Abstract
gal-1 (galectin-1) mediates cell–cell and cell–extracellular matrix adhesion, essentially by interacting with β-galactoside-containing glycans of cell-surface glycoconjugates. Although most structural studies with gal-1 have investigated its binding to simple carbohydrates, in particular lactose and N-acetyl-lactosamine, this view is limited, because gal-1 functions at the cell surface by interacting with more complex glycans that are heterogeneous in size and composition. In the present study we used NMR spectroscopy to investigate the interaction of human gal-1 with a large (120 kDa) complex glycan, GRG (galactorhamnogalacturonate glycan), that contains non-randomly distributed mostly terminal β(1→4)-linked galactose side chains. We used 15N–1H-HSQC (heteronuclear single quantum coherence) NMR experiments with 15N-enriched gal-1 to identify the GRG-binding region on gal-1 and found that this region covers a large surface area on gal-1 that includes the quintessential lactose-binding site and runs from that site through a broad valley or cleft towards the dimer interface. HSQC and pulsed-field-gradient NMR diffusion experiments also show that gal-1 binds GRG with a gal-1:GRG stoichiometry of about 5:1 (or 6:1) and with average macroscopic and microscopic equilibrium dissociation constants (Kd) of 8×10−6 M and 40×10−6 M (or 48×10−6 M) respectively, indicating stronger binding than to lactose (Kd=520×10−6 M). Although gal-1 may bind GRG in various ways, the glycan can be competed for by lactose, suggesting that there is one major mode of interaction. Furthermore, even though terminal motifs on GRG are Gal-β(1→4)-Gal rather than the traditional Gal-β(1→4)-Glc/GlcNAc (where GlcNAc is N-acetylglucosamine), we show that the disaccharide Gal-β(1→4)-Gal can bind gal-1 at the lactose-binding domain. In addition, gal-1 binding to GRG disrupts inter-glycan interactions and decreases glycan-mediated solution viscosity, a glycan decongestion effect that may help explain why gal-1 promotes membrane fluidity and lateral diffusion of glycoconjugates within cell membranes. Overall, our results provide an insight into the function of galectin in situ and have potential significant biological consequences.
Keywords: carbohydrate-binding domain, cell surface, galectin–glycan interactions, heteronuclear single quantum coherence nuclear magnetic resonance (HSQC NMR), NMR diffusion spectroscopy, simple saccharide
Abbreviations: CRD, carbohydrate-recognition domain; ERK, extracellular-signal-regulated kinase; gal-1, galectin-1; GRG, galactorhamnogalacturonate glycan [α-(1→2)-l-rhamnosyl-α-(1→4)-D-galacturonosyl]; HSQC, heteronuclear single quantum coherence; Lamp, lysosomal-membrane-associated protein; PFG, pulsed-field gradient
INTRODUCTION
Traditionally, galectins have been defined as a subfamily of lectins that selectively bind β-galactosides, and all galectins share significant amino acid sequence conservation within their CRD (carbohydrate-recognition domain) [1]. Although galectins in general can have intracellular functions (e.g. modulating proliferation, apoptosis and pre-mRNA splicing), they are best known for their extracellular activities in mediating cell–cell and cell–matrix adhesion and migration by interacting with various glycan groups of cell-surface glycoproteins and/or glycolipids [2]. gal-1 (galectin-1) interacts with various glycoconjugate ligands of the extracellular matrix [e.g. laminin, fibronectin, the β1 subunit of integrins, ganglioside GM1 and the Lamps (lysosomal-membrane-associated proteins) Lamp 1 and Lamp 2)], as well as those on endothelial cells {e.g. integrins αvβ3 and αvβ5, ROBO4 [roundabout homologue 4, magic roundabout (Drosophila)], CD36, and CD13} and on T-lymphocytes (e.g. CD7, CD43, and CD45), where it is known to induce apoptosis [2]. Their binding to glycans of cell-surface glycoconjugates can also trigger intracellular activity. For example, gal-1 interacts with the α5β1 fibronectin receptor to restrict carcinoma cell growth via induction of p21 and p27 {Ras–MEK–ERK [Ras GTPase–MAPK (mitogen-activated protein kinase)/ERK (extracellular-signal-regulated kinase) kinase–ERK]} pathway [3].
The disaccharide lactose (Gal-β(1→4)-Glc) is the simplest carbohydrate to which galectins bind and one that has been used most often to delineate galectin function and structure–activity relationships in vitro [1]. High-resolution structures of glycan–galectin interactions have been limited to galectin bound to lactose, N-acetyl-lactosamine, trisaccharide and an N-acetyl-lactosamine octasaccharide [4]. In all these cases, a β-galactoside-containing disaccharide moiety is shown to bind galectin in a similar fashion at its classical CRD, i.e. the quintessential lactose-binding domain, and even the largest one, N-acetyl-lactosamine octasaccharide, has the remaining six saccharide units jutting out from the galectin CRD into solution. Although providing a structural insight into gal-1 saccharide binding, previous studies have provided a biased picture of how galectins interact with, and affect, the properties of complex, heterogeneous cell-surface glycans. Actual galectin-binding glycans of various glycoconjugates in situ have far more varied saccharide compositions and complicated structures than the simple saccharides mentioned above, as they are larger, heterogeneous in size and composition, densely packed and therefore self-associating, and it is likely that gal-1 interacts to some extent with saccharide units other than β-galactose in those glycans [5]. The relatively low intrinsic binding affinity (micro- to milli-molar) of simple saccharides to galectins is significantly increased by multivalent interactions with larger glycan ligands. Moreover, gal-1, like most galectins, is a dimer with two CRDs that can mediate intermolecular glycan cross-linking [6]. All this and more could determine and/or differentiate galectin structure–function relationships.
In the present paper we report on NMR experiments with human gal-1 and its interactions with GRG (galactorhamnogalacturonate glycan), a relatively large heterogeneous glycan derived from citrus pectins. Citrus pectins are branched heteropolysaccharides of various molecular masses ranging from about 40 kDa to several million Da, and contain multiple branches of oligosaccharides comprised of one to 20 or more moieties of galacturonate, arabinose, galactose, glucose, xylose, mannose etc. [7]. These complex glycans can also include single residues or chains of uronic acid, and their carboxylic groups can be esterified to the extent of 2–30%. Owing to their large size, these glycans tend to form gels in solution [8]. Following processing, the GRG has a weight-average molecular mass of about 120 kDa [9] and can be characterized as having an irregular structure with five principal components: rhamnose, fucose, arabinose, galactose and uronate, which by weight account for about 4.3, 3.7, 19, 37 and 36% respectively [10,11]. The backbone of the GRG polymer is mainly composed of α-(1→2)-L-rhamnosyl-α-(1→4)-D-galacturonosyl sections. As with most complex glycans, precise structural details for GRG are lacking. However, it is known that the galactose side-chains in GRG are randomly distributed via β(1→4) linkages, most of which are at terminal positions [7,10,12], suggesting the potential for interactions with galectins.
In this regard, pectin-derived polysaccharides have been reported to interact with gal-3 [13], as well as to inhibit human cancer cell growth and metastasis in mice [14] and to enhance apoptosis in human prostrate-cancer cells [15] and multiple myeloma cells in vitro, probably also by interacting with, and antagonizing, the function of gal-3 and possibly that of other galectins [16]. GRG is currently being investigated and developed as a therapeutic in the oncology arena. Here, we demonstrate that GRG interacts with gal-1 over a large surface area on the protein that includes the quintessential lactose-binding site and runs from that site through a broad valley or cleft towards the dimer interface. We also show that binding of gal-1 to GRG disrupts inter-glycan interactions and decreases glycan-mediated solution viscosity, which may help explain previous findings that gal-1 promotes increased membrane fluidity and lateral diffusion of glycoconjugates within cell membranes. Overall, our results provide a novel picture of gal-1 binding to a relatively complex glycan, as well as an insight into galectin structure–function relationships with potential significant biological consequences.
MATERIALS AND METHODS
Gal-1 preparation
Uniformly 15N-labelled gal-1 was expressed in Escherichia coli BL21(DE3) competent cells (Novagen) grown in minimal medium. It was purified over a β-lactose affinity column, and further fractionated on a gel-filtration column as described previously [17]. Typically, 44 mg of purified protein were obtained from 1 litre of cell culture. The purity of the final sample was quantified by using the Bio-Rad protein assay and was checked for purity by SDS/PAGE. The functional activity of the purified protein was assessed by using a T-cell death assay.
Glycan preparation
GRG was produced from crude citrus pectins that were decreased in size to increase solubility and expose galactose residues for potentially better interactions with gal-1. The glycan size-reduction protocol used controlled conditions (temperature, reaction time, catalysts) with hydrogen peroxide and L-ascorbate to catalytically hydrolyse glycosidic linkages in the polysaccharide backbone and oligosaccharide side chains into smaller polysaccharide molecules [10,11]. This chemical modification was aimed at limited depolymerization of the backbone of the polymer, mainly the α-(1→2)-L-rhamnosyl-α-(1→4)-D-galacturonosyl sections, accompanied by de-methoxylation and de-acetylation of carboxylic groups of the polymer, which also decreased hydrophobicity, and hence increased aqueous solubility. Briefly, the crude commercially available citrus pectin (provided as solid crude powder of a kind typically used in food industry) was dissolved in water to pH 10 with 3 M NaOH. After incubation at 50 °C for 30 min, 20% (v/v) ethanol was added, and the partially purified polysaccharide was precipitated to remove proteins and pigments. The polysaccharide was then dissolved to 20 g/litre in water, followed by addition of trifluoroacetic acid to a final concentration of 0.5 M for controlled de-polymerization in which the galactorhamnogalacturonan backbone is cleaved to the desired size. After incubation for 24 h at 60 °C, the solution pH was adjusted to 4. The solution was cooled to 4 °C and centrifuged (15000 g for 60 min) to remove insoluble matter. The supernatant was then neutralized to a final pH of 8.0 with 1 M NaOH, and 20% ethanol was added to recover soluble polysaccharide. The resulting soluble branched polysaccharide product (GRG) was washed with 70% ethanol or with 100% acetone to provide a final dry powder.
HSQC (heteronuclear single quantum coherence) NMR spectroscopy
Uniformly 15N-labelled gal-1 was dissolved at a concentration of 0.3 mM in 20 mM potassium phosphate buffer, pH 7.0, and 0.8 mM dithiothreitol, made up using a 1H2O/2H2O (9:1) mixture. Using uniformly 15N-enriched gal-1, we performed HSQC NMR experiments to investigate binding of gal-1 to GRG [17]. 1H and 15N resonance assignments for the gal-1 have already been reported [17].
All NMR experiments were carried out at 30 °C on a Varian Unity Inova 600 MHz spectrometer equipped with an H/C/N triple-resonance probe and x/y/z triple-axis PFG (pulsed-field gradient) unit. A gradient sensitivity-enhanced version of two-dimensional 1H–15N HSQC was applied with 256 (t1) x 2048 (t2) complex data points in nitrogen and proton dimensions respectively. Raw data were converted and processed by using NMRPipe [18] and were analysed by using NMRview [19].
PFG NMR self-diffusion measurements
For NMR measurements, gal-1 was dissolved in 0.6 ml of 20 mM potassium phosphate buffer, pH 7.0, and 0.8 mM dithiothreitol, made up using 2H2O, and the pH was adjusted by adding microlitre quantities of NaO2H or 2HCl. PFG NMR self-diffusion measurements were made on a Varian INOVA-600 spectrometer using a GRASP™ (gradient-accelerated spectroscopy) unit as previously described [20]. NMR spectra for measurement of diffusion coefficients, D, were acquired using a 5 mm triple-resonance probe equipped with an actively shielded z-gradient coil. The maximum magnitude of the gradient was calibrated using the manufacturer's standard procedure based on the frequency spread of the applied gradient and was found to be 100 G/cm. This was consistent with the value of 98 G/cm obtained from analysis of PFG data on water using its known diffusion constant [20]. The linearity of the gradient was checked by performing diffusion measurements on water over different ranges of the gradient. The PFG longitudinal eddy-current delay pulse sequence [20] was used for self-diffusion measurements, which were performed in 20 mM potassium phosphate buffer, pH 7.0, and 0.8 mM dithiothreitol, made up using 2H2O at a temperature of 30 °C.
For unrestricted diffusion of a molecule in an isotropic liquid, the PFG NMR signal amplitude, A, normalized to the signal obtained in the absence of gradient pulses, is related to D by eqn (1):
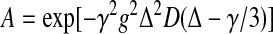 |
where γ is the gyromagnetic ratio of the observed nucleus, g and δ are the magnitude and duration of the magnetic field gradient pulses respectively, and Δ is the time between the gradient pulses [20]. For these experiments, δ=4 ms, g=1–75 G/cm, Δ=34.2 ms, and the longitudinal eddy-current delay (Te) was 100 ms. Each D value was determined from a series of 12 one-dimensional PFG spectra acquired using different g values. Measurements of D were calibrated by performing the PFG NMR self-diffusion measurements on dextran standards and standard proteins [9,21].
RESULTS
Gal-1 binds to GRG
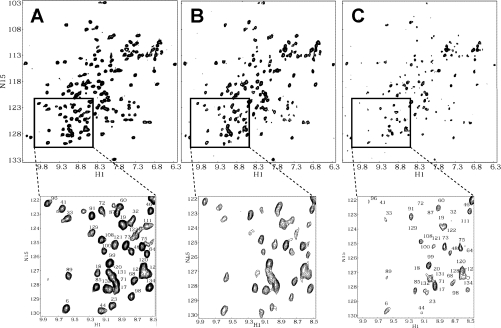
Figure 1(A) shows a 1H–15N HSQC spectrum of uniformly 15N-enriched gal-1, with cross-peaks labelled as assigned previously [17]. At this concentration (1 mg/ml; 70×10−6 M), gal-1 with a monomer↔dimer equilibrium dissociation constant (Kd) of about 2×10−6 M [4], is >99% dimer. As the concentration of GRG is increased, gal-1 resonances are differentially broadened, as exemplified by the 15N-enriched gal-1 HSQC spectra acquired at gal-1/GRG molar ratios of 20:1 and 5:1 (Figures 1B and 1C respectively). Differential broadening is better appreciated in the HSQC spectral expansions shown below each of the full spectra in Figure 1. Because all three HSQC spectra were collected, processed and plotted the same way, it is apparent that some cross-peaks have disappeared, some are greatly decreased in intensity and others appear relatively less affected. These data alone indicate that gal-1 interacts with GRG. Although the increased weight-average molecular mass(es) of the complex(es) formed between gal-1 and GRG likely contribute(s) somewhat to the observed broadening of gal-1 resonances, most of this effect is the result of exchange between or among gal-1 and binding sites on GRG that is occurring on the intermediate chemical-shift time scale [22].
Figure 1. 1H-15N HSQC spectra for gal-1 with and without GRG.
HSQC spectra are shown for 15N-enriched gal-1 (1 mg/ml) alone (A) and at gal-1/GRG molar ratios of 20:1 (B) and 5:1 (C). Spectral expansions are provided below each plot to visualize better the observed resonance broadening as GRG is added to the gal-1 solution. Resonances in the expansion plot to (A) are labelled with assignments reported by Nesmelova et al. [17]. H1, 1H; N15, 15N.
These HSQC data also indicate that the folded structure of gal-1 dimer is not significantly perturbed by binding to GRG. We reached this conclusion because even though 1H–15N resonances are differentially broadened (intensities decreased) during the titration, the chemical shifts of resonances remaining during the titration are mostly unchanged.
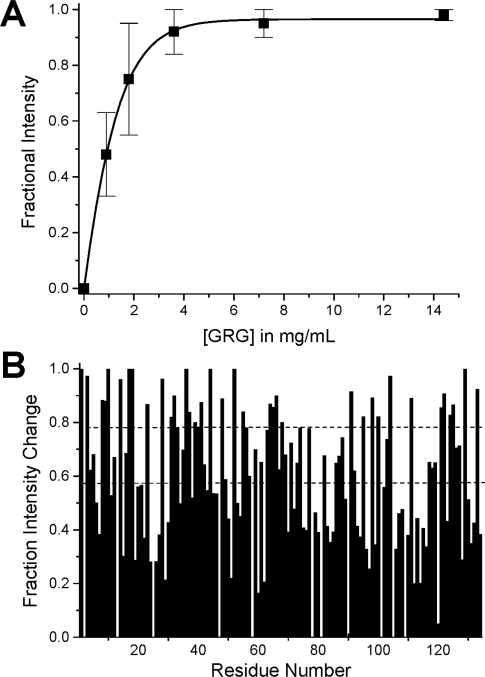
Using our HSQC data, we could also estimate the gal-1–GRG binding affinity and stoichiometry by plotting the fractional change in gal-1 HSQC resonance intensities as a function of the concentration (mg/ml) of GRG (Figure 2A). Fractional changes were calculated by subtracting from 1 the intensity of a given HSQC cross-peak divided by that in pure gal-1 at the same protein concentration and with NMR data collected and processed the same way. The average of fractional changes for the 40 most broadened resonances is plotted as a function of the concentration of GRG in Figure 2(A). S.D. values are indicated by error bars. The continuous line represents a sigmoidal (Boltzmann) fit using the general equation:
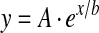 |
(where y is fractional intensity, A is the signal amplitude, x is the concentration of gRG and A and b are variable fitting parameters) through the average value at each concentration point. The χ2 value for the fit was <0.0005. An apparent Kd value was estimated at the point on this curve where 50% of the gal-1 molecules are bound to GRG. In this regard, the average fractional intensity change of 0.5 (50% bound) occurs at a GRG concentration of 1 mg/ml, which corresponds to an apparent macroscopic Kd value of about 8×10−6 M when a weight-average molecular mass of 120 kDa was used for GRG [9]. Furthermore, because gal-1 at 1 mg/ml has about 70×10−6 M in gal-1 CRD equivalents (14500 Da), each average GRG molecule should saturate about five gal-1 CRDs. This binding stoichiometry of 5:1 (gal-1/GRG) would yield an average microscopic binding constant of about 40×10−6 M/site.
Figure 2. Gal-1–GRG binding curve and HSQC resonance broadening mapping.
(A) The average of fractional changes for the 40 most broadened resonances is plotted against the concentration (mg/ml) of GRG. Error bars represent S.D. The continuous line represents a sigmoidal (Boltzmann) fit using the general equation y=A·ex/b through the average value at each concentration point. The χ2 value for the fit was <0.0005. (B) This plot shows initial fractional changes in gal-1 resonance intensities observed at lower glycan/gal-1 molar ratios (where most gal-1 resonances are still apparent) against the amino acid sequence of gal-1 for GRG. A value of 1 indicates that the resonance associated with that particular residue is no longer apparent, and a value of zero indicates no change in resonance intensity.
GRG binding site on gal-1
Using these HSQC data, we also gained insight into where GRG interacts on the surface of gal-1. We do this in a way that is similar to HSQC chemical-shift mapping [23], which is performed when binding interactions occur in the fast or slow exchange regimes on the chemical-shift time scale. In these instances, resonances are chemically shifted, and little broadened, during the titration with ligand. In our case, gal-1 resonances initially may be shifted somewhat by interaction with GRG, but are primarily broadened due to the exchange process which falls in the intermediate exchange regime on the NMR chemical-shift time scale [22]. There are a number of factors that can contribute to a system falling into a particular NMR exchange regime. However, the general tendency is that as the lifetime of a complex is increased (i.e. binding becomes relatively stronger), the exchange regime on the chemical-shift time scale goes from fast to intermediate to slow. Because interactions occurring on the intermediate exchange time scale may not show discrete resonances, the way in which we present observed broadening effects is different from the way in which we would show 1H and 15N weighted average chemical-shift changes for a system in the fast or slow exchange regimes.
In the intermediate exchange regime, we show differential broadening at a molar ratio where most resonances are still observed, but are at lower intensities, owing to the broadening. We refer to this as ‘HSQC resonance broadening mapping’ in order to distinguish it from ‘HSQC chemical-shift mapping’. The interpretation is essentially the same as with HSQC chemical-shift mapping, i.e. those resonances that are initially broadened the most are associated with that site(s) on gal-1 that interacts with GRG. We create this map by taking the initial fractional changes in gal-1 HSQC resonance intensities observed at a low GRG/gal-1 molar ratio (where most gal-1 resonances are still apparent) and by plotting them against the amino acid sequence of gal-1. This is illustrated in Figure 2(B), where a value of 1 indicates that the resonance associated with that particular residue is no longer apparent (highly broadened), and a value of 0 indicates no change in resonance intensity (not broadened). From these data it is apparent which gal-1 residues are initially more affected than others.
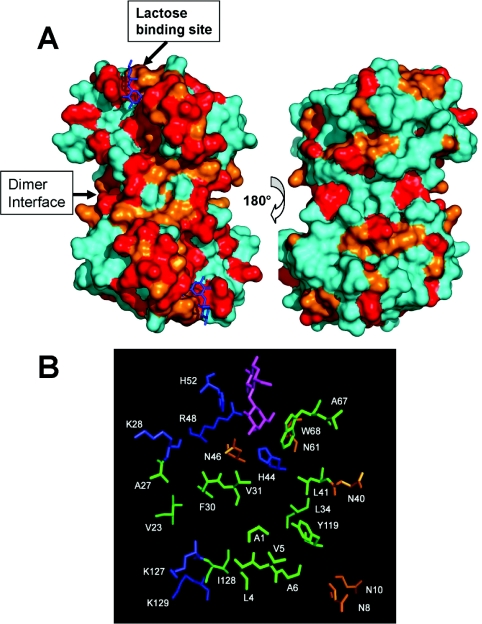
Figure 3(A) highlights the most affected residues on the folded structure of gal-1 dimer. Resonances whose fractional intensity decrease is more than 0.8, or between 0.6 and 0.8 (Figure 2B), are highlighted in red and orange respectively. The image on the left illustrates the dimer surface with the lactose-binding sites (one per monomer) oriented at the top left and bottom right. For reference, lactose molecules are shown in blue at their previously determined binding sites [24]. Clearly, gal-1 residues at the quintessential lactose-binding site are affected by binding to GRG. However, many other gal-1 residues on this same surface are also significantly affected, indicating that GRG interacts with a larger area on this surface of gal-1. By contrast, GRG binding also affects a few residues on the opposite surface of the gal-1 dimer (Figure 3A, right-hand image). Although the reason for this is unclear, it may be that GRG interacts to some extent with these residues, or, more likely, that GRG binding on the front surface (Figure 3A, left-hand image) perturbs NH groups on the opposite face through the β-sheet sandwich. Furthermore, this novel extended glycan-binding domain on gal-1 could be explained by various modes of interaction with binding sites on heterogeneous GRG. However, when we add a large molar excess of lactose (200 mM), the NMR spectral effects on gal-1 from GRG binding can be reversed, indicating that lactose competes with GRG for binding to gal-1. This suggests that we are dealing with only one major mode of interaction.
Figure 3. The GRG binding site on gal-1.
(A) Residues on the folded structure of gal-1 that have been most affected by binding to GRG are highlighted in red and orange as discussed in the text. The X-ray structure of lactose-bound human gal-1 has been used in this Figure (Protein Data Base access code: 1 gzw) [24]. The orientation at the left shows the face of the dimer where GRG binds. Bound lactose molecules are shown in blue on the gal-1 dimer structure. The gal-1 dimer interface is also indicated. The orientation at the right shows the opposite side of the dimer where lactose binds. (B) Illustration of gal-1 residues in the GRG-binding domain. Polar, positively charged and hydrophobic residues are colored in orange, blue, and green respectively. For reference, the lactose molecule in its binding site is shown in purple. The one-letter code for amino acids is used (e.g. K28=Lys28 etc.).
The GRG-binding domain includes residues from the quintessential lactose binding site (H44, N46, R48, H52, N61, W68, using the one-letter amino acid code), as well as others that extend out from that site down towards the gal-1 dimer interface. These residues are shown for one gal-1 subunit in Figure 3(B), along with the lactose molecule to illustrate proportional size of the binding domain to the disaccharide. This amino acid composition is similar to that of the lactose-binding site and suggests the potential for similar types of protein–glycan interactions, namely hydrogen bonding and hydrophobic interactions, as well as electrostatic interactions between or among positively charged lysine residues (K28, K127 and K129) from gal-1 and negatively charged galactouronate carboxylate groups from the backbone of GRG.
HSQC mapping with lactose
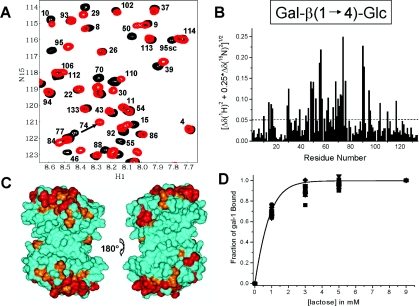
Lactose is the quintessential ligand for any galectin. For this reason, we performed the same gal-1 15N HSQC titration experiment with lactose. As lactose was titrated (1–10 mM) into solution, we observed that a number of gal-1 HSQC cross-peaks were significantly chemically shifted by the presence of lactose, with saturation being achieved by about 5 mM lactose. Figure 4(A) overlays two HSQC spectral expansions for pure gal-1 (black) and gal-1 plus 5 mM lactose (red). 15N–1H weighted chemical-shift changes against the amino acid sequence of gal-1 are plotted in Figure 4(B). The simple average over all changes is 0.05 p.p.m. Although a good number of gal-1 residues are affected by lactose binding, those residues that are affected most (mean+ 1 S.D.) are highlighted in red on the surface of the gal-1 dimer in Figure 4(C). (Here we used the same surface orientation as illustrated for GRG binding in Figure 3.) Other gal-1 residues that fall above the simple average are highlighted in orange. As expected, the most affected residues are around the lactose-binding site identified previously from X-ray-crystallographic studies [24], and these include many of the same residues affected by GRG binding to gal-1.
Figure 4. HSQC chemical-shift mapping for lactose binding to gal-1.
(A) Two 15N–1H HSQC spectra of gal-1 (1 mg/ml) are overlayed, one without lactose (black cross-peaks) and one with 5 mM lactose (red cross-peaks). Some of the cross-peaks have been labelled, as discussed in the text. Resonances are labelled with assignments reported by Nesmelova et al [17]. H1, 1H; N15, 15N. (B) 15N and 1H weighted chemical-shift changes between pure gal-1 and gal-1 upon addition of 5 mM lactose are plotted against the amino acid sequence of gal-1. The broken line indicates the simple average over all values. (C) Residues on the folded structure of gal-1 that have been most shifted by binding to lactose are highlighted in red and orange as discussed in the text. The X-ray structure of lactose-bound human gal-1 has been used in this Figure (Protein Data Base access code: 1gzw) [24]. The orientation at the left shows the face of the dimer where lactose binds. The orientation at the right shows the opposite side of the dimer where lactose binds. (D) The fractional change in gal-1 HSQC weighted chemical shifts is plotted against the concentration of lactose. The continuous line represents a sigmoidal (Boltzmann) fit using the general equation y=A·ex/b through the average value at each concentration point. The χ2 value for the fit was <0.0005.
Interestingly, lactose binding also elicits an effect on residues at the back side of gal-1 (see Figure 4C), and these are among those also affected by GRG binding (see Figure 3A, right-hand image). Because these residues are relatively well removed from the lactose-binding site, it appears that lactose binding induces conformational changes, however minor, in other regions of the folded protein. This observation with lactose supports the idea that GRG binding to the front face of gal-1 (Figure 3A) does the same.
To determine the Kd for lactose binding to gal-1, we plotted the fractional changes in 15N–1H weighted chemical shifts for the 15 most shifted resonances as a function of the lactose concentration (Figure 4D). The continuous line represents a sigmoidal (Boltzmann) fit using the general equation y=A·ex/b, through the average value at each concentration point. The χ2 value for the fit was <0.0005. A microscopic Kd value of 520×10−6 M was determined from the lactose concentration at the fractional change of 0.5 (50% bound) and a statistical correction for the number of lactose binding sites, i.e. two in the dimer. Our Kd value is similar to that of 330×10−6 M reported from calorimetric studies for lactose binding to human gal-1 at 30 °C [24] and somewhat higher than that reported for lactose binding to bovine gal-1 (170×10−6 M at 30 °C) [25]. Nevertheless, these values are all in the same range, and any of these Kd values for lactose binding to gal-1 is considerably greater than the average value we found for gal-1 binding to GRG. The difference in Kd values is consistent with the significant resonance broadening we observed with gal-1 binding to GRG, placing the interaction in the intermediate exchange regime on the chemical-shift time scale [22]. In the case of lactose binding to gal-1, we observed that gal-1 resonances were merely chemically shifted during the titration with lactose, which is what one would expect with fast exchange on the chemical-shift time scale.
Gal-β((1→4)-Gal binds gal-1
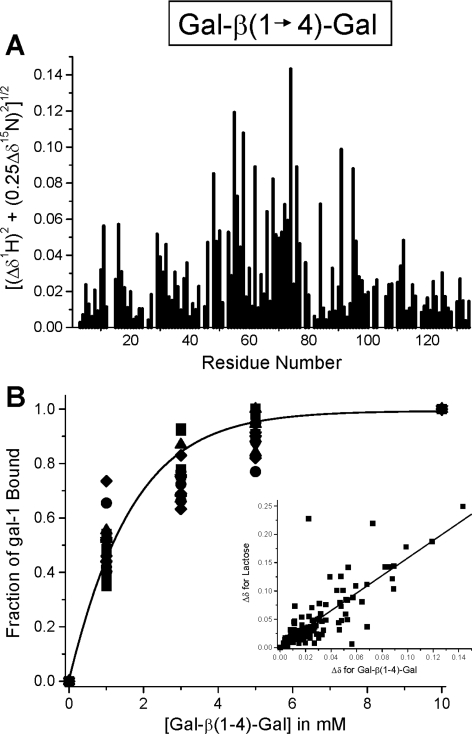
Even though we demonstrated that gal-1 binds GRG, the terminal motifs of GRG to which gal-1 most likely interacts are Gal-β(1→4)-Gal. Galectin/saccharide-binding dogma, however, would say that gal-1 would not bind to a Gal-β(1→4)-Gal motif, primarily because of the axial C4 hydroxy group on the reducing-end Gal unit. In lactose [Gal-β(1→4)-Glc], the Glc C4 hydroxy group is equatorial. For this reason, we performed the gal-1 15N HSQC titration with the disaccharide Gal-β(1→4)-Gal and found that gal-1 HSQC cross-peaks are chemically shifted as they were upon titration with lactose, indicating interaction of Gal-β(1→4)-Gal with gal-1.
Figure 5(A) indicates which residues in gal-1 are most affected upon binding Gal-β(1→4)-Gal. If we correlate these chemical-shift changes with those from the binding of lactose (Figure 4B), we get a linear regression coefficient of 0.82 (Figure 5B, inset), which indicates that Gal-β(1→4)-Gal interacts primarily at the same site on gal-1 as lactose. In Figure 5(B) we plot the fraction bound for the 20 most chemically shifted gal-1 resonances against the concentration of the disaccharide. For this, we assumed that saturation occurs by about 10 mM Gal-β(1→4)-Gal, and set this value as a bound fraction of 1.0. The continuous line represents a sigmoidal (Boltzmann) fit using the general equation y=A·ex/b through the average value at each concentration point. The χ2 value for the fit was <0.0005. A microscopic Kd value of 1200×10−6 M was determined at the fractional change of 0.5 (50% bound). This Kd value is significantly greater than that for lactose (520×10−6 M), indicating weaker binding of Gal-β(1→4)-Gal to gal-1.
Figure 5. 1H-15N HSQC of gal-1 with and without Gal-β(1→4)-Gal.
(A) The histogram shows fractional changes in gal-1 (1 mg/ml) HSQC resonance intensities observed at a Gal-β(1→4)-Gal/gal-1 molar ratio of 4:1 (where most gal-1 resonances are still apparent) plotted against the amino acid sequence of gal-1. A value of 1 indicates that the resonance associated with that particular residue is no longer apparent, and a value of zero indicates no change in resonance intensity. (B) The fractional change in gal-1 HSQC resonance broadening is plotted against the concentration (mg/ml) of Gal-β(1→4)-Gal. Variation among these fractional values arises because of differential broadening, in that some resonances are affected more than others at different points during the titration. The continuous line represents a sigmoidal (Boltzmann) fit using the general equation y=A·ex/b, through the average value at each concentration point. The χ2 value for the fit was <0.0005. The inset to (B) correlates HSQC chemical-shift changes of gal-1 resonances for binding of lactose and Gal-β(1→4)-Gal with gal-1. The continuous line represents a linear fit to these points, with a regression coefficient of 0.82.
From the glycan side
In our NMR experiments we actually observe an average effect from GRG glycans that vary in size [9] and from which we cannot get structural detail. Therefore, we performed PFG NMR diffusion experiments to derive D values for insight into what occurs from the perspective of the glycan when it binds gal-1. For the most part, changes in D values reflect changes in molecular size (apparent molecular mass and hydrodynamic radius) and/or solution viscosity due to intermolecular interactions among glycans. A smaller D value indicates an increase in molecular size and/or viscosity and vice versa.
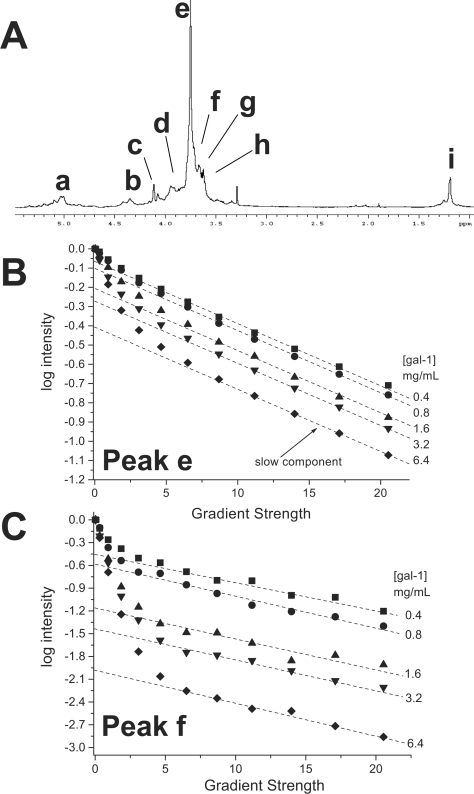
For these diffusion experiments we used a fixed concentration of GRG (4.6 mg/ml) and titrated gal-1 into the glycan solution. During the titration, D values were acquired for the most intense GRG resonances, as identified in the 1H NMR spectral trace of the glycan preparation shown in Figure 6(A). If this were a pure single-size glycan, individual resonances would represent different chemical groups within the GRG molecule and D values derived from any given resonance should be the same or very nearly the same (e.g. differences in internal motions of glycan side chains could affect D). Here, this is not the case, because our GRG preparation is composed of heterogeneous galactorhamnogalacturonates, with a distribution of molecular sizes and a weight-average molecular mass of about 120 kDa [9]. Moreover, even though chemical shift is not a function of molecular mass, it is a function of, e.g. chemical composition, folding and supramolecular structure, i.e. anything that affects the chemical environment of a particular saccharide residue. Therefore, depending upon which GRG resonance, or even upon which point within a resonance envelope, D values can and will vary because of both these factors and experimental error. It is for this reason that we measured diffusion-mediated decay curves for several of the GRG resonances.
Figure 6. Diffusion decay curves for the titration of gal-1 with GRG.
Diffusion decay curves for GRG resonances e (B) and f (C) are shown. The glycan concentration was held constant at 4.6 mg/ml, and gal-1 was titrated into solution. Broken lines show extrapolation to the Y-intercept of the slow component of the decay curve, as discussed in the text. A 1H-NMR spectral trace for GRG is shown above the plot (A), with resonances labelled as discussed in the text.
To show data quality and give a feel for experimental error, Figures 6(B) and 6(C) show diffusion decay curves for two GRG resonances (e and f) as a function of gal-1 concentration. In the absence of gal-1, diffusion decay curves for these resonances appear linear (albeit with different slopes; results not shown), indicating the presence of either a single molecular-size component for each or any number of different glycan components with nearly the same molecular size. Decay curves become increasingly curvilinear as gal-1 is titrated into the GRG solution. If we deconvolute each decay curve into two components simply by approximating slopes for initial and final parts of the decay curves, we can estimate D values for the fast and slow decay components. Broken lines are shown through the final six to eight points of the curves to indicate the slow decay components. Linear fits to these data points are very good, with regression coefficients greater than 0.9. In this regard, differences in D values are primarily the result of differences in apparent size of the glycan's internal flexibility and solution viscosity, as opposed to experimental error. The fractional contribution of these slow components to the full decay curves was estimated from the Y-intercept of the linearly extrapolated curves (broken lines).
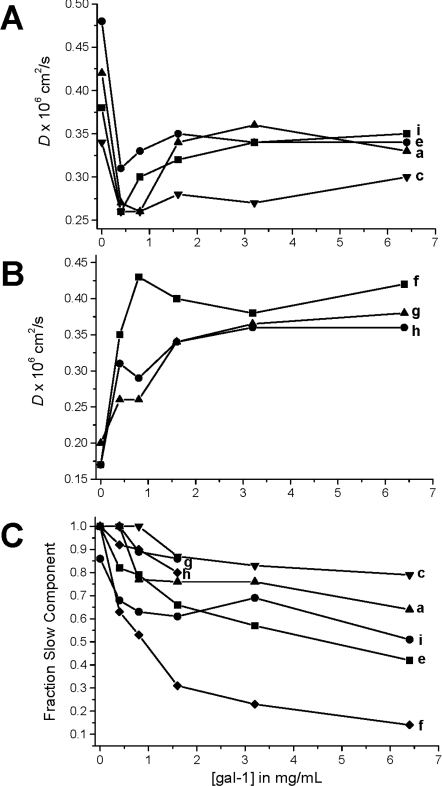
In Figure 7, D values derived from the slow decay components are plotted as a function of gal-1 concentration. Two types of trends are observed. For some resonances (a, c, e, and i), D values initially decrease upon addition of gal-1, and then they increase somewhat and remain essentially constant (Figure 7A). In the second set (f, g and h), D values merely increase and plateau upon addition of gal-1 (Figure 7B). For the first set, the initial decrease in D indicates an increase in molecular size of GRG upon binding gal-1, as opposed to an increase in solution viscosity. If solution viscosity were responsible for the initial decrease in D, the second set would have behaved similarly, and it did not. If anything, the immediate increase in D values in the second set indicates that solution viscosity is decreased and/or internal flexibility is decreased. Moreover, this trend in the second set suggests that GRG glycans associated with D values derived from resonances f, g and h either do not bind gal-1 or respond differently to interactions with gal-1.
Figure 7. D values for GRG.
(A) Some D values for the slow component of the deconvoluted diffusion decay curves are plotted against gal-1 concentration. (B) Other D values for the slow component of the deconvoluted diffusion decay curves are plotted against gal-1 concentration. (C) Estimation of the fraction of the slow decay component by linearly extrapolating the slow component line to the Y-intercept is shown. These fractions yield the fraction of gal-1-bound glycan.
The initial decrease in D occurs at gal-1 concentrations below 1 mg/ml, where gal-1 binding sites on GRG (4.6 mg/ml) are not fully occupied (vis-à-vis Figure 2). As the gal-1 concentration is increased above 1 mg/ml, more and more binding sites on GRG become occupied such that the apparent molecular size of the complex should increase, and yet we observe an increase in D values, indicating the opposite. There are two likely explanations for this apparent conundrum: (1) gal-1 binding disrupts the extensive inter-glycan networks that contribute to the normally high solution viscosity of these polysaccharide solutions; and (2) the hydrodynamic radius of the glycan is decreased by gal-1 binding. In either case, gal-1 binding appears to alter GRG conformation and perturb inter-glycan interactions. Note that, upon gal-1 binding, the fraction of the GRG slow decay component is decreased and tends towards a limiting value (Figure 7C), consistent with saturation of the gal-1-binding sites on GRG (Figures 2 and 7A/7B). Some slow-component fractions are decreased more than others, suggesting different extents of the effect(s) from gal-1 binding.
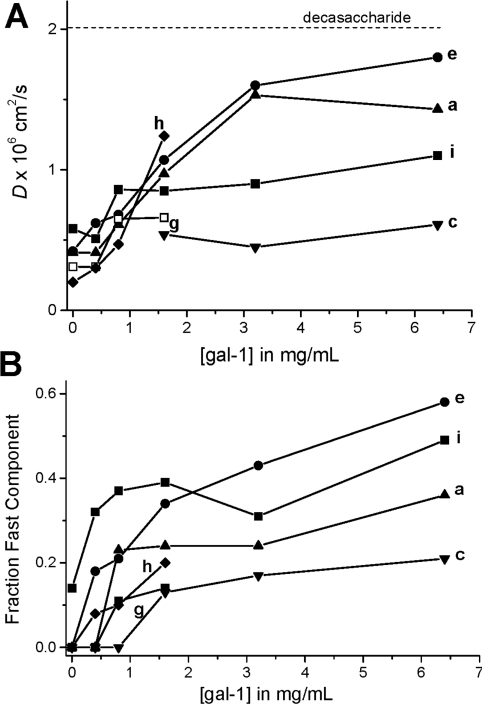
This gal-1-induced re-organization or conformational change of the GRG glycan network apparently leads to release of lower-molecular-size glycan species, as evidenced by trends in D values derived from the fast components of the decay curves (Figure 8A). If we assume that only two glycan components are represented in each curvilinear decay curve, then the fraction of the fast component can be estimated by subtracting from one the fraction of the slow component (Figure 7B), as plotted in Figure 8(B). During the titration with gal-1, the fraction of the fast component increases rapidly up to the gal-1 saturation point (about 1 mg/ml) and then tends to level off at anywhere from about 20 to 50% of the total glycan species reflected in D values derived from that resonance. On the basis of apparent D values, we estimate that molecular sizes of galectin-mediated GRG-released glycans range from about 20 to 200 saccharide units, compared with 500 to 2000 units prior to addition of gal-1 [9]. The dotted line labelled ‘decasaccharide’ in Figure 8(A) was drawn as a point of reference, because a decasaccharide in dilute solution would have a D value around 2×10−6 cm2/s [9]. It is important to note here, however, that because our GRG glycans are not in dilute solution, actual D values will be lower, and changes in D values correlate better with changes in hydrodynamic radii than with molecular mass [9]. It is for this reason that we have provided a very wide range in our estimates of the possible number of saccharide units stated above. Nevertheless, these ranges are reasonable. Furthermore, because gal-1 at these concentrations is a dimer with a molecular mass of 29 kDa, it is unlikely that gal-1 binds to the smallest glycans with D values near to, or larger than, that for the gal-1 dimer (D=1.04×10−6 cm2/s). If gal-1 were to bind to these GRG glycans, the D values would be significantly lower by the end of the titration, and they are not.
Figure 8. D values for GRG fast component.
(A) D values for the fast component of the deconvoluted diffusion decay curves are plotted against gal-1 concentration. The dotted line labelled ‘decasaccharide’ was drawn as a point of reference, because a decasaccharide in dilute solution would have a D value of about 2×10−6 cm2/s [9]. (B) Estimation of the fraction of the fast decay component by linearly extrapolating the slow component line to the Y-intercept is shown. This estimation assumes that the decay curve is composed of only two glycan fractions.
On the basis of these data, we estimate a binding stiochiometry of 6:1 (gal-1/GRG) by assuming that saturation of GRG (37×10−6 M) with gal-1 occurs at a gal-1 concentration of about 240×10−6 M (Figures 7 and 8). This stiochiometry is essentially the same as that derived from our HSQC binding curve (Figure 2A), which indicated a binding stiochiometry of about 5:1 (gal-1/GRG). Nevertheless, the average microscopic dissociation constants for gal-1 binding to GRG derived either way would be essentially the same, i.e. 48×10−6 M at 6:1 compared with 40×10−6 M at 5:1. This indicates that gal-1 binds to sites on GRG about 10-fold more strongly than it does to lactose.
DISCUSSION
We report here several findings that increase our understanding of how gal-1 interacts with larger, more complex, glycans, in the present case with a pectin-derived GRG, galactorhamnogalacturonate. First of all, we found that gal-1 binds relatively strongly to GRG glycans (microscopic Kd≈40×10−6 M), more so than it does to lactose (Kd≈520×10−6 M). The multivalent nature of GRG (five or six gal-1 CRDs per average glycan molecule) contributes to this greater affinity or avidity for gal-1 (e.g. [6,26,27]), something that is observed with many cell-surface glycoconjugates that bear multiple galectin-binding sites [28]. The corresponding free energies for binding of GRG glycans and lactose are 6.08 kcal/mol and 4.54 kcal/mol (1 kcal=4.184 kJ) respectively, which (assuming a single mode of interaction) reflect the greater surface area on gal-1 with which GRG interacts, approx. 1600 Å2 (1 Å=0.1 nm) compared with 700 Å2 for the much smaller disaccharide lactose.
The larger binding region on gal-1 GRG not only includes residues of the lactose-binding domain, but extends from there through a broad valley or cleft towards the dimer interface. From the overall dimensions of the protein surface, we estimate that this binding domain could accommodate about eight to ten saccharide units, a number that is consistent with the up to 20 or so saccharide units present in any of the multiple oligosaccharide branches of GRG. Moreover, the amino acid composition of the binding domain is consistent with promotion of protein–glycan interactions, as it contains a significant number of polar residues that could potentially form hydrogen bonds with polar groups from the glycan. In addition, negatively charged groups are absent from this domain, whereas positively charged amino acid residues are present for potential electrostatic interactions with negatively charged galacturonate carboxylate groups in the backbone of GRG, as has been reported to occur with the binding of sulfated glycosaminoglycans to platelet factor-4 or sulfated galactosaminoglycans to other galectins [29,30].
Although we do not know which specific groups on GRG are involved in interactions with gal-1, galectins are generally known for their ability to bind to β-galactosides [1]. GRG is composed of about one-third galactose units, which are in either α(1→4) or β(1→4) anomeric linkages. However, most of these are found in the glycan backbone, mainly as sections of α-(1→2)-L-rhamnosyl-α-(1→4)-D-galacturonosyl, and it is unlikely that these galactose units would be accessible to bind to gal-1, as, for example, branched saccharide side chains would likely interfere sterically with the interaction. On the other hand, GRG contains terminal β(1→4)-galactose units, which we propose are likely to be involved in interactions with gal-1. This suggestion, however, poses a problem in that these terminal units exist in a Gal-β(1→4)-Gal motif to which gal-1 should not bind because, unlike lactose [Gal-β(1→4)-Glc], the reducing-end saccharide (galactose) has an axial (not equatorial) C4 hydroxy group. Nevertheless, our HSQC data demonstrate that Gal-β(1→4)-Gal does bind at the gal-1 lactose-binding site, albeit not as strongly. Manual docking of Gal-β(1→4)-Gal into the lactose-binding site on gal-1 in silico suggests that this galactose C4 axial hydroxy group would be sterically hindered only by the side chain of R48 (Arg48) from gal-1, and that a conformational change in the R48 side chain could alleviate most of the unfavourable steric hindrance. In support of this, Leffler et al., who have done extensive work with galectin-binding oligosaccharides (see, e.g., [31,32]) reported that the gal-1 carbohydrate-binding domain can tolerate many chemical modifications and extensions, both linear and branched. Our observation may also be relevant to mammalian glycobiology, as β-D-galactose is the terminal saccharide unit found in 23% of all mammalian glycans [33], and their terminal β-D-galactose units would be linked in a similar fashion.
Even if terminal β(1→4)-galactose on GRG were part of the gal-1-binding epitope, GRG is a heterogeneous glycan, and there could be multiple modes of interaction. Four points, however, support the idea that there is one major mode of interaction: (1) during the gal-1 HSQC titration with GRG, resonance broadening for all residues followed essentially the same titration curve; (2) most residues affected by the presence of GRG are located over a contiguous surface of the folded protein; (3) the binding strength, i.e. Kd, that we observe would likely not be as great if there were numerous smaller binding patches interacting in multiple ways; and (4) lactose competes with gal-1 for binding to GRG. Nevertheless, although we can not exclude the possibility of more than one mode of binding, the more important message from our data is that GRG (and likely other larger glycans) can interact with the surface of gal-1 over a larger region than previously thought from studies with small simple saccharides [34–38].
We also found that gal-1 interactions with GRG attenuate inter-glycan interactions. By forming large intermolecular networks, glycans generally exhibit highly increased solution viscosity. For example, Daas et al. reported that, because of their high hydrodynamic volume, polysaccharides, even at low concentration, can form viscous solutions [39]. We demonstrated this previously with GRG by measuring D as a function of GRG concentration, such that, as the concentration was lowered, D values increased [9]. In the present study we observed that the D value for GRG initially decreases in some instances, simply increases in others, but nevertheless eventually increases in all cases as gal-1 is titrated into the GRG solution. Since we know that gal-1 interacts with GRG, the most reasonable explanation for increasing D values is that gal-1 binding decreases the apparent solution viscosity, which most likely occurs via attenuation of inter-glycan interactions. Although it is unclear how this occurs at the molecular level, we suggest that gal-1 binds saccharide groups that would otherwise be involved in intermolecular inter-glycan interactions and that gal-1 binding somehow re-conforms glycan molecules to promote their dissociation. A common analogy would be decongesting a traffic jam or nasal blockage.
Gal-1-mediated glycan decongestion may be biologically relevant. Glycans, in one form or another (glycoproteins or glycolipids), are a major component of any cell-surface micro-environment, and the glycan concentration within micro-environments can be quite high. In fact, we can estimate the glycan concentration in situ. At least for engineered glycan-containing mucins in a plasma membrane, cell-surface packing density is about 50 molecules/μm2 [40,41]. Assuming that the glycan moieties of each mucin extend about 100×10−10 m from the membrane surface to define the volume of each molecule, we would have 8×10−23 mol per 1×10−17 litre, or a concentration of about 8×10−6 M. Considering that there are numerous glycoproteins on a typical cell surface, this estimate seems reasonable and may even be low, because glycoproteins are not generally homogeneously dispersed on the cell surface (as assumed in our estimation) and are often found in plasma-membrane microdomains [42,43]. Therefore, the concentration of some glycoproteins (and their associated glycans) can be relatively high, possibly in the micro- to milli-molar range, as used here in vitro with GRG.
The functional relevance of galectin-mediated glycan decongestion is further supported by the observation that gal-1 interactions with cell-surface glycans increase membrane fluidity. Gupta et al. used EPR spectroscopy to demonstrate that gal-1 (in a concentration-dependent manner from 1.1 to 4.4 μM) increases erythrocyte membrane fluidity up to a factor of about 3-fold [44]. In this regard, cell-surface viscosity should be decreased, as fluidity is inversely correlated with viscosity. Increased membrane fluidity may be necessary for cell-surface glycoproteins to re-organize within the plasma membrane, as is required, for example, for cell adhesion [45]. Confocal microscopy has demonstrated that, upon exposure to gal-1, CD45 and CD43 receptors cluster on the cell surface of MOLT-4 cells (leucocytes) within 20 min upon exposure to gal-1 [43]. A similar observation has been made with gal-3, which associates and forms clusters when it interacts with cell-surface glycans, thereby explaining its ability to mediate reorganization of cell-surface glycoproteins [46]. In these studies, cell-surface glycoprotein re-organization and clustering are triggered by gal-1–glycan (glycoconjugate) binding and, given the limited time scale over which this process occurs, the phenomenon is likely primarily physically driven. A number of other studies have also proposed that binding of various galectins (i.e. gal-1, -3, -4 and -9) to specific cell-surface glycoproteins contributes to plasma-membrane microdomain assembly and/or maintenance [47–52].
In the light of our present findings, we propose that cell-surface re-organization of glycoconjugates into plasma-membrane microdomains may be promoted by galectin-mediated glycan decongestion. Even though the nature of inter-glycan interactions (specific or non-specific) among various glycoconjugates on the cell surface is unknown, these interactions do occur and would likely attenuate lateral diffusion of glycoproteins and glycolipids within the cell membrane. Galectin interactions with cell-surface glycoproteins may then promote glycan decongestion, increase membrane fluidity and thereby allow glycoconjugate re-organization within the cell membrane to occur.
Conclusions
In the present study we used NMR spectroscopy to investigate interactions between gal-1 and a heterogeneous galactorhamnogalacturonate, GRG. We found that GRG binds to gal-1 more strongly than lactose, and over a broader area on the protein surface, which runs from the quintessential lactose-binding site through a broad valley towards the dimer interface. This information expands our view of how galectins in general may interact with glycans in situ, and this may play a role in determining and/or differentiating galectin function. Moreover, we found that gal-1 binding to the glycan acts to decongest inter-glycan interactions. Because of this finding, our cell-surface glycan decongestion hypothesis may be biologically relevant in that glycan decongestion could promote re-organization of cell-surface glycoconjugates into plasma-membrane microdomains. Overall, our results provide an insight into galectin function in situ and may help explain how gal-1 mediates cell–cell and cell–matrix adhesion and migration.
AUTHOR CONTRIBUTION
Michelle Miller designed, conducted and analysed the HSQC NMR and diffusion experiments and wrote the paper. Irina Nesmelova performed the HSQC NMR experiment with gal-1 and lactose and helped write the paper. David Platt and Anatole Klyosov helped prepare the glycan GRG and write the paper. Kevin Mayo designed and analysed the NMR experiments, supervised the overall work and wrote the paper.
ACKNOWLEDGEMENTS
We are most grateful and indebted to Professor Linda Baum and Ms Mabel Pang of the Department of Pathology and Laboratory Medicine, University of California Los Angeles School of Medicine, Los Angeles, CA, U.S.A., for providing us with isotopically enriched recombinant human gal-1, as well as for reading the manuscript and for many helpful comments and suggestions. We are also indebted to Dr Eliezer Zomer of Pro-Pharmaceuticals for help in preparing this manuscript.
FUNDING
This work was supported by the National Cancer Institute [National Institutes of Health grant number CA096090 (to K. H. M.)]; National Institutes of Health [grant number T32 CA009138 (Cancer Biology Training grant to M.M.)]; National Science Foundation [grant number BIR-961477 (funds for NMR instrumentation)]; University of Minnesota Graduate School (funds for NMR instrumentation); Minnesota Medical Foundation (funds for NMR instrumentation).
References
- 1.Barondes S. H., Castronovo V., Cooper D. N., Cummings R. D., Drickamer K., Feizi T., Gitt M. A., Hirabayashi J., Hughes C., Kasai K., et al. Galectins: a family of animal β-galactoside-binding lectins. Cell. 1994;76:597–598. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- 2.Liu F. T., Rabinovich G. A. Galectins as modulators of tumour progression. Nat. Rev. Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 3.Fischer C., Sanchez-Ruderisch H., Welzel M., et al. Gal-1 interacts with the α5β1 fibronectin receptor to restrict carcinoma cell growth via induction of p21 and p27. J. Biol. Chem. 2005;280:37266–37277. doi: 10.1074/jbc.M411580200. [DOI] [PubMed] [Google Scholar]
- 4.Nesmelova I. V., Dings R. P. M., Mayo K. H. Understanding galectin structure–function relationship to design effective antagonists. In: Klyosov A. A., Witczak Z. J., Platt D., editors. Galectins. New York: John Wiley and Sons; 2008. pp. 33–69. [Google Scholar]
- 5.Klyosov A. A. Galectins and their functions in plain language. In: Klyosov A. A., Witczak Z. J., Platt D., editors. Galectins. New York: John Wiley and Sons; 2008. pp. 9–31. [Google Scholar]
- 6.Collins B. E., Paulson J. C. Cell surface biology mediated by low affinity multivalent protein–glycan interactions. Curr. Opin. Chem. Biol. 2004;8:617–625. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Ridley B. L., O'Neill M. A., Mohnen D. Pectins: structure, biosynthesis and oligogalacturonide-related signaling. Phytochemistry. 2001;57:929–967. doi: 10.1016/s0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- 8.Mikkonen K. S., Rita H., Helen H., Talja R. A., Hyvoenen L., Tenkanen M. Effect of polysaccharide structure on mechanical and thermal properties of galactomannan-based films. Biomacromolecules. 2007;8:3198–3205. doi: 10.1021/bm700538c. [DOI] [PubMed] [Google Scholar]
- 9.Miller M., Klyosov A., Platt D., Mayo K. H. Using pulse field gradient NMR diffusion measurements to define molecular weight distributions in glycan preparations. Carbohydr. Res. 2009;344:1205–1212. doi: 10.1016/j.carres.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Klyosov A., Platt D. Galactose-pronged polysaccharides in a formulation for antifibrotic therapies. 20080107622. U.S. Pat. 2008 USPTO (United States Patent and Trademark Office) pending.
- 11.Tapie N., Malhiac C., Hucher N., Grisel M. Determination of galactose and mannose residues in natural galactomannans using a fast and efficient high-performance liquid chromatography/UV detection. J. Chromatogr. A. 2008;1181:45–50. doi: 10.1016/j.chroma.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 12.Voragen A. G. J., Pilnik W., Thibault J. F., Axelos M. A., Renard C. M. G. C. Isolation and structural characterization of rhamnogalacturonan oligmers. In: Stephen A.M., editor. Food Polysaccharides and their Applications. New York: Food Science and Technology Series, Marcel Dekker; 1995. pp. 287–340. [Google Scholar]
- 13.Sathisha U. V., Jayaram S., Nayaka M. A. H., Dharmesh S. M. Inhibition of galectin-3 mediated cellular interactions by pectic polysaccharides from dietary sources. Glycoconj. J. 2007;24:497–507. doi: 10.1007/s10719-007-9042-3. [DOI] [PubMed] [Google Scholar]
- 14.Nangia-Makker P., Hogan V., Honjo Y., Baccarini S., Tait L., Bresalier R., Raz A. Inhibition of human cancer cell growth and metastasis in nude mice by oral intake of modified citrus pectin. J. Natl. Cancer Inst. 2002;94:1854–1862. doi: 10.1093/jnci/94.24.1854. [DOI] [PubMed] [Google Scholar]
- 15.Jackson C. L., Dreaden T. M., Theobald L. K., Tran N. M., Beal T. L., Eid M., Gao M. Y., Shirley R. B., Stoffel M. T., Kumar M. V., Mohnen D. Pectin induces apoptosis in human prostrate cancer cells: correlation of apoptotic function with pectin structure. Glycobiology. 2007;17:805–819. doi: 10.1093/glycob/cwm054. [DOI] [PubMed] [Google Scholar]
- 16.Chauhan D., Li G., Podar K., Hideshima T., Neri P., He D., Mitsiades N., Richardson P., Chang Y., Schindler J., et al. A novel carbohydrate-based therapeutic GCS-100 overcomes bortezomib resistance and enhances dexamethasone- induced apoptosis in multiple myeloma cells. Cancer Res. 2005;65:8350–8358. doi: 10.1158/0008-5472.CAN-05-0163. [DOI] [PubMed] [Google Scholar]
- 17.Nesmelova I. V., Pang M., Baum L. G., Mayo K. H. 1H, 13C and 15N backbone and side-chain chemical shift assignments for the 29 kDa human gal-1 protein dimer. Biol. NMR Assign. 2008;2:203–205. doi: 10.1007/s12104-008-9121-9. [DOI] [PubMed] [Google Scholar]
- 18.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 19.Johnson B. A., Blevins R. A. NMR View: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 20.Mayo K. H., Ilyina E., Park H. A recipe for designing water-soluble, β-sheet-forming peptides. Protein Sci. 1996;5:1301–1315. doi: 10.1002/pro.5560050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilyina E., Roongta V., Pan H., Woodward C., Mayo K. H. A pulsed-field gradient NMR study of bovine pancreatic trypsin inhibitor self-association. Biochemistry. 1997;36:3383–3388. doi: 10.1021/bi9622229. [DOI] [PubMed] [Google Scholar]
- 22.Keeler J. New York: Wiley and Sons; 2005. Understanding NMR Spectroscopy. [Google Scholar]
- 23.Rajagopal P., Waygood E. B., Reizer J., Saier M. H., Klevit R. E. Demonstration of protein–protein interaction specificity by NMR chemical shift mapping. Protein Sci. 1997;6:2624–2627. doi: 10.1002/pro.5560061214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Lucendo M. F., Solis D., Andre S., Hirabayashi J., Kasai K., Kaltner H., Gabius H.-J., Romero A. Growth-regulatory human gal-1: crystallographic characterisation of the structural changes induced by single-site mutations and their impact on the thermodynamics of ligand binding. J. Mol. Biol. 2004;343:957–970. doi: 10.1016/j.jmb.2004.08.078. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz F. P., Ahmed H., Bianchet M. A., Amzel L. M., Vasta G. R. Thermodynamics of bovine spleen gal-1 binding to disaccharides: correlation with structure and its effect on oligomerization at the denaturation temperature. Biochemistry. 1998;37:5867–5877. doi: 10.1021/bi9716478. [DOI] [PubMed] [Google Scholar]
- 26.Brewer C. F., Miceli M. C., Baum L. G. Clusters, bundles, arrays and lattices: novel mechanisms for lectin–saccharide-mediated cellular interactions. Curr. Opin. Struct. Biol. 2002;12:616–623. doi: 10.1016/s0959-440x(02)00364-0. [DOI] [PubMed] [Google Scholar]
- 27.Dam T. K., Gabius H.-J., andre S., Kaltner H., Lensch M., Brewer C. F. Galectins bind to the multivalent glycoprotein asialofetuin with enhanced affinities and a gradient of decreasing binding constants. Biochemistry. 2005;44:12564–12571. doi: 10.1021/bi051144z. [DOI] [PubMed] [Google Scholar]
- 28.Daniels M. A., Hogquist K. A., Jameson S. C. Sweet ‘n’ sour: the impact of differential glycosylation on T cell responses. Nat. Immunol. 2002;3:903–910. doi: 10.1038/ni1002-903. [DOI] [PubMed] [Google Scholar]
- 29.Mikhailov D., Young H. C., Linhardt R. J., Mayo K. H. Heparin dodecasaccharide binding to platelet factor-4 and growth-related protein-α. Induction of a partially folded state and implications for heparin-induced thrombocytopenia. J. Biol. Chem. 1999;274:25317–25329. doi: 10.1074/jbc.274.36.25317. [DOI] [PubMed] [Google Scholar]
- 30.Iwaki J., Minamisawa T., Tateno H., Kominami J., Suzuki K., Nishi N., Nakamura T., Hirabayashi J. Desulfated galactosaminoglycans are potential ligands for galectins: evidence from frontal affinity chromatography. Biochem. Biophys. Res. Commun. 2008;373:206–212. doi: 10.1016/j.bbrc.2008.05.190. [DOI] [PubMed] [Google Scholar]
- 31.Sörme P., Kahl-Knutsson B., Wellmar U., Magnusson B.-G., Leffler H., Nilsson U. J. Design and synthesis of galectin inhibitors. Methods Enzymol. 2003;363:157–169. doi: 10.1016/S0076-6879(03)01050-4. [DOI] [PubMed] [Google Scholar]
- 32.Carlsson S., Oberg C. T., Carlsson M. C., Sundin A., Nilsson U. J., Smith D., Cummings R. D., Almkvist J., Karlsson A., Leffler H. Affinity of galectin-8 and its carbohydrate recognition domains for ligands in solution and at the cell surface. Glycobiology. 2007;17:663–676. doi: 10.1093/glycob/cwm026. [DOI] [PubMed] [Google Scholar]
- 33.Werz D. B., Ranzinger R., Herget S., Adibekian A., von der Lieth C.-W., Seeberger P. H. Exploring the structural diversity of mammalian carbohydrates (“glycospace”) by statistical databank analysis. Chem. Biol. 2007;2:685–691. doi: 10.1021/cb700178s. [DOI] [PubMed] [Google Scholar]
- 34.Bourne Y., Bolgiano B., Liao D. I., Strecker G., Cantau P., Herzberg O., Feizi T., Cambillau C. Crosslinking of mammalian lectin (gal-1) by complex biantennary saccharides. Nat. Struct. Biol. 1994;1:863–870. doi: 10.1038/nsb1294-863. [DOI] [PubMed] [Google Scholar]
- 35.Leonidas D. D., Vatzaki E. H., Vorum H., Celis J. E., Madsen P., Acharya K. R. Structural basis for the recognition of carbohydrates by human galectin-7. Biochemistry. 1998;37:13930–13940. doi: 10.1021/bi981056x. [DOI] [PubMed] [Google Scholar]
- 36.Liao D. I., Kapadia G., Ahmed H., Vasta G. R., Herzberg O. Structure of S-lectin, a developmentally regulated vertebrate β-galactoside-binding protein. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1428–1432. doi: 10.1073/pnas.91.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagae M., Nishi N., Murata T., Usui T., Nakamura T., Wakatsuki S., Kato R. Crystal structure of the galectin-9 N-terminal carbohydrate recognition domain from Mus musculus reveals the basic mechanism of carbohydrate recognition. J. Biol. Chem. 2006;281:35884–35893. doi: 10.1074/jbc.M606648200. [DOI] [PubMed] [Google Scholar]
- 38.Walser P. J., Haebel P. W., Kunzler M., Sargent D., Kues U., Aebi M., Ban N. Structure and functional analysis of the fungal galectin CGL2. Structure. 2004;12:689–702. doi: 10.1016/j.str.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Daas P. J. H., Schols H. A., de Jongh H. H. J. On the galactosyl distribution of commercial galactomannans. Carbohydr. Res. 2000;329:609–619. doi: 10.1016/s0008-6215(00)00209-3. [DOI] [PubMed] [Google Scholar]
- 40.Rabuka D., Forstner M. B., Groves J. T., Bertozzi C. R. Noncovalent cell surface engineering: incorporation of bioactive synthetic glycopolymers into cellular membranes. J. Am. Chem. Soc. 2008;130:5947–5953. doi: 10.1021/ja710644g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabuka D., Parthasarathy R., Lee G. S., Chen X., Groves J. T., Bertozzi C. R. Hierarchical assembly of model cell surfaces: synthesis of mucin mimetic polymers and their display on supported bilayers. J. Am. Chem. Soc. 2007;129:5462–5471. doi: 10.1021/ja067819i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernandez J. D., Nguyen J. T., He J., Wang W., Ardman B., Green J. M., Fukuda M., Baum L. G. Gal-1 binds different CD43 glycoforms to cluster CD43 and regulate T cell death. J. Immunol. 2006;177:5328–5336. doi: 10.4049/jimmunol.177.8.5328. [DOI] [PubMed] [Google Scholar]
- 43.Pace K. E., Lee C., Stewart L. G., Baum L. G. Restricted receptor segregation into membrane microdomains occurs on human T cells during apoptosis induced by gal-1. J. Immunol. 1999;163:3801–3811. [PubMed] [Google Scholar]
- 44.Gupta R. K., Pande A. H., Gulla K. C., Gabius H.-J., Hajela K. Carbohydrate-induced modulation of cell membrane. VIII. Agglutination with mammalian lectin gal-1 increases osmofragility and membrane fluidity of trypsinized erythrocytes. FEBS Lett. 2006;580:1691–1695. doi: 10.1016/j.febslet.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Taylor M. E., Drickamer K. Paradigms for glycan-binding receptors in cell adhesion. Curr. Opin. Cell Biol. 2007;19:572–577. doi: 10.1016/j.ceb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 46.Nieminen J., Kuno A., Hirabayashi J., Sato S. Visualization of galectin-3 oligomerization on the surface of neutrophils and endothelial cells using fluorescence resonance energy transfer. J. Biol. Chem. 2007;282:1374–1383. doi: 10.1074/jbc.M604506200. [DOI] [PubMed] [Google Scholar]
- 47.Chung C. D., Patel V. P., Moran M., Lewis L. A., Miceli M. C. Gal-1 induces partial TCR ζ-chain phosphorylation and antagonizes processive TCR signal transduction. J. Immunol. 2000;165:3722–3729. doi: 10.4049/jimmunol.165.7.3722. [DOI] [PubMed] [Google Scholar]
- 48.Demetriou M., Granovsky M., Quaggin S., Dennis J. W. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature. 2001;409:733–739. doi: 10.1038/35055582. [DOI] [PubMed] [Google Scholar]
- 49.Partridge E. A., Le Roy C., Di Guglielmo G. M., Pawling J., Cheung P., Granovsky M., Nabi I. R., Wrana J. L., Dennis J. W. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306:120–124. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- 50.Lau K. S., Partridge E. A., Grigorian A., Silvescu C. I., Reinhold V. N., Demetriou M., Dennis J. W. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 51.Braccia A., Villani M., Immerdal L., Niels-Christiansen L. L., Nystrøm B. T., Hansen G. H., Danielsen E. M. Microvillar membrane microdomains exist at physiological temperature. Role of galectin-4 as lipid raft stabilizer revealed by “superrafts”. J. Biol. Chem. 2003;278:15679–15684. doi: 10.1074/jbc.M211228200. [DOI] [PubMed] [Google Scholar]
- 52.Ohtsubo K., Takamatsu S., Minowa M. T., Yoshida A., Takeuchi M., Marth J. D. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell. 2005;123:1307–1321. doi: 10.1016/j.cell.2005.09.041. [DOI] [PubMed] [Google Scholar]