Abstract
RTKs (receptor tyrosine kinases) play important roles in cellular proliferation and differentiation. In addition, RTKs reveal oncogenic potential when their kinase activities are constitutively enhanced by point mutation, amplification or rearrangement of the corresponding genes. The ALK (anaplastic lymphoma kinase) RTK was originally identified as a member of the insulin receptor subfamily of RTKs that acquires transforming capability when truncated and fused to NPM (nucleophosmin) in the t(2;5) chromosomal rearrangement associated with ALCL (anaplastic large cell lymphoma). To date, many chromosomal rearrangements leading to enhanced ALK activity have been described and are implicated in a number of cancer types. Recent reports of the EML4 (echinoderm microtubule-associated protein like 4)–ALK oncoprotein in NSCLC (non-small cell lung cancer), together with the identification of activating point mutations in neuroblastoma, have highlighted ALK as a significant player and target for drug development in cancer. In the present review we address the role of ALK in development and disease and discuss implications for the future.
Keywords: anaplastic lymphoma kinase (ALK), anaplastic large cell lymphoma (ALCL), extracellular-signal-regulated kinase (ERK), inflammatory myofibroblastic tumour (IMT), midkine, neuroblastoma, non-small cell lung cancer (NSLCL), pleiotrophin
Abbreviations: ALCL, anaplastic large cell lymphoma; ALK, anaplastic lymphoma kinase; ALO17, ALK lymphoma oligomerization partner on chromosome 17; ATIC, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase; BCR-Abl, breakpoint cluster region-Abl; CARS, cysteinyl-tRNA synthetase; Cdc42, cell division cycle 42; C/EBPβ, CCAAT/enhancer-binding protein β; CLTC, clathrin heavy chain; CML, chronic myeloid leukaemia; CNS, central nervous system; dALK, Drosophila ALK; DLBCL, diffuse large B-cell lymphoma; Dpp, decapentaplegic; DRG, dorsal root ganglia; EGFR, epidermal growth factor receptor; EML4, echinoderm microtubule-associated protein like 4; ERK, extracellular-signal-regulated kinase; FOXO3a, forkhead box O 3a; FRS2, fibroblast growth factor receptor substrate 2; GIST, gastrointestinal stromal tumour; Grb2, growthfactor-receptor-bound protein 2; HEK, human embryonic kidney; Hen-1, hesitation-1; IL-3, interleukin-3; IMT, inflammatory myofibroblastic tumour; IR, insulin receptor; IRS-1, IR substrate-1; JAK, Janus kinase; Jeb, jelly belly; JNK, c-Jun N-terminal kinase; LDLa, low-density lipoprotein class A; LTK, leucocyte tyrosine kinase; MAM, meprin, A5 protein and receptor protein tyrosine phosphatase mu; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; MK, midkine; MSN, moesin; mTOR, mammalian target of rapamycin; MUC-1, mucin-1; MYH9, non-muscle myosin heavy chain; NF-κB, nuclear factor κB; NIPA, nuclear interacting partner of ALK; NPM, nucleophosmin; NSCLC, non-small cell lung cancer; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B; PLCγ, phospholipase Cγ; PTN, pleiotrophin; RANBP2, Ran-binding protein 2; RPTP, receptor protein tyrosine phosphatase; RTK, receptor tyrosine kinase; SCC, squamous cell carcinoma; SCD-2, suppressor of constitutive dauer 2; SEC31L1, SEC31 homologue A; SH2, Src homology 2; Shc, Src homology and collagen homology; SHH, sonic hedghog; Shp1, SH2 domain-containing phosphatase 1; STAT, signal transducer and activator of transcription; TFG, TRK-fused gene; TGFβ, transforming growth factor β; TPM, tropomyosin; UCN-01, unco-ordinated 1
DISCOVERY OF THE ALK (ANAPLASTIC LYMPHOMA KINASE) RTK (RECEPTOR TYROSINE KINASE)
ALK was originally described as a novel tyrosine phosphoprotein in ALCL (anaplastic large cell lymphoma) cell lines in 1994 [1,2]. The identity of this protein was revealed as a chimaeric protein created via a translocation event between chromosomes (2;5)(p23:q35), generating a previously uncharacterized fusion protein, NPM (nucleophosmin)–ALK. NPM–ALK contains the N-terminal portion of the NPM protein and the kinase domain of a then novel tyrosine kinase protein that was named ALK after the disease [1].
STRUCTURAL FEATURES OF ALK
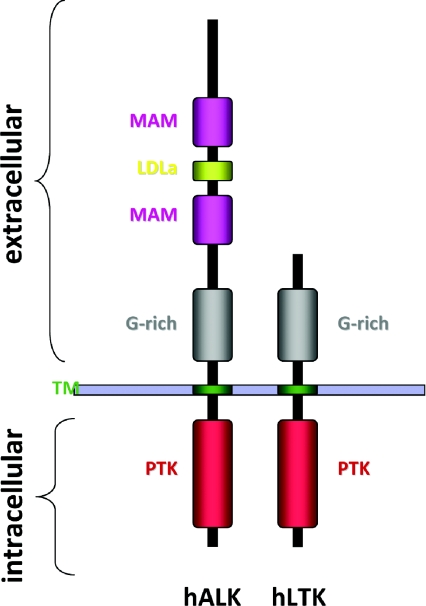
The intriguing characteristics of the full-length ALK RTK were not revealed until 1997, when they were simultaneously reported by two groups [3,4]. ALK displays the classical structural features of a RTK, with an extracellular ligand-binding domain, a transmembrane-spanning region and an intracellular tyrosine kinase domain. Based on overall homology, ALK is grouped with LTK (leucocyte tyrosine kinase), and together they form their own subgroup within the IR (insulin receptor) superfamily [3,4]. The human ALK gene encodes a protein of 1620 amino acids giving rise to a protein of approx. 180 kDa. However, as a result of post-translational modifications such as N-linked glycosylations, ALK migrates at approx. 220 kDa on SDS/PAGE [3,4]. The ALK extracellular region contains a unique combination of domains among the RTKs, exhibiting an N-terminal signal peptide, followed by two MAM (meprin, A5 protein and receptor protein tyrosine phosphatase mu) domains, an LDLa (low-density lipoprotein class A) motif and a large glycine-rich region proximal to the membrane [3–6] (Figure 1).
Figure 1. Domain structure of human ALK and human LTK.
The N-terminal region of human ALK (hALK) comprises two MAM domains (amino acids 264–427 and 480–626), one LDLa domain (amino acids 453–471) and a glycine rich (G-rich) region (amino acids 816–940). A transmembrane (TM)-spanning segment, connects the extracellular region with the protein tyrosine kinase (PTK), domain (amino acids 1116–1383)-containing intracellular region. The closest family member, LTK [hLTK (human LTK)], is depicted with the corresponding regions denoted. The signal peptide (amino acids 1–16), the glycine rich, G-rich, domain (amino acids 63–334) and the kinase domain (amino acids 510–777) located in the intracellular C-terminal region of the protein.
Within ALK, the LDLa domain has an unknown function. However, this module mediates the binding between the LDL-receptor and LDL [7,8], suggesting a potential role in ligand binding for this domain of ALK. MAM domains are thought to participate in cell–cell interactions [9], but their significance for ALK function is unclear. The importance of the MAM domain is nevertheless emphasized in studies from Drosophila in which a point mutation altering a highly conserved aspartic acid residue in the MAM domain to arginine renders dALK (Drosophila ALK) inactive [10]. The functional significance of the glycine-rich domain has also been reported in Drosophila, where several loss-of-function dALK mutants display point mutations that convert a single glycine residue within the glycine-rich region into an acidic amino acid [10]. The domain organization of ALK is conserved throughout evolution, with the highest conservation found in the kinase domains. In fact, mouse and human ALK show 87% overall homology at the protein level, and within the kinase domain these differ at only four amino acids. Although mouse and human ALK are highly similar, it should be noted that human ALK contains one extra tyrosine residue, Tyr1604, which has been implicated in tumour progression [11] (Figure 2). Within the activation loop of the kinase domain, ALK contains a YxxxYY motif in common with the IR. It has been reported that phosphorylation of the first tyrosine residue (Tyr1278) of this YxxxYY motif is predominant in the autoactivation of the ALK kinase domain. Phosphorylation of Tyr1278 appears to be determined in part by the intervening RAS amino acid triplet in the activation loop (Y'RAS'YY), immediately following Tyr1278 of ALK, which differs from the activation loop of the IR RTK [12,13].
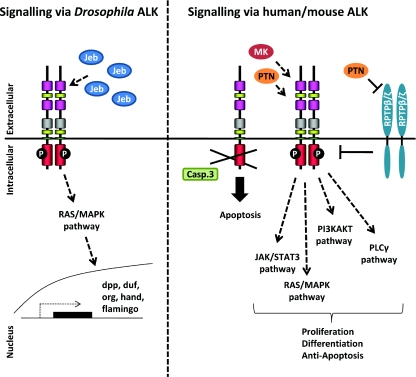
Figure 2. Signalling via ALK.
Signalling via dALK occurs via binding of the ligand Jeb, downstream activation of ERK and transcription of downstream target genes. Signalling via mammalian ALK is thought to occur via ligand-mediated dimerization in response to the MK and PTN ligands. ALK mediates signalling via the JAK/STAT, RAS/MAPK, PI3K and PLCγ pathways. Activation of ALK via RPTPβ/ζ, independently of direct ALK–ligand interactions has also been proposed. Lastly, ALK is proposed to function as a dependency receptor which is cleaved by caspase 3 (Casp. 3) in the absence of ligand, thereby promoting apoptosis.
ALK FUNCTION IN MODEL ORGANISMS
dALK
The In vivo function of ALK has been most thoroughly studied in Drosophila melanogaster where ALK was initially shown to drive ERK (extracellular-signal-regulated kinase) activation In vivo [5]. During embryonic development of the fruitfly, dALK plays a vital role in the formation of the visceral musculature of the gut [10]. In the absence of dALK, Drosophila embryos hatch into gut-less larvae, which consequently die. This phenotype is due to a lack of specification of a particular cell type, the founder cell, in the developing gut musculature of Drosophila embryos deficient in dALK signalling [14–16]. Activation of the dALK signal transduction pathway in wild-type flies is initiated by binding of the Jeb (jelly belly) ligand to a specific set of cells in the embryonic visceral mesoderm, which thereby are specified as founder cells. Founder cells then fuse with fusion competent myoblasts to give rise to the multinucleated visceral musculature of the gut [14–18]. Since dALK signalling specifies founder cells, loss of dALK results in an absence of founder cells, as well as muscle cell fusion, which as a consequence leads to defective assembly of a functional gut musculature in dALK mutant flies.
The Jeb protein is now well-established as an activating ligand for dALK, and as such is also required for founder-cell specification [14–16]. Jeb is a secreted protein of approx. 61 kDa containing a secretory signal and an LDLa domain [19]. The interaction of Jeb and dALK appears to be mediated via the LDLa domain in Jeb, since a Jeb mutant protein lacking the LDLa domain is unable to bind dALK [15]. Activation of the Jeb/dALK signalling pathway leads to ERK activation and further downstream transcription of target molecules such as Duf (dumbfounded)/Kirre (kin of irregular chiasm) [14–16], Org [15], Hand [20] and Dpp (decapentaplegic) [21] in the fruitfly (Figure 2).
The Jeb/dALK signalling pathway is also critical for development of the Drosophila embryonic endoderm in an indirect manner, since dALK activity in the visceral mesoderm is required for Dpp [TGFβ (transforming growth factor β)] transcription and subsequent signalling in the adjacent endoderm [21]. Moreover, dALK and Jeb play a central role acting as an anterograde signalling pathway mediating neuronal circuit assembly in the Drosophila visual system. In the developing Drosophila eye, dALK is expressed in, and required for, the targeting of neurons in the optic lobe, whereas Jeb is primarily produced by photoreceptor axons, functioning to control target selection of R1–R6 axons in the lamina and R8 axons in the medulla. Lack of either protein results in mistargeting of the R8 axons during later maturation of the optic lobe neuropile [22].
Caenorhabditis elegans ALK [SCD-2 (suppressor of constitutive dauer-2)]
In C. elegans, ALK has been implicated in neuronal control of entry into dauer and synapse stabilization [23,24]. C. elegans ALK (T10H9.2), now formally known as SCD-2, was identified as an effector through which the F-box protein Fsn-1 (F-box protein at the synapse-1) stabilizes synapse formation in GABAergic neuromuscular junctions [23]. More recently, SCD-2 mutants were characterized which display a temperature-sensitive adaptation to dauer entry [24]. Epistatic analysis in C. elegans suggests that SCD-2 modulates the TGFβ pathway upstream of Daf3 (abnormal dauer formation 3; Smad)/Daf5 (Sno/Ski) [24]. The SCD-2 ligand has been identified as Hen-1 (hesitation-1) [25], which like Drosophila Jeb is a secreted ligand containing an LDLa domain, and the genetically mapped signal transduction pathway employs the adaptor SOC-1 (suppressor of Clr-1) and the MAPK (mitogen-activated protein kinase) SMA-5 (small body size-5) [24].
Zebrafish LTK/ALK
Elegant studies in the zebrafish Danio rerio have recently demonstrated an In vivo role for LTK. During zebrafish embryogenesis LTK signalling leads to specification of iridophores from the neural crest linage, and mutants in LTK (known as shady) display defects in pigmentation patterns [26]. The LTK/ALK family in zebrafish comprises two genes: LTK and ALK, both of which contain MAM domains [26]. This is in contrast with mouse and human LTK which both lack MAM domains. Although zebrafish LTK has no reported ligand to date, its expression in the developing neural crest is of particular interest given the recent reports of human ALK activating mutations in neuroblastoma (see below).
MAMMALIAN ALK
Mammalian ALK has been postulated to play a role in the normal development and function of the nervous system, a hypothesis arising from the extensive expression of ALK mRNA throughout the nervous system during mouse embryogenesis [3,4,27]. Furthermore, this pattern of ALK mRNA expression is recapitulated in the developing CNS (central nervous system) of the chicken where ALK localizes to a subset of spinal motor neurons, the sympathetic ganglia and DRG (dorsal root ganglia) [28]. A similar pattern of expression in a subtype of DRG neurons during DRG development has also been reported in the rat [29]. In mice, the intensity of the ALK transcript and protein diminishes after birth, reaching minimum levels at 3 weeks of age, and is thereafter maintained at low levels in the adult animal [3]. Immunohistochemical studies of adult human tissue reveal a pattern consistent with that previously reported for mouse ALK, with a weak ALK signal observed in the CNS [30]. ALK mRNA transcripts of different sizes have also been reported in the testis, small intestine, colon, prostate and brain of adult human material, suggesting that differential splicing of ALK may occur [1].
in vitro studies also support a role for ALK in neuronal development. For instance, substitution of the extracellular region of ALK with the mouse IgG Fc domain, resulting in the creation of a constitutively active membrane-bound ALK–IgG Fc hybrid protein, has the capacity to induce neuronal differentiation of PC12 cells. Inhibition of MEK (MAPK/ERK kinase) abolishes ALK–IgG Fc-induced PC12 cell neurite outgrowth, implying that neurite outgrowth activity is mediated via the MAPK pathway [31]. Other groups have also reported a role for ALK in neurite outgrowth in cell culture [32–35]. Moreover, employing antibodies to activate ALK, it has been shown that Shc (Src homology and collagen homology) association with ALK is required for downstream ERK1/2 activation and neurite extension, further strengthening the hypothesis that neuronal differentiation induced by ALK is mediated via the MAPK pathway [33,36]. FRS2 (fibroblast growth factor receptor substrate 2)/SNT has also been reported to bind both ALK and NPM-ALK [36,37]. Since prolonged activation of ERK is associated with differentiation of PC12 cells, FRS2 recruitment may contribute to the neuronal differentiation of PC12 cells stimulated by activated ALK [36]. Experiments replacing the extracellular domain of ALK with the extracellular region of the EGFR (epidermal growth factor receptor), leads to the phosphorylation of ALK and subsequent activation of PLCγ (phospholipase Cγ) and PI3K (phosphoinositide 3-kinase). Activation of chimaeric EGFR–ALK efficiently transforms NIH 3T3 cells, further illustrating the potential oncogenic properties of ALK when deregulated [38].
LIGANDS OF MAMMALIAN ALK
In mammals PTN (pleiotrophin) also known as HB-GAM (heparin-binding growth-associated molecule) [39], OSF-1 (osteoblast-specific factor-1) [40], HARP (heparin affinity regulatory peptide) [41] and HBNF (heparin-binding neurotrophic factor) [42]; and MK (midkine) [43], also known as RIHB (retinoic acid-inducible heparin-binding protein) [44], have been postulated to be the activating ligands for ALK [6,45]. MK and PTN are small, heparin-binding growth factors implicated in diverse processes such as neural development, cell migration and angiogenesis [46,47]. The observation that PTN could function as a ligand for ALK arises from the isolation of a small portion of the extracellular region of ALK which was identified upon screening a human foetal brain phage display cDNA library for PTN-binding partners [6]. Subsequently, the PTN-related protein MK was identified as an ALK ligand. Furthermore, antibodies directed toward the ALK extracellular domain could inhibit the in vitro ligand–receptor interaction, suggesting that MK and PTN bind ALK [45].
MK and PTN are conserved throughout evolution and are found in species ranging from Drosophila to humans [46]. The subject of receptors for MK and PTN is a complex one since, in addition to ALK, a number of other proposed receptors exist. To date, MK and PTN have been shown to bind and signal via RPTPβ/ζ (receptor protein tyrosine phosphatase β/ζ) [48,49] and N-syndecan [50,51], whereas MK can also bind LRP (LDL-related protein) [52] as well as the α4β1- and α6β1-integrins [53].
Enhanced proliferation has been reported upon ALK activation via PTN [6,54], a function that seems to be dependent on PKB (protein kinase B)/Akt activation, thus implicating the PI3K pathway as a target of PTN–ALK signalling [54,55]. Moreover, a role for ALK in opposing apoptosis via the MAPK pathway in NIH 3T3 cells has been suggested [54]. In agreement, ribozyme-mediated reduction of ALK in glioblastoma cell lines results in increased apoptosis [55]. MK–ALK signalling has also been reported to be important for proliferation [45,56]. MK-mediated activation of ALK leads to IRS-1 (IR substrate-1) and Shc interaction, stimulating downstream signalling and ultimately activation of NF-κB (nuclear factor κB), thereby inducing cell growth, which is abrogated upon knockdown of the p65 subunit of NF-κB [56].
Although several groups have shown that PTN and MK activate ALK [6,45,54–57] some questions remain, since a number of groups have reported contradictory findings [32,33,58–60]. Agonist monoclonal ALK antibodies mediate efficient ERK1/2 phosphorylation and differentiation of PC12 cells; however, this could not be reproduced with PTN which failed to induce phosphorylation of ALK [32]. Furthermore, treatment with independently developed ALK-activating antibodies stimulates neuronal differentiation of SK-N-SH cells, which can be blocked by the MEK inhibitor PD98059, thus implicating the MAPK pathway in this process. However, this activation of ALK was not reproduced when MK and PTN were employed [33].
Interestingly, two different species of PTN, PTN15 and PTN18, have been described. PTN15 has been reported to promote glioblastoma growth via ALK, whereas PTN18 promoted glioblastoma migration in an RPTPβ/ζ-dependent manner [57]. The existence of two different PTN isoforms has been hypothesized to explain the discrepancy in reports concerning the ability of PTN to activate ALK. However, some investigators have been unable to reproduce the effects of either PTN15 or PTN18 on ALK [59]. Thus the physiological significance of ALK activation via PTN and MK is still a matter of debate and investigation within the field.
One hypothesis concerning ALK activation via PTN comes from the observation that PTN can indirectly lead to phosphorylation of ALK via binding to, and inactivation of, the phosphatase RPTPβ/ζ [61]. In this scenario, phosphorylation of ALK is independent of the ALK extracellular region, since a membrane-bound construct containing the ALK intracellular region is phosphorylated as effectively as the full-length protein [61].
MK and PTN display no obvious homology toward the dALK ligand Jeb, or the C. elegans ligand Hen-1 [19,25]. Jeb harbours a signal peptide and an LDLa domain [19], whereas MK and PTN are composed of two domains, one N-terminal N-domain and one C-terminal C-domain [62,63], which contain heparin-binding modules important for their activity [63,64]. In Drosophila the homologues for MK and PTN are the orphan ligands Miple1 and Miple2 (where Miple is Midkine and Pleiotrophin) [65]. Although the combined expression pattern of Miple1 and Miple2 complement the expression pattern of dALK, suggesting that a role as activating ligands for dALK is possible, this has yet to be tested at the gene level [65].
Finally, a novel dependence receptor function for ALK has recently been reported, suggesting that ALK may have an activation independent function [60]. In this study [60], cleavage of ALK by caspase 3 was found to expose a pro-apoptotic intracellular domain of ALK, resulting in increased apoptosis in Jurkat and 13.S.1.24 cells treated with apoptosis-inducing agents. Moreover, this pro-apoptotic function was counteracted by activation of ALK (Figure 2). The relevance of these intriguing results in an In vivo context remains to be explored.
ALK IN THE MOUSE
Throughout the literature a number of comments have been made regarding ALK mutant mice generated by the Morris group, stating that they are viable without any gross alterations [66]. This is in agreement with observations from the Palmer and Hallberg groups, who have also generated ALK mutant mice (E. Vernersson, R.H. Palmer and B. Hallberg, unpublished work). A recent study describes a third independently generated mouse ALK knockout, which displays increased hippocampal progenitor proliferation, increased performance in hippocampal-associated tasks, as well as increased levels of dopamine within the basal cortex [67]. Interestingly, the authors observed an increase in the number of calretinin-positive cells within the hippocampus, a phenotype also noted in MK-knockout animals [67,68].
MK and PTN exhibit similar expression patterns in rodents [69–72], and studies of MK mutant mice embryos, which display a strong up-regulation of PTN expression, suggest that PTN and MK are functionally redundant [73]. Indeed, the MK/PTN double mutant shows a more severe phenotype than either of the single knockouts. These phenotypes include female infertility [74] and hearing impairments [75]. The closest ALK relative, LTK, is expressed in pre-B-cells and adult neurons in the hippocampus and cerebral cortex [76]. Given that the ALK mutant phenotype is hippocampus related, the idea that LTK might compensate for ALK loss is intriguing and relevant [67]. It is known that LTK is able to promote neuronal differentiation of PC12 cells, underscoring the fact that ALK and LTK may potentially compensate for one another [77]. Thus far, however, mouse LTK appears only to be expressed in the adult and not during development [76], suggesting a clear distinction from ALK, which is extensively expressed during embryogenesis [3,4,27]. Furthermore, whereas LTK and ALK share a highly similar intracellular tyrosine kinase domain, there are notable differences in the extracellular region, with LTK lacking several domains found in ALK, such as the MAM and LDLa domains (Figure 1). To date, there is no published mouse LTK knockout to suggest a role for this RTK In vivo, and therefore the question of whether or not LTK might substitute for loss of ALK must await future investigation.
ALK IN HUMANS
The human ALK appears to exist as a 140 kDa protein, as well as the 220 kDa full-length ALK species [3]. The 140 kDa ALK species is thought to be the result of a cleavage within the extracellular region of full-length ALK, generating an 80 kDa form of unknown function [32]. The 140 kDa ALK protein is phosphorylated in response to activation of ALK [33,36,59]. In addition, the presence of an 85 kDa ALK protein which is phosphorylated in response to activating ALK antibodies has been observed in NIH 3T3 cells expressing full-length ALK [33]. Interestingly, a recent report suggests that an as yet unidentified factor secreted by Schwann cells can prevent ALK cleavage [29]. As yet no physiological function has been reported for these shorter ALK variants, and it is possible that the shorter forms of ALK are generated after activation of the receptor as part of down-regulation/degradation processes within the cell. However, future studies should lead to the definition of the cleavage sites, as well as the functional relevance of these forms of ALK In vivo.
ALK IN DISEASE
Since the initial discovery of the NPM–ALK fusion protein in patients suffering from ALCL [1,2], ALK fusion proteins have also been described in IMTs (inflammatory myofibroblastic tumours) [78], NSCLC (non-small cell lung cancer) [79,80], DLBCLs (diffuse large B-cell lymphomas) [81], and SCC (squamous cell carcinoma) of the oesophagus [82,83]. Furthermore, full-length ALK has also been reported to be expressed in cell lines and tumours suggesting oncogenic progression via overexpression [30,55,58,84–92] or gain-of-function mutations, as has recently has been reported in cases of neuroblastoma [93–97].
ALCL
ALK has been most extensively studied in the context of ALCL, a disease first described in 1985 [98]. ALCL is a non-Hodgkin's lymphoma arising from T-cells, and is characterized by the expression of CD30. ALK-positive ALCL is most commonly observed in young adults and children, accounting for 3% of adult non-Hodgkin's lymphoma and 10–30% of all non-Hodgkin's lymphoma in children [99–101]. ALK expression is an important prognostic factor used to predict clinical outcome for patients presenting with ALCL, since ALK-positive patients have a significantly higher 5-year survival rate as compared with ALK-negative cases [101–105]. ALK-positive ALCL occurs to a higher extent in children and young adults [101]; however, ALK expression in ALCL is a positive prognostic factor independent of patient age [103]. Currently, combinatorial chemotherapy treatment, CHOP [cyclophosphamide, hydroxydaunorubicin (doxorubicin), oncovin (vincristine) and prednisone] is applied for treatment of ALCL patients as a first approach. In addition, radiation therapy can be employed as an important complement to CHOP therapy [103,105,106]. Currently, there is no treatment for ALK-positive ALCL aimed at directly targeting ALK.
Besides ALK, active caspase 3 expression is also used as a prognostic indicator for favourable outcome in ALCL. In fact, caspase 3 activity is strongly correlated with the expression of ALK [107]. The presence of STAT3 (signal transducer and activator of transcription 3) in both ALK-positive and ALK-negative ALCL has led to the suggestion that activated STAT3 may be a negative prognostic factor independent of ALK expression in ALCL [108]. Survivin [109] and MUC-1 (mucin-1) [110] are two additional markers that indicate a poorer outcome in ALCL regardless of ALK status. Within ALK-positive ALCL, expression of MUC-1 is indicative of a poorer prognosis with a decrease in overall survival [110]. The exact mechanism of MUC-1 in the modulation of ALK-positive ALCL is not fully understood. However, MUC-1 is commonly overexpressed in oncogenic processes, and via its adhesive properties is thought to modulate both metastatic capacity and provide hindrance for immune cells trying to access the tumour cells [111].
There have been a number of intriguing reports concerning expression of ALK-fusion proteins in healthy individuals. Indeed, NPM–ALK transcripts have been detected in blood from healthy donors [112], as well as in healthy lymphoid tissue via RT (reverse transcriptase)–PCR [113]. Such reports raise the relevant question of whether the fusion transcript on its own is enough for inducing oncogenic transformation or whether secondary events are also required.
ALK fusion proteins in ALCL
In ALCL the kinase domain of ALK is fused to NPM, creating the constitutively expressed fusion protein NPM–ALK [1]. NPM is a multifunctional protein which serves as a molecular chaperone involved in the shuttling of pre-ribosome subunits from the nucleus to the cytoplasm during ribosome biogenesis, in addition to playing a role in DNA repair, transcription and genomic stability [114]. In the NPM–ALK fusion protein, oligomerization mediated via NPM juxtaposes two ALK tyrosine kinase domains, resulting in autophosphorylation and activation of ALK kinase activity [115,116]. The subcellular localization of NPM–ALK, which is present both in the cytoplasm and the nucleus, seems not to be critical for NPM–ALK-mediated transformation [116]. Besides the dimerization ability of NPM, the kinase activity of NPM–ALK is an absolute requirement for transforming activity, since mutation of the ATP-binding site (K219R) renders NPM–ALK kinase dead and eliminates transformation [116].
In addition to NPM, numerous other fusion partners exist for ALK in ALCL namely ALO17 (ALK lymphoma oligomerization partner on chromosome 17) [117], TFG (TRK-fused gene) [118,119], MSN (moesin) [120], TPM3 and 4 (tropomyosin 3 and 4) [121–123], ATIC (5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase) [124–126], MYH9 (non-muscle myosin heavy chain) [127] and CLTC (clathrin heavy chain) [128] (Table 1). These fusion partner proteins of ALK share several common characteristics: (i) the transcription of the fusion protein is driven via the promoter of the ALK partner protein; (ii) the localization of the fusion protein is determined by the ALK partner protein and; (iii) oligomerization via the ALK partner protein induces autophosphorylation and thereby activation of the ALK kinase domain.
Table 1. Fusion partners of ALK in disease.
| Disease | Fusion protein | Chromosomal abnormality | References |
|---|---|---|---|
| ALCL | NPM–ALK | t(2;5)(p23;q35) | [1,2] |
| ALCL | ALO17–ALK | t(2;17)(p23;q25) | [117] |
| ALCL | TFG–ALK | t(2;3)(p23;q21) | [118,119] |
| ALCL | MSN–ALK | t(2;X)(p23;q11-12) | [120] |
| ALCL | TPM3–ALK | t(1;2)(q25;p23) | [121,122] |
| ALCL | TPM4–ALK | t(2;19)(p23;p13) | [123] |
| ALCL | ATIC–ALK | inv(2)(p23;q35) | [124–126] |
| ALCL | MYH9–ALK | t(2;22)(p23;q11.2) | [127] |
| ALCL | CLTC–ALK | t(2;17)(p23;q23) | [128] |
| IMT | TPM4–ALK | t(2;19)(p23;p13) | [174] |
| IMT | TPM3–ALK | t(1;2)(q25;p23) | [174] |
| IMT | CLTC–ALK | t(2;17)(p23;q23) | [176,177] |
| IMT | ATIC–ALK | inv(2)(p23;q35) | [175] |
| IMT | SEC31L1–ALK | t(2;4)(p23;q21) | [180] |
| IMT | RANBP2–ALK | t(2;2)(p23;q13) inv(2)(p23;q11-13) | [179] |
| IMT | CARS–ALK | t(2;11;2)(p23;p15;q31) | [117,178] |
| NSCLC | EML4–ALK | inv(2)(p21;p23) | [79,80] |
| NSCLC | TFG–ALK | t(2;3)(p23;q21) | [80] |
| DLBCL | NPM–ALK | t(2;5)(p23;q35) | [204] |
| DLBCL | CLTC–ALK | t(2;17)(p23;q23) | [237] |
| SCC | TPM4–ALK | t(2;19)(p23;p13) | [82,83] |
As discussed above, NPM is responsible for the dimerization essential for autophosphorylation of, and downstream signalling via the NPM–ALK tyrosine kinase domain [115,116]. The TPM3 and TFG fusion partners contain dimeric α-coiled-coil structures that are hypothesized to mediate the dimerization of TPM3–ALK [121] and TFG–ALK [119] respectively. Since ATIC exists as a homodimer [129], this property is assumed to be responsible for the activation of ATIC–ALK [124–126]. For MSN, MYH9 and CLTC, the means of dimerization seems to be more complex. It is believed that clathrin coat formation activates the kinase domain of ALK via the close proximity of the CLTC–ALK fusion proteins, since CLTC is a component of clathrin-coated vesicles. In agreement with this suggestion, CLTC–ALK is localized in a punctuated pattern within the cell, consistent with clathrin-coated vesicles [128]. The theory of ‘close proximity’ as a means of activating the ALK tyrosine kinase domain also applies to MSN–ALK, which is thought to be activated via the ability of MSN to crosslink the plasma membrane with the actin cytoskeleton [120]. Myosin heavy chain is also known to form a dimer, however the MYH9 sequence that is fused to ALK is missing the dimerization domain present in the full-length protein. In spite of this, the MYH9–ALK fusion protein is phosphorylated In vivo, indicating either an uncharacterized dimerization domain or that MYH9 potentially interacts with other proteins leading to multimerization and consequent activation of ALK [127].
Oncogenic signalling via ALK-fusion proteins
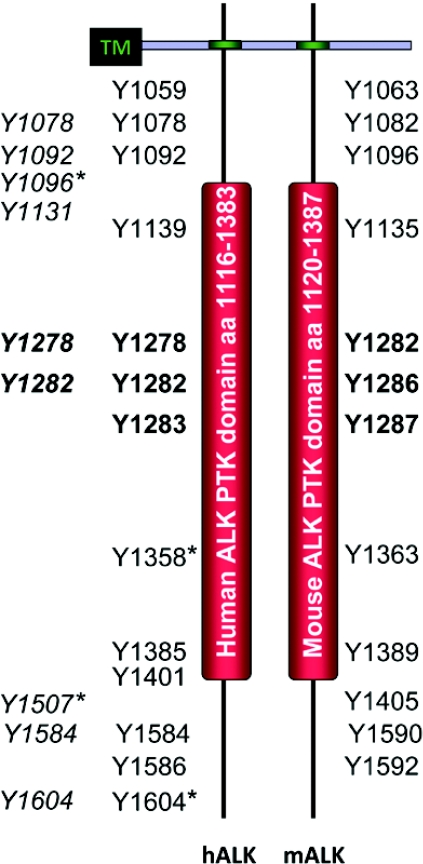
The most studied ALK-fusion protein, NPM–ALK, signals via the PLCγ, PI3K, RAS/MAPK and JAK (Janus kinase)/STAT pathways. PLCγ has been shown to directly interact with NPM–ALK via its SH2 (Src homology 2) domain. This interaction is of importance for the transforming potential of NPM–ALK, since mutation of the NPM–ALK/PLCγ interaction site abrogates transformation in transfected cell culture models [11]. A systematic analysis of NPM–ALK tyrosine to phenylalanine mutants (Figure 3), has identified Tyr664 (corresponding to Tyr1604 in human full-length ALK; Tyr1604 is present in human ALK but not in mouse ALK) as responsible for the PLCγ interaction. Interestingly, all other mutants (except the kinase dead Y338F, Y342F and Y343F mutants) retained their ability to confer IL-3 (interleukin-3)-independent growth of Ba/F3 cells [11], indicating a key role for the PLCγ pathway in ALK-dependent transformation.
Figure 3. Tyrosine residues phosphorylated in the intracellular regions of human and mouse ALK.
The intracellular regions of human and mouse ALK (hALK and mALK respectively) contain the PTK domain. Potential autophosphorylation sites are shown within human and mouse ALK [11]. Note that there is no equivalent tyrosine residue in mouse ALK for human Tyr1604. Tyrosine residues within the activation loop are shown in bold. In italics is a profiling of putative phosphorylated tyrosine residues in NPM–ALK from ALCL cancer cells [166]. Four tyrosine sites, marked with*, have been tested by mutagenesis in NPM–ALK for interaction with signalling proteins such as PLC-γ (Tyr1604 in hALK) [11], Shc (Tyr1507 in hALK) [115], Src (Tyr1358 in hALK) [157], IRS-1 (Tyr1096 in hALK) [115] and SNT (Tyr1096 and Tyr1507 in hALK) [37]. Localization of tyrosine residues within the intracellular region is not to scale. TM, transmembrane domain.
NPM–ALK interacts with PI3K thereby activating the catalytic subunit of PI3K, and leading to the phosphorylation of PKB/Akt and subsequent downstream signalling events [130]. This interaction has been reported to occur via both the SH2 and SH3 domains of the p85 subunit of PI3K [130,131]. Furthermore, an indirect interaction of p85 and NPM–ALK via other adaptor molecules must also be considered. Inactivation of the PI3K pathway induces apoptosis in NPM–ALK-positive cells [130,132], indicating a role for the PI3K/PKB/Akt pathway in anti-apoptotic signalling. Moreover, PKB/Akt activation appears to be critical for transformation by NPM–ALK, since mice inoculated with NPM–ALK-positive cells expressing a dominant-negative PKB/Akt display impaired oncogenic growth and delayed tumour formation [132].
NPM–ALK regulates the FOXO3a (forkhead box O 3a) transcription factor via the PI3K/PKB/Akt pathway. Active PKB/Akt phosphorylates FOXO3a, leading to retention of FOXO3a in the cytoplasm. This has been shown to result in alterations at the level of transcription of several FOXO3a target genes in NPM–ALK-expressing lymphoma, including cyclin D2, Bin-1 and p27kip1[133]. In agreement, inactivation of PKB/Akt activity in ALCL cell lines, employing small molecule inhibitors, results in up-regulation of p27kip1 levels and induction of cell-cycle arrest [134]. An additional target for PI3K/PKB/Akt signalling in ALK-positive ALCL cell lines is mTOR (mammalian target of rapamycin), which displays reduced levels of phosphorylation in response to inhibition of PKB/Akt [135]. In addition, the RAS/MAPK pathway has also been reported to be important for mTOR activation in NPM–ALK-expressing cells [136].
NPM–ALK interacts with IRS-1, Shc and Grb2 (growth-factor-receptor-bound protein 2), thus implicating the RAS/MAPK pathway as a downstream target of NPM–ALK activity [115]. Interaction with IRS-1 and Shc are non-essential for transformation, given that NPM–ALK mutants unable to interact with Shc and IRS-1 are still able to transform NIH 3T3 cells [115]. Previous reports suggest that NPM–ALK may be able to activate MEK directly [137,138]. Furthermore, simultaneous depletion of ERK1 and ERK2 impairs cell proliferation, whereas ERK1 depletion alone induces apoptosis in an ALCL-derived ALK-positive cell line, indicating that ERK1/2 are involved in survival and pro-apoptotic functions [138].
The last major pathway engaged by NPM–ALK is the JAK/STAT pathway. Several reports have demonstrated a correlation between NPM–ALK expression and STAT3 phosphorylation status and activation [139–141]. In agreement, inactivation of NPM–ALK with small molecule ALK inhibitors results in reduced STAT3 phosphorylation [142–144]. The observed interaction of NPM–ALK with JAK3, the receptor-associated tyrosine kinase responsible for STAT3 activation [140,145], provides a possible explanation for the effect of NPM–ALK on STAT3, given that inhibition of JAK3 leads to a reduction of STAT3 activation by NPM–ALK together with increased cellular apoptosis [145,146]. Although the precise mechanism of NPM–ALK induced activation of STAT3 is still a matter of investigation, it is clear that JAK3 activity is strongly associated with ALK expression and STAT3 phosphorylation In vivo [147]. It is also clear that down-regulation of active STAT3 is incompatible with the ALK-positive ALCL-transforming phenotype, as STAT3 compromised cells display increased apoptosis and cell-cycle arrest [143,145,148,149]. This finding is further supported by gene-targeting experiments which support a role for STAT3 in NPM–ALK-induced tumour growth In vivo [150].
Several groups have provided additional information concerning the importance of the JAK/STAT pathway in ALCL. Loss of the JAK/STAT pathway negative regulator Shp1 (SH2 domain-containing phosphatase 1) as a result of DNA methylation has been reported in ALK-positive ALCL [151]. Indeed, restoring Shp1 levels in NPM–ALK-expressing cell lines inactivates the JAK/STAT pathway and blocks cell-cycle progression [149]. In addition, protein phosphatase 2A, a STAT3-interacting protein necessary for sustained STAT3 phosphorylation, is overexpressed in ALK-positive ALCL [141].
Another member of the STAT family, STAT5, plays a less clear role in the pathogenesis of ALK-positive ALCL. Although several reports have been unable to detect activation of STAT5 in ALK-expressing cell lines [140,141], an interaction between STAT5B and NPM–ALK has been observed by others [152]. Furthermore, expression of dominant-negative STAT5B induces apoptosis and cell-cycle arrest in ALCL-derived cell lines expressing NPM–ALK [152]. In ALK-positive ALCL cell lines, STAT5A is silenced via methylation in a STAT3-dependent manner. Up-regulation of STAT5A by inhibition of methylation, results in decreased transcription of NPM–ALK due to the ability of STAT5A to interact with the NPM–ALK promoter region, indicating a tumour suppressor function for STAT5A in ALK-positive ALCL-derived cell lines [153]. Future investigations should clarify the role of STAT5A and B in NPM–ALK-induced tumorigenesis.
Besides the above mentioned players there are a number of other proteins implicated in NPM–ALK-mediated malignancy. The small GTPases Rac1 (ras-related C3 botulinium toxin substrate 1) and Cdc42 (cell division cycle 42) are regulated by NPM–ALK in ALCL and other cell lines [154,155]. Furthermore, upon depletion of Cdc42, cell-cycle arrest and apoptosis are induced [154]. In addition, loss of p130Cas (Crk-associated substrate) modifies cell shape and inhibits cellular transformation by NPM–ALK. This dependency on p130Cas has been suggested to be modulated via Grb2 in NPM–ALK-positive ALCL cells [154]. Moreover, knockdown of Shp2 reduces the migratory capacity of cells expressing NPM–ALK [156] and Src kinases, in particular pp60Src, have been suggested to be important for the proliferative capacity of NPM–ALK-positive ALCL cells [157].
Yeast two-hybrid screening has led to the identification of NIPA (nuclear interacting partner of ALK) as a novel downstream target of NPM–ALK that has been shown to interact with NPM–ALK, as well as other ALK fusions, in a tyrosine kinase-dependent manner. NIPA has been characterized as an F-box-containing protein that defines the multisubunit ubiquitin E3 ligase complex SCFNIPA (where SCF is stem cell factor), which targets nuclear cyclin B1 for ubiquitination during interphase, thus contributing to the timing of mitotic entry [158]. Overexpression of NIPA protects Ba/F3 cells from apoptosis induced by IL-3 withdrawal. Furthermore, apoptosis triggered by wortmannin treatment in NPM–ALK-transformed Ba/F3 cells is enhanced by overexpression of dominant-negative NIPA mutants, implying an anti-apoptotic role for NIPA in NPM–ALK-mediated signalling events [159].
Activation of JNK (c-Jun N-terminal kinase) in ALK-transformed cells has also been reported. Utilizing the Vav promoter to drive NPM–ALK expression in mice results in development of lymphomas which display a robust increase in JNK phosphorylation relative to controls [160]. Subsequent work has reported activation of JNK and c-Jun in ALCL cell lines as well as primary tumour cells. Similar activation was also observed upon introduction of NPM–ALK, but not a kinase-dead mutant NPM–ALK, into HEK (human embryonic kidney)-293T cells, suggesting that NPM–ALK is indeed capable of activating JNK [161].
Elevation of SHH (sonic hedghog) expression in ALK-positive ALCL has recently been reported [162]. This increase of SHH expression is apparently dependent on NPM–ALK-induced PI3K activity, since inhibition of PI3K leads to a concentration-dependent decrease of SHH protein levels. Inhibition of SHH pathway activity results in decreased cell viability, colony formation and cell-cycle arrest in ALK-positive ALCL cell lines [162].
A number of studies have used high-throughput approaches to identify novel ALK targets [80,137,163–168]. Immunoprecipitation of phosphotyrosine peptides from ALCL cell extracts followed by LC–MS/MS (liquid chromatography tandem MS) analysis identifies ALK as the sole tyrosine kinase phosphorylated within the activation loop, together with tyrosine phosphorylated signalling molecules such as dok2 (docking protein 2), IRS-1, Shc, Crk, CrkL and STAT3 among others [166]. A similar global survey of phosphotyrosine signalling in lung cancer found up-regulation and activation of full-length ALK, and independently identified the EML4 (echinoderm microtubule-associated protein like 4)–ALK oncogene, as well as identifying a number of potential ALK downstream signalling proteins [80]. Immunoprecipitation of NPM–ALK from the Karpas 299 cell lines with monoclonal and polyclonal antibodies led to the identification of 36 NPM–ALK binding partners, including both known and novel ALK-interacting proteins [137]. Recent results employing a proteomics approach with NPM–ALK have moreover defined a set of phosphorylated proteins, including VASP (vasodilator-stimulated phosphoprotein) and ATIC, as ALK targets [163]. In addition, analysis of the transcriptomes of ALCL cell lines has established a number of ALK-regulated genes, including the transcription factor C/EBPβ (CCAAT/enhancer-binding protein β) and the anti-apoptotic protein BCL2A1 (B-cell lymphoma 2A1) which are required to sustain the growth and survival of ALK-positive ALCL cells [164]. A further study comparing ALK-positive and ALK-negative ALCL led to the identification of a number of ALK transcriptional targets, of which BCL-6, serpinA1 and C/EBPβ were also confirmed at the protein level [165]. Finally, using a tandem-affinity purification approach several novel interaction partners for ALK were isolated including MCM6, MSH2, Nup98, Importin 8, 82Fip and Bag2 [168].
None of the other known ALK-fusion proteins have been studied as extensively as NPM–ALK with respect to downstream signalling. Nevertheless they are assumed to function in a similar manner as NPM–ALK. This assumption is supported by studies on the ATIC–ALK and TFG–ALK fusion proteins, in which ATIC–ALK was shown to associate with Grb2 and Shc [125], and TFG–ALK with Grb2, Shc and PLCγ [118].
An extensive evaluation of the oncogenic potential of NPM–ALK, TPM3–ALK, TFG–ALK, CLTLC–ALK and ATIC–ALK both In vivo and in vitro suggests that TPM3–ALK-expressing cells display a higher migratory and invasive capacity in vitro as compared with the other ALK-fusion proteins examined. However, when grafting transfected NIH 3T3 cells subcutaneously into nude mice, NPM–ALK- and TFG–ALK-mediated tumours were readily detected 6 days after inoculation, whereas TPM3–ALK-induced tumours were detected a few days later [169]. The invasive properties of the ALK-fusion proteins were further evaluated by an In vivo lung metastasis assay, in which TPM3–ALK displayed an increased capacity to form metastases in comparison with other ALK-fusion proteins [169]. However, whether this increased ability of TPM3–ALK-transfected cells to induce metastases has clinical relevance is currently unclear. In patient samples, the observation that activated STAT3 is expressed in 84% of ALK-positive ALCL cases has been speculated to be due to differences in signalling via the different ALK-fusion proteins [108]. Indeed, this hypothesis is supported by reports of efficient STAT3 phosphorylation in cells by NPM–ALK, TPM3–ALK, TFG–ALK and ATIC–ALK, but not by CLTLC–ALK [169,170].
IMT
IMT is a benign malignancy of mesenchymal origin with a prominent inflammatory component consisting of plasma cells and lymphocytes [171]. Although IMT mostly affects young individuals, it can develop at all ages. First described in the lung, these ‘inflammatory pseudotumours’ were considered a post-inflammatory condition rather than a malignant process [172]. IMTs can manifest in almost any anatomical site [171], although they are most common in tissues such as the lung, abdomen and retroperitoneum [173]. In 1999, Griffin et al. [78] reported an association between the 2p23 chromosomal rearrangement and expression of ALK with IMTs; the first indication that IMTs, or a subpopulation of IMTs, were neoplastic in origin rather than reactive [78]. Further studies have confirmed the presence of several different ALK-fusion proteins in IMTs, all containing the kinase domain of ALK together with one of a variety of partner proteins responsible for dimerization (Table 1). Those fusion partners described to date are TMP3–ALK [174], TMP4–ALK[174], ATIC–ALK [175], CLTC–ALK [176,177], CARS (cysteinyl-tRNA synthetase)–ALK [117,178], RANBP2 (Ran-binding protein 2)–ALK [179] and SEC31L1 (SEC31 homologue A)–ALK [180]. In fact, 35–60% of all IMTs appear to exhibit ALK rearrangements [181,182], with ALK-positive IMTs preferentially affecting young individuals [174,181,182]. Interestingly, ALK-positive IMT reoccurs in approx. 50% of cases, although no metastasis was detected in this group [183]. This pattern of occurrence is similar to that noted for ALK-positive ALCL. Moreover the prognosis appears to be better for ALK-positive IMTs [184], as is the case for ALK-positive ALCL. However, no obvious ALCL prognostic factors correlate with IMT [183] and the difference between ALK-positive and ALK-negative IMTs is still unclear. Thus we do not presently understand the functional relevance and consequences of ALK signalling in ALK-positive IMT.
NSCLC
Lung cancer is the most common cause of cancer death in the world. Of the annual 1.3 million cases worldwide, NSCLC accounts for approx. 80% of all lung cancers [185,186]. In 2007 a novel gene fusion between ALK and EML4 was reported in NSCLC [79,80]. Studies from over 1900 NSCLC cases suggest a frequency of EML4–ALK fusion in the range 0.1–7.9%, encompassing a number of different translocation variants [79,80,187–193]. Extrapolation would suggest that approx. 3.5% of all NSCLC cases contain an EML4–ALK translocation, equivalent to over 45000 patients worldwide. Furthermore, the EML4–ALK translocation seems to be uninfluenced by ethnic differences, in contrast with point mutations in the EGFR observed in NSCLC [194]. EML4 does not appear to be exclusive as the fusion partner for ALK in NSCLC, since Rikova et al. [80], also identified TFG as an ALK-fusion partner in one NSCLC sample [80]. Also unclear is the issue of whether EML4–ALK is causative in NSCLC, and whether EML4–ALK will be an effective clinical marker/therapeutic target. However, cells expressing oncogenic variants of ALK or EML4–ALK fusion proteins show reduced growth upon treatment with ALK inhibitors, such as PF-2341066 or NVP-TAE684 [195–198]. Furthermore, mice overexpressing EML4–ALK (variant 1) develop tumours with malignant characteristics, which are treatable with administration of a small specific ALK inhibitor, NVP-TAE684 [142], confirming the potent oncogenic activity of the fusion kinase [195].
Although an attractive diagnostic marker, some caution should be employed when considering EML4–ALK as a diagnostic marker for NSCLC. A recent study by Martelli et al. [193] observed a subset (7.5%) of EML4–ALK translocations in NSCLC samples from European patients. However, the presence of EML4–ALK was also detected in non-tumour lung samples, where the EML4–ALK transcript was not detected in matching tumour samples from the same patient [193]. This observation raises some important questions concerning the role of EML4–ALK in the development of NSCLC, and further work is required to clarify this important issue.
ALK-positive DLBCL
Among diffuse DLBCLs there is an ALK-positive subpopulation, which shows features of plasmablastic morphology. ALK-positive DLBCL usually expresses markers such as EMA (epithelial membrane antigen), CD138 and cytoplasmic Ig, together with ALK. Furthermore, ALK-positive DLBCL lacks expression of B-cell markers (CD20, CD79a), the T-cell marker CD3 and is typically negative for CD30 expression [85,199]. To date, 41 cases of ALK-positive DLBCL have been reported, spanning all age groups and displaying an overall predominance in males (male/female ratio, 5:1). In addition, ALK-positive DLBCL is correlated with an aggressive clinical outcome as well as a poor response to treatment [170,199–202]. The most frequent chromosomal rearrangement in ALK-positive DLBCL is the t(2;17)(p23;q23) translocation which generates CLTLC–ALK, although a few cases of NPM–ALK fusions have also been described [203,204]. Recently, a third variant of DLBCL was reported with a cryptic insertion of the 3′-ALK sequence at chromosome 4q22-24, displaying immunohistochemical staining characteristic of a DLBCL and focal granular cytoplasmatic staining of ALK [200], although the ALK-fusion partner in this case has not been identified. As previously discussed NPM–ALK translocation induces activation of STAT3 [140,141], and this also seems to occur in CLTC–ALK-driven DLBCL, but not in ALK-negative DLBCL [170]. These observations again indicate a close connection between deregulated ALK and phosphorylation of STAT3, suggesting that STAT3 inhibitors might be useful therapies for DLBCL.
Overexpression of ALK in cancer
Several groups have examined overexpression of ALK in different tumour materials, reporting excessive ALK expression in thyroid carcinoma, NSCLC, breast cancer, melanoma, neuroblastoma, glioblastoma, astrocytoma, retinoblastoma, ewing sarcoma and rhabdomyosarcoma [58]. Besides these tumour types, ALK expression has also been described in leiomyosarcoma and malignant peripheral nerve sheath tumours, as well as malignant fibrous histocytoma [86]. Some of these, NSCLC, glioblastoma and neuroblastoma, have also been evaluated with respect to ALK activation. However, the significance of ALK overexpression in many of these cancer types is uncharacterized at the molecular level.
In the case of rhabdomyosarcoma, leiomyosarcoma and malignant fibrous histocytomas, increased copy number of the chromosomal region 2p23 may lead to the overexpression of ALK [86].
PTN has been reported to induce ALK phosphorylation and subsequent downstream PKB/Akt activation in glioblastoma. In agreement, glioblastoma cell lines depleted of ALK grow at a reduced rate compared with parental cell lines [55]. Similar observations have also been reported in U87MG glioblastoma cell lines stimulated with MK [45]. Combined targeting of ALK and PTN in U87 glioblastoma cells significantly impairs tumour growth in an In vivo xenograft model [205].
A clear role for ALK in breast cancer has not been firmly established; however, several lines of evidence suggest a role for ALK in this disease. First, ALK is strongly expressed in several different subtypes of human breast cancers, in a pattern not consistent with normal tissue [84]. Secondly, PTN, the proposed mammalian ALK ligand, is extensively expressed in breast cancer [206,207], and expression of truncated PTN in a human breast cancer cell line abrogates tumour formation in nude mice [208]. Thirdly, the PTN receptor RPTPβ/ζ is strongly expressed in different subtypes of human breast cancer [61]. Taken together with the hypothesis that ALK is indirectly activated via PTN/RPTPβ/ζ signalling [61], it is possible that ALK harbours oncogenic potential in breast cancer development.
ALK gain-of-function mutations in neuroblastoma
Neuroblastoma is derived from neural crest cells of the sympaticoadrenal lineage and can therefore arise throughout the sympathetic nervous system. It is the most common solid tumour in childhood and accounts for 15% of all paediatric oncology deaths [209]. Neuroblastoma tumours show heterogeneous biological and clinical features and a subset may undergo spontaneous differentiation or regression with little or no therapy, whereas the majority are difficult to cure with current regimes. The most common genetic features of neuroblastoma are amplification of the proto-oncogene MYCN (v-Myc myelocytomatosis viral-related oncogene, neuroblastoma derived), deletions of parts of chromosome arms 1p and 11q, gain of parts of 17q and triploidy [210]. Expression of full-length ALK in neuroblastoma was first demonstrated in 2000 [88], and is supported by subsequent studies showing that the ALK locus is amplified in neuroblastoma cell lines, as well as in primary patient samples [89,90]. A physical association between ALK and ShcC has also been demonstrated [89]. In agreement, silencing of ALK in NB-39-nu and Nagai neuroblastoma cell lines results in the down-regulation of ShcC phosphorylation, as well as PKB/Akt activation, with concomitant apoptosis [90].
During 2008, five groundbreaking studies reported the presence of activating ALK mutations in both familiar [96,97] and sporadic [93–97] cases of neuroblastoma. Interestingly, all mutations (except one) are localized within the kinase domain of ALK and are assumed to be activating in nature (Table 2). Furthermore, neuroblastoma patients who are positive for ALK mutations appear to have a worse prognosis [93–97]. In line with an oncogenic role for ALK in neuroblastoma, a number of neuroblastoma cell lines were also shown to harbour activating ALK mutations, and knockdown of ALK in these cell lines resulted in an inhibition of proliferation [97]. Two of these activating ALK mutants, F1174L and K1062M, can indeed induce rapid formation of subcutaneous tumours in nude mice, thus displaying transforming potential In vivo [94].
Table 2. Activating mutations within the ALK kinase domain in neuroblastoma patients.
| Nucleotide changes | Amino acid mutation | Targeted region | References |
|---|---|---|---|
| 3260 C→T | T1087I | Juxtamembrane domain | [94] |
| 3271 G→A | D1091N | Juxtamembrane domain | [97] |
| 3383 G→C | G1128A | P loop | [97] |
| 3452 C→T | T1151M | Kinase domain | [95] |
| 3497 T→G | M1166R | C helix | [97] |
| 3512 T→A | I1171N | C helix | [97] |
| 3520 T→A | F1174I | End of C helix | [93,97] |
| 3521 T→G | F1174C | End of C helix | [94,96] |
| 3520 T→G | F1174V | End of C helix | [94] |
| 3522 C→A/G | F1174L | End of C helix | [93–95,97] |
| 3575 G→C | R1192P | β4 strand | [97] |
| 3700 G→A | A1234T | Catalytic loop? | [95] |
| 3733 T→G | F1245V | Catalytic loop | [97] |
| 3733 T→A | F1245I | Catalytic loop | [93] |
| 3735 C→A/G | F1245L | Catalytic loop | [93,94] |
| 3734 T→G | F1245C | Catalytic loop | [95,97] |
| 3749 T→C | I1250T | Catalytic loop | [97] |
| 3824 G→T | R1275L | Activation loop | [96] |
| 3824 G→A | R1275Q | Activation loop | [93–97] |
| 3833 A→C | Y1278S | Activation loop? | [96] |
The work described above highlights the relevance of understanding ALK-mediated signalling in a physiological context, since ALK ligands, as well as mutations in downstream ALK signalling pathway components, are obvious candidates with potential roles in the progression of neuroblastoma. The identification of a role for a drug targetable RTK, such as ALK in neuroblastoma development, provides a real hope for future therapeutic treatments for this devastating disease.
MOUSE MODELS FOR ALK-DRIVEN ALCL
NPM–ALK has been established as the causative protein in ALCL by a number of groups both in vitro and In vivo. Bone marrow-expressing human NPM–ALK is able to induce lymphoid malignancies in lethally irradiated mice, providing In vivo support for NPM–ALK as an oncogenic agent [211,212]. Moreover, transgenic animals expressing NPM–ALK under the control of the CD4 promoter develop CD30-positive NPM–ALK T-cell lymphomas, as well as plasma cell tumours [213]. Utilization of the haematopoietic cell-specific Vav promoter [160] and the Lck promoter [214] to overexpress NPM–ALK in mice further confirm a role in the development of lymphomas. Additional strategies of investigating NPM–ALK in the development of ALCL using animal models have been reported and recently reviewed [215].
POTENTIAL TREATMENT STRATEGIES FOR ALK-POSITIVE CANCERS
The development of tyrosine kinase inhibitors for use in cancer therapy has proven effective in several cases. Most well-known is Gleevec which targets the BCR-Abl (breakpoint cluster region-Abl) fusion protein in CML (chronic myeloid leukaemia) [216,217]. In addition to inactivating Abl, Gleevec also targets the c-Kit RTK, as well as the PDGFR (platelet-derived growth factor receptor) [218,219]. c-Kit is highly expressed in GISTs (gastrointestinal stromal tumours) [220] and clinical trials have established Gleevec as a treatment for GIST patients [221–223]. Other examples of RTK inhibitors are Gefitinib and Erlotinib, monoclonal antibodies which selectively inhibit EGFR (ErbB1), and are used in treatment of NSCLC [224].
A screen of more than 600 different human cancer-derived cell lines with the ALK inhibitor TAE684 identified a number of ALK-positive ALCL, neuroblastoma and NSCLC cell lines in which proliferation was inhibited [198]. TAE684 is a 5-chloro-2,4-diaminophenylpyrimidine which targets the ATP-binding pocket of ALK thereby blocking ATP binding [142]. Furthermore, administration of TAE684 in mice after injection of NPM–ALK-positive ALCL cells (Karpas 299 cell line) prevented development of disease and induced regression of pre-induced ALCL [142].
Neuroblastoma cell lines harbouring different activating ALK mutations appear to respond differently to inhibition of ALK via the inhibitor TAE684. For example, the ALK mutant F1174L observed in SH-SY5Y and KELLY neuroblastoma cell lines is sensitive to TAE684, whereas the R1275W ALK mutant observed in the SMS-KCNR cell line is unaffected. ALK-silencing experiments in these cell lines support the previous inhibitor TAE684 results, since growth inhibition and increased apoptosis were noted in SH-SY5Y and KELLY cells, but not in SMS-KCNR cells [95,97]. It should be noted that these ALK mutants were also tested in a Ba/BF3 cells, where both mutants exhibited transforming ability and were sensitive to TAE684 [95]. The reasons behind this differential response to TAE684 are presently unclear, but may indicate differences in genetic background, or reflect complex genetic predispositions acquired by these cells.
An additional ALK inhibitor is PF-02341066, also an ATP competitor, which targets both c-Met and ALK [196]. PF-02341066 has been shown to reduce ALK-positive ALCL development in an animal model [197], and is currently in phase I trails for patients with advanced anaplastic large cell lymphomas (www.ClinicalTrials.gov). UCN-01 (unco-ordinated 1), a staurosporine derivative, which inhibits PKC (protein kinase C) [225] and chk1 [226], has been reported to cause disease regression in one patient with ALK-positive ALCL [227]. However, others have been unable to confirm a direct relationship between UCN-01 and ALK [228]. Presently a phase I clinical trial using UCN-01 in patients with relapsed or refractory systemic ALCL or mature T-cell lymphoma is underway (www.ClinicalTrials.gov). Although several PTK inhibitors are used successfully in the clinic, ALK inhibitors are not yet so well developed. This active area of research should hopefully produce effective and clinically relevant compounds [229,230].
Besides small-molecule chemical inhibitors, ALK has also been the target of a number of strategies aimed at reducing mRNA levels, thus depleting the ALK protein. In glioblastoma, PTN and ALK appear to be up-regulated and functionally relevant, since ALK and PTN ribozyme-expressing glioblastoma cell lines show reduced signalling potential. Moreover, in xenograft models, silencing of either ALK or PTN results in reduced tumour growth [55,231]. A double knockdown of PTN and ALK completely inhibited xenograft tumour growth over a 60-day experimental period [205]. Similar ribozyme-mediated silencing was observed for NPM–ALK in ALCL and Hodgkin's lymphoma-derived cell lines [232]. Finally, Piva et al. [233] have reported induction of cell death in ALK-positive ALCL cell lines by ALK silencing in vitro as well as in In vivo mouse models of ALCL tumour growth. In this case silencing was achieved by injection of adenovirus-expressing an shRNA (small hairpin RNA)-targeting ALK [233].
Interestingly, DNA vaccination of animals against human ALK prior to challenge with ALK-expressing lymphoma cells provides protection against disease. Furthermore, in already diseased animals ALK vaccination in combination with standard treatment increased the cure rate [234].
Although the direct targeting of ALK may be preferential for treatment of ALK-positive malignancies, other approaches may provide a beneficial complement. By targeting different ALK-interacting proteins, several reports have demonstrated reduced viability and tumour-forming capacity of NPM–ALK-positive cell lines. For example, NPM–ALK interacts with the chaperone Hsp90 (heat-shock protein 90), and inactivation of Hsp90 with 17-AAG (17-allyl-amino-demethoxygeldanamycin) results in degradation of NPM–ALK and subsequent apoptosis in ALCL cell lines [235]. Another example of possible downstream targets in ALK-positive ALCL is PKB/Akt. Injection of mice with cells expressing NPM–ALK, together with dominant-negative PKB/Akt results in a severely impaired tumour-forming capacity as compared with animals receiving control NPM–ALK-expressing cells [132].
Inhibitory antibodies are also a feasible alternative to small molecules. One such example is Trastuzumab (Herceptin), a monoclonal antibody, which binds to the extracellular juxtamembrane domain of HER2 and prolongs life in patients with human epithelial cancer [236]. Inhibitory antibodies to human ALK have been described, which reduce the level of phosphorylation of ALK, and consequently also the basal activity of MAPK in constitutively active ALK-expressing HEK-293 cells. Furthermore, these blocking antibodies are able to reduce the basal differentiation of ALK-transfected PC12 cells [32]. The authors suggest that this monoclonal antibody may function by blocking dimerization of ALK receptors, since stimulation with activating antibodies is abrogated by the inhibitory antibody. Whether this antibody indeed blocks ligand(s)-mediated receptor dimerization of ALK remains to be seen, and it is possible that this blocking antibody against ALK will display inhibitory activity towards the recently discovered activating ALK mutants in neuroblastoma [93–97]. Such blocking antibodies are potential therapeutics for cancers displaying overexpression or activating mutations of the full-length ALK RTK, and not the translocation-generated fusion oncogenes, and may indeed have the capacity to prolong life and increase the therapeutic effects of other treatments.
OUTSTANDING QUESTIONS IN ALK RESEARCH: SPECULATION FOR THE FUTURE
One of the most disputed areas in the ALK research field at the present time is the issue of PTN and MK as ligands of ALK In vivo. To date, there is no genetic evidence to support this from model systems, in spite of the fact that a number of organisms in which ALK signalling has been studied (such as Drosophila and zebra fish) have clear homologues. The possibility exists that different ligands for ALK are utilized in different developmental processes, and that indeed PTN/MK function as ALK ligands, but that as yet unidentified ‘Jeb-like’ vertebrate ligands also exist. In the absence of ‘Jeb-like’ vertebrate ligands, the spatial and temporal relevance of the PTN/MK–ALK interaction In vivo needs to be addressed. The issue of ALK ligands is of particular relevance in neuroblastoma, since it follows that any misregulation of an ALK ligand may influence neuroblastoma progression even in the absence of activating ALK mutations. Structural information regarding ALK is also lacking, and would be an invaluable treasure trove of information. The unique organization of ALK in the extracellular region suggests that important information will be gleaned from solving the ALK structure, with or without PTN/MK. A simpler task would be elucidation of the crystal structure of the intracellular domain of ALK, in the presence or absence of inhibitors, or indeed in the active or inactive conformation. In this regard it would be important to consider crystallization of the human ALK in order to understand the relevance, if any, of the Tyr1604 residue which is lacking in the mouse ALK.
Much work is ongoing concerning generation of specific ALK inhibitors, which would surely be simplified by intricate knowledge of the ALK kinase domain structure. This is an obvious area of importance to be pursued over the coming years. A number of different ALK inhibitors may be needed for therapeutic use, if the paradigm of BCR-Abl and CML inhibitors will also apply to ALK-driven neoplasms. Further development and exploration of monoclonal antibodies to ALK may also provide important therapeutic approaches, particularly in neuroblastoma, and potentially offers the opportunity to interrupt the interaction between ALK and its ligand(s) In vivo. While frustrating for those who have painstakingly developed and analysed the ALK-knockout mice, the lack of obvious phenotypes observed in these animals offers hope that side effects of ALK inhibition may be minimal.
Although a wealth of information exists concerning NPM–ALK signalling, it is less clear whether this information can be directly translated over to the many other ALK fusion proteins. Even less clear is the signalling mediated by the full-length ALK receptor. Further work will hopefully address these issues. We are currently unclear on the mechanisms of down-regulation of full-length ALK, and also of the physiological relevance of the different ALK cleavage products. The functional relevance of the potential ALK-dependence receptor In vivo also needs to be clarified. While considering the In vivo importance of the full-length ALK, it should be noted that RTK loss-of-function is often associated with developmental syndromes, and whereas no connection has been made to date, future work may implicate ALK in a human syndrome perhaps involving the nervous system.
A large number of studies have identified ALK-interacting proteins as well as molecules which are transcriptionally modulated in response to ALK activation. Only a handful of these have been investigated in detail. The difficult task of functional characterization and identification of biologically and clinically relevant molecules in these data sets lies in the future. However, the value of these data sets will not be realised without corresponding biological context.
Further insight into ALK function, in terms of signalling pathways and developmental context, may come from genetic screens and analysis in the model organism systems such as C.elegans, Drosophila, zebrafish and mouse. The function of the second zebrafish ALK family member is of interest, as is the question of a function for the MK/PTN homologues in the fruitfly.
Many clinically relevant questions are also outstanding, such as whether ALK is capable of driving tumours alone in humans, or whether there are particular molecular partners that function with ALK in tumour progression. Difficult issues such as genetic predisposition seem to be especially important in neuroblastoma development. It is intriguing to consider why the ALK genetic locus is so sensitive to translocation events, and, in this regard, very little is known. Bearing in mind the recent gain-of-function ALK mutations in neuroblastoma, it is obvious to wonder whether these gain-of-function mutant ALK RTKs are ligand independent, either partially or completely. Furthermore, will we in the future find mutations in ALK ligands implicated in neuroblastoma progression? One area of research which we will most surely hear more of in the near future is the development of mouse models to investigate ALK-driven neuroblastoma, which will allow a deeper analysis of ALK action in neuroblastoma in a more In vivo context.
CONCLUDING REMARKS
Currently, there are no clinically approved treatments for oncogenic malignancies caused by aberrant ALK activation that directly target ALK or the downstream ALK-activated signalling pathways. The accumulating body of work concerning ALK signal transduction pathways in both physiological and pathological states has provided a number of very real candidate targets for the development of clinical therapeutics. Outstanding questions surrounding the ALK ligand(s) and their mode of action In vivo are issues which need to be addressed. The future holds great hope for more tailoured therapeutic approaches for those suffering from ALK-driven cancers. Such ALK-directed treatments should benefit a number of different cancer patient populations, from ALCL patients to children with neuroblastoma, as well as NSCLC sufferers.
ACKNOWLEDGEMENTS
The authors would like to thank Margret Shirinian for comments and critical reading of the review prior to submission. We apologize to collegues whose work could not be cited owing to space limitations.
FUNDING
The author's work is supported by the Swedish Research Council [grant number 621-2003-3399 (to R. H. P.), 08-0597 (to B. H.)]; the Swedish Cancer Research Foundation [grant number 07/959 (to B. H.)]; and the Swedish Childhood Cancer Foundation [grant number 08/074 (to R. H. P.), 08/084 (to B. H.)]. R. H. P. is a Swedish Cancer Foundation Research Fellow.
References
- 1.Morris S. W., Kirstein M. N., Valentine M. B., Dittmer K. G., Shapiro D. N., Saltman D. L., Look A. T. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 2.Shiota M., Fujimoto J., Semba T., Satoh H., Yamamoto T., Mori S. Hyperphosphorylation of a novel 80 kDa protein-tyrosine kinase similar to Ltk in a human Ki-1 lymphoma cell line, AMS3. Oncogene. 1994;9:1567–1574. [PubMed] [Google Scholar]
- 3.Iwahara T., Fujimoto J., Wen D., Cupples R., Bucay N., Arakawa T., Mori S., Ratzkin B., Yamamoto T. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14:439–449. doi: 10.1038/sj.onc.1200849. [DOI] [PubMed] [Google Scholar]
- 4.Morris S. W., Naeve C., Mathew P., James P. L., Kirstein M. N., Cui X., Witte D. P. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin's lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK) Oncogene. 1997;14:2175–2188. doi: 10.1038/sj.onc.1201062. [DOI] [PubMed] [Google Scholar]
- 5.Loren C. E., Scully A., Grabbe C., Edeen P. T., Thomas J., McKeown M., Hunter T., Palmer R. H. Identification and characterization of DAlk: a novel Drosophila melanogaster RTK which drives ERK activation In vivo. Genes Cells. 2001;6:531–544. doi: 10.1046/j.1365-2443.2001.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoica G. E., Kuo A., Aigner A., Sunitha I., Souttou B., Malerczyk C., Caughey D. J., Wen D., Karavanov A., Riegel A. T., Wellstein A. Identification of anaplastic lymphoma kinase as a receptor for the growth factor pleiotrophin. J. Biol. Chem. 2001;276:16772–16779. doi: 10.1074/jbc.M010660200. [DOI] [PubMed] [Google Scholar]
- 7.Daly N. L., Scanlon M. J., Djordjevic J. T., Kroon P. A., Smith R. Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6334–6338. doi: 10.1073/pnas.92.14.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fass D., Blacklow S., Kim P. S., Berger J. M. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature. 1997;388:691–693. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- 9.Beckmann G., Bork P. An adhesive domain detected in functionally diverse receptors. Trends Biochem. Sci. 1993;18:40–41. doi: 10.1016/0968-0004(93)90049-s. [DOI] [PubMed] [Google Scholar]
- 10.Loren C. E., Englund C., Grabbe C., Hallberg B., Hunter T., Palmer R. H. A crucial role for the Anaplastic lymphoma kinase receptor tyrosine kinase in gut development in Drosophila melanogaster. EMBO Rep. 2003;4:781–786. doi: 10.1038/sj.embor.embor897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai R. Y., Dieter P., Peschel C., Morris S. W., Duyster J. Nucleophosmin-anaplastic lymphoma kinase of large-cell anaplastic lymphoma is a constitutively active tyrosine kinase that utilizes phospholipase C-γ to mediate its mitogenicity. Mol. Cell. Biol. 1998;18:6951–6961. doi: 10.1128/mcb.18.12.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donella-Deana A., Marin O., Cesaro L., Gunby R. H., Ferrarese A., Coluccia A. M., Tartari C. J., Mologni L., Scapozza L., Gambacorti-Passerini C., Pinna L. A. Unique substrate specificity of anaplastic lymphoma kinase (ALK): development of phosphoacceptor peptides for the assay of ALK activity. Biochemistry. 2005;44:8533–8542. doi: 10.1021/bi0472954. [DOI] [PubMed] [Google Scholar]
- 13.Tartari C. J., Gunby R. H., Coluccia A. M., Sottocornola R., Cimbro B., Scapozza L., Donella-Deana A., Pinna L. A., Gambacorti-Passerini C. Characterization of some molecular mechanisms governing autoactivation of the catalytic domain of the anaplastic lymphoma kinase. J. Biol. Chem. 2008;283:3743–3750. doi: 10.1074/jbc.M706067200. [DOI] [PubMed] [Google Scholar]
- 14.Englund C., Loren C. E., Grabbe C., Varshney G. K., Deleuil F., Hallberg B., Palmer R. H. Jeb signals through the Alk receptor tyrosine kinase to drive visceral muscle fusion. Nature. 2003;425:512–516. doi: 10.1038/nature01950. [DOI] [PubMed] [Google Scholar]
- 15.Lee H. H., Norris A., Weiss J. B., Frasch M. Jelly belly protein activates the receptor tyrosine kinase Alk to specify visceral muscle pioneers. Nature. 2003;425:507–512. doi: 10.1038/nature01916. [DOI] [PubMed] [Google Scholar]
- 16.Stute C., Schimmelpfeng K., Renkawitz-Pohl R., Palmer R. H., Holz A. Myoblast determination in the somatic and visceral mesoderm depends on Notch signalling as well as on milliways [mili(Alk)] as receptor for Jeb signalling. Development. 2004;131:743–754. doi: 10.1242/dev.00972. [DOI] [PubMed] [Google Scholar]
- 17.Klapper R., Stute C., Schomaker O., Strasser T., Janning W., Renkawitz-Pohl R., Holz A. The formation of syncytia within the visceral musculature of the Drosophila midgut is dependent on duf, sns and mbc. Mech. Dev. 2002;110:85–96. doi: 10.1016/s0925-4773(01)00567-6. [DOI] [PubMed] [Google Scholar]
- 18.Martin B. S., Ruiz-Gomez M., Landgraf M., Bate M. A distinct set of founders and fusion-competent myoblasts make visceral muscles in the Drosophila embryo. Development. 2001;128:3331–3338. doi: 10.1242/dev.128.17.3331. [DOI] [PubMed] [Google Scholar]
- 19.Weiss J. B., Suyama K. L., Lee H. H., Scott M. P. Jelly belly: a Drosophila LDL receptor repeat-containing signal required for mesoderm migration and differentiation. Cell. 2001;107:387–398. doi: 10.1016/s0092-8674(01)00540-2. [DOI] [PubMed] [Google Scholar]
- 20.Varshney G. K., Palmer R. H. The bHLH transcription factor Hand is regulated by Alk in the Drosophila embryonic gut. Biochem. Biophys. Res. Commun. 2006;351:839–846. doi: 10.1016/j.bbrc.2006.10.117. [DOI] [PubMed] [Google Scholar]
- 21.Shirinian M., Varshney G., Loren C. E., Grabbe C., Palmer R. H. Drosophila anaplastic lymphoma kinase regulates Dpp signalling in the developing embryonic gut. Differentiation. 2007;75:418–426. doi: 10.1111/j.1432-0436.2006.00148.x. [DOI] [PubMed] [Google Scholar]
- 22.Bazigou E., Apitz H., Johansson J., Loren C. E., Hirst E. M., Chen P. L., Palmer R. H., Salecker I. Anterograde Jelly belly and Alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell. 2007;128:961–975. doi: 10.1016/j.cell.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 23.Liao E. H., Hung W., Abrams B., Zhen M. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430:345–350. doi: 10.1038/nature02647. [DOI] [PubMed] [Google Scholar]
- 24.Reiner D. J., Ailion M., Thomas J. H., Meyer B. J. C elegans anaplastic lymphoma kinase ortholog SCD-2 controls dauer formation by modulating TGF-β signaling. Curr. Biol. 2008;18:1101–1109. doi: 10.1016/j.cub.2008.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishihara T., Iino Y., Mohri A., Mori I., Gengyo-Ando K., Mitani S., Katsura I. HEN-1, a secretory protein with an LDL receptor motif, regulates sensory integration and learning in Caenorhabditis elegans. Cell. 2002;109:639–649. doi: 10.1016/s0092-8674(02)00748-1. [DOI] [PubMed] [Google Scholar]
- 26.Lopes S. S., Yang X., Muller J., Carney T. J., McAdow A. R., Rauch G. J., Jacoby A. S., Hurst L. D., Delfino-Machin M., Haffter P., et al. Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet. 2008;4:e1000026. doi: 10.1371/journal.pgen.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vernersson E., Khoo N. K., Henriksson M. L., Roos G., Palmer R. H., Hallberg B. Characterization of the expression of the ALK receptor tyrosine kinase in mice. Gene Expr. Patterns. 2006;6:448–461. doi: 10.1016/j.modgep.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Hurley S. P., Clary D. O., Copie V., Lefcort F. Anaplastic lymphoma kinase is dynamically expressed on subsets of motor neurons and in the peripheral nervous system. J. Comp. Neurol. 2006;495:202–212. doi: 10.1002/cne.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degoutin J., Brunet-de Carvalho N., Cifuentes-Diaz C., Vigny M. ALK (Anaplastic Lymphoma Kinase) expression in DRG neurons and its involvement in neuron-Schwann cells interaction. Eur. J. Neurosci. 2009;29:275–286. doi: 10.1111/j.1460-9568.2008.06593.x. [DOI] [PubMed] [Google Scholar]
- 30.Pulford K., Lamant L., Morris S. W., Butler L. H., Wood K. M., Stroud D., Delsol G., Mason D. Y. Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)-ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood. 1997;89:1394–1404. [PubMed] [Google Scholar]
- 31.Souttou B., Carvalho N. B., Raulais D., Vigny M. Activation of anaplastic lymphoma kinase receptor tyrosine kinase induces neuronal differentiation through the mitogen-activated protein kinase pathway. J. Biol. Chem. 2001;276:9526–9531. doi: 10.1074/jbc.M007333200. [DOI] [PubMed] [Google Scholar]
- 32.Moog-Lutz C., Degoutin J., Gouzi J. Y., Frobert Y., Brunet-de Carvalho N., Bureau J., Creminon C., Vigny M. Activation and inhibition of anaplastic lymphoma kinase receptor tyrosine kinase by monoclonal antibodies and absence of agonist activity of pleiotrophin. J. Biol. Chem. 2005;280:26039–26048. doi: 10.1074/jbc.M501972200. [DOI] [PubMed] [Google Scholar]
- 33.Motegi A., Fujimoto J., Kotani M., Sakuraba H., Yamamoto T. ALK receptor tyrosine kinase promotes cell growth and neurite outgrowth. J. Cell Sci. 2004;117:3319–3329. doi: 10.1242/jcs.01183. [DOI] [PubMed] [Google Scholar]
- 34.Yang H. L., Eriksson T., Vernersson E., Vigny M., Hallberg B., Palmer R. H. The ligand Jelly Belly (Jeb) activates the Drosophila Alk RTK to drive PC12 cell differentiation, but is unable to activate the Mouse ALK RTK. J. Exp. Zool. B Mol. Dev. Evol. 2007;308:269–282. doi: 10.1002/jez.b.21146. [DOI] [PubMed] [Google Scholar]
- 35.Gouzi J. Y., Moog-Lutz C., Vigny M., Brunet-de Carvalho N. Role of the subcellular localization of ALK tyrosine kinase domain in neuronal differentiation of PC12 cells. J. Cell Sci. 2005;118:5811–5823. doi: 10.1242/jcs.02695. [DOI] [PubMed] [Google Scholar]
- 36.Degoutin J., Vigny M., Gouzi J. Y. ALK activation induces Shc and FRS2 recruitment: Signaling and phenotypic outcomes in PC12 cells differentiation. FEBS Lett. 2007;581:727–734. doi: 10.1016/j.febslet.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 37.Chikamori M., Fujimoto J., Tokai-Nishizumi N., Yamamoto T. Identification of multiple SNT-binding sites on NPM-ALK oncoprotein and their involvement in cell transformation. Oncogene. 2007;26:2950–2954. doi: 10.1038/sj.onc.1210095. [DOI] [PubMed] [Google Scholar]
- 38.Piccinini G., Bacchiocchi R., Serresi M., Vivani C., Rossetti S., Gennaretti C., Carbonari D., Fazioli F. A ligand-inducible epidermal growth factor receptor/anaplastic lymphoma kinase chimera promotes mitogenesis and transforming properties in 3T3 cells. J. Biol. Chem. 2002;277:22231–22239. doi: 10.1074/jbc.M111145200. [DOI] [PubMed] [Google Scholar]
- 39.Merenmies J., Rauvala H. Molecular cloning of the 18-kDa growth-associated protein of developing brain. J. Biol. Chem. 1990;265:16721–16724. [PubMed] [Google Scholar]
- 40.Tezuka K., Takeshita S., Hakeda Y., Kumegawa M., Kikuno R., Hashimoto-Gotoh T. Isolation of mouse and human cDNA clones encoding a protein expressed specifically in osteoblasts and brain tissues. Biochem. Biophys. Res. Commun. 1990;173:246–251. doi: 10.1016/s0006-291x(05)81048-4. [DOI] [PubMed] [Google Scholar]
- 41.Courty J., Dauchel M. C., Caruelle D., Perderiset M., Barritault D. Mitogenic properties of a new endothelial cell growth factor related to pleiotrophin. Biochem. Biophys. Res. Commun. 1991;180:145–151. doi: 10.1016/s0006-291x(05)81267-7. [DOI] [PubMed] [Google Scholar]
- 42.Kovesdi I., Fairhurst J. L., Kretschmer P. J., Bohlen P. Heparin-binding neurotrophic factor (HBNF) and MK, members of a new family of homologous, developmentally regulated proteins. Biochem. Biophys. Res. Commun. 1990;172:850–854. doi: 10.1016/0006-291x(90)90753-a. [DOI] [PubMed] [Google Scholar]
- 43.Tomomura M., Kadomatsu K., Matsubara S., Muramatsu T. A retinoic acid-responsive gene, MK, found in the teratocarcinoma system. Heterogeneity of the transcript and the nature of the translation product. J. Biol. Chem. 1990;265:10765–10770. [PubMed] [Google Scholar]
- 44.Vigny M., Raulais D., Puzenat N., Duprez D., Hartmann M. P., Jeanny J. C., Courtois Y. Identification of a new heparin-binding protein localized within chick basement membranes. Eur. J. Biochem. 1989;186:733–740. doi: 10.1111/j.1432-1033.1989.tb15267.x. [DOI] [PubMed] [Google Scholar]
- 45.Stoica G. E., Kuo A., Powers C., Bowden E. T., Sale E. B., Riegel A. T., Wellstein A. Midkine binds to anaplastic lymphoma kinase (ALK) and acts as a growth factor for different cell types. J. Biol. Chem. 2002;277:35990–35998. doi: 10.1074/jbc.M205749200. [DOI] [PubMed] [Google Scholar]
- 46.Kadomatsu K., Muramatsu T. Midkine and pleiotrophin in neural development and cancer. Cancer Lett. 2004;204:127–143. doi: 10.1016/S0304-3835(03)00450-6. [DOI] [PubMed] [Google Scholar]
- 47.Muramatsu T. Midkine and pleiotrophin: two related proteins involved in development, survival, inflammation and tumorigenesis. J. Biochem. (Tokyo) 2002;132:359–371. doi: 10.1093/oxfordjournals.jbchem.a003231. [DOI] [PubMed] [Google Scholar]
- 48.Meng K., Rodriguez-Pena A., Dimitrov T., Chen W., Yamin M., Noda M., Deuel T. F. Pleiotrophin signals increased tyrosine phosphorylation of β-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase β/ζ. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2603–2608. doi: 10.1073/pnas.020487997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maeda N., Nishiwaki T., Shintani T., Hamanaka H., Noda M. 6B4 proteoglycan/phosphacan, an extracellular variant of receptor-like protein-tyrosine phosphatase ζ/RPTPβ, binds pleiotrophin/heparin-binding growth-associated molecule (HB-GAM) J. Biol. Chem. 1996;271:21446–21452. doi: 10.1074/jbc.271.35.21446. [DOI] [PubMed] [Google Scholar]
- 50.Nakanishi T., Kadomatsu K., Okamoto T., Ichihara-Tanaka K., Kojima T., Saito H., Tomoda Y., Muramatsu T. Expression of syndecan-1 and -3 during embryogenesis of the central nervous system in relation to binding with midkine. J. Biochem. 1997;121:197–205. [PubMed] [Google Scholar]
- 51.Raulo E., Chernousov M. A., Carey D. J., Nolo R., Rauvala H. Isolation of a neuronal cell surface receptor of heparin binding growth-associated molecule (HB-GAM). Identification as N-syndecan (syndecan-3) J. Biol. Chem. 1994;269:12999–13004. [PubMed] [Google Scholar]
- 52.Muramatsu H., Zou K., Sakaguchi N., Ikematsu S., Sakuma S., Muramatsu T. LDL receptor-related protein as a component of the midkine receptor. Biochem. Biophys. Res. Commun. 2000;270:936–941. doi: 10.1006/bbrc.2000.2549. [DOI] [PubMed] [Google Scholar]
- 53.Muramatsu H., Zou P., Suzuki H., Oda Y., Chen G. Y., Sakaguchi N., Sakuma S., Maeda N., Noda M., Takada Y., Muramatsu T. α4β1- and α6β1-integrins are functional receptors for midkine, a heparin-binding growth factor. J. Cell Sci. 2004;117:5405–5415. doi: 10.1242/jcs.01423. [DOI] [PubMed] [Google Scholar]
- 54.Bowden E. T., Stoica G. E., Wellstein A. Anti-apoptotic signaling of pleiotrophin through its receptor, anaplastic lymphoma kinase. J. Biol. Chem. 2002;277:35862–35868. doi: 10.1074/jbc.M203963200. [DOI] [PubMed] [Google Scholar]
- 55.Powers C., Aigner A., Stoica G. E., McDonnell K., Wellstein A. Pleiotrophin signaling through anaplastic lymphoma kinase is rate-limiting for glioblastoma growth. J. Biol. Chem. 2002;277:14153–14158. doi: 10.1074/jbc.M112354200. [DOI] [PubMed] [Google Scholar]
- 56.Kuo A. H., Stoica G. E., Riegel A. T., Wellstein A. Recruitment of insulin receptor substrate-1 and activation of NF-κB essential for midkine growth signaling through anaplastic lymphoma kinase. Oncogene. 2007;26:859–869. doi: 10.1038/sj.onc.1209840. [DOI] [PubMed] [Google Scholar]
- 57.Lu K. V., Jong K. A., Kim G. Y., Singh J., Dia E. Q., Yoshimoto K., Wang M. Y., Cloughesy T. F., Nelson S. F., Mischel P. S. Differential induction of glioblastoma migration and growth by two forms of pleiotrophin. J. Biol. Chem. 2005;280:26953–26964. doi: 10.1074/jbc.M502614200. [DOI] [PubMed] [Google Scholar]
- 58.Dirks W. G., Fahnrich S., Lis Y., Becker E., MacLeod R. A., Drexler H. G. Expression and functional analysis of the anaplastic lymphoma kinase (ALK) gene in tumor cell lines. Int. J. Cancer. 2002;100:49–56. doi: 10.1002/ijc.10435. [DOI] [PubMed] [Google Scholar]
- 59.Mathivet T., Mazot P., Vigny M. In contrast to agonist monoclonal antibodies, both C-terminal truncated form and full length form of Pleiotrophin failed to activate vertebrate ALK (anaplastic lymphoma kinase)? Cell. Signalling. 2007;19:2434–2443. doi: 10.1016/j.cellsig.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 60.Mourali J., Benard A., Lourenco F. C., Monnet C., Greenland C., Moog-Lutz C., Racaud-Sultan C., Gonzalez-Dunia D., Vigny M., Mehlen P., et al. Anaplastic lymphoma kinase is a dependence receptor whose proapoptotic functions are activated by caspase cleavage. Mol. Cell. Biol. 2006;26:6209–6222. doi: 10.1128/MCB.01515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez-Pinera P., Zhang W., Chang Y., Vega J. A., Deuel T. F. Anaplastic lymphoma kinase is activated through the pleiotrophin/receptor protein-tyrosine phosphatase β/ζ signaling pathway: an alternative mechanism of receptor tyrosine kinase activation. J. Biol. Chem. 2007;282:28683–28690. doi: 10.1074/jbc.M704505200. [DOI] [PubMed] [Google Scholar]
- 62.Fabri L., Nice E. C., Ward L. D., Maruta H., Burgess A. W., Simpson R. J. Characterization of bovine heparin-binding neurotrophic factor (HBNF): assignment of disulfide bonds. Biochem. Int. 1992;28:1–9. [PubMed] [Google Scholar]
- 63.Muramatsu H., Inui T., Kimura T., Sakakibara S., Song X. J., Maruta H., Muramatsu T. Localization of heparin-binding, neurite outgrowth and antigenic regions in midkine molecule. Biochem. Biophys. Res. Commun. 1994;203:1131–1139. doi: 10.1006/bbrc.1994.2300. [DOI] [PubMed] [Google Scholar]
- 64.Kilpelainen I., Kaksonen M., Avikainen H., Fath M., Linhardt R. J., Raulo E., Rauvala H. Heparin-binding growth-associated molecule contains two heparin-binding β-sheet domains that are homologous to the thrombospondin type I repeat. J. Biol. Chem. 2000;275:13564–13570. doi: 10.1074/jbc.275.18.13564. [DOI] [PubMed] [Google Scholar]
- 65.Englund C., Birve A., Falileeva L., Grabbe C., Palmer R. H. Miple1 and miple2 encode a family of MK/PTN homologues in Drosophila melanogaster. Dev. Genes Evol. 2006;216:10–18. doi: 10.1007/s00427-005-0025-8. [DOI] [PubMed] [Google Scholar]
- 66.Pulford K., Morris S. W., Turturro F. Anaplastic lymphoma kinase proteins in growth control and cancer. J. Cell Physiol. 2004;199:330–358. doi: 10.1002/jcp.10472. [DOI] [PubMed] [Google Scholar]
- 67.Bilsland J. G., Wheeldon A., Mead A., Znamenskiy P., Almond S., Waters K. A., Thakur M., Beaumont V., Bonnert T. P., Heavens R., et al. Behavioral and neurochemical alterations in mice deficient in anaplastic lymphoma kinase suggest therapeutic potential for psychiatric indications. Neuropsychopharmacology. 2008;33:685–700. doi: 10.1038/sj.npp.1301446. [DOI] [PubMed] [Google Scholar]
- 68.Nakamura E., Kadomatsu K., Yuasa S., Muramatsu H., Mamiya T., Nabeshima T., Fan Q. W., Ishiguro K., Igakura T., Matsubara S., et al. Disruption of the midkine gene (Mdk) resulted in altered expression of a calcium binding protein in the hippocampus of infant mice and their abnormal behaviour. Genes Cells. 1998;3:811–822. doi: 10.1046/j.1365-2443.1998.00231.x. [DOI] [PubMed] [Google Scholar]
- 69.Fan Q. W., Muramatsu T., Kadomatsu K. Distinct expression of midkine and pleiotrophin in the spinal cord and placental tissues during early mouse development. Dev. Growth Differ. 2000;42:113–119. doi: 10.1046/j.1440-169x.2000.00497.x. [DOI] [PubMed] [Google Scholar]
- 70.Matsumoto K., Wanaka A., Mori T., Taguchi A., Ishii N., Muramatsu H., Muramatsu T., Tohyama M. Localization of pleiotrophin and midkine in the postnatal developing cerebellum. Neurosci. Lett. 1994;178:216–220. doi: 10.1016/0304-3940(94)90762-5. [DOI] [PubMed] [Google Scholar]
- 71.Mitsiadis T. A., Salmivirta M., Muramatsu T., Muramatsu H., Rauvala H., Lehtonen E., Jalkanen M., Thesleff I. Expression of the heparin-binding cytokines, midkine (MK) and HB-GAM (pleiotrophin) is associated with epithelial-mesenchymal interactions during fetal development and organogenesis. Development. 1995;121:37–51. doi: 10.1242/dev.121.1.37. [DOI] [PubMed] [Google Scholar]
- 72.Vanderwinden J. M., Mailleux P., Schiffmann S. N., Vanderhaeghen J. J. Cellular distribution of the new growth factor pleiotrophin (HB-GAM) mRNA in developing and adult rat tissues. Anat. Embryol. 1992;186:387–406. doi: 10.1007/BF00185989. [DOI] [PubMed] [Google Scholar]
- 73.Herradon G., Ezquerra L., Nguyen T., Silos-Santiago I., Deuel T. F. Midkine regulates pleiotrophin organ-specific gene expression: evidence for transcriptional regulation and functional redundancy within the pleiotrophin/midkine developmental gene family. Biochem. Biophys. Res. Commun. 2005;333:714–721. doi: 10.1016/j.bbrc.2005.05.160. [DOI] [PubMed] [Google Scholar]
- 74.Muramatsu H., Zou P., Kurosawa N., Ichihara-Tanaka K., Maruyama K., Inoh K., Sakai T., Chen L., Sato M., Muramatsu T. Female infertility in mice deficient in midkine and pleiotrophin, which form a distinct family of growth factors. Genes Cells. 2006;11:1405–1417. doi: 10.1111/j.1365-2443.2006.01028.x. [DOI] [PubMed] [Google Scholar]
- 75.Zou P., Muramatsu H., Sone M., Hayashi H., Nakashima T., Muramatsu T. Mice doubly deficient in the midkine and pleiotrophin genes exhibit deficits in the expression of β-tectorin gene and in auditory response. Lab. Invest. 2006;86:645–653. doi: 10.1038/labinvest.3700428. [DOI] [PubMed] [Google Scholar]
- 76.Bernards A., de la Monte S. M. The ltk receptor tyrosine kinase is expressed in pre-B lymphocytes and cerebral neurons and uses a non-AUG translational initiator. EMBO J. 1990;9:2279–2287. doi: 10.1002/j.1460-2075.1990.tb07399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamada S., Nomura T., Takano K., Fujita S., Miyake M., Miyake J. Expression of a chimeric CSF1R-LTK mediates ligand-dependent neurite outgrowth. NeuroRep. 2008;19:1733–1738. doi: 10.1097/WNR.0b013e3283186bf8. [DOI] [PubMed] [Google Scholar]
- 78.Griffin C. A., Hawkins A. L., Dvorak C., Henkle C., Ellingham T., Perlman E. J. Recurrent involvement of 2p23 in inflammatory myofibroblastic tumors. Cancer Res. 1999;59:2776–2780. [PubMed] [Google Scholar]
- 79.Soda M., Choi Y. L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H., et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 80.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 81.Arber D. A., Sun L. H., Weiss L. M. Detection of the t(2;5)(p23;q35) chromosomal translocation in large B-cell lymphomas other than anaplastic large cell lymphoma. Hum. Pathol. 1996;27:590–594. doi: 10.1016/s0046-8177(96)90167-7. [DOI] [PubMed] [Google Scholar]
- 82.Du X. L., Hu H., Lin D. C., Xia S. H., Shen X. M., Zhang Y., Luo M. L., Feng Y. B., Cai Y., Xu X., et al. Proteomic profiling of proteins dysregulted in Chinese esophageal squamous cell carcinoma. J. Mol. Med. 2007;85:863–875. doi: 10.1007/s00109-007-0159-4. [DOI] [PubMed] [Google Scholar]
- 83.Jazii F. R., Najafi Z., Malekzadeh R., Conrads T. P., Ziaee A. A., Abnet C., Yazdznbod M., Karkhane A. A., Salekdeh G. H. Identification of squamous cell carcinoma associated proteins by proteomics and loss of β tropomyosin expression in esophageal cancer. World J. Gastroenterol. 2006;12:7104–7112. doi: 10.3748/wjg.v12.i44.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perez-Pinera P., Chang Y., Astudillo A., Mortimer J., Deuel T. F. Anaplastic lymphoma kinase is expressed in different subtypes of human breast cancer. Biochem. Biophys. Res. Commun. 2007;358:399–403. doi: 10.1016/j.bbrc.2007.04.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Delsol G., Lamant L., Mariame B., Pulford K., Dastugue N., Brousset P., Rigal-Huguet F., al Saati T., Cerretti D. P., Morris S. W., Mason D. Y. A new subtype of large B-cell lymphoma expressing the ALK kinase and lacking the 2; 5 translocation. Blood. 1997;89:1483–1490. [PubMed] [Google Scholar]
- 86.Cessna M. H., Zhou H., Sanger W. G., Perkins S. L., Tripp S., Pickering D., Daines C., Coffin C. M. Expression of ALK1 and p80 in inflammatory myofibroblastic tumor and its mesenchymal mimics: a study of 135 cases. Mod. Pathol. 2002;15:931–938. doi: 10.1097/01.MP.0000026615.04130.1F. [DOI] [PubMed] [Google Scholar]
- 87.Pillay K., Govender D., Chetty R. ALK protein expression in rhabdomyosarcomas. Histopathology. 2002;41:461–467. doi: 10.1046/j.1365-2559.2002.01534.x. [DOI] [PubMed] [Google Scholar]
- 88.Lamant L., Pulford K., Bischof D., Morris S. W., Mason D. Y., Delsol G., Mariame B. Expression of the ALK tyrosine kinase gene in neuroblastoma. Am. J. Pathol. 2000;156:1711–1721. doi: 10.1016/S0002-9440(10)65042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miyake I., Hakomori Y., Shinohara A., Gamou T., Saito M., Iwamatsu A., Sakai R. Activation of anaplastic lymphoma kinase is responsible for hyperphosphorylation of ShcC in neuroblastoma cell lines. Oncogene. 2002;21:5823–5834. doi: 10.1038/sj.onc.1205735. [DOI] [PubMed] [Google Scholar]
- 90.Osajima-Hakomori Y., Miyake I., Ohira M., Nakagawara A., Nakagawa A., Sakai R. Biological role of anaplastic lymphoma kinase in neuroblastoma. Am. J. Pathol. 2005;167:213–222. doi: 10.1016/S0002-9440(10)62966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Corao D., Biegel J. A., Coffin C. M., Barr F. G., Wainwright L. M., Ernst L. M., Choi J. K., Zhang P. J., Pawel B. ALK expression in rhabdomyosarcomas: correlation with histologic subtype and fusion status. Pediatr. Dev. Pathol. 2008 doi: 10.2350/08-03-0434.1. doi: 10.2350/08-03-0434.1. [DOI] [PubMed] [Google Scholar]
- 92.Li X. Q., Hisaoka M., Shi D. R., Zhu X. Z., Hashimoto H. Expression of anaplastic lymphoma kinase in soft tissue tumors: an immunohistochemical and molecular study of 249 cases. Hum. Pathol. 2004;35:711–721. doi: 10.1016/j.humpath.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 93.Caren H., Abel F., Kogner P., Martinsson T. High incidence of DNA mutations and gene amplifications of the ALK gene in advanced sporadic neuroblastoma tumours. Biochem. J. 2008;416:153–159. doi: 10.1042/bj20081834. [DOI] [PubMed] [Google Scholar]
- 94.Chen Y., Takita J., Choi Y. L., Kato M., Ohira M., Sanada M., Wang L., Soda M., Kikuchi A., Igarashi T., et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 95.George R. E., Sanda T., Hanna M., Frohling S., Luther W., 2nd, Zhang J., Ahn Y., Zhou W., London W. B., McGrady P., et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Janoueix-Lerosey I., Lequin D., Brugieres L., Ribeiro A., de Pontual L., Combaret V., Raynal V., Puisieux A., Schleiermacher G., Pierron G., et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 97.Mosse Y. P., Laudenslager M., Longo L., Cole K. A., Wood A., Attiyeh E. F., Laquaglia M. J., Sennett R., Lynch J. E., Perri P., et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stein H., Mason D. Y., Gerdes J., O'Connor N., Wainscoat J., Pallesen G., Gatter K., Falini B., Delsol G., Lemke H., et al. The expression of the Hodgkin's disease associated antigen Ki-1 in reactive and neoplastic lymphoid tissue: evidence that Reed-Sternberg cells and histiocytic malignancies are derived from activated lymphoid cells. Blood. 1985;66:848–858. [PubMed] [Google Scholar]
- 99.Stein H., Foss H. D., Durkop H., Marafioti T., Delsol G., Pulford K., Pileri S., Falini B. CD30+ anaplastic large cell lymphoma: a review of its histopathologic, genetic, and clinical features. Blood. 2000;96:3681–3695. [PubMed] [Google Scholar]
- 100.Amin H. M., Lai R. Pathobiology of ALK+ anaplastic large-cell lymphoma. Blood. 2007;110:2259–2267. doi: 10.1182/blood-2007-04-060715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Swerdlow S. H., Campo E., Harris N. L., Jaffe E. S., Pileri S. A., Stein H., Thiele J., Vardiman J. W. Lyon: International Agency for Research on Cancer; 2008. WHO Classification of Tumours of Haematopoietic and Lymphiod Tissues. [Google Scholar]
- 102.Savage K. J., Harris N. L., Vose J. M., Ullrich F., Jaffe E. S., Connors J. M., Rimsza L., Pileri S. A., Chhanabhai M., Gascoyne R. D., et al. ALK anaplastic large-cell lymphoma is clinically and immunophenotypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: report from the International Peripheral T-Cell Lymphoma Project. Blood. 2008;111:5496–5504. doi: 10.1182/blood-2008-01-134270. [DOI] [PubMed] [Google Scholar]
- 103.Gascoyne R. D., Aoun P., Wu D., Chhanabhai M., Skinnider B. F., Greiner T. C., Morris S. W., Connors J. M., Vose J. M., Viswanatha D. S., et al. Prognostic significance of anaplastic lymphoma kinase (ALK) protein expression in adults with anaplastic large cell lymphoma. Blood. 1999;93:3913–3921. [PubMed] [Google Scholar]
- 104.Shiota M., Nakamura S., Ichinohasama R., Abe M., Akagi T., Takeshita M., Mori N., Fujimoto J., Miyauchi J., Mikata A., et al. Anaplastic large cell lymphomas expressing the novel chimeric protein p80NPM/ALK: a distinct clinicopathologic entity. Blood. 1995;86:1954–1960. [PubMed] [Google Scholar]
- 105.Falini B., Pileri S., Zinzani P. L., Carbone A., Zagonel V., Wolf-Peeters C., Verhoef G., Menestrina F., Todeschini G., Paulli M., et al. ALK+ lymphoma: clinico-pathological findings and outcome. Blood. 1999;93:2697–2706. [PubMed] [Google Scholar]
- 106.Brugieres L., Quartier P., Le Deley M. C., Pacquement H., Perel Y., Bergeron C., Schmitt C., Landmann J., Patte C., Terrier-Lacombe M. J., et al. Relapses of childhood anaplastic large-cell lymphoma: treatment results in a series of 41 children–a report from the French Society of Pediatric Oncology. Ann. Oncol. 2000;11:53–58. doi: 10.1023/a:1008352726155. [DOI] [PubMed] [Google Scholar]
- 107.ten Berge R. L., Meijer C. J., Dukers D. F., Kummer J. A., Bladergroen B. A., Vos W., Hack C. E., Ossenkoppele G. J., Oudejans J. J. Expression levels of apoptosis-related proteins predict clinical outcome in anaplastic large cell lymphoma. Blood. 2002;99:4540–4546. doi: 10.1182/blood.v99.12.4540. [DOI] [PubMed] [Google Scholar]
- 108.Khoury J. D., Medeiros L. J., Rassidakis G. Z., Yared M. A., Tsioli P., Leventaki V., Schmitt-Graeff A., Herling M., Amin H. M., Lai R. Differential expression and clinical significance of tyrosine-phosphorylated STAT3 in ALK+ and ALK- anaplastic large cell lymphoma. Clin. Cancer Res. 2003;9:3692–3699. [PubMed] [Google Scholar]
- 109.Schlette E. J., Medeiros L. J., Goy A., Lai R., Rassidakis G. Z. Survivin expression predicts poorer prognosis in anaplastic large-cell lymphoma. J. Clin. Oncol. 2004;22:1682–1688. doi: 10.1200/JCO.2004.10.172. [DOI] [PubMed] [Google Scholar]
- 110.Rassidakis G. Z., Goy A., Medeiros L. J., Jiang Y., Thomaides A., Remache Y., Cabanillas F., Sarris A. H., Gilles F. Prognostic significance of MUC-1 expression in systemic anaplastic large cell lymphoma. Clin. Cancer Res. 2003;9:2213–2220. [PubMed] [Google Scholar]
- 111.Hattrup C. L., Gendler S. J. Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- 112.Trumper L., Pfreundschuh M., Bonin F. V., Daus H. Detection of the t(2;5)-associated NPM/ALK fusion cDNA in peripheral blood cells of healthy individuals. Br. J. Haematol. 1998;103:1138–1144. doi: 10.1046/j.1365-2141.1998.01097.x. [DOI] [PubMed] [Google Scholar]
- 113.Maes B., Vanhentenrijk V., Wlodarska I., Cools J., Peeters B., Marynen P., de Wolf-Peeters C. The NPM-ALK and the ATIC-ALK fusion genes can be detected in non-neoplastic cells. Am. J. Pathol. 2001;158:2185–2193. doi: 10.1016/S0002-9440(10)64690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Okuwaki M. The structure and functions of NPM1/Nucleophsmin/B23, a multifunctional nucleolar acidic protein. J. Biochem. 2008;143:441–448. doi: 10.1093/jb/mvm222. [DOI] [PubMed] [Google Scholar]
- 115.Fujimoto J., Shiota M., Iwahara T., Seki N., Satoh H., Mori S., Yamamoto T. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5) Proc. Natl. Acad. Sci. U.S.A. 1996;93:4181–4186. doi: 10.1073/pnas.93.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bischof D., Pulford K., Mason D. Y., Morris S. W. Role of the nucleophosmin (NPM) portion of the non-Hodgkin's lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol. Cell. Biol. 1997;17:2312–2325. doi: 10.1128/mcb.17.4.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cools J., Wlodarska I., Somers R., Mentens N., Pedeutour F., Maes B., De Wolf-Peeters C., Pauwels P., Hagemeijer A., Marynen P. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2002;34:354–362. doi: 10.1002/gcc.10033. [DOI] [PubMed] [Google Scholar]
- 118.Hernandez L., Bea S., Bellosillo B., Pinyol M., Falini B., Carbone A., Ott G., Rosenwald A., Fernandez A., Pulford K., et al. Diversity of genomic breakpoints in TFG-ALK translocations in anaplastic large cell lymphomas: identification of a new TFG-ALK(XL) chimeric gene with transforming activity. Am. J. Pathol. 2002;160:1487–1494. doi: 10.1016/S0002-9440(10)62574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hernandez L., Pinyol M., Hernandez S., Bea S., Pulford K., Rosenwald A., Lamant L., Falini B., Ott G., Mason D. Y., et al. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood. 1999;94:3265–3268. [PubMed] [Google Scholar]
- 120.Tort F., Pinyol M., Pulford K., Roncador G., Hernandez L., Nayach I., Kluin-Nelemans H. C., Kluin P., Touriol C., Delsol G., et al. Molecular characterization of a new ALK translocation involving moesin (MSN-ALK) in anaplastic large cell lymphoma. Lab. Invest. 2001;81:419–426. doi: 10.1038/labinvest.3780249. [DOI] [PubMed] [Google Scholar]
- 121.Lamant L., Dastugue N., Pulford K., Delsol G., Mariame B. A new fusion gene TPM3-ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood. 1999;93:3088–3095. [PubMed] [Google Scholar]
- 122.Siebert R., Gesk S., Harder L., Steinemann D., Grote W., Schlegelberger B., Tiemann M., Wlodarska I., Schemmel V. Complex variant translocation t(1;2) with TPM3-ALK fusion due to cryptic ALK gene rearrangement in anaplastic large-cell lymphoma. Blood. 1999;94:3614–3617. [PubMed] [Google Scholar]
- 123.Meech S. J., McGavran L., Odom L. F., Liang X., Meltesen L., Gump J., Wei Q., Carlsen S., Hunger S. P. Unusual childhood extramedullary hematologic malignancy with natural killer cell properties that contains tropomyosin 4–anaplastic lymphoma kinase gene fusion. Blood. 2001;98:1209–1216. doi: 10.1182/blood.v98.4.1209. [DOI] [PubMed] [Google Scholar]
- 124.Ma Z., Cools J., Marynen P., Cui X., Siebert R., Gesk S., Schlegelberger B., Peeters B., De Wolf-Peeters C., Wlodarska I., Morris S. W. Inv(2)(p23q35) in anaplastic large-cell lymphoma induces constitutive anaplastic lymphoma kinase (ALK) tyrosine kinase activation by fusion to ATIC, an enzyme involved in purine nucleotide biosynthesis. Blood. 2000;95:2144–2149. [PubMed] [Google Scholar]
- 125.Trinei M., Lanfrancone L., Campo E., Pulford K., Mason D. Y., Pelicci P. G., Falini B. A new variant anaplastic lymphoma kinase (ALK)-fusion protein (ATIC-ALK) in a case of ALK-positive anaplastic large cell lymphoma. Cancer Res. 2000;60:793–798. [PubMed] [Google Scholar]
- 126.Colleoni G. W., Bridge J. A., Garicochea B., Liu J., Filippa D. A., Ladanyi M. ATIC-ALK: a novel variant ALK gene fusion in anaplastic large cell lymphoma resulting from the recurrent cryptic chromosomal inversion, inv(2)(p23q35) Am. J. Pathol. 2000;156:781–789. doi: 10.1016/S0002-9440(10)64945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lamant L., Gascoyne R. D., Duplantier M. M., Armstrong F., Raghab A., Chhanabhai M., Rajcan-Separovic E., Raghab J., Delsol G., Espinos E. Non-muscle myosin heavy chain (MYH9): a new partner fused to ALK in anaplastic large cell lymphoma. Genes Chromosomes Cancer. 2003;37:427–432. doi: 10.1002/gcc.10232. [DOI] [PubMed] [Google Scholar]
- 128.Touriol C., Greenland C., Lamant L., Pulford K., Bernard F., Rousset T., Mason D. Y., Delsol G. Further demonstration of the diversity of chromosomal changes involving 2p23 in ALK-positive lymphoma: 2 cases expressing ALK kinase fused to CLTCL (clathrin chain polypeptide-like) Blood. 2000;95:3204–3207. [PubMed] [Google Scholar]
- 129.Beardsley G. P., Rayl E. A., Gunn K., Moroson B. A., Seow H., Anderson K. S., Vergis J., Fleming K., Worland S., Condon B., Davies J. Structure and functional relationships in human pur H. Adv. Exp. Med. Biol. 1998;431:221–226. doi: 10.1007/978-1-4615-5381-6_43. [DOI] [PubMed] [Google Scholar]
- 130.Bai R. Y., Ouyang T., Miething C., Morris S. W., Peschel C., Duyster J. Nucleophosmin-anaplastic lymphoma kinase associated with anaplastic large-cell lymphoma activates the phosphatidylinositol 3-kinase/Akt antiapoptotic signaling pathway. Blood. 2000;96:4319–4327. [PubMed] [Google Scholar]
- 131.Polgar D., Leisser C., Maier S., Strasser S., Ruger B., Dettke M., Khorchide M., Simonitsch I., Cerni C., Krupitza G. Truncated ALK derived from chromosomal translocation t(2;5)(p23;q35) binds to the SH3 domain of p85-PI3K. Mutat. Res. 2005;570:9–15. doi: 10.1016/j.mrfmmm.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 132.Slupianek A., Nieborowska-Skorska M., Hoser G., Morrione A., Majewski M., Xue L., Morris S. W., Wasik M. A., Skorski T. Role of phosphatidylinositol 3-kinase-Akt pathway in nucleophosmin/anaplastic lymphoma kinase-mediated lymphomagenesis. Cancer Res. 2001;61:2194–2199. [PubMed] [Google Scholar]
- 133.Gu T. L., Tothova Z., Scheijen B., Griffin J. D., Gilliland D. G., Sternberg D. W. NPM-ALK fusion kinase of anaplastic large-cell lymphoma regulates survival and proliferative signaling through modulation of FOXO3a. Blood. 2004;103:4622–4629. doi: 10.1182/blood-2003-03-0820. [DOI] [PubMed] [Google Scholar]
- 134.Rassidakis G. Z., Feretzaki M., Atwell C., Grammatikakis I., Lin Q., Lai R., Claret F. X., Medeiros L. J., Amin H. M. Inhibition of Akt increases p27Kip1 levels and induces cell cycle arrest in anaplastic large cell lymphoma. Blood. 2005;105:827–829. doi: 10.1182/blood-2004-06-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vega F., Medeiros L. J., Leventaki V., Atwell C., Cho-Vega J. H., Tian L., Claret F. X., Rassidakis G. Z. Activation of mammalian target of rapamycin signaling pathway contributes to tumor cell survival in anaplastic lymphoma kinase-positive anaplastic large cell lymphoma. Cancer Res. 2006;66:6589–6597. doi: 10.1158/0008-5472.CAN-05-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marzec M., Kasprzycka M., Liu X., El-Salem M., Halasa K., Raghunath P. N., Bucki R., Wlodarski P., Wasik M. A. Oncogenic tyrosine kinase NPM/ALK induces activation of the rapamycin-sensitive mTOR signaling pathway. Oncogene. 2007;26:5606–5614. doi: 10.1038/sj.onc.1210346. [DOI] [PubMed] [Google Scholar]
- 137.Crockett D. K., Lin Z., Elenitoba-Johnson K. S., Lim M. S. Identification of NPM–ALK interacting proteins by tandem mass spectrometry. Oncogene. 2004;23:2617–2629. doi: 10.1038/sj.onc.1207398. [DOI] [PubMed] [Google Scholar]
- 138.Marzec M., Kasprzycka M., Liu X., Raghunath P. N., Wlodarski P., Wasik M. A. Oncogenic tyrosine kinase NPM/ALK induces activation of the MEK/ERK signaling pathway independently of c-Raf. Oncogene. 2007;26:813–821. doi: 10.1038/sj.onc.1209843. [DOI] [PubMed] [Google Scholar]
- 139.Amin H. M., McDonnell T. J., Ma Y., Lin Q., Fujio Y., Kunisada K., Leventaki V., Das P., Rassidakis G. Z., Cutler C., et al. Selective inhibition of STAT3 induces apoptosis and G(1) cell cycle arrest in ALK-positive anaplastic large cell lymphoma. Oncogene. 2004;23:5426–5434. doi: 10.1038/sj.onc.1207703. [DOI] [PubMed] [Google Scholar]
- 140.Zamo A., Chiarle R., Piva R., Howes J., Fan Y., Chilosi M., Levy D. E., Inghirami G. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene. 2002;21:1038–1047. doi: 10.1038/sj.onc.1205152. [DOI] [PubMed] [Google Scholar]
- 141.Zhang Q., Raghunath P. N., Xue L., Majewski M., Carpentieri D. F., Odum N., Morris S., Skorski T., Wasik M. A. Multilevel dysregulation of STAT3 activation in anaplastic lymphoma kinase-positive T/null-cell lymphoma. J. Immunol. 2002;168:466–474. doi: 10.4049/jimmunol.168.1.466. [DOI] [PubMed] [Google Scholar]
- 142.Galkin A. V., Melnick J. S., Kim S., Hood T. L., Li N., Li L., Xia G., Steensma R., Chopiuk G., Jiang J., et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc. Natl. Acad. Sci. U.S.A. 2007;104:270–275. doi: 10.1073/pnas.0609412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Marzec M., Kasprzycka M., Ptasznik A., Wlodarski P., Zhang Q., Odum N., Wasik M. A. Inhibition of ALK enzymatic activity in T-cell lymphoma cells induces apoptosis and suppresses proliferation and STAT3 phosphorylation independently of Jak3. Lab. Invest. 2005;85:1544–1554. doi: 10.1038/labinvest.3700348. [DOI] [PubMed] [Google Scholar]
- 144.Wan W., Albom M. S., Lu L., Quail M. R., Becknell N. C., Weinberg L. R., Reddy D. R., Holskin B. P., Angeles T. S., Underiner T. L., et al. Anaplastic lymphoma kinase activity is essential for the proliferation and survival of anaplastic large-cell lymphoma cells. Blood. 2006;107:1617–1623. doi: 10.1182/blood-2005-08-3254. [DOI] [PubMed] [Google Scholar]
- 145.Amin H. M., Medeiros L. J., Ma Y., Feretzaki M., Das P., Leventaki V., Rassidakis G. Z., O'Connor S. L., McDonnell T. J., Lai R. Inhibition of JAK3 induces apoptosis and decreases anaplastic lymphoma kinase activity in anaplastic large cell lymphoma. Oncogene. 2003;22:5399–5407. doi: 10.1038/sj.onc.1206849. [DOI] [PubMed] [Google Scholar]
- 146.Shi X., Franko B., Frantz C., Amin H. M., Lai R. JSI-124 (cucurbitacin I) inhibits Janus kinase-3/signal transducer and activator of transcription-3 signalling, downregulates nucleophosmin-anaplastic lymphoma kinase (ALK), and induces apoptosis in ALK-positive anaplastic large cell lymphoma cells. Br. J. Haematol. 2006;135:26–32. doi: 10.1111/j.1365-2141.2006.06259.x. [DOI] [PubMed] [Google Scholar]
- 147.Lai R., Rassidakis G. Z., Lin Q., Atwell C., Medeiros L. J., Amin H. M. Jak3 activation is significantly associated with ALK expression in anaplastic large cell lymphoma. Hum. Pathol. 2005;36:939–944. doi: 10.1016/j.humpath.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 148.Qiu L., Lai R., Lin Q., Lau E., Thomazy D. M., Calame D., Ford R. J., Kwak L. W., Kirken R. A., Amin H. M. Autocrine release of interleukin-9 promotes Jak3-dependent survival of ALK+ anaplastic large-cell lymphoma cells. Blood. 2006;108:2407–2415. doi: 10.1182/blood-2006-04-020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Han Y., Amin H. M., Franko B., Frantz C., Shi X., Lai R. Loss of SHP1 enhances JAK3/STAT3 signaling and decreases proteosome degradation of JAK3 and NPM-ALK in ALK+ anaplastic large-cell lymphoma. Blood. 2006;108:2796–2803. doi: 10.1182/blood-2006-04-017434. [DOI] [PubMed] [Google Scholar]
- 150.Chiarle R., Simmons W. J., Cai H., Dhall G., Zamo A., Raz R., Karras J. G., Levy D. E., Inghirami G. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat. Med. 2005;11:623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 151.Khoury J. D., Rassidakis G. Z., Medeiros L. J., Amin H. M., Lai R. Methylation of SHP1 gene and loss of SHP1 protein expression are frequent in systemic anaplastic large cell lymphoma. Blood. 2004;104:1580–1581. doi: 10.1182/blood-2004-03-1151. [DOI] [PubMed] [Google Scholar]
- 152.Nieborowska-Skorska M., Slupianek A., Xue L., Zhang Q., Raghunath P. N., Hoser G., Wasik M. A., Morris S. W., Skorski T. Role of signal transducer and activator of transcription 5 in nucleophosmin/anaplastic lymphoma kinase-mediated malignant transformation of lymphoid cells. Cancer Res. 2001;61:6517–6523. [PubMed] [Google Scholar]
- 153.Zhang Q., Wang H. Y., Liu X., Wasik M. A. STAT5A is epigenetically silenced by the tyrosine kinase NPM1-ALK and acts as a tumor suppressor by reciprocally inhibiting NPM1-ALK expression. Nat. Med. 2007;13:1341–1348. doi: 10.1038/nm1659. [DOI] [PubMed] [Google Scholar]
- 154.Ambrogio C., Voena C., Manazza A. D., Martinengo C., Costa C., Kirchhausen T., Hirsch E., Inghirami G., Chiarle R. The anaplastic lymphoma kinase controls cell shape and growth of anaplastic large cell lymphoma through Cdc42 activation. Cancer Res. 2008;68:8899–8907. doi: 10.1158/0008-5472.CAN-08-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Colomba A., Courilleau D., Ramel D., Billadeau D. D., Espinos E., Delsol G., Payrastre B., Gaits-Iacovoni F. Activation of Rac1 and the exchange factor Vav3 are involved in NPM-ALK signaling in anaplastic large cell lymphomas. Oncogene. 2008;27:2728–2736. doi: 10.1038/sj.onc.1210921. [DOI] [PubMed] [Google Scholar]
- 156.Voena C., Conte C., Ambrogio C., Boeri Erba E., Boccalatte F., Mohammed S., Jensen O. N., Palestro G., Inghirami G., Chiarle R. The tyrosine phosphatase Shp2 interacts with NPM-ALK and regulates anaplastic lymphoma cell growth and migration. Cancer Res. 2007;67:4278–4286. doi: 10.1158/0008-5472.CAN-06-4350. [DOI] [PubMed] [Google Scholar]
- 157.Cussac D., Greenland C., Roche S., Bai R. Y., Duyster J., Morris S. W., Delsol G., Allouche M., Payrastre B. Nucleophosmin-anaplastic lymphoma kinase of anaplastic large-cell lymphoma recruits, activates, and uses pp60c-src to mediate its mitogenicity. Blood. 2004;103:1464–1471. doi: 10.1182/blood-2003-04-1038. [DOI] [PubMed] [Google Scholar]
- 158.Bassermann F., von Klitzing C., Munch S., Bai R. Y., Kawaguchi H., Morris S. W., Peschel C., Duyster J. NIPA defines an SCF-type mammalian E3 ligase that regulates mitotic entry. Cell. 2005;122:45–57. doi: 10.1016/j.cell.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 159.Ouyang T., Bai R. Y., Bassermann F., von Klitzing C., Klumpen S., Miething C., Morris S. W., Peschel C., Duyster J. Identification and characterization of a nuclear interacting partner of anaplastic lymphoma kinase (NIPA) J. Biol. Chem. 2003;278:30028–30036. doi: 10.1074/jbc.M300883200. [DOI] [PubMed] [Google Scholar]
- 160.Turner S. D., Tooze R., Maclennan K., Alexander D. R. Vav-promoter regulated oncogenic fusion protein NPM-ALK in transgenic mice causes B-cell lymphomas with hyperactive Jun kinase. Oncogene. 2003;22:7750–7761. doi: 10.1038/sj.onc.1207048. [DOI] [PubMed] [Google Scholar]
- 161.Leventaki V., Drakos E., Medeiros L. J., Lim M. S., Elenitoba-Johnson K. S., Claret F. X., Rassidakis G. Z. NPM–ALK oncogenic kinase promotes cell cycle progression through activation of JNK/cJun signaling in anaplastic large cell lymphoma. Blood. 2007;110:1621–1630. doi: 10.1182/blood-2006-11-059451. [DOI] [PubMed] [Google Scholar]
- 162.Singh R. R., Cho-Vega J. H., Davuluri Y., Ma S., Kasbidi F., Milito C., Lennon P. A., Drakos E., Medeiros L. J., Luthra R., Vega F. Sonic hedgehog signaling pathway is activated in ALK-positive anaplastic large cell lymphoma. Cancer Res. 2009;69:2550–2558. doi: 10.1158/0008-5472.CAN-08-1808. [DOI] [PubMed] [Google Scholar]
- 163.Boccalatte F. E., Voena C., Riganti C., Bosia A., d'Amico L., Riera L., Cheng M., Ruggeri B., Jensen O. N., Goss V. L., et al. The enzymatic activity of 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC) is enhanced by NPM-ALK: new insights in ALK-mediated pathogenesis and the treatment of ALCL. Blood. 2008;113:2776–2790. doi: 10.1182/blood-2008-06-161018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Piva R., Pellegrino E., Mattioli M., Agnelli L., Lombardi L., Boccalatte F., Costa G., Ruggeri B. A., Cheng M., Chiarle R., et al. Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and BCL2A1 as critical target genes. J. Clin. Invest. 2006;116:3171–3182. doi: 10.1172/JCI29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lamant L., de Reynies A., Duplantier M. M., Rickman D. S., Sabourdy F., Giuriato S., Brugieres L., Gaulard P., Espinos E., Delsol G. Gene-expression profiling of systemic anaplastic large-cell lymphoma reveals differences based on ALK status and two distinct morphologic ALK+ subtypes. Blood. 2007;109:2156–2164. doi: 10.1182/blood-2006-06-028969. [DOI] [PubMed] [Google Scholar]
- 166.Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 167.Sjostrom C., Seiler C., Crockett D. K., Tripp S. R., Elenitoba Johnson K. S., Lim M. S. Global proteome profiling of NPM/ALK-positive anaplastic large cell lymphoma. Exp. Haematol. 2007;35:1240–1248. doi: 10.1016/j.exphem.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 168.Wu F., Wang P., Young L. C., Lai R., Li L. Proteome-wide identification of novel binding partners to the oncogenic fusion gene protein, NPM-ALK, using tandem affinity purification and mass spectrometry. Am. J. Pathol. 2009;174:361–370. doi: 10.2353/ajpath.2009.080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Armstrong F., Duplantier M. M., Trempat P., Hieblot C., Lamant L., Espinos E., Racaud-Sultan C., Allouche M., Campo E., Delsol G., Touriol C. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene. 2004;23:6071–6082. doi: 10.1038/sj.onc.1207813. [DOI] [PubMed] [Google Scholar]
- 170.Momose S., Tamaru J., Kishi H., Mikata I., Mori M., Toyozumi Y., Itoyama S. Hyperactivated STAT3 in ALK-positive diffuse large B-cell lymphoma with clathrin-ALK fusion. Hum. Pathol. 2009;40:75–82. doi: 10.1016/j.humpath.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 171.Gleason B. C., Hornick J. L. Inflammatory myofibroblastic tumours: where are we now? J. Clin. Pathol. 2008;61:428–437. doi: 10.1136/jcp.2007.049387. [DOI] [PubMed] [Google Scholar]
- 172.Umiker W. O., Iverson L. Postinflammatory tumors of the lung; report of four cases simulating xanthoma, fibroma, or plasma cell tumor. J. Thorac. Surg. 1954;28:55–63. [PubMed] [Google Scholar]
- 173.Coffin C. M., Watterson J., Priest J. R., Dehner L. P. Extrapulmonary inflammatory myofibroblastic tumor (inflammatory pseudotumor). A clinicopathologic and immunohistochemical study of 84 cases. Am. J. Surg. Pathol. 1995;19:859–872. doi: 10.1097/00000478-199508000-00001. [DOI] [PubMed] [Google Scholar]
- 174.Lawrence B., Perez-Atayde A., Hibbard M. K., Rubin B. P., Dal Cin P., Pinkus J. L., Pinkus G. S., Xiao S., Yi E. S., Fletcher C. D., Fletcher J. A. TPM3–ALK and TPM4–ALK oncogenes in inflammatory myofibroblastic tumors. Am. J. Pathol. 2000;157:377–384. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Debiec-Rychter M., Marynen P., Hagemeijer A., Pauwels P. ALK–ATIC fusion in urinary bladder inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2003;38:187–190. doi: 10.1002/gcc.10267. [DOI] [PubMed] [Google Scholar]
- 176.Patel A. S., Murphy K. M., Hawkins A. L., Cohen J. S., Long P. P., Perlman E. J., Griffin C. A. RANBP2 and CLTC are involved in ALK rearrangements in inflammatory myofibroblastic tumors. Cancer Genet. Cytogenet. 2007;176:107–114. doi: 10.1016/j.cancergencyto.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 177.Bridge J. A., Kanamori M., Ma Z., Pickering D., Hill D. A., Lydiatt W., Lui M. Y., Colleoni G. W., Antonescu C. R., Ladanyi M., Morris S. W. Fusion of the ALK gene to the clathrin heavy chain gene, CLTC, in inflammatory myofibroblastic tumor. Am. J. Pathol. 2001;159:411–415. doi: 10.1016/S0002-9440(10)61711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Debelenko L. V., Arthur D. C., Pack S. D., Helman L. J., Schrump D. S., Tsokos M. Identification of CARS-ALK fusion in primary and metastatic lesions of an inflammatory myofibroblastic tumor. Lab. Invest. 2003;83:1255–1265. doi: 10.1097/01.lab.0000088856.49388.ea. [DOI] [PubMed] [Google Scholar]
- 179.Ma Z., Hill D. A., Collins M. H., Morris S. W., Sumegi J., Zhou M., Zuppan C., Bridge J. A. Fusion of ALK to the Ran-binding protein 2 (RANBP2) gene in inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2003;37:98–105. doi: 10.1002/gcc.10177. [DOI] [PubMed] [Google Scholar]
- 180.Panagopoulos I., Nilsson T., Domanski H. A., Isaksson M., Lindblom P., Mertens F., Mandahl N. Fusion of the SEC31L1 and ALK genes in an inflammatory myofibroblastic tumor. Int. J. Cancer. 2006;118:1181–1186. doi: 10.1002/ijc.21490. [DOI] [PubMed] [Google Scholar]
- 181.Coffin C. M., Patel A., Perkins S., Elenitoba-Johnson K. S., Perlman E., Griffin C. A. ALK1 and p80 expression and chromosomal rearrangements involving 2p23 in inflammatory myofibroblastic tumor. Mod. Pathol. 2001;14:569–576. doi: 10.1038/modpathol.3880352. [DOI] [PubMed] [Google Scholar]
- 182.Cook J. R., Dehner L. P., Collins M. H., Ma Z., Morris S. W., Coffin C. M., Hill D. A. Anaplastic lymphoma kinase (ALK) expression in the inflammatory myofibroblastic tumor: a comparative immunohistochemical study. Am. J. Surg. Pathol. 2001;25:1364–1371. doi: 10.1097/00000478-200111000-00003. [DOI] [PubMed] [Google Scholar]
- 183.Coffin C. M., Hornick J. L., Fletcher C. D. Inflammatory myofibroblastic tumor: comparison of clinicopathologic, histologic, and immunohistochemical features including ALK expression in atypical and aggressive cases. Am. J. Surg. Pathol. 2007;31:509–520. doi: 10.1097/01.pas.0000213393.57322.c7. [DOI] [PubMed] [Google Scholar]
- 184.Chun Y. S., Wang L., Nascimento A. G., Moir C. R., Rodeberg D. A. Pediatric inflammatory myofibroblastic tumor: anaplastic lymphoma kinase (ALK) expression and prognosis. Ped. Blood Cancer. 2005;45:796–801. doi: 10.1002/pbc.20294. [DOI] [PubMed] [Google Scholar]
- 185.Jemal A., Clegg L. X., Ward E., Ries L. A., Wu X., Jamison P. M., Wingo P. A., Howe H. L., Anderson R. N., Edwards B. K. Annual report to the nation on the status of cancer, 1975–2001, with a special feature regarding survival. Cancer. 2004;101:3–27. doi: 10.1002/cncr.20288. [DOI] [PubMed] [Google Scholar]
- 186.Kamangar F., Dores G. M., Anderson W. F. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J. Clin. Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 187.Perner S., Wagner P. L., Demichelis F., Mehra R., Lafargue C. J., Moss B. J., Arbogast S., Soltermann A., Weder W., Giordano T. J., et al. EML4–ALK fusion lung cancer: a rare acquired event. Neoplasia. 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Inamura K., Takeuchi K., Togashi Y., Nomura K., Ninomiya H., Okui M., Satoh Y., Okumura S., Nakagawa K., Soda M., et al. EML4–ALK fusion is linked to histological characteristics in a subset of lung cancers. J. Thorac. Oncol. 2008;3:13–17. doi: 10.1097/JTO.0b013e31815e8b60. [DOI] [PubMed] [Google Scholar]
- 189.Fukuyoshi Y., Inoue H., Kita Y., Utsunomiya T., Ishida T., Mori M. EML4-ALK fusion transcript is not found in gastrointestinal and breast cancers. Br. J. Cancer. 2008;98:1536–1539. doi: 10.1038/sj.bjc.6604341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shinmura K., Kageyama S., Tao H., Bunai T., Suzuki M., Kamo T., Takamochi K., Suzuki K., Tanahashi M., Niwa H., et al. EML4–ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer. 2008;61:163–169. doi: 10.1016/j.lungcan.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 191.Koivunen J. P., Mermel C., Zejnullahu K., Murphy C., Lifshits E., Holmes A. J., Choi H. G., Kim J., Chiang D., Thomas R., et al. EML4–ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin. Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wong D. W., Leung E. L., So K. K., Tam I. Y., Sihoe A. D., Cheng L. C., Ho K. K., Au J. S., Chung L. P., Pik Wong M. The EML4–ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS, Cancer. 2009 doi: 10.1002/cncr.24181. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 193.Martelli M. P., Sozzi G., Hernandez L., Pettirossi V., Navarro A., Conte D., Gasparini P., Perrone F., Modena P., Pastorino U., et al. EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am. J. Pathol. 2009;174:661–670. doi: 10.2353/ajpath.2009.080755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Paez J. G., Janne P. A., Lee J. C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F. J., Lindeman N., Boggon T. J., et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 195.Soda M., Takada S., Takeuchi K., Choi Y. L., Enomoto M., Ueno T., Haruta H., Hamada T., Yamashita Y., Ishikawa Y., et al. A mouse model for EML4-ALK-positive lung cancer. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zou H. Y., Li Q., Lee J. H., Arango M. E., McDonnell S. R., Yamazaki S., Koudriakova T. B., Alton G., Cui J. J., Kung P. P., et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 197.Christensen J. G., Zou H. Y., Arango M. E., Li Q., Lee J. H., McDonnell S. R., Yamazaki S., Alton G. R., Mroczkowski B., Los G. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol. Cancer Ther. 2007;6:3314–3322. doi: 10.1158/1535-7163.MCT-07-0365. [DOI] [PubMed] [Google Scholar]
- 198.McDermott U., Iafrate A. J., Gray N. S., Shioda T., Classon M., Maheswaran S., Zhou W., Choi H. G., Smith S. L., Dowell L., et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–3395. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- 199.Reichard K. K., McKenna R. W., Kroft S. H. ALK-positive diffuse large B-cell lymphoma: report of four cases and review of the literature. Mod. Pathol. 2007;20:310–319. doi: 10.1038/modpathol.3800742. [DOI] [PubMed] [Google Scholar]
- 200.Stachurski D., Miron P. M., Al-Homsi S., Hutchinson L., Harris N. L., Woda B., Wang S. A. Anaplastic lymphoma kinase-positive diffuse large B-cell lymphoma with a complex karyotype and cryptic 3′ ALK gene insertion to chromosome 4 q22–24. Hum. Pathol. 2007;38:940–945. doi: 10.1016/j.humpath.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 201.Lee H. W., Kim K., Kim W., Ko Y. H. ALK-positive diffuse large B-cell lymphoma: report of three cases. Hematol. Oncol. 2008;26:108–113. doi: 10.1002/hon.841. [DOI] [PubMed] [Google Scholar]
- 202.Choung H. S., Kim H. J., Kim W. S., Kim K., Kim S. H. [Cytomorphology and molecular characterization of CLTC-ALK rearrangement in 2 cases of ALK-positive diffuse large B-cell lymphoma with extensive bone marrow involvement] Korean J. Lab. Med. 2008;28:89–94. doi: 10.3343/kjlm.2008.28.2.89. [DOI] [PubMed] [Google Scholar]
- 203.Onciu M., Behm F. G., Downing J. R., Shurtleff S. A., Raimondi S. C., Ma Z., Morris S. W., Kennedy W., Jones S. C., Sandlund J. T. ALK-positive plasmablastic B-cell lymphoma with expression of the NPM-ALK fusion transcript: report of 2 cases. Blood. 2003;102:2642–2644. doi: 10.1182/blood-2003-04-1095. [DOI] [PubMed] [Google Scholar]
- 204.Adam P., Katzenberger T., Seeberger H., Gattenlohner S., Wolf J., Steinlein C., Schmid M., Muller-Hermelink H. K., Ott G. A case of a diffuse large B-cell lymphoma of plasmablastic type associated with the t(2;5)(p23;q35) chromosome translocation. Am. J. Surg. Pathol. 2003;27:1473–1476. doi: 10.1097/00000478-200311000-00012. [DOI] [PubMed] [Google Scholar]
- 205.Grzelinski M., Steinberg F., Martens T., Czubayko F., Lamszus K., Aigner A. Enhanced antitumorigenic effects in glioblastoma on double targeting of pleiotrophin and its receptor ALK. Neoplasia. 2009;11:145–156. doi: 10.1593/neo.81040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Garver R. I., Jr, Radford D. M., Donis-Keller H., Wick M. R., Milner P. G. Midkine and pleiotrophin expression in normal and malignant breast tissue. Cancer. 1994;74:1584–1590. doi: 10.1002/1097-0142(19940901)74:5<1584::aid-cncr2820740514>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 207.Riegel A. T., Wellstein A. The potential role of the heparin-binding growth factor pleiotrophin in breast cancer. Breast Cancer Res. Treatment. 1994;31:309–314. doi: 10.1007/BF00666163. [DOI] [PubMed] [Google Scholar]
- 208.Zhang N., Zhong R., Wang Z. Y., Deuel T. F. Human breast cancer growth inhibited In vivo by a dominant negative pleiotrophin mutant. J. Biol. Chem. 1997;272:16733–16736. doi: 10.1074/jbc.272.27.16733. [DOI] [PubMed] [Google Scholar]
- 209.Maris J. M., Hogarty M. D., Bagatell R., Cohn S. L. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 210.Brodeur G. M. Neuroblastoma: biological insights into a clinical enigma. Nat. Rev. Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 211.Kuefer M. U., Look A. T., Pulford K., Behm F. G., Pattengale P. K., Mason D. Y., Morris S. W. Retrovirus-mediated gene transfer of NPM–ALK causes lymphoid malignancy in mice. Blood. 1997;90:2901–2910. [PubMed] [Google Scholar]
- 212.Lange K., Uckert W., Blankenstein T., Nadrowitz R., Bittner C., Renauld J. C., van Snick J., Feller A. C., Merz H. Overexpression of NPM-ALK induces different types of malignant lymphomas in IL-9 transgenic mice. Oncogene. 2003;22:517–527. doi: 10.1038/sj.onc.1206076. [DOI] [PubMed] [Google Scholar]
- 213.Chiarle R., Gong J. Z., Guasparri I., Pesci A., Cai J., Liu J., Simmons W. J., Dhall G., Howes J., Piva R., Inghirami G. NPM-ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood. 2003;101:1919–1927. doi: 10.1182/blood-2002-05-1343. [DOI] [PubMed] [Google Scholar]
- 214.Jager R., Hahne J., Jacob A., Egert A., Schenkel J., Wernert N., Schorle H., Wellmann A. Mice transgenic for NPM-ALK develop non-Hodgkin lymphomas. Anticancer Res. 2005;25:3191–3196. [PubMed] [Google Scholar]
- 215.Turner S. D., Alexander D. R. What have we learnt from mouse models of NPM-ALK-induced lymphomagenesis? Leukemia. 2005;19:1128–1134. doi: 10.1038/sj.leu.2403797. [DOI] [PubMed] [Google Scholar]
- 216.Druker B. J., Tamura S., Buchdunger E., Ohno S., Segal G. M., Fanning S., Zimmermann J., Lydon N. B. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 217.Druker B. J. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 218.Buchdunger E., Cioffi C. L., Law N., Stover D., Ohno-Jones S., Druker B. J., Lydon N. B. Abl protein-tyrosine kinase inhibitor STI571 inhibits in vitro signal transduction mediated by c-kit and platelet-derived growth factor receptors. J. Pharmacol. Exp. Ther. 2000;295:139–145. [PubMed] [Google Scholar]
- 219.Buchdunger E., Zimmermann J., Mett H., Meyer T., Muller M., Regenass U., Lydon N. B. Selective inhibition of the platelet-derived growth factor signal transduction pathway by a protein-tyrosine kinase inhibitor of the 2-phenylaminopyrimidine class. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2558–2562. doi: 10.1073/pnas.92.7.2558. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 220.Hirota S., Isozaki K., Moriyama Y., Hashimoto K., Nishida T., Ishiguro S., Kawano K., Hanada M., Kurata A., Takeda M., et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 221.Demetri G. D., von Mehren M., Blanke C. D., Van den Abbeele A. D., Eisenberg B., Roberts P. J., Heinrich M. C., Tuveson D. A., Singer S., Janicek M., et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 222.van Oosterom A. T., Judson I., Verweij J., Stroobants S., Donato di Paola E., Dimitrijevic S., Martens M., Webb A., Sciot R., Van Glabbeke M., et al. Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: a phase I study. Lancet. 2001;358:1421–1423. doi: 10.1016/s0140-6736(01)06535-7. [DOI] [PubMed] [Google Scholar]
- 223.Joensuu H., Roberts P. J., Sarlomo-Rikala M., Andersson L. C., Tervahartiala P., Tuveson D., Silberman S., Capdeville R., Dimitrijevic S., Druker B., Demetri G. D. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N. Engl. J. Med. 2001;344:1052–1056. doi: 10.1056/NEJM200104053441404. [DOI] [PubMed] [Google Scholar]
- 224.Arora A., Scholar E. M. Role of tyrosine kinase inhibitors in cancer therapy. J. Pharmacol. Exp. Ther. 2005;315:971–979. doi: 10.1124/jpet.105.084145. [DOI] [PubMed] [Google Scholar]
- 225.Takahashi I., Saitoh Y., Yoshida M., Sano H., Nakano H., Morimoto M., Tamaoki T. UCN-01 and UCN-02, new selective inhibitors of protein kinase C. II. Purification, physico-chemical properties, structural determination and biological activities. J. Antibiotics. 1989;42:571–576. doi: 10.7164/antibiotics.42.571. [DOI] [PubMed] [Google Scholar]
- 226.Graves P. R., Yu L., Schwarz J. K., Gales J., Sausville E. A., O'Connor P. M., Piwnica-Worms H. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J. Biol. Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 227.Sausville E. A., Arbuck S. G., Messmann R., Headlee D., Bauer K. S., Lush R. M., Murgo A., Figg W. D., Lahusen T., Jaken S., et al. Phase I trial of 72-hour continuous infusion UCN-01 in patients with refractory neoplasms. J. Clin. Oncol. 2001;19:2319–2333. doi: 10.1200/JCO.2001.19.8.2319. [DOI] [PubMed] [Google Scholar]
- 228.Gunby R. H., Tartari C. J., Porchia F., Donella-Deana A., Scapozza L., Gambacorti-Passerini C. An enzyme-linked immunosorbent assay to screen for inhibitors of the oncogenic anaplastic lymphoma kinase. Haematologica. 2005;90:988–990. [PubMed] [Google Scholar]
- 229.Li R., Xue L., Zhu T., Jiang Q., Cui X., Yan Z., McGee D., Wang J., Gantla V. R., Pickens J. C., et al. Design and synthesis of 5-aryl-pyridone-carboxamides as inhibitors of anaplastic lymphoma kinase. J. Med. Chem. 2006;49:1006–1015. doi: 10.1021/jm050824x. [DOI] [PubMed] [Google Scholar]
- 230.Li R., Morris S. W. Development of anaplastic lymphoma kinase (ALK) small-molecule inhibitors for cancer therapy. Med. Res. Rev. 2008;28:372–412. doi: 10.1002/med.20109. [DOI] [PubMed] [Google Scholar]
- 231.Grzelinski M., Bader N., Czubayko F., Aigner A. Ribozyme-targeting reveals the rate-limiting role of pleiotrophin in glioblastoma. Int. J. Cancer. 2005;117:942–951. doi: 10.1002/ijc.21276. [DOI] [PubMed] [Google Scholar]
- 232.Hubinger G., Wehnes E., Xue L., Morris S. W., Maurer U. Hammerhead ribozyme-mediated cleavage of the fusion transcript NPM-ALK associated with anaplastic large-cell lymphoma. Exp. Hematol. 2003;31:226–233. doi: 10.1016/s0301-472x(02)01084-6. [DOI] [PubMed] [Google Scholar]
- 233.Piva R., Chiarle R., Manazza A. D., Taulli R., Simmons W., Ambrogio C., d'Escamard V., Pellegrino E., Ponzetto C., Palestro G., Inghirami G. Ablation of oncogenic ALK is a viable therapeutic approach for anaplastic large-cell lymphomas. Blood. 2006;107:689–697. doi: 10.1182/blood-2005-05-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chiarle R., Martinengo C., Mastini C., Ambrogio C., d'Escamard V., Forni G., Inghirami G. The anaplastic lymphoma kinase is an effective oncoantigen for lymphoma vaccination. Nat. Med. 2008;14:676–680. doi: 10.1038/nm1769. [DOI] [PubMed] [Google Scholar]
- 235.Bonvini P., Gastaldi T., Falini B., Rosolen A. Nucleophosmin-anaplastic lymphoma kinase (NPM-ALK), a novel Hsp90-client tyrosine kinase: down-regulation of NPM-ALK expression and tyrosine phosphorylation in ALK+ CD30+ lymphoma cells by the Hsp90 antagonist 17-allylamino,17-demethoxygeldanamycin. Cancer Res. 2002;62:1559–1566. [PubMed] [Google Scholar]
- 236.Hudis C. A. Trastuzumab: mechanism of action and use in clinical practice. N. Engl. J. Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 237.De Paepe P., Baens M., van Krieken H., Verhasselt B., Stul M., Simons A., Poppe B., Laureys G., Brons P., Vandenberghe P., et al. ALK activation by the CLTC-ALK fusion is a recurrent event in large B-cell lymphoma. Blood. 2003;102:2638–2641. doi: 10.1182/blood-2003-04-1050. [DOI] [PubMed] [Google Scholar]