Abstract
DNA polymerases catalyze template-dependent genome replication. The assembly of a high affinity ternary complex between these enzymes, the double strand-single strand junction of their DNA substrate, and the deoxynucleoside triphosphate (dNTP) complementary to the first template base in the polymerase active site is essential to this process. We present a single molecule method for iterative measurements of DNA-polymerase complex assembly with high temporal resolution, using active voltage control of individual DNA substrate molecules tethered noncovalently in an α-hemolysin nanopore. DNA binding states of the Klenow fragment of Escherichia coli DNA polymerase I (KF) were diagnosed based upon their ionic current signature, and reacted to with sub-millisecond precision to execute voltage changes that controlled exposure of the DNA substrate to KF and dNTP. Precise control of exposure times allowed measurements of DNA-KF complex assembly on a time scale that superimposed with the rate of KF binding. Hundreds of measurements were made with a single tethered DNA molecule within seconds, and dozens of molecules can be tethered within a single experiment. This approach allows statistically robust analysis of the assembly of complexes between DNA and RNA processing enzymes and their substrates at the single molecule level.
The Klenow fragment of Escherichia coli DNA polymerase I (KF) is a model for studying DNA polymerase structure and function.1,2 Crystal structures of these enzymes, which catalyze template-dependent DNA replication, reveal a highly conserved catalytic domain that resembles a partially open right hand.3 The palm subdomain contains residues essential for catalysis, the thumb subdomain positions the primer/template DNA duplex in the active site, and the fingers subdomain is essential for binding incoming deoxynucleoside triphosphate (dNTP) substrates. The KF catalytic cycle has been characterized in kinetic studies.2,4 In an ordered assembly mechanism, KF recognizes the double-strand/single-strand junction of its primer/template substrate to form a binary complex, to which incoming dNTP binds to form a ternary complex. Binary and ternary complex crystal structures of A-family polymerases closely related to KF reveal a large conformational transition between these two functional states.5–7 In polymerase-DNA-dNTP ternary complexes, the fingers subdomain rotates toward the active site to achieve a tight steric fit with the nascent base pair. In this closed ternary complex, the affinity of KF for its DNA substrate is increased by ~5 to 8-fold.8
In this study we present a nanopore-based technique for detecting and manipulating individual KF molecules as they bind to DNA. Nanopores are an established tool for single molecule analysis that have been employed recently to study protein/DNA interactions.8–10 While other single molecule techniques such as optical trapping permit measurements at higher spatial resolution than have thus far been achieved with nanopores,11 these methods generally require more preparative steps. A greater number of molecules can be analyzed over a common time period using nanopores, making it possible to examine population distributions. For example, this capability has enabled statistically robust analysis of the effect of force on DNA-protein dissociation times.10
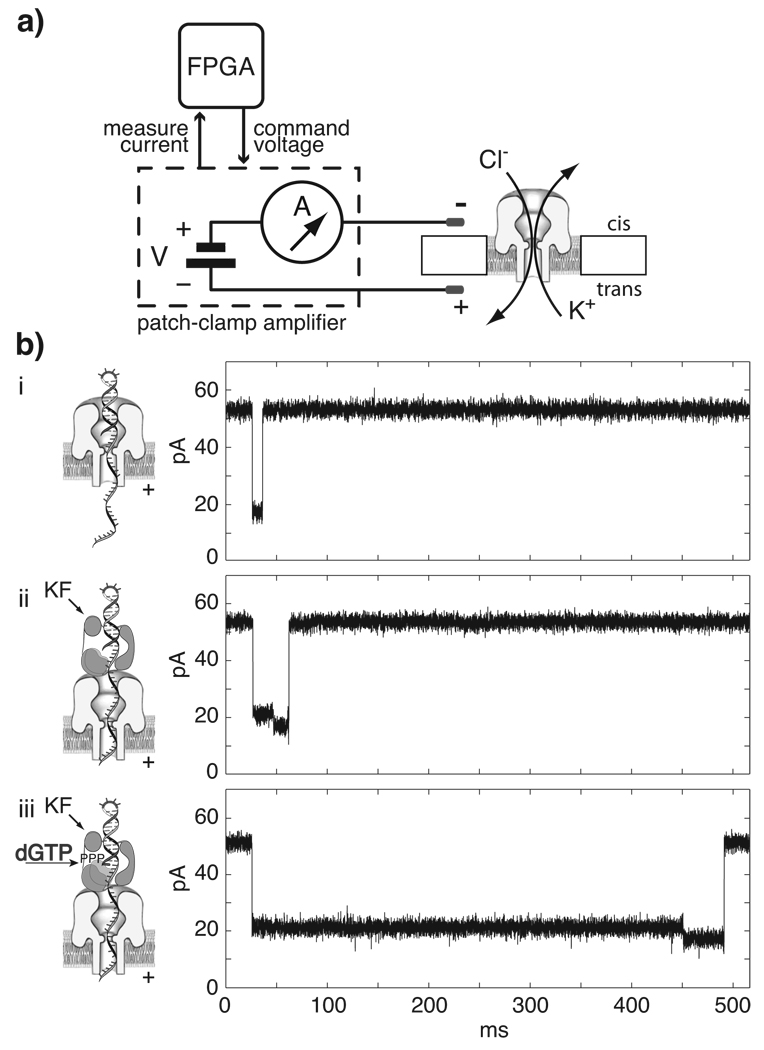
A schematic of the nanopore device used in our experiments is shown in Figure 1a. A single α-hemolysin (α-HL) nanopore is inserted into a lipid bilayer that separates two compartments (cis and trans) containing electrolyte solution. On the cis side the pore opening has a vestibule with a diameter of 3.6 nm that can accommodate double-stranded DNA (dsDNA).12 The pore tapers to a limiting aperture with a 1.5 nm diameter,13 just wide enough to accommodate single-stranded DNA (ssDNA). A voltage potential of 160 mV applied across the membrane generates an open channel ionic current of 53 pA in buffer containing 0.3M KCl. DNA molecules are captured and driven through the pore to the trans side, with each molecule causing a current blockade for a discrete time. This dwell time, and the amplitude for each current state, characterize events in nanopore experiments.
Figure 1. KF binding to individual DNA molecules captured in a nanopore.
a) Schematic of the nanopore device. A patch-clamp amplifier supplies voltage and measures ionic current through a single α-hemolysin channel inserted in a ~25 µm diameter lipid bilayer. Field programmable gate array (FPGA) hardware connected to the amplifier monitors current levels with high temporal resolution (~200 kHz) and allows rapid execution of voltage control logic. b) Illustrations and representative current traces for nanopore capture events of (i) unbound DNA hairpin, (ii) DNA-KF binary complex, and (iii) DNA-KF-dNTP ternary complex.
We recently characterized the current signatures of KF binary and ternary complexes captured in the nanopore.9 We used DNA template or hairpin substrates bearing a 2′–3′ dideoxycytidine (ddC) terminus, which permit ternary complex formation without catalytic turnover. While DNA substrates absent KF yield fast events (Figure 1b, i), capture events for KF-DNA binary complexes (Figure 1b, ii) or KF-DNA-dNTP ternary complexes (Figure 1b, iii), are longer and display two current levels. The initial current level corresponds to the enzyme bound state (EBS) of the captured DNA, with the complex perched atop the pore vestibule and the single-stranded template threaded through the nanopore lumen (Figure 2a, i). This first EBS segment is longer for ternary complexes than for binary complexes (Figure 1b, ii,iii). The second current segment results upon voltage-promoted enzyme dissociation, when the duplex DNA drops into the nanopore vestibule (Figure 2a, ii) followed by duplex unzipping and translocation through the pore (Figure 2a, iii). The duration of this terminal current step is the same as the duration of capture events for the DNA substrate alone, as a function of both applied voltage and duplex length.9
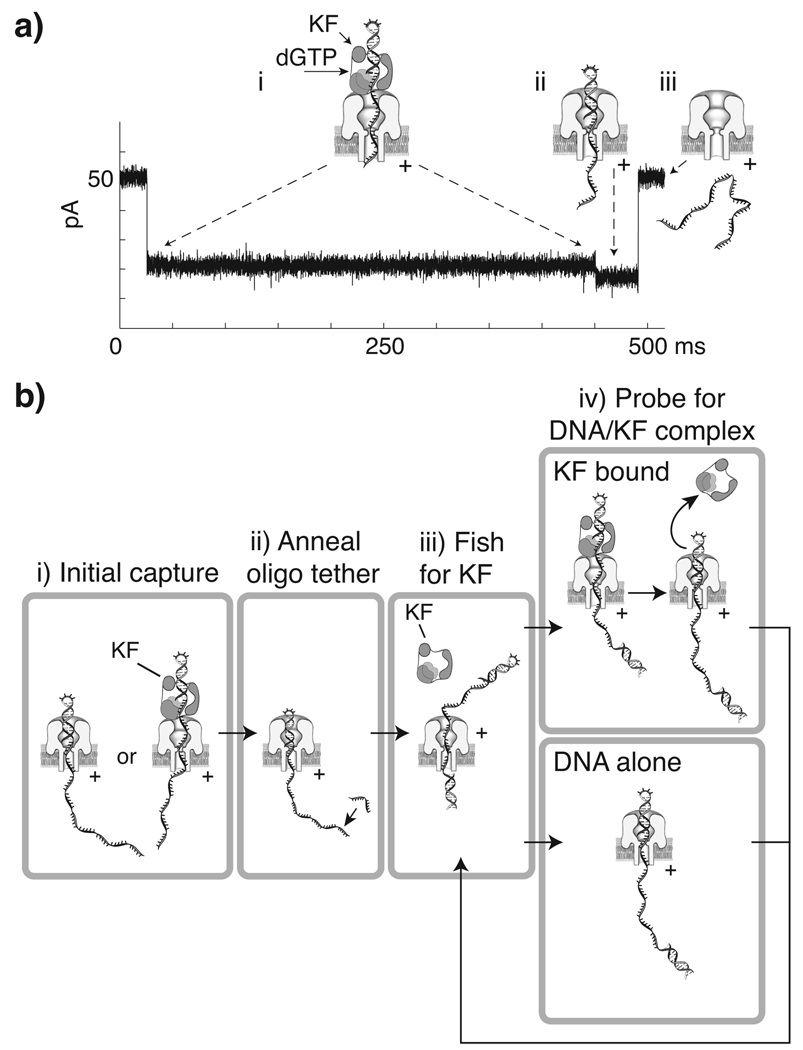
Figure 2. Use of the two step current signature of DNA-KF and DNA-KF-dNTP complexes to control KF association and dissociation from a DNA substrate tethered in the nanopore.
a) Representative current trace and illustration of corresponding molecular events during nanopore capture of a DNA-KF-dGTP ternary complex. The initial ~21.5 pA blockade (i) arises from capture of the enzyme bound DNA, with the duplex held atop the pore vestibule bound to KF. The shorter ~18.5 pA terminal step (ii) occurs upon KF dissociation, when duplex DNA is pulled into the nanopore vestibule, followed by (iii) duplex unzipping and translocation of the DNA through the nanopore, and return to the open channel current. b) Strategy for control of iterative KF association and dissociation from a nanopore-tethered DNA molecule. (i) A DNA hairpin, either KF bound or unbound, is captured under applied voltage. When the amplitude level that distinguishes capture of DNA alone or the terminal step of KF bound events is detected, (ii) the FSM lowers the voltage to hold the hairpin duplex in the pore vestibule long enough to anneal an oligonucleotide to the single-stranded end protruding into the trans chamber. With the DNA tethered in the pore by duplexes at both ends, a negative voltage is applied (iii) to expose the DNA to KF in the cis chamber (‘fishing’). After a programmed time period, the voltage is reversed (iv) to draw the duplex back to the pore and diagnose whether it is KF bound based upon amplitude (‘probing’). Detection of unbound DNA prompts a return to the fishing voltage. Diagnosis of the KF bound state results in continued application of the probing voltage until the terminal step is detected, which then prompts a return to fishing.
In this study, we employed this two step current signature for DNA-KF complexes to develop repetitive, real-time detection and control of KF association and dissociation from single DNA molecules. Individual primer/template DNA molecules were topologically tethered with their ss-DNA segment threaded through the nanopore and duplex segments on either side of the pore (Figure 2b). We applied finite state machine (FSM) logic implemented on a field programmable gate array (FPGA) (Figure 1a) to monitor current levels and control voltage with sub-millisecond precision in response to current changes. We exploited the complementarity of the α-HL nanopore structure to the ds-ssDNA junction to achieve control of voltage promoted DNA-KF dissociation. By varying the duration of exposure to KF and dNTP, we probed the assembly of DNA-KF complexes, and the effect of dGTP on the rate of assembly.
Results and Discussion
Our objective was to measure and control iterative KF dissociation and re-association to an individual DNA molecule tethered in the α-HL nanopore. To achieve this, we exploited the current change that accompanies KF dissociation from its DNA substrate (Figure 2a). Using an FSM implemented on an FPGA to exert active voltage control,9,10,14 we reasoned that we could diagnose the EBS of captured complexes. We could then monitor the current level to detect and react to the terminal current step with a voltage reversal executed after KF dissociation, but before DNA translocation through the nanopore.
Specifically, our strategy was to capture a 23 base-pair DNA hairpin substrate bearing a 2′–3′ dideoxy terminus and a 5′ overhang from the cis chamber, and promptly lower the voltage to a level sufficient to hold the duplex in the nanopore vestibule with its single-stranded end protruding into the trans chamber (Figure 2b, i). This allows a 20-mer oligonucleotide to anneal to the 5′ end of the DNA, tethering it in the nanopore by duplex regions at both ends (Figure 2 b, ii). In a step termed ‘fishing’, a small negative voltage is then applied for a programmed time interval to expose the double-strand/single-strand junction of the DNA to the bulk phase in the cis chamber (Figure 2b, iii). The cis chamber contained either DNA alone, DNA and KF, or DNA, KF, and dGTP (the complement to the template base at n=0). In a step termed ‘probing’, a positive voltage is applied next, drawing the double-strand/single-strand junction to the nanopore (Figure 2b, iv). By monitoring the ionic current, the FSM determines if KF is bound to the DNA. If KF binding is not detected, the FSM reverses the voltage to fish again. If KF binding is diagnosed, the probing voltage is maintained until detection of the terminal step that reports enzyme dissociation. A voltage reversal can then be executed before the DNA molecule translocates through the nanopore in order to fish again (Figure 2b, iii).
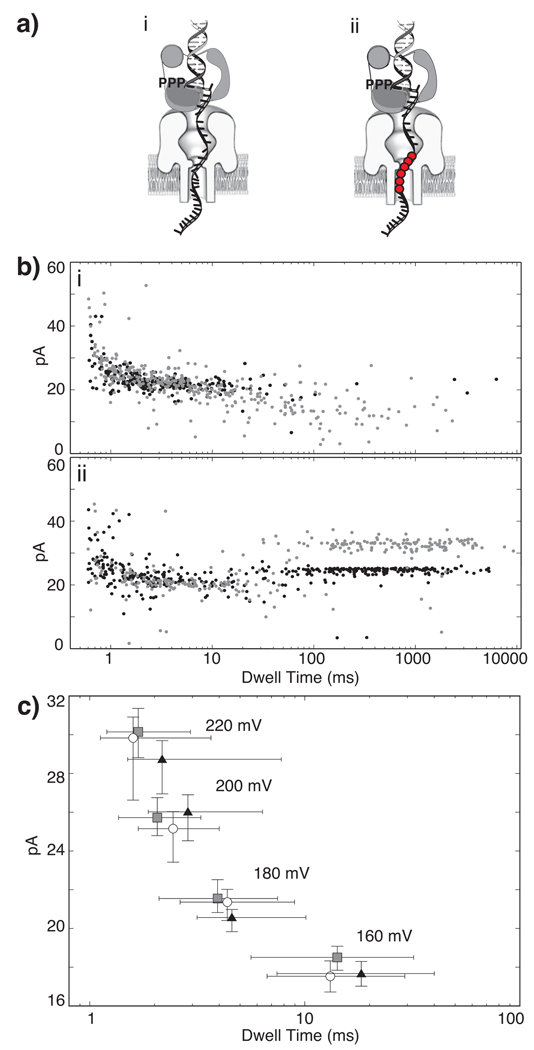
The duration of the first current segment of DNA-KF binary events is extended by the presence of dNTP complementary to the template base in the enzyme active site (Figure 1b, ii,iii), consistent with assignment of this segment to the EBS.9 Because the tethered probing experiments rely on the accuracy of this assignment, we performed an additional test. We designed a template bearing a block of 6 abasic (1′, 2′-dideoxy) residues. Using the known dimensions of the nanopore13 and the length of single-stranded DNA, the abasic residues were placed so that when the template is hybridized to a 23 mer primer, they span residues +12 to +17 relative to position n = 0 in the KF catalytic site. If DNA is perched atop the nanopore vestibule when it is bound to KF, captured complexes formed with this substrate should position the abasic segment in the nanopore lumen (Figure 3a, ii), where it is predicted to impede ionic current through the pore less than standard DNA residues. Thus the current level of the initial segment of the events should be higher than for an all DNA substrate. In contrast, when duplex DNA, absent KF, is captured in the pore vestibule, the abasic residues would be displaced into the trans compartment, where they would be predicted to have minimal effect on current levels. When KF and dGTP were present with the primer/template substrate bearing 6 abasic residues, the median amplitude of the EBS of capture events at 180 mV was 33.1 pA, compared to 24.8 pA for the control substrate absent abasic residues (Figure 3b, ii). The median amplitude of primer/template capture events, absent KF and dGTP, was 21.6 pA for the substrate containing abasic residues, and 22.3 pA for the DNA substrate not containing abasic residues (Figure 3b, i). Thus the effect of the abasic residues is exerted in the first current segment, verifying that it corresponds to the EBS of capture events.
Figure 3. Characterization of molecular events that cause the two step current signature of DNA-KF complexes in the nanopore.
a) Diagram of nanopore capture of a DNA-KF-dGTP ternary complex with (i) the all DNA template, and (ii) the 6 abasic residue-containing template. Abasic residues are in template positions +12 to +17, and are shown as red circles. b) Superimposed plots showing amplitude vs. dwell time for capture events for DNA substrates with abasic-containing template (black dots) or the all DNA template (grey dots). In (i) 1 µM DNA is present but KF and dGTP are absent; in (ii) 1 µM DNA, 1 µM KF and 200 µM dGTP are present. Offline data analysis described in Methods was used to extract the EBS and terminal steps from each event in b(ii). This experiment was conducted at 180 mV applied voltage. c) Median amplitude vs. median dwell time plot for DNA alone (grey squares), or the terminal current steps of binary (white circles) or ternary complexes (black triangles) captured from the bulk phase in the cis chamber at the indicated constant voltage levels (error bars are defined by first and third quartiles). Each data point was obtained from analysis of 181 to 552 events.
Diagnosis of the state of complexes captured in the nanopore requires characterization of their current signatures at an appropriate probing voltage. This voltage must yield sufficient ionic current for reliable discrimination between KF bound and unbound states. It must also support dwell times long enough to allow subsequent diagnosis by the FSM. We compared current patterns for DNA alone or KF bound events with a 23 base-pair all DNA hairpin substrate at 220, 200, 180, and 160 mV to identify an optimal voltage for real-time detection and control. Consistent with previous results,9 at each voltage examined, values for the amplitude and duration of the terminal step scaled with values for DNA alone (Figure 3c). At 160 mV, the median amplitude and dwell time for DNA alone events were 18.5 pA and 14.7 ms, respectively, while corresponding values for the terminal step of DNA-KF complexes were 17.7 pA and 13.1 ms. The duration of unbound DNA events or terminal steps at 160 mV exceeds the ~5 ms settling time for the capacitive transient superimposed on the current signal immediately following a voltage change, allowing reliable FSM reaction to this state prior to translocation. Using online filtering of the current signal to mitigate noise, the ~4 pA difference between the amplitude of the enzyme bound state (21.5 pA) and the terminal current step is sufficient for accurate event diagnosis.
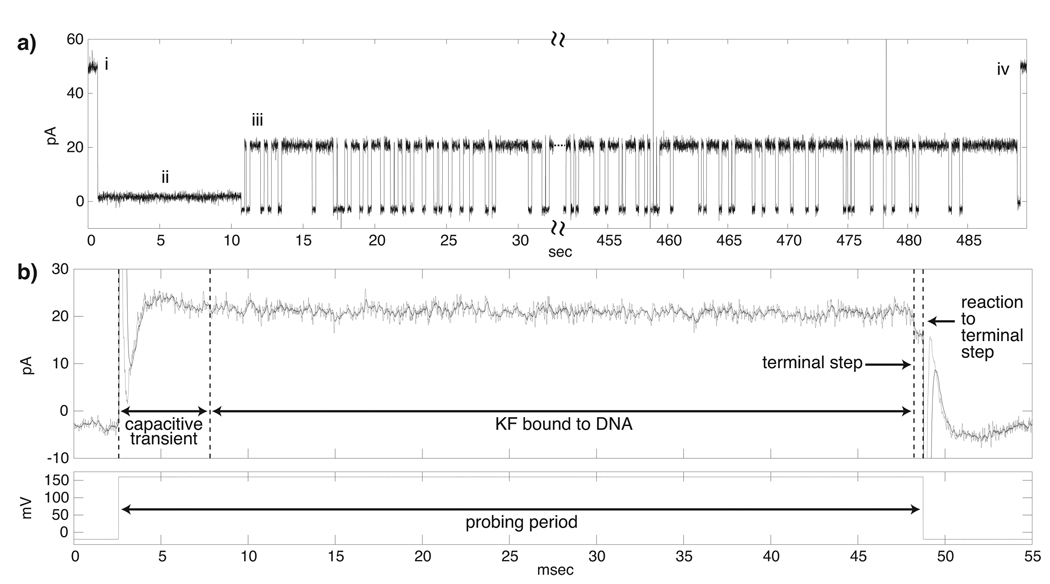
We conducted fishing experiments using this 23 base-pair all DNA hairpin, which bears a 50 nucleotide single-stranded overhang. Figure 4 shows an experiment in which a single DNA substrate molecule was tethered in the nanopore and ~500 fishing and probing cycles were achieved in the presence of KF and dGTP. Current traces show the repeated fishing/probing events (Figure 4a, iii), and an expanded view of a single ternary complex capture event (Figure 4b) with the corresponding levels of applied voltage shown below the expanded current trace. Starting from the open channel current (Figure 4a, i), a DNA hairpin molecule is captured at 160 mV and detected, and the FSM logic lowers the voltage to 50 mV to hold the DNA for 10 seconds (Figure 4a, ii). This period is sufficient to anneal an oligonucleotide complementary to the last 20 nucleotides at the 5′ end of the substrate without promoting translocation. Once the DNA strand is tethered in the nanopore by duplex regions at both ends, the FSM applies −20 mV for a programmed fishing time interval (250 ms in the experiment shown), to expose the tethered DNA to the bulk phase in the cis chamber containing DNA, KF, and dGTP. After a voltage reversal to 160 mV, the state of the captured DNA molecule is identified as either enzyme bound or unbound by the current level. If the DNA is unbound, the −20 mV fishing voltage is reapplied. If the DNA is KF bound (Figure 4b), the probing voltage is maintained until detection of the terminal step that reports KF dissociation, after which the −20 mV fishing voltage is reapplied. Eventual translocation of the DNA, detected as a return to the open channel current (Figure 4a, iv), prompts the FSM to await detection of another capture event at 160 mV and then return to the oligonucleotide annealing step (Figure 4a, ii) to begin the process again.
Figure 4. Fishing for KF with DNA tethered in the nanopore.
a) Representative current trace for (i) DNA capture, (ii) DNA tethering via annealing of trans side oligonucleotide, (iii) ~500 cycles of fishing and probing and (iv) DNA translocation and return to open channel current. In this experiment the fishing interval was 250 ms, with 1 µM DNA, 2 µM KF, and 400 µM dGTP in the cis chamber. b) Expanded current trace and corresponding applied voltage levels (below) during a single fishing and probing cycle. The trace shows the ~5 ms capacitive transient that follows the change from the −20 mV fishing voltage to the 160 mV probing voltage. The filtered signal (black trace) mitigates noise present in the raw signal (grey trace) to avoid false detection of terminal steps. This event was diagnosed as enzyme bound since its amplitude was outside [14.75, 18.75] pA, the range employed to diagnose DNA alone events and terminal steps. The 160 mV probing voltage was maintained until detection of the current drop to within [14.75, 18.75] pA, followed by an additional 0.5 ms to ensure accurate diagnosis. A voltage reversal to −20mV to fish again is then applied.
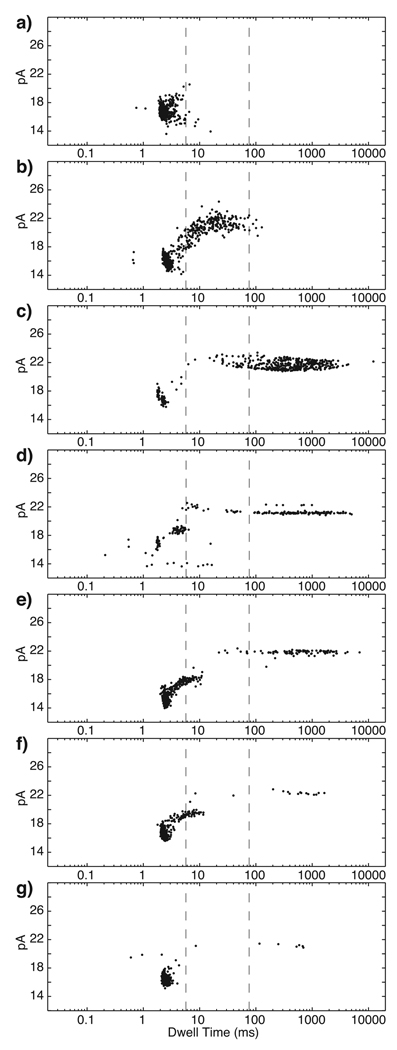
Our next goal was to use this electronic control capability to probe and control the assembly of KF complexes by varying the duration of the fishing interval (Figure 5; Table 1). We first examined events for a long fishing interval (500 ms). When only the DNA hairpin (1 µM) was present in the nanopore chamber, a population of probing events with a median amplitude of 16.8 pA and median dwell time of 2.1 ms was detected (Figure 5a). The majority of the events in the presence of DNA alone (98%) had dwell times below 5.61 ms (Table 1, line 1). Upon addition of 2 µM KF, a new population of events emerged at ~21 pA. Under these conditions, the dwell times for ~38% of the total population were longer than 5.61 ms (Figure 5b; Table 1, line 2), indicating that at least 38% of events are attributable to binary complexes. Addition of 400 µM dGTP yielded an additional population of events, with ~75% of the total population lasting longer than 75.22 ms (Figure 5c; Table 1, line 3). Therefore, at least 75% of the total events were characteristic of ternary complexes.
Figure 5. Control of complex assembly by varying fishing duration.
Plots of amplitude vs. dwell time for events detected in the probing step after a 500 ms fishing interval with a) DNA alone (1 µM), b) DNA and KF (2 µM), or c) DNA, KF and dGTP (400 µM) present in the bulk phase in the nanopore cis chamber. With DNA, KF, and dGTP present, the fishing time interval was reduced to d) 50 ms, e) 10 ms, f) 7.5 ms, or g) 5 ms. The two dashed vertical lines through the plots indicate the upper dwell time limits for DNA alone (5.61 ms) and KF-DNA binary complexes (75.22 ms), determined as described in Table 1. The upper dwell time limit for DNA alone is shorter than the median dwell time for DNA alone (14.7 ms) in Figure 3c because in fishing experiments these events are truncated by a voltage reversal (Figure 2b).
Table 1.
Percentage of total fishing events in specified dwell time ranges
| Components in chambera |
Fishing time, ms |
Total events |
Events < 5.61 msb |
Events ≥ 5.61, < 75.22 msc |
Events ≥ 75.22 ms |
|---|---|---|---|---|---|
| DNA | 500 | 609 | 98.01% | 1.99% | 0% |
| DNA, KF | 500 | 853 | 61.14% | 38.04% | 0.82% |
| DNA, KF, dGTP | 500 | 536 | 14.99% | 9.51% | 75.5% |
| DNA, KF, dGTP | 100 | 537 | 14.63% | 9.01% | 76.36% |
| DNA, KF, dGTP | 50 | 269 | 39.42% | 11.3% | 49.28% |
| DNA, KF, dGTP | 10 | 1052 | 82.36% | 7.15% | 10.49% |
| DNA, KF, dGTP | 7.5 | 670 | 91.55% | 5.54% | 2.92% |
| DNA, KF, dGTP | 5 | 668 | 97.96% | 0.73% | 1.31% |
The concentration of each component in this experiment, when present as indicated, was: 1 µM DNA, 2 µM KF, and 400 µM dGTP.
This upper dwell time limit for unbound DNA events was calculated as the time below which 98% of events occurred when the DNA hairpin was in the cis chamber absent KF and dGTP (Figure 5a).
An upper dwell time limit (75.22 ms) for binary complex events was defined as the time below which 98% of the events that exceed the first boundary of 5.61 ms occur, when both DNA and KF were present (Figure 5b).
We used these three data sets determined at a 500 ms fishing interval to designate two dwell time boundaries (Table 1). The first boundary (5.61 ms) assigns an upper dwell time limit for events attributable to capture of unbound DNA (Figure 5a). The second boundary (75.22 ms) assigns an upper dwell time limit for binary complex events (Figure 5b). Thus, events lasting longer than 5.61 ms were attributed to KF bound DNA, and events lasting longer than 75.22 ms, when DNA, KF, and dGTP were present in the chamber (Figure 5c), were considered to be due to ternary complexes.
We then examined whether shifts in the event populations occurred as the fishing interval was shortened, in the presence of all components required for ternary complex formation (1 µM DNA, 2 µM KF, and 400 µM dGTP). At this high dGTP concentration we expect that for all fishing intervals, almost every time that a DNA-KF complex is drawn back to the pore, the dwell time measured during the probing phase will be characteristic of a ternary complex. This is because regardless of whether the complex is binary or ternary at the moment of capture from the bulk phase, dGTP can bind, unbind, and rebind to the complex during the probing period, extending its lifetime atop the pore.15 Thus, in the presence of 400 µM dGTP, as fishing time is changed, variation in the percentage of EBS events is dictated primarily by the rate of KF binding to the tethered DNA.
When the fishing time was reduced from 500 ms (Figure 5c) to 100 ms, the percentage of total events within the designated dwell time boundaries was virtually indistinguishable (Table 1, lines 3 and 4), suggesting that equilibration with the the bulk phase occurs between these time intervals. When the fishing time was reduced to 50 ms, the number of events longer than 75.22 ms decreased to ~50% of the total population, with a concomitant increase in putative DNA events (<5.61 ms) to ~40% (Figure 5d; Table 1, line 5). No significant increase in events with dwell times between 5 ms and 75 ms was observed. Further reductions of the fishing time to 10 ms and 7.5 ms shifted the populations further (Figure 5e and f; Table 1, lines 6 and 7). At 5 ms fishing time, ~98% of events were of an amplitude (~16 pA) and duration characteristic of unbound DNA (Figure 5g; Table 1, line 8).
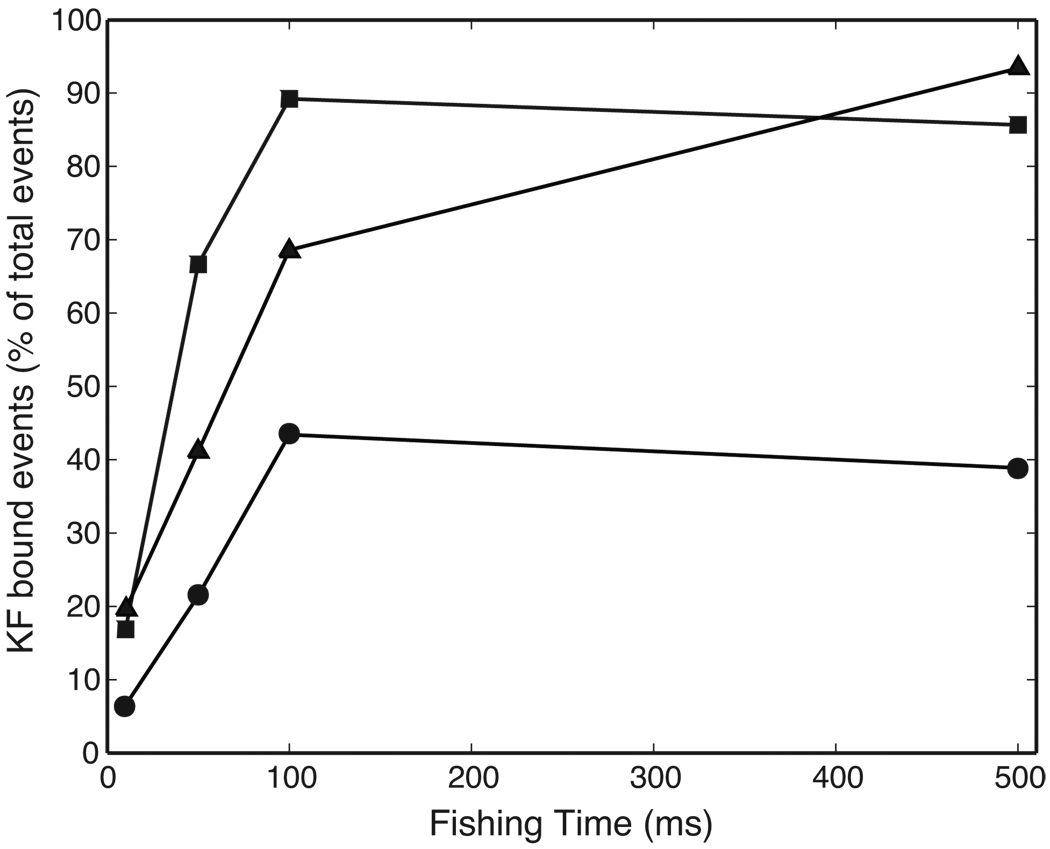
Figure 6 shows the effect of dGTP on the percentage of EBS events as a function of fishing time. This representation of the data does not distinguish among different EBS events based upon dwell time. It is therefore sensitive to the effect of dGTP in the bulk phase on the probability of drawing a KF-bound complex back to the pore, but is independent of the effect of dGTP binding and rebinding to KF-bound complexes while they reside atop the pore during the probing period, which causes a dGTP-concentration dependent increase in dwell time.15
Figure 6. Effect of dGTP on DNA-KF complex assembly.
Plot of the percentage of total events diagnosed as KF-bound when the fishing interval was varied in the presence of 1 µM DNA, 2 µM KF, either absent dGTP (circles), or in the presence of 50 µM dGTP (triangles), or 400 µM dGTP (squares). The percentage of EBS events was determined as described in Methods. Percentage values for each plotted point were determined from at least 255 and up to 7828 events (Table 2).
With a 500 ms fishing interval, 39% of events detected absent dGTP were enzyme bound (Figure 6, Table 2), compared to 93% and 86% in the presence of either 50 or 400 µM dGTP, respectively. This reflects the increased affinity of KF for DNA conferred by the presence of dNTP.4 At this long fishing interval, EBS events converge to ~90% for the dGTP concentrations examined (Figure 6, Table 2). This behavior is predicted by the value of Kd ~5 µM for bulk phase dNTP binding to DNA-KF complexes.15,16 Thus at dGTP concentrations above ~10 µM, the ability of dNTP to increase the affinity of KF for DNA at equilibrium is saturated.
Table 2.
Percentage of total fishing events diagnosed as enzyme bound
| dGTP concentration, µM |
Fishing time, ms |
Total events | EBS eventsa | Percentage of events with EBS |
|---|---|---|---|---|
| 0 | 10 | 7828 | 511 | 6.5% |
| 0 | 50 | 6781 | 1454 | 21.4% |
| 0 | 100 | 997 | 433 | 43.4% |
| 0 | 500 | 854 | 332 | 38.9% |
| 50 | 10 | 1635 | 320 | 19.6% |
| 50 | 50 | 749 | 308 | 41.4% |
| 50 | 100 | 653 | 448 | 68.6% |
| 50 | 500 | 1031 | 963 | 93.4 % |
| 400 | 10 | 1050 | 177 | 16.9% |
| 400 | 50 | 255 | 170 | 66.7% |
| 400 | 100 | 520 | 464 | 89.2% |
| 400 | 500 | 531 | 455 | 85.7 % |
Number of events with dwell times longer than the upper dwell time limit for unbound DNA. The upper limit is the time below which 98% of events occurred when the DNA hairpin was in the cis chamber absent KF and dGTP.
At fishing intervals shorter than 500 ms, decreases in the percentage of EBS events are observed. The extent of these decreases varied with dGTP concentration. Thus the slopes of the curves for the percentage of KF-bound events as a function of fishing time increased as dGTP concentration was increased from 0 to 50 to 400 µM (Figure 6). This suggests that the rate of approach to equilibrium is dependent upon dGTP concentration. That is, for fishing intervals within the approach to equilibrium, increasing dGTP concentration increased the probability of the DNA molecule being KF-bound at the moment it was drawn back to the pore.
Experiments in which the fishing interval is varied thus probe complex assembly on a time scale that superimposes with the rate of KF binding to DNA (Figure 5), and with the time scale of dGTP binding to binary complexes (Figure 6). Each fishing and probing event uses an individual DNA molecule, known to be unbound by enzyme at the moment that it is exposed to KF and dNTP in the bulk phase. This yields the potential to measure pre-steady state complex assembly at the single molecule level. This approach is statistically robust, as hundreds to thousands of events can be analyzed in each experiment. Finer temporal resolution using additional fishing time intervals, in combination with mathematical modeling,15 should permit the determination of binding rate constants.
Methods
Materials
DNA oligonucleotides were synthesized by Midland Certified Reagent Company or the Stanford University Protein and Nucleic Acid Facility and purified by denaturing PAGE. Sequences of the oligonucleotides used in this study are shown below. The sequences of the 6 abasic residue-containing 79-mer template and the all DNA control are identical except for the region replaced by abasic residues, each indicated by an X. The 3′ terminal residue of the 23 base pair hairpin (bphp) and 23-mer primer was 2′–3′ dideoxycytidine (ddC), enabling ternary complex formation without catalytic turnover. The n = 0 template base for all substrates was C; thus the complementary incoming nucleotide was dGTP.
-
23 bphp:
5′-CTCACCTATCCTTCCACTCATTCCTTACCATTCATTCACATCTCACTATCG CATTCTCATGCAGGTCGTAGCCTTTTGGCTACGACCTGCATGAGAATGddC 3′
-
20-mer tethering oligonucleotide:
5′-TGAGTGGAAGGATAGGTGAG-3′
-
23-mer primer:
5′-GGCTACGACCTGCATGAGAATGddC- 3′
-
All DNA control 79-mer template:
5′-CTCACCTATCCTTCCACTCATTCCAATTAATTACCATTCATTCAGATCTCAC TATCGCATTCTCATGCAGGTCGTAGCC-3′
-
6 abasic residue-containing 79-mer template
5′-CTCACCTATCCTTCCACTCATTCCAATTAATTACCATTXXXXXXGATCTCAC TATCGCATTCTCATGCAGGTCGTAGCC-3′
Prior to use, the 23 bp hairpin was denatured at 95°C for 2.5 min and quickly annealed in ice water to prevent intermolecular hybridization. The 23-mer primer was annealed to each 79-mer template as previously described.9 Klenow fragment (exo-, 100,000 U/ml, 20,000 U/mg) was obtained from New England Biolabs.
General Nanopore Methods
Experiments were conducted at 23°C in 10 mM HEPES/KOH, pH 8.00 ±0.05, 0.3 M KCl, 5 mM MgCl2, which are conditions shown to support KF catalytic function.9 In all experiments 1 µM DNA substrate was present in the cis chamber. KF and dGTP were used, when indicated, at 2 µM and 400 µM, respectively, unless otherwise indicated in figure legends. Single α-HL channels were formed as described.12 A patch-clamp amplifier (AxoPatch 200B, Molecular Devices, Sunnyvale, CA) was used to apply trans-membrane voltage and measure ionic current, with the 4-pole Bessel filter set at 5 kHz bandwidth. A digitizer (Digidata 1440A, Molecular Devices) stored data sampled at 200 kHz.
Active Voltage Control Experiments
Active voltage control logic was implemented as a finite state machine (FSM), a logic construct where program execution is broken into a series of individual states, as described previously.9 Measurements determine which state the logic is in, and therefore the sequence of executed commands over time.17 The FSM was programmed with LabVIEW software (Version 8, National Instruments, Austin, TX) and implemented on an FPGA system (PCI-7831R, National Instruments), a reconfigurable hardware platform.18 The FPGA was connected to the Axopatch 200B and permitted trans-membrane voltage control and ionic current measurements at 5.3 µs updates. To improve the signal-to-noise ratio, the ionic current signal was filtered on the FPGA using a single-pole low pass filter with 1.58 kHz cutoff frequency and a 10%–90% rise time of ~0.2 ms.
The FSM initialized tethering by detecting capture of a DNA molecule at 160 mV, and reducing the voltage to 50 mV for 10 s. With the 20-mer tethering oligonucleotide present in the trans chamber at 4 µM, this period yielded 70% oligonucleotide annealing efficiency. The FSM then triggered the −20 mV fishing voltage, which was sustained for the time periods specified in the figures and table. During each 160 mV probing period, the FSM logic diagnosed the DNA as unbound if the low-pass filtered current remained within an amplitude range for 0.5 ms. The range was calibrated at the beginning of each experiment, by monitoring bound and unbound tethered DNA probing event amplitudes, and was not varied during a given experiment at a set voltage and KF and dGTP concentrations. Experiment-to-experiment variations in the range was due largely to evaporation-induced drift in the open channel current by 1–3 pA, over the course of several experiments. Example ranges used are [14.25, 18.25] pA (for 0 µM and 50 µM dGTP at all reported fishing times) and [15.5, 19.5] pA (for 400 µM dGTP at 500 ms fishing time). These ranges for unbound DNA diagnosis were consistent with the observed amplitude separation between enzyme bound (21.5 pA) and unbound (18.5 pA) DNA in constant 160 mV experiments. If the low-pass filtered current remained outside the amplitude range, the DNA was diagnosed as enzyme bound. Upon diagnosis of unbound DNA, the FSM re-triggered and sustained the fishing voltage period.
In nanopore systems using patch-clamp technology, a step change in voltage induces a capacitive transient superimposed on the ionic current measurement. The transient masks information in the measured current for a time proportional to the magnitude of the net voltage change. The change from the −20 mV fishing voltage to the +160 mV probing voltage excited a capacitive transient that lasted ~5 ms. Since this transient time exceeds the filter settling time (0.2 ms), initial diagnosis of the tethered DNA as enzyme bound or unbound was dictated by the transient settling time. In experiments with untethered DNA at a constant voltage of 160 mV, 50% of binary complex bound states lasted longer than 5.8 ms. Therefore up to 50% of binary complexes captured may be diagnosed as unbound DNA events by the FSM control logic because the EBS was masked by the transient. At 50 µM and 400 µM dGTP the EBS duration is extended, significantly reducing the likelihood that the transient would mask an EBS and subsequent terminal step. When any EBS lasted longer than the transient settling time, KF dissociation was diagnosed by the FSM within ~0.7 ms of when the terminal step occurred, dictated by the sum of the filter rise time (0.2 ms) and the control logic diagnosis time (0.5 ms).
Offline Data Processing
Recorded current blockades reported in the figures were tabulated offline using MATLAB (2007a, The MathWorks, Natick, MA) with software developed in our lab. In experiments with untethered DNA, an event was identified when the open channel current (~53 pA at 160 mV) dropped by at least 8 pA for at least 0.2 ms. In the experiments with tethered DNA, the dwell time of each event was defined as the period during which the FSM applied the probing voltage.
To identify and quantify terminal steps in constant voltage experiments with untethered DNA (Fig. 3), a baseline amplitude was calculated as the mean of the first 15% of the event amplitude for all events longer than 1 ms. Events that ended with a segment of at least 3.5 pA below the baseline amplitude were identified as having a terminal step, and the dwell time and mean amplitude of each end segment were used to quantify the terminal step. Offline analysis was used to estimate the performance of the FSM control logic in tethered DNA experiments. On average, 96% of terminal steps were detected and reacted to within 2 ms.
In Figure 5, the mean amplitude of each probing event was computed from the time that the transient crossed 12 pA (1–2 ms after the start of probing) to the end of the probing period. To compute the percentage of EBS events in an experiment (Figure 6, Table 2), a probing event with a dwell time longer than the 98%-cutoff time for unbound DNA was diagnosed as enzyme bound. The 0 µM and 400 µM dGTP experiments were performed on one nanopore station (with a corresponding unbound DNA cutoff time of 5.61 ms (Figure 5a)), and the 50 µM dGTP experiments were performed on another nanopore station (with a corresponding unbound DNA cutoff time of 9.22 ms). The transient pattern dictated the unbound DNA diagnosis time; thus, differences in the transient duration on the two stations determined the different 98%-cutoff times characteristic of unbound DNA.
Acknowledgement
We are grateful to Seico Benner for help in the early stages of the experiments, and to David Deamer for advice and discussion. This work was supported by National Institutes of Health grants GM23950-444071 (M.A.) and HG004035-03 (W.D.).
References
- 1.Joyce CM, Steitz TA. Function and Structure Relationships in DNA Polymerases. Anal. Rev. Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 2.Joyce CM, Benkovic SJ. DNA Polymerase Fidelity: Kinetics, Structure, and Checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 3.Brautigam CA, Steitz TA. Structural and Functional Insights Provided by Crystal Structures of DNA Polymerases and their Substrate Complexes. Current Opinion in Structural Biology. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 4.Joyce CM, Potapova O, Delucia A, Huang X, Basu V, Grindley NDF. Fingers-Closing and Other Rapid Conformational Changes in DNA Polymerase I (Klenow Fragment) and Their Role in Nucleotide Selectivity. Biochemistry. 2008;47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 5.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal Structure of a Bacteriophage T7 DNA Replication Complex at 2.2 Å Resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 6.Johnson SJ, Taylor JS, Beese LS. Processive DNA Synthesis Observed in a Polymerase Crystal Suggests a Mechanism for the Prevention of Frameshift Mutations. Proc. Natl. Acad. Sci. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Korolev S, Waksman G. Crystal Structures of Open and Closed Forms of Binary and Ternary Complexes of the Large Fragment of Thermus Aquaticus DNA Polymerase I: Structural Basis for Nucleotide Incorporationpen and Closed Forms of Binary and Ternary Complexes of the Large Fragment of Thermus Aquaticus DNA Polymerase I: Structural Basis for Nucleotide Incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cockroft SL, Chu J, Amorin M, Ghadiri MR. A Single-Molecule Nanopore Device Detects DNA Polymerase Activity with Single-Nucleotide Resolution. J. Am. Chem. Soc. 2008;130:818–820. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benner S, Chen RJA, Wilson NA, Abu-Shumays R, Hurt N, Lieberman KR, Deamer DW, Dunbar WB, Akeson M. Sequence-Specific Detection of Individual DNA Polymerase Complexes in Real Time Using a Nanopore. Nat. Nanotechnology. 2007;2:718–724. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornblower B, Coombs A, Whitaker R, Kolomeisky A, Picone S, Meller A, Akeson M. Single-Molecule Analysis of DNA-Protein Complexes Using Nanopores. Nat. Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 11.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct Observation of Base-Pair Stepping by RNA Polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vercoutere W, Akeson M. Biosensors for DNA Sequence Detection. Current Opinion in Chemical Biology. 2002;6:816–822. doi: 10.1016/s1367-5931(02)00395-2. [DOI] [PubMed] [Google Scholar]
- 13.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of Staphylococcal Alpha-Hemolysin, a Heptameric Transmembrane Pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 14.Tropini C, Marziali A. Multi-Nanopore Force Spectroscopy for DNA Analysis. Biophysical Journal. 2007;92:1632–1637. doi: 10.1529/biophysj.106.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurt N, Wang H, Akeson M, Lieberman KR. Specific Nucleotide Binding and Rebinding to Individual DNA Polymerase Complexes Captured on a Nanopore. J. Am. Chem. Soc. 2009 doi: 10.1021/ja809663f. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahlberg ME, Benkovic SJ. Kinetic Mechanism of DNA Polymerase I (Klenow Fragment): Identification of a Second Conformational Change and Evaluation of the Internal Equilibrium Constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 17.Gill A. Introduction to the Theory of Finite-state Machines. McGraw-Hill; 1962. [Google Scholar]
- 18.Trimberger SM. Field-Programmable Gate Array Technology. Springer; 1994. [Google Scholar]