late S-phase, were acquired with various time-lapse intervals in twenty six sessions. For double labeling studies, twenty seven cells in nine sessions were documented. 570, 260 and 590 ChrD were measured for early-, mid- and late- S labeled ChrD, respectively.
A computer software program was developed for the calculation of the ChrD linear movement length. In the first step, ChrD were segmented using an in-house developed, spot-based algorithm (Samarabandu, et al. 1995). As a second step, translation and rotation of the cell in x-y plane in subsequent time points were corrected using in house software running a modified algorithm of Jahne (Jahne 2002). Finally, the linear distance was measured between centers of gravity of corresponding ChrD.
Acknowledgements
The authors are grateful to Alan J. Siegel for extensive discussions. These studies were supported by the National Institutes of Health (NIH) grant GM 072131 awarded to R.B.
Abbreviations List
- ChrD
Chromatin Domains
- DRB
5,6-dichloro-1-β-D-ribobenzimidazole
- TS
Transcription Site(s)
References
- Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J Cell Biol. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R. Regulating the mammalian genome: the role of nuclear architecture. Adv Enzyme Regul. 2002;42:39–52. doi: 10.1016/s0065-2571(01)00041-3. [DOI] [PubMed] [Google Scholar]
- Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys J. 1999;77:2871–2886. doi: 10.1016/S0006-3495(99)77119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12:439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Edelmann P, Bornfleth H, Zink D, Cremer T, Cremer C. Morphology and dynamics of chromosome territories in living cells. Biochim Biophys Acta. 2001;1551:M29–M39. doi: 10.1016/s0304-419x(01)00023-3. [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Galli-Stauber C, Raho G, Rossi D, Corona DF, Pirola B, Bonaglia MC, Zuffardi O, Sorrentino V. Genomic structure and chromosomal location of the human TGFbeta-receptor interacting protein-1 (TRIP-1) gene to 1p34.1. FEBS Lett. 1998;426:279–282. doi: 10.1016/s0014-5793(98)00361-5. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Goldman MA, Holmquist GP, Gray MC, Caston LA, Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahne B. Digital Image Processing. Springer. 2002 [Google Scholar]
- Kamei Y, Tsutsumi O, Taketani Y, Watanabe K. cDNA cloning and chromosomal localization of neural adhesion molecule NB-3 in human. J Neurosci Res. 1998;51:275–283. doi: 10.1002/(SICI)1097-4547(19980201)51:3<275::AID-JNR1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Koberna K, Ligasova A, Malinsky J, Pliss A, Siegel AJ, Cvackova Z, Fidlerova H, Masata M, Fialova M, Raska I, Berezney R. Electron microscopy of DNA replication in 3-D: evidence for similar-sized replication foci throughout S-phase. J Cell Biochem. 2005;94:126–138. doi: 10.1002/jcb.20300. [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132:929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Clemson CM. Gene associations: true romance or chance meeting in a nuclear neighborhood? J Cell Biol. 2008;182:1035–1038. doi: 10.1083/jcb.200808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Sudlow G, Belmont AS. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J Cell Biol. 1998;140:975–989. doi: 10.1083/jcb.140.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol. 2002;159:753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyavantham KS, Bhattacharya S, Alonso WD, Acharya R, Berezney R. Spatio-temporal dynamics of replication and transcription sites in the mammalian cell nucleus. Chromosoma. 2008a;117:553–567. doi: 10.1007/s00412-008-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyavantham KS, Bhattacharya S, Barbeitos M, Mukherjee L, Xu J, Fackelmayer FO, Berezney R. Identifying functional neighborhoods within the cell nucleus: proximity analysis of early S-phase replicating chromatin domains to sites of transcription, RNA polymerase II, HP1gamma, matrin 3 and SAF-A. J Cell Biochem. 2008b;105:391–403. doi: 10.1002/jcb.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi T, Kishimoto T, Makino Y, Shimada M, Nakadai T, Aoki T, Kawata T, Niwa S, Tamura T. Isolation of cDNA, chromosome mapping, and expression of the human TBP-like protein. Biochem Biophys Res Commun. 1999;255:137–142. doi: 10.1006/bbrc.1999.0159. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Rouquier S, Trask BJ, Taviaux S, van den Engh G, Diriong S, Lennon GG, Giorgi D. Direct selection of cDNAs using whole chromosomes. Nucleic Acids Res. 1995;23:4415–4420. doi: 10.1093/nar/23.21.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoni N, Langer S, Fauth C, Bernardi G, Cremer T, Turner BM, Zink D. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J Cell Biol. 1999;146:1211–1226. doi: 10.1083/jcb.146.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarabandu J, Ma H, Acharya R, Cheng PC, Berezney R. Image analysis techniques for visualizing the spatial organization of DNA replication sites in the mammalian cell nucleus using multi-channel confocal microscopy. SPIE. 1995:370–375. [Google Scholar]
- Schubeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Sobell HM. Actinomycin and DNA transcription. Proc Natl Acad Sci U S A. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen PD, Wintero AK, Fredholm M. Chromosomal assignments of 19 porcine cDNA sequences by FISH. Mamm Genome. 1998;9:394–396. doi: 10.1007/s003359900779. [DOI] [PubMed] [Google Scholar]
- Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol. 2004;14:166–172. doi: 10.1016/j.cub.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Belmont AS. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat Cell Biol. 2001;3:134–139. doi: 10.1038/35055033. [DOI] [PubMed] [Google Scholar]
- Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont AS, van Driel R. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, Trowsdale J, Sheer D. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Qiu QQ, Seufert W, Taguchi T, Testa JR, Whitmore SA, Callen DF, Welsh D, Shenk T, Deuel TF. Molecular cloning of the cDNA and chromosome localization of the gene for human ubiquitin-conjugating enzyme 9. J Biol Chem. 1996;271:24811–24816. doi: 10.1074/jbc.271.40.24811. [DOI] [PubMed] [Google Scholar]
- Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–1506. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- Wiesmeijer K, Krouwels IM, Tanke HJ, Dirks RW. Chromatin movement visualized with photoactivable GFP-labeled histone H4. Differentiation. 2008;76:83–90. doi: 10.1111/j.1432-0436.2007.00234.x. [DOI] [PubMed] [Google Scholar]
- Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Yokoyama M, Kobayashi E, Takai S, Hino O. Mapping and determination of the cDNA sequence of the Erc gene preferentially expressed in renal cell carcinoma in the Tsc2 gene mutant (Eker) rat model. Biochem Biophys Res Commun. 2000;275:134–140. doi: 10.1006/bbrc.2000.3280. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zink D, Cremer T, Saffrich R, Fischer R, Trendelenburg MF, Ansorge W, Stelzer EH. Structure and dynamics of human interphase chromosome territories in vivo. Hum Genet. 1998;102:241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]