Abstract
RAP (receptor-associated protein) is a three domain 38 kDa ER (endoplasmic reticulum)-resident protein that is a chaperone for the LRP (low-density lipoprotein receptor-related protein). Whereas RAP is known to compete for binding of all known LRP ligands, neither the location, the number of binding sites on LRP, nor the domains of RAP involved in binding is known with certainty. We have systematically examined the binding of each of the three RAP domains (D1, D2 and D3) to tandem and triple CRs (complement-like repeats) that span the principal ligand-binding region, cluster II, of LRP. We found that D3 binds with low nanomolar affinity to all (CR)2 species examined. Addition of a third CR domain increases the affinity for D3 slightly. A pH change from 7.4 to 5.5 gave only a 6-fold increase in Kd for D3 at 37 °C, whereas temperature change from 22 °C to 37 °C has a similar small effect on affinity, raising questions about the recently proposed D3-destabilization mechanism of RAP release from LRP. Surprisingly, and in contrast to literature suggestions, D1 and D2 also bind to most (CR)2 and (CR)3 constructs with nanomolar affinity. Although this suggested that there might be three high-affinity binding sites in RAP for LRP, studies with intact RAP showed that only two binding sites are available in the intact chaperone. These findings suggest a new model for RAP to function as a folding chaperone and also for the involvement of YWTD domains in RAP release from LRP in the Golgi.
Keywords: chaperone, ligand release, low-density lipoprotein receptor (LDLR), LDLR-associated protein (LRP), receptor-associated protein (RAP), YWTD domain
Abbreviations: CR, complement-like repeat; (CR)x LRP fragment containing x CR domains; CRxy, LRP fragment containing domains CRx and CRy; CRxyz, LRP fragment containing domains CRx, CRy and CRz; D1, D2 and D3, first, second and third domains of RAP; ER, endoplasmic reticulum; GST, glutathione transferase; IPTG, isopropyl β-D-thiogalactoside; ITC, isothermal titration calorimetry; 2-ME, 2-mercaptoethanol; LA34, third and fourth CR domains from the ligand-binding cluster of LDLR; (V)LDLR, (very-) low-density lipoprotein receptor; LRP, low-density lipoprotein receptor-related protein; RAP, receptor-associated protein; TEV, tobacco etch virus
INTRODUCTION
Members of the LDLR (low-density lipoprotein receptor) family are involved in binding and internalizing a wide range of structurally unrelated proteins [1,2]. Binding of ligands occurs to regions of the receptors that are built from clusters of non-identical copies of the CR (complement-like repeat) domain, which is an ∼40 residue module that derives its structural integrity from the presence of three disulfides and a calcium co-ordination site. Probably because of the presence of such a high density of disulfides in these ligand-binding clusters of LDLR family members, a chaperone is necessary for efficient folding of these receptors. For VLDLR (very-LDLR), LRP (LDLR-associated protein) and some other members of the family, though perhaps not LDLR itself [3], this chaperone is the 38 kDa ER (endoplasmic reticulum)-resident protein RAP (receptor-associated protein) [4–8], which consists of three roughly equal-sized domains [9,10] and terminates with an HNEL ER retention sequence [11]. It has been proposed that RAP serves two critical functions with respect to these receptors, the first to ensure correct folding of their CR domains and the second to prevent premature tight binding of other protein ligands to the CR clusters within the ER [9,12].
Although an early report on the number and location of binding sites within RAP for LRP suggested that there are two such sites [13], later quantitative studies, which compared the binding of the third RAP domain (D3) with that of intact RAP found apparently similar affinities for D3 and intact RAP, implying that the other two RAP domains contributed little to binding [9]. This conclusion was reinforced by a study that used ITC (isothermal titration calorimetry) to examine the binding of the CR tandem construct CR56 (where CRxy is the LRP fragment containing domains CRx and CRy) from ligand binding cluster II of LRP to RAP domains D1, D2 and D3 that found moderately high affinity (0.2 μM) for D3, but much weaker affinity for D1 (3 μM) and D2 (19 μM) [14]. Similarly, low affinity was found in a study that examined the binding of CR56 to RAP D1 [15]. However, there are potential problems with these studies in that the CR56 construct was either immobilized or fused to ubiquitin, which may account for none of the affinities approaching those reported for either D3 or RAP for intact LRP (1–5 nM) [9].
To provide more complete and less ambiguous data on the binding of RAP to LRP, we have systematically examined the binding of tandem and triple CR domain constructs that cover the portion of cluster II of LRP to which protein ligands are known to bind (CR3–CR9) to each of the three domains of RAP on their own, as well as in the context of intact RAP. Our results from the present study provide evidence for tight binding sites on each of the three domains of RAP, only two of which can be simultaneously engaged in intact RAP. Of significance for the functioning of RAP is that each of these high-affinity binding sites is in the low nanomolar range, making it likely that both are engaged in vivo. In addition, whereas there is a pH-dependence of the binding of D3, it seems to be of too small a magnitude to be the principal mechanism of dissociation of RAP from folded LRP as the latter progresses from the ER to the lower pH environment of the Golgi. Furthermore, no such pH-dependence is observed for D1 or D2. This also has implications for binding and displacement of protein ligands other than RAP. Based on these findings, we propose a new model for how RAP may both function to promote correct folding of LRP and subsequently dissociate as the folded LRP is transported from the ER to the Golgi, with concomitant lowering of the pH. The latter also has implications for the release of protein ligands from LRP subsequent to their receptor-mediated internalization.
MATERIALS AND METHODS
Expression, purification and refolding of CR constructs
All CR constructs (see Supplementary Table S1 at http://www.BiochemJ.org/bj/421/bj4210273add.htm) were expressed in 2YT [1.6% (w/v) tryptone, 1% (w/v) yeast extract and 0.5% (w/v) NaCl] medium. CR34, CR56, CR78 and CR567 (where CRxyz is the LRP fragment containing domains CRx, CRy and CRz) were cloned in pGEX-2T, modified to contain a TEV (tobacco etch virus) cleavage site, and expressed as GST (glutathione transferase)-fusion proteins in BL21 cells. Cells were grown to D600=0.6–1.0 before induction with 1 mM IPTG (isopropyl β-D-thiogalactoside), and the cells were harvested after 5–6 h incubation at 37 °C. The GST-fusion proteins were purified from cleared cell lysate by GSH–Sepharose chromatography, and the GST-tag was removed by TEV cleavage during overnight dialysis against 4 litres of 20 mM Tris/HCl, pH 8.0, 50 mM NaCl and 4 mM EDTA, containing 14 mM 2-ME (2-mercaptoethanol). GST and uncleaved GST–CR fusion proteins were removed by passage through the GSH-column.
CR89, CR345, CR456 and CR678 were cloned in pQE-30, modified to contain a GB1 fusion partner and a TEV cleavage site, and CR45 was cloned in pQE-30, modified to contain a NusA fusion partner and a TEV cleavage site. These His6 (hexahistidine)-tagged fusion proteins were expressed in SG13009 cells containing the plasmid pRARE. The CR fusion proteins were purified from cell lysate by Ni2+ or TALON chromatography, and the fusion partner was removed by TEV cleavage during overnight dialysis against PBS containing either 14 or 7 mM 2-ME.
Before refolding, all (CR)x (LRP fragment containing x CR domains) species were further purified by Q-Sepharose HP chromatography, using a gradient of 0–1000 mM NaCl in 20 mM Tris/HCl, pH 8.0, and 6 M urea. The denatured (CR)x species were diluted to 0.1 mg/ml with 50 mM Tris/HCl, pH 8.5, 50 mM NaCl and 10 mM CaCl2, and refolded by dialysis against buffer containing 14 mM 2-ME and 8 mM 2-hydroxyethyl disulfide. For increased folding efficiency, all (CR)x species except CR78 were then mixed with the chaperone GST–RAP as a GST-fusion protein [GST-RAP(IAA)] at a 1:1.25 ratio (CR:RAP) [16]. After 24 h at room temperature (22 °C) with N2 bubbling, the dialysis was continued for 24 h at 4 °C without N2. Finally, the mixture was dialysed against 2×4 litres of 20 mM Tris/HCl, pH 7.8, 50 mM NaCl and 1 mM CaCl2 at 4 °C. The refolding mixture was loaded on to GSH–Sepharose, and folded CR constructs, i.e. those capable of binding to RAP and therefore retained on the column, were eluted with 40 mM Tris/HCl, pH 8.0, 100 mM NaCl and 8 mM EDTA. Folded (CR)x species were further purified by Q-Sepharose HP chromatography (50–1000 mM NaCl in 20 mM Tris/HCl, pH 8.0, and 0.1 mM CaCl2). If additional purification was needed, calcium was removed by EDTA and the (CR)x species was passed through a Superdex 75 size-exclusion column equilibrated in 20 mM Tris/HCl, pH 7.8, 150 mM NaCl and 4 mM EDTA.
As a quality control between each step of purification, samples were analysed by reducing and non-reducing SDS/PAGE and non-denaturing PAGE. The presence of a single band on SDS/PAGE with a reduction in mobility under reducing conditions indicated homogeneous samples, with formed disulfide bridges. A single band on non-denaturing PAGE indicated the presence of a single folding product. The final products were judged to be more than 95% pure by SDS/PAGE.
Expression and purification of RAP
RAP cDNA, cloned in pGEX-2T, was a gift from Dr Dudley Strickland (University of Maryland School of Medicine, Baltimore, MA, U.S.A.). GST–RAP was expressed in BL21 cells, induced with 1 mM IPTG at a D600=0.6–1.0, and harvested after 5 h incubation at 25 °C. GST–RAP was purified by GSH–Sepharose chromatography. GST–RAP that was to be used for CR refolding was then dialysed overnight against 4 litres of 20 mM Tris/HCl, pH 8.0, 50 mM NaCl and 4 mM EDTA, containing 14 mM 2-ME. Free cysteines in the GST moiety were blocked by treatment with 20 mM iodoacetamide for 1 h at room temperature and then dialysed against 4 litres of 20 mM Tris/HCl, pH 8.0, 50 mM NaCl and 4 mM EDTA, followed by dialysis against 2×4 litres of refolding buffer (50 mM Tris/HCl, pH 8.5, 50 mM NaCl and 10 mM CaCl2).
For GST-free RAP used in binding assays, a TEV cleavage site was introduced between the GST and RAP moieties. GST-free RAP was prepared by removing the N-terminal GST-tag with TEV protease after the GSH–Sepharose step. This was performed in the presence of TEV protease (1:10000) during an overnight dialysis at 4 °C against 4 litres of 20 mM Tris/HCl, pH 8.0, 50 mM NaCl and 4 mM EDTA supplemented with 14 mM 2-ME, after the initial GSH–Sepharose step. The mixture was then reapplied to the GSH column and GST-free RAP was collected in the flow-through. Pooled RAP was then dialysed against 2×1 litre of 50 mM sodium acetate, pH 5.0, and 50 mM NaCl at room temperature, and subjected to a final SP (sulfopropyl)-Sepharose step eluting pure RAP with a 50–750 mM NaCl gradient. The final RAP sample, with a purity greater than 95% as verified by SDS/PAGE, was prepared by dialysis into the assay buffer (20 mM Tris/HCl, pH 7.4, 150 mM NaCl and 1 mM CaCl2).
Expression and purification of RAP domains
RAP domains 1, 2 and 3 (see Supplementary Table S1) were cloned in pQE30, modified to contain a TEV cleavage site, and expressed in SG13009 cells. Cells were grown at 25 °C in 2YT media to D600=0.6–1.0 before induction with 1 mM IPTG, and harvested after 5 h. The protein was purified from cleared cell lysate by nickel affinity chromatography. The N-terminal His6-tag was removed by TEV cleavage and the sample was reapplied to the nickel affinity column, collecting tag-free RAP domains in the flow-through. If further purification was needed, a size-exclusion chromatography step on a Superdex 75 column was used. Final sample purity of more than 95% was verified by SDS/PAGE.
Purification of RAP-CR56 and RAP-CR345 complexes
RAP was mixed with a 4.8 molar excess of CR56 or CR345 and applied directly to a 200 ml Superdex 75 size-exclusion column equilibrated in 20 mM Tris/HCl, pH 7.4, 150 mM NaCl and 1 mM CaCl2. Fractions eluted at about 70 ml, corresponding to a molecular mass higher than RAP alone, contained the complex. The amount of protein applied to the column was based on two criteria: first, to ensure saturation of the complex, by keeping the concentration of each component well above 3 μM; secondly, to make the final absorbance at 280 nm in directly collected fractions suitable for analytical ultracentrifugation analysis without further concentration.
An estimate of the stoichiometry of the complex was obtained by integrating the absorbance at 280 nm of the elution profile and using the individual extinction coefficients, the initial molar ratio of proteins, and the assumptions that all RAP is complexed and that the late-eluting peak contained only CR protein. Both assumptions were verified by SDS/PAGE.
Fluorescence spectroscopy
Both fluorescence emission spectra and binding experiments were performed on a PTI Quantamaster spectrofluorimeter, equipped with double monochromators on both the excitation and emission sides. All experiments were performed in a quartz cuvette in 1200 μl of 20 mM Tris/HCl, pH 7.4, 150 mM NaCl and 1 mM CaCl2 supplemented with 0.1% (w/v) PEG [poly(ethylene glycol)] of 8000. The temperature of cuvette and sample was controlled by a water bath set to 22 °C except for the 37 °C experiments. For the low pH experiments, Tris was replaced by 20 mM Mes adjusted to pH 5.5.
Fluorescence spectra were recorded with an excitation wave-length of 295 nm, scanning the emission wavelength region of 300–450 nm in 4 nm steps. As all proteins examined contain tryptophan, the reference spectra of the individual components were recorded and subtracted from complex spectra to obtain the difference perturbation spectrum for the binding of each CR. Protein concentrations of 100–2000 nM were used.
Binding experiments with RAP domains 1, 2 and 3 to (CR)x species were performed with a fixed amount of CR of between 100 and 4000 nM depending on the affinity. Additions of 2 μl up to 40–60 μl of ligand were made to a final ratio of ∼3.5:1 (350–12500 nM). The fluorescence change at 338 nm was measured for an average of 60 s at each titration step, and the subsequent titration curve was corrected for ligand without CR. It should be noted that, although the fluorescence perturbation may come from either or both component, this is not relevant to the use of the perturbation to follow the binding process. The resulting curve was then fitted to a standard 1:1 binding isotherm with floating Fmax, Kd and CR concentration by non-linear least-squares fit. The fitted CR concentrations provided an independent estimate of the individual CR folding efficiencies, which were typically 50–100%. For binding experiments at pH 5.5 and 37 °C, the titration was reversed, with CR56 titrated into RAP. This was done to accommodate pH or temperature effects on the activity of the RAP domains.
To determine the stoichiometry of complex formation with full-length RAP, a fixed concentration of RAP (100–1000 nM) was titrated with (CR)x species up to 3.5 μM and the data fitted to various possible binding ratios of (CR)x species to RAP.
Analytical ultracentrifugation
All sedimentation velocity experiments were performed in a Beckman XL-I analytical ultracentrifuge. Samples were extensively dialysed into buffer (20 mM Tris/HCl, pH 7.4, 150 mM NaCl and 1 mM CaCl2) and loaded into a dual sector charcoal-filled Epon centrepiece. The samples were centrifuged at 50000 rev./min in an An60-Ti rotor and sedimentation was monitored by absorbance at 280 nm. Data were analysed using the program SEDFIT, which generates a continuous c(s) distribution for the sedimenting species [17]. Solvent densities and viscosities were calculated with SEDNTERP [17].
For multisignal analysis of protein complexes, sedimentation was followed measuring two signals from the proteins simultaneously. Data were acquired using laser interferometry and absorbance spectrophotometry at a wavelength of 280 nm. Data were analysed according to the multisignal ck(s) method implemented in the program SEDPHAT [18]. Briefly, the extinction coefficients of the individual proteins were predetermined in separate experiments with each protein component alone. Global modelling of data acquired at the different signals permits spectral distinction of the different sedimenting protein components and allows for separate deconvolution of the sedimentation coefficient distributions ck(s) of species containing protein component k. Integration of a ck(s) peak yields the concentration of a given component in that peak and comparison of the two concentrations yields the stoichiometry of the proteins in that peak.
ITC
Binding studies were performed on a MicroCal VP isothermal titrational calorimeter at 25 °C. All protein samples were dialysed into the same buffer containing 20 mM Tris/HCl, pH 7.4, 150 mM NaCl and 1 mM CaCl2. CR56, diluted in dialysis buffer, was titrated with 7–12 μl additions of D1, D2 or D3 at 25–55 μM. Experiments with full-length RAP were performed with 3 μM RAP in the cell, titrated with 7 μl injections of 45 μM CR56. Injection peaks were integrated using Origin software provided by the manufacturer. Integrated and progressively-summed heats were used to follow binding progress. Data were fitted by nonlinear least-squares analysis to a one binding-site model for the single RAP domains, and a two independent site model for intact RAP, using the program Scientist (MicroMath). Control experiments of titrant into buffer were performed for all titrants.
SDS/PAGE and non-denaturing PAGE
SDS/PAGE was performed using Novex 10–20% gels (Invitrogen) with standard Laemmli buffers with or without reducing agent (2-ME) in the loading buffer. Non-denaturing PAGE was performed using Novex 8% gels, pre-run for 3.5 h at 15 mA in 375 mM Tris/HCl, pH 8.8, and 5 mM CaCl2 to introduce calcium into the gel. Gels were run in a standard Tris-glycine buffer system, supplemented with 5 mM CaCl2 at 15 mA for 2.5 h. For gel-shift experiments, a fixed amount of RAP or RAP fragments was incubated with the indicated molar ratio of CR for 10 min at room temperature prior to loading.
RESULTS
Domain D3 binds tightly to all (CR)2 species
It has previously been shown qualitatively that all of the two domain constructs from cluster II of LRP, except CR9 and CR10, can bind with similar affinity to domain D3 from RAP when they are expressed as fusion proteins with ubiquitin [19]. Quantitative SPR (surface plasmon resonance) and calorimetric data from the same laboratory on the binding of D3 to the single tandem construct ubiquitin–CR56 gave an affinity of approx. 170 nM [14]. We have now determined the affinities of tandem pairs from CR34 to CR89, expressed as non-conjugated species, for RAP D3, using perturbation of tryptophan fluorescence (Figure 1). This has the advantage over previous studies that there are no possible complications from attached tags or from steric constraints imposed by surface immobilization. In keeping with the qualitative findings of Andersen et al. [19], all of these pairs could bind to D3 (Table 1). However, whereas the Kd of ubiquitin–CR56 to D3 was reported to be 170–700 nM [14], we now find a value 5–19-fold tighter (Kd=37 nM) for tag-free CR56, and Kd values for the other tandem domains that range from 55 nM for CR78 to as tight as 3.5 nM for CR34. Unlike the previously reported affinities, these are similar in magnitude to those reported for binding of D3 to intact LRP [20].
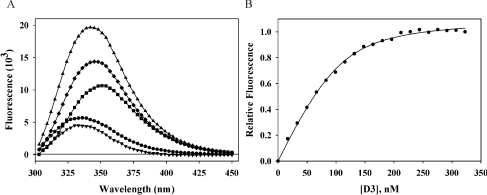
Figure 1. Fluorescence determination of affinities of CR fragments for RAP D3.
Examples of emission spectra and fluorescence determination of affinities of CR fragments for RAP and its fragments. (A) Emission spectra of intrinsic tryptophan fluorescence excited at 295 nm of (■) 200 nM CR45, (●) 200 nM RAP-D3, (▲) mixture, (♦) difference between mixture and RAP-D3 and (▼) CR45 bound–unbound difference spectrum. (B) Change in tryptophan fluorescence followed at 338 nm for 200 nM CR45 titrated with 0–350 nM RAP-D3, and corrected for D3 fluorescence. Normalized raw data (n=3) are shown, with a best fit to a 1:1 binding model.
Table 1. Affinities of (CR)2 and (CR)3 species for RAP fragments D1, D2, and D3.
| D1 | D2 | D3 | ||||
|---|---|---|---|---|---|---|
| CR species | Kd (nM) | ΔG0 (kJ·mol−1) | Kd (nM) | ΔG0 (kJ·mol−1) | Kd (nM) | ΔG0 (kJ·mol−1) |
| CR34 | 38±11 | −41.9±0.7 | 19±12 | −43.6±1.8 | 3.5±0.4 | −47.7±0.3 |
| CR45 | 24±6 | −43.0±0.6 | 30±11 | −42.4±0.9 | 17±3 | −43.8±0.4 |
| CR56 | 20±3 | −43.4±0.4 | 54±15 | −41.0±0.7 | 37±3 | −41.9±0.2 |
| CR78 | 927±254 | −34.0±0.7 | 116±68 | −39.1±1.6 | 55±20 | −41.0±0.9 |
| CR89 | 35±6 | −42.1±0.4 | 42±17 | −41.6±0.5 | 20±5 | −43.4±0.6 |
| CR345 | 23±2 | −43.1±0.2 | 23±2 | −43.1±0.2 | 7±3 | −46.0±1.1 |
| CR456 | 6±1 | −46.4±0.4 | 6±2 | −46.4±0.8 | 8±1 | −45.7±0.3 |
| CR567 | 15±2 | −44.1±0.3 | 18±5 | −43.7±0.7 | 5.3±0.8 | −46.7±0.4 |
| CR678 | 859±518 | −34.2±1.6 | 44±16 | −41.5±0.9 | 26±10 | −42.8±1.0 |
Two CR domains provide most of the binding energy to D3
The only X-ray structure of CR domains bound to any part of RAP is of two CR domains from LDLR bound to RAP D3 [21]. In this structure, the two CR domains lie at an acute angle to the long axis of the elongated D3, but span less than two-thirds of the length of D3. Thus there is space on the surface of D3 for an additional CR domain to bind. We therefore examined whether (CR)3 species might bind with a significantly higher affinity than (CR)2 species to D3. Although most of the (CR)3 species examined had increased affinity to D3, with Kd values reduced to single digit nanomolar values, the increase represented only a 3–11% increase in binding energy (Table 1). The only exception was CR678, which bound with a Kd of 26 nM. Thus most of the binding energy of binding to D3 arises from interactions with two CR domains, and presumably mimics the interactions seen in the X-ray structure of D3 with LA34 (where LA3 and LA4 are the third and fourth CR domains from the ligand-binding cluster of LDLR) [21].
Domains D1 and D2 can also bind to LRP fragments with very high affinity
A previous study that examined the binding of D1 or D2 to fragments of LRP concluded that the affinities are low, with Kd of 2.8 μM and 19 μM respectively to ubiquitin-conjugated CR56 [14]. Given the much higher affinities that we found for D3 binding to LRP fragments than found by others, we wanted to repeat binding experiments on D1 and D2, but using the whole range of two and three domain species we had used for D3 binding. Somewhat surprisingly we found that, at least to fragments that did not contain both CR7 and CR8 domains together, binding was as tight or even tighter than to D3 (Table 1). The tightness of binding is also seen in gel-shift assays, where both D1 and D2 give clear shifts that are saturable (Figure 2). Even for the two-domain constructs, Kd values were in the 19–54 nM range and tightened to 6–18 nM for three domain constructs. Only binding that involved CR78, either alone or in the longer construct CR678, was significantly weaker, with Kd values of 44–930 nM. However, even these are much tighter than reported elsewhere for either of these RAP domains [14]. Together with the results on binding of D3, this implies that there might be three tight binding sites in RAP for two or three domain fragments of LRP.
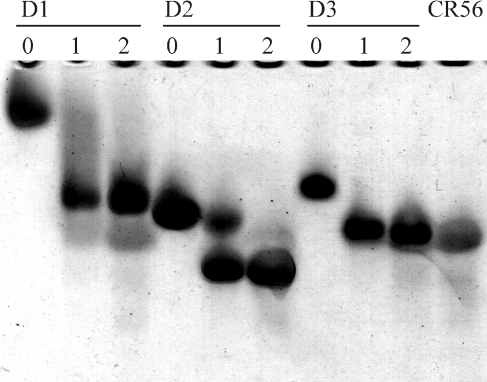
Figure 2. Gel-shift assay of binding of D1, D2 and D3 to CR56.
Binding of CR56 to RAP-D1, D2, and D3 by non-denaturing gel-shift assay. The Figure shows the migration of 2μg of the indicated RAP-domain (D1, D2 or D3) incubated with 0, 1 or 2 molar ratio of CR56 on an 8% non-denaturing PAGE. The migration of 3 μg of CR56 alone is shown in the far-right lane.
Influence of temperature on binding
It has been suggested that the principal mechanism of dissociation of ligands from RAP D3 depends on promoting the unfolding of this domain by ionization of histidines as the pH is lowered upon movement of LRP–RAP from the ER to the Golgi [22,23]. Since it has marginal stability at physiological temperature (Tm∼42 °C) [10], one might expect that such a destabilization mechanism would also be apparent at constant pH upon increasing the temperature. We therefore examined the effect of increasing the temperature from 22 °C to 37 °C at pH 7.4 on the binding of CR56 (which binds tightly to all three RAP domains at 22 °C) to D1, D2 and D3. Surprisingly, only the most stable domain, D1, showed a reduction in affinity, and then by less than a factor of 2, whereas D2 and the least stable D3 actually increased their affinity 2- and 5-fold respectively (Table 2).
Table 2. Temperature and pH dependence of affinities of D1, D2 and D3 for CR56.
| D1 | D2 | D3 | |||||
|---|---|---|---|---|---|---|---|
| Temperature (°C) | pH | Kd (nM) | ΔG0 (kJ·mol−1) | Kd (nM) | ΔG0 (kJ·mol−1) | Kd (nM) | ΔG0 (kJ·mol−1) |
| 22 | 7.4 | 20±3 | −43.4±0.4 | 54±15 | −41.0±0.7 | 37±3 | −41.9±0.2 |
| 37 | 7.4 | 29±7 | −44.6±0.6 | 22±11 | −45.3±1.4 | 7±2 | −48.3±0.8 |
| 37 | 5.5 | 26±11 | −44.9±1.2 | 46±9 | −43.4±0.5 | 41±11 | −43.7±0.7 |
Effect of pH on binding to CR56
The effect of lowering the pH from 7.4 to 5.5 was examined at 37 °C for binding of CR56 to each of the three RAP domains. Both D2 and D3 showed a reduction in affinity, though this was only 2-fold for D2 and 6-fold for D3 (Table 2). In contrast, the affinity of D1 was unaffected. Although this suggests that histidine ionization might destabilize D3, the effect is not large, resulting in only a 10% reduction in free the energy of binding.
Intact RAP possesses only two high-affinity sites for LRP fragments
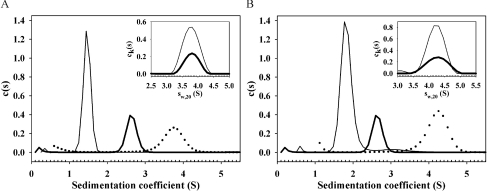
Given that each of D1, D2 and D3 can bind tightly to most tandem and triple CR fragments from cluster II of LRP, we next examined how many such CR fragments could bind to intact RAP. This was examined in several ways. First, we examined binding by gel-shift assay. Both (CR)2 and (CR)3 species gave clear shifts in the position of RAP that seemed to saturate at approximately two equivalents (Figure 3). The second approach was more qualitative and used titration of CR species into RAP followed using the same fluorescence perturbation technique as used for determining individual affinities above. Binding of CR456 (Figure 4) or CR345 to RAP resulted in saturable binding, which could be well fitted to two tight binding sites (Table 3). CR56 also gave saturable binding that could be fitted to two tight binding sites (Table 3). Given that both binding sites appear to be quite tight it is difficult to obtain accurate values for both Kd values simultaneously. However, fixing one Kd at the value for binding of the CR fragment to D1 gave the second, weaker Kd in the range 36–56 nM, indicating that the presence of the first fragment did not antagonize binding of the second in a major way. Finally, we purified complexes of RAP with both CR56 and CR345, by adding excess CR56 or CR345 to RAP and fractionating by size, and then examined their properties by sedimentation velocity measurements. Size-exclusion chromatography gave clear separation of complex from unbound (CR)x fragments (Figure 5). A stoichiometry of 1:2.1 (RAP:CR56) and 1:1.8 (RAP:CR345) was estimated by integrating the elution profiles. Sedimentation velocity gave sedimentation coefficients (s20,w) for the complexes of 3.79 and 4.26 compared with 2.7 for RAP alone (Table 4 and Figure 6). Best fits to the data gave molecular masses of 53 kDa for the RAP–CR56 complex, 66 kDa for the RAP–CR345 complex and 38 kDa for RAP alone. These are consistent with the larger species being 2:1 complexes of either CR56 (8.9 kDa) or CR345 (13.6 kDa) with RAP (38 kDa). This stoichiometry is also supported by multisignal analysis of the sedimentation velocity data, which independently gives ratios for the complexes of 1:2.6 (RAP:CR56) and 1:2.3 (RAP:CR345) (see insets of Figure 6). The frictional ratios of 1.47 for RAP, 1.38 for the RAP–CR56 complex, and 1.38 for the RAP–CR345 complex are consistent with an elongated shape for each species, though with an increase in the thickness of the complexes, presumably reflecting the association of two CR56 or CR345 fragments along the long axis of the RAP molecule.
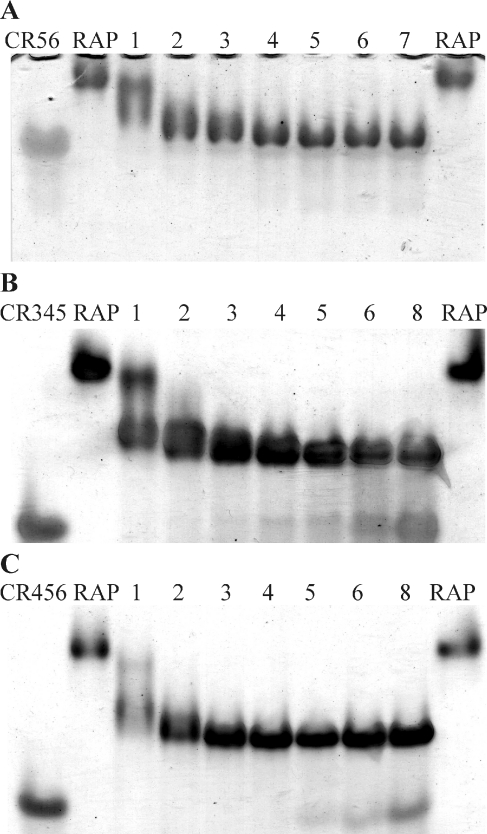
Figure 3. Gel-shift assay of binding of CRx to intact RAP.
Binding titration of CR56, CR345 and CR456 to RAP by non-denaturing gel-shift assay. The Figure show the migration of 2.5 μg of RAP either alone (RAP) or incubated with the indicated molar ratios of CR by 8% non-denaturing PAGE. (A) RAP incubated with 1–7 molar ratios of CR56. (B) RAP incubated with 1–8 molar ratios of CR345. (C) RAP incubated with 1–8 molar ratios of CR456. The migration of 7 μg of the indicated CR alone is shown in the far-left lane of each respective gel.
Figure 4. Fluorescence titration of CR into RAP.
Change in intrinsic tryptophan fluorescence followed at 338 nm. (A) RAP (100 nM) titrated with 0–500 nM CR56 and (B) 1 μM RAP titrated with 0–2.7μM CR456. Normalized raw data with a best fit to a two-binding site model is shown.
Table 3. Affinities of binding sites in intact RAP for (CR)2 and (CR)3 fragments.
| Kd1* (nM) | Kd2 (nM) | |
|---|---|---|
| CR56 | 20 | 56±8 |
| CR345 | 20 | 42±11 |
| CR456 | 10 | 36±5 |
*The fit to two tight binding sites was made by fixing the first Kd at a value close to that found for binding of the same fragment to D1 alone.
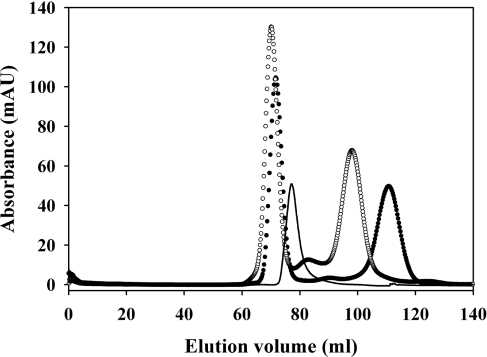
Figure 5. Size-exclusion chromatographic separation of (CR)x–RAP complexes.
Purification of RAP–CR56 and RAP–CR345 complexes by size-exclusion chromatography. The profiles show the separation of RAP-CR56 (●) and RAP-CR345 (○) complexes from 1:4.8 RAP:CR mixtures. As confirmed by SDS/PAGE (results not shown), RAP–CR complexes elute close to 70 ml, whereas un-complexed CR56 and CR345 elute at 112 ml and 98 ml respectively. RAP alone in an equivalent amount is shown as a solid line.
Table 4. Analytical ultracentrifugation of RAP, CR56, CR345 and their complexes.
| Species | Mr (kDa)* | Mr (kDa)† | sw,20‡ | f/f0§ | a/b¶ |
|---|---|---|---|---|---|
| RAP | 38±2 | 37.8 | 2.7±0.1 | 1.47±0.02 | 6.9±0.3 |
| CR56 | 7.8±0.3 | 8.9 | 1.49±0.02 | 1.46±0.04 | 5.5±0.5 |
| RAP–(CR56)2 | 53±3 | 55.6 | 3.79±0.02 | 1.38±0.04 | 4.7±0.6 |
| CR345 | 14.2±0.1 | 13.6 | 1.87±0.01 | 1.37±0.01 | 4.4±0.1 |
| RAP–(CR345)2 | 66±3 | 65 | 4.26±0.01 | 1.38±0.05 | 4.8±0.7 |
*Molecular mass calculated from sedimentation data.
†Molecular mass calculated from sequence and stoichiometry of 1:2 for RAP:(CR)x complexes.
†Temperature-corrected sedimentation coefficient at 20 °C in water.
§Frictional ratio.
¶Shape factor expressed as ratio of long to short axes (a/b) for a prolate ellipsoid.
Figure 6. Sedimentation velocity analysis of (CR)x–RAP complexes.
Analysis of RAP–CR56 and RAP–CR345 complexes by analytical ultracentrifugation. Overlay of sedimentation velocity profiles for CR (thin line), RAP (thick line), and purified RAP–CR complex (●) are shown. (A) CR56. (B) CR345. The respective global fits for extinction coefficient-based stoichiometry determination are shown as small insets in top right corner of (A and B) [CR (thin line), RAP (thick line)].
ITC analysis of binding of CR56
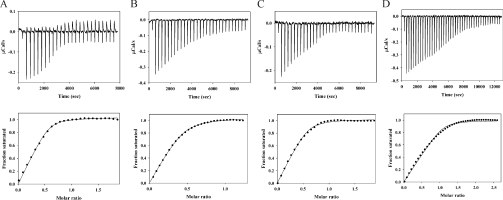
Although our fluorescence approach to measuring Kd values for the various RAP–CR interactions has an advantage over ITC of being able to be carried out at much lower protein concentrations and is therefore better for accurate determination of tight interactions, we nevertheless wanted some independent quantitative corroboration of our principal findings that (i) D1, D2 and D3 could each bind tightly to CR fragments, and (ii) that only two high-affinity binding sites are present in intact RAP. We chose to examine the binding of CR56, since this is the LRP fragment for which ITC data exist for binding to RAP, albeit for ubiquitin-conjugated CR56 [14,15]. Using a low μM concentration of CR56. we found that D1, D2 and D3 could all bind quite tightly (Figures 7A–7C). The Kd values obtained were 62 nM, 270 nM and 77 nM respectively. Although these are somewhat weaker than found above by fluorescence (20 nM, 54 nM and 37 nM), they clearly demonstrate tight binding of all three isolated RAP domains to CR56 that has no attached protein tag. The somewhat lower affinities obtained by ITC probably reflect the need to use higher protein concentration and the significant correction to the raw heat data that must always be made for the control titration of titrant into buffer. Our ITC values, however, stand in marked contrast to those reported previously for ubiquitin–CR56 binding to the same RAP fragments. Thus for D1, our value of 62 nM compares with a reported value of 2650 nM, for D2 270 nM compares with 16700 nM and for D3 77 nM compares with 126 nM.
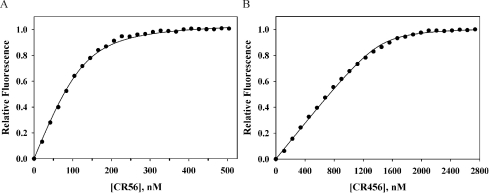
Figure 7. ITC analysis of binding of CR56 to RAP and RAP fragments.
(A) Titration of D1 into 1.8 μM CR56; (B) titration of D2 into 4.6 μM CR56; (C) titration of D3 into 1.8 μM CR56; (D) titration of CR56 into 3 μM RAP. The upper part of each panel gives the raw ITC data for the titration prior to correction for dilution of titrant, and the lower portion gives the best fit of the data to a single site model (A–C) or a two-site model (D).
We also examined binding of CR56 to intact RAP and found binding that corresponded to a stoichiometry of two CR56 to one RAP (Figure 7D), further supporting the fluorescence, gel-shift, size-exclusion chromatography and sedimentation velocity analyses reported above. One benefit of the ITC analysis was that it also provided ΔH values for all binding processes. These were −77.0 kJ·mol−1, −36.8 kJ·mol−1 and −71.6 kJ·mol−1 respectively for D1, D2 and D3, and the overall ΔH for binding of two equivalents of CR56 to intact RAP was −164.1 kJ·mol−1, which is more consistent with binding of CR56 to D1 and D3 than with binding to D2 and either D1 or D3.
DISCUSSION
By fluorescence, gel-shift assay, ITC, analytical ultracentrifugation and size-exclusion chromatography, we have demonstrated that all three RAP domains bind tightly to nearly all fragments from LRP cluster II. Kd values were in the low to mid nM range. This contrasts with an earlier study that found extremely weak binding (μM) for D1 and D2, and binding for D3 that, though tighter, was orders of magnitude weaker than for D3 to intact LRP [14]. This high affinity but low specificity is not surprising given the chaperone function of RAP and the mode of binding seen in the D3–LA34 complex [21], which involves co-ordination of an exposed lysine by the calcium-binding region that is common to all CR domains [24–27]. The source of the disagreement is unclear, but may result from use of ubiquitin-conjugated CR fragments, immobilization of ligands, use of CR domains from LDLR, for which RAP is not thought to be the in vivo chaperone [23], or the lower accuracy of ITC for measuring tight interactions when very high protein concentrations are used.
For the first time, binding data for (CR)3 fragments are reported. These show that a third CR domain contributes very much less to binding than the average contribution of each of the first two domains. This suggests that, although there are likely to be two ‘hot spot’ lysines on each of D1, D2 and D3, each of which engages with a CR domain via its calcium-binding region, there is no third such interaction. Importantly, we have shown that only two of the three binding sites are available in intact RAP, though they retain the very high affinities seen for isolated RAP domains to the respective (CR)x species. This is in agreement with a study that identified one high-affinity binding site in the N-terminal half of RAP and one in the C-terminal half [13]. It is also in keeping with an earlier study from this laboratory that found two binding sites for CR78 to intact RAP [28]. With the benefit of hindsight, it was unfortunate that we chose CR78, since this is the ‘odd-man-out’ in having relatively low affinity for D1 and D2. Nevertheless, the conclusion reached there of two binding sites is the same as reached here with a more extensive range of much tighter binding species.
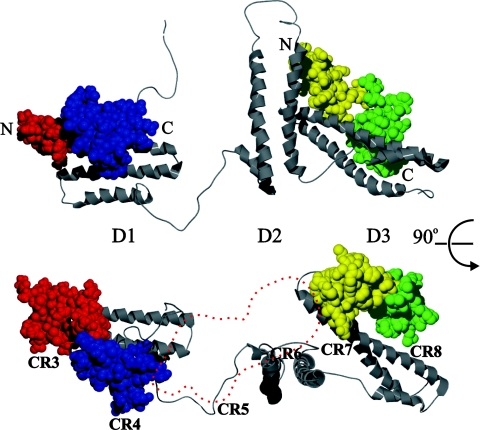
Based on the existence of two high-affinity binding sites, on a model for RAP derived from NMR structures of each of the three RAP domains [28a], and on two structures of RAP domain complexes with (CR)2 fragments, one from X-ray [21] and one from NMR and modelling [15], we propose a model for the binding of RAP to a contiguous stretch of CR domains. In this model the locations of the two (CR)2 fragments from the structures of D1 and D3 in separate complexes have been superimposed on one of the ensemble of 20 energy-minimized structures of RAP (Figure 8). Model 2 was chosen as being one with lowest energy and one that best fitted the elongated structure of RAP implied by the high frictional ratio obtained here from sedimentation velocity data. The orientation of D3 relative to D1 would allow a contiguous stretch of CR domains to bind in the same manner as in the two complexes of isolated domains. It is intriguing that, when two (CR)2 fragments are positioned in this way, there is just sufficient space between their C and N-terminal ends to accommodate an additional pair of domains, although in a location that does not give direct additional contact with the globular parts of either D1 or D3. This could explain how two (CR)3 fragments bind to intact RAP with only slightly higher affinity than two (CR)2 fragments. A final pleasing aspect of the model is that D2 is approximately orthogonal to the long axis that runs along D1 and D3. If the binding site on D2 for a (CR)2 fragment is parallel to the long axis of the domain, as it is in D1 and D3, it might not be possible to simultaneously engage all three binding sites in intact RAP without interference. This would explain the stoichiometry of 2:1 rather than 3:1 for (CR)x binding to intact RAP. Furthermore, the ΔH values obtained for binding CR56 to intact RAP compared with isolated domains favour binding to D1 and D3 in intact RAP rather than to D2 in combination with either D1 or D3. Although our binding experiments using a single type of (CR)x species precluded being able to simultaneously bind different fragments of cluster II to RAP, it is clear from Table 1 that RAP could tightly engage CR3–CR8, with D1 binding to CR345 (Kd of 23 nM) and D3 binding to CR678 (Kd of 26 nM).
Figure 8. Model of RAP–cluster II complex.
Model of RAP binding to CR stretches. The Low energy solution 2 from a RAP structure modelling study by Lee et al. [28a], using individual NMR structures of RAP domain 1, 2 and 3 (PDB ID 2P01), is shown in a grey ribbon diagram. D1 is aligned with the D1 from a D1-CR56 complex based on NMR data by Jensen et al. [15] (PDB ID 2FYL). D3 is aligned with D3 from the X-ray crystal structure of D3 in complex with LA34 from the LDLR by Fisher et al. [21] (PDB ID 2FCW). CR5 and CR6 are shown in red and blue space-fill respectively. LA3 and LA4 are shown in yellow and green space-fill respectively. N- and C-termini of the CR and LA tandem repeats and the identity of the RAP domains are indicated. Top and bottom structures are related by a 90° rotation around a horizontal axis through the middle of the Figure. An outline trace of the CR56 space-filling structure in the bottom structure has been positioned between the end of the tandem domain bound to D1 and the tandem domain bound to D3 (shown in red dashes), to represent a possible location of an additional CR tandem repeat. The numbering CR3 through CR8 below the CR domains in the bottom representation indicates how the (CR)6 species CR3–CR8 might bind.
The presence of two such high-affinity binding sites within RAP should have profound consequences for the chaperone function of RAP. A landmark study showed that a receptor–ligand complex between a synthetic trivalent vancomycin and a trivalent D-Ala-D-Ala ligand had binding energy that was approximately equal to the sum of the three separate binding energies of the monovalent ligand and receptor [29], resulting in an astonishing 40 aM (4×10−17 M) Kd for the trivalent system. More intriguingly, since a monovalent ligand could compete for any of the individual interactions in the trivalent complex, the dissociation kinetics of the trivalent complex using a monovalent competitive ligand reflected the off rate for the individual interactions rather than the much slower dissociation of the whole trimer. This system thus combined ultra high affinity with the ability to compete off parts of the whole ligand at relatively fast rates [30]. This has previously been exploited in the design of retractable, but high affinity, bivalent inhibitors of thrombin [31].
The consequences for RAP–LRP are two-fold. The first is that, with two binding sites on RAP for up to 3 CR domains each, with individual Kd value of ∼25 nM, RAP should bind to cluster II with an overall Kd of ∼0.6 fM. The second is that, even though the overall affinity of RAP for cluster II should be ultra-high, each of the two binding sites should be displaceable by a ligand with affinity equal only to that of the individual site. This could explain the apparent conflict between the present proposal for ultra-high-affinity binding of RAP to LRP and the many literature reports of binding of RAP to LRP only in the low-nanomolar range, since most such binding studies infer an affinity by competition with a ligand (whether bivalent RAP or monovalent D3) that could bind at either of the two sites on RAP that is bound to LRP. Such ultra-high affinity of RAP for folded cluster II would facilitate its function as a folding chaperone, while still allowing rapid dissociation in two steps by competition with ligands that separately target each of the two binding sites on RAP. An obvious candidate for such a competing ligand are the YWTD ‘propeller’ domains that flank the ligand-binding clusters in LRP and other LDLR family members [32]. LRP has eight YWTD domains, with four flanking cluster II. In a beautiful structure of the extracellular portion of LDLR at low pH, it was shown that the single YWTD domain present interacted with CR modules 4 and 5 of the lipoprotein-binding region [33]. It was proposed that several histidines at the interface between the YWTD domain and the CR domains might promote binding of YWTD to CR at low pH and thereby displace lipoprotein bound at, or close to, that same site. In support of this, it was recently shown that displacement of bound lipoprotein depends on these histidines in the YWTD domain in the expected pH-dependent manner [34]. By extension, displacement of RAP at the pH of the Golgi might occur by a similar mechanism. The main difference is that, since RAP has two tight binding sites on cluster II, two such YWTD domains would be needed, one for each binding site. However, there are quite sufficient YWTD domains available, flanking each end of the cluster. YWTD modules could then effectively displace a single RAP, even though the latter bound with overall ultra-high affinity, by individually displacing each of the two binding sites on RAP.
This proposal for the basis of pH-dependent release of RAP from LRP is at variance with previous studies that suggested that pH-dependent unfolding of RAP D3, resulting from histidine protonation, is the cause of RAP dissociation [22,23]. There are, however, a number of concerns with these studies. One is that, even if D3 dissociated in this manner, D1 should remain tightly bound, based on the pH-independence of the high-affinity interaction of D1 with CR56 found here. The second is that, although we also found that lower pH reduces the affinity of CR56 for D3, the reduction is small (from 7 nM at pH 7.4 to 41 nM at pH 5.5). Indeed, in previous competition studies between wild-type D3 and D3 with mutated histidines, a similar relatively small reduction in affinity (∼7–9 fold) was found [22]. A final concern that applies to cell-based studies on LRP ‘mini-receptors’ is that, to the best of our understanding, none of these truncated receptors contains any YWTD domains [6,22,35]. If these domains do play a dominant role in RAP release, and hence in trafficking of receptor to the cell surface, the results obtained could be misleading, by focusing on a second-rank event such as modest weakening of D3 affinity, rather than the principal player.
Online data
AUTHOR CONTRIBUTION
Jan Jensen carried out experiments and analysed data, Klavs Dolmer helped with protein expression, Christine Schar carried out and analysed analytical the ultracentrifugation experiments and Peter Gettins designed experiments, analysed data and wrote the manuscript.
ACKNOWLEDGEMENTS
We thank Steven Olson for helpful comments on the manuscript.
FUNDING
This work was supported by the National Institutes of Health [grant numbers R01 GM54414 and HL79430 (to P. G. W. G.)] and by fellowships (to J. K. J.) from the Danish Research Council and the Carlsberg Foundation. The Beckman XL-I analytical ultracentrifuge was purchased with funds from a shared instrumentation grant from the National Institutes of Health [grant number S10 RR22361].
References
- 1.Hussain M. M., Strickland D. K., Bakillah A. The mammalian low-density lipoprotein receptor family. Annu. Rev. Nutr. 1999;19:141–172. doi: 10.1146/annurev.nutr.19.1.141. [DOI] [PubMed] [Google Scholar]
- 2.Herz J., Strickland D. K. LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willnow T. E., Armstrong S. A., Hammer R. E., Herz J. Functional expression of low density lipoprotein receptor-related protein is controlled by receptor-associated protein in vivo. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4537–4541. doi: 10.1073/pnas.92.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orlando R. A. The low-density lipoprotein receptor-related protein associates with calnexin, calreticulin, and protein disulfide isomerase in receptor-associated-protein- deficient fibroblasts. Exp. Cell Res. 2004;294:244–253. doi: 10.1016/j.yexcr.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Willnow T. E., Rohlmann A., Horton J., Otani H., Braun J. R., Hammer R. E., Herz J. RAP, a specialized chaperone, prevents ligand-induced ER retention and degradation of LDL receptor-related endocytic receptors. EMBO J. 1996;15:2632–2639. [PMC free article] [PubMed] [Google Scholar]
- 6.Bu G., Rennke S. Receptor-associated protein is a folding chaperone for low density lipoprotein receptor-related protein. J. Biol. Chem. 1996;271:22218–22224. doi: 10.1074/jbc.271.36.22218. [DOI] [PubMed] [Google Scholar]
- 7.Herz J., Marschang P. Coaxing the LDL receptor family into the fold. Cell. 2003;112:289–292. doi: 10.1016/s0092-8674(03)00073-4. [DOI] [PubMed] [Google Scholar]
- 8.Bu G., Marzolo M. P. Role of RAP in the biogenesis of lipoprotein receptors. Trends Cardiovasc. Med. 2000;10:148–155. doi: 10.1016/s1050-1738(00)00045-1. [DOI] [PubMed] [Google Scholar]
- 9.Obermoeller L. M., Warshawsky I., Wardell M. R., Bu G. Differential functions of triplicated repeats suggest two independent roles for the receptor-associated protein as a molecular chaperone. J. Biol. Chem. 1997;272:10761–10768. doi: 10.1074/jbc.272.16.10761. [DOI] [PubMed] [Google Scholar]
- 10.Lazic A., Dolmer K., Strickland D. K., Gettins P. G. W. Structural organization of the receptor associated protein. Biochemistry. 2003;42:14913–14920. doi: 10.1021/bi035779e. [DOI] [PubMed] [Google Scholar]
- 11.Bu G., Rennke S., Geuze H. J. ERD2 proteins mediate ER retention of the HNEL signal of LRP's receptor associated protein (RAP) J. Cell Sci. 1997;110:65–73. doi: 10.1242/jcs.110.1.65. [DOI] [PubMed] [Google Scholar]
- 12.Bu G., Geuze H. J., Strous G. J., Schwartz A. L. 39 kDa receptor-associated protein is an ER resident protein and molecular chaperone for LDL receptor-related protein. EMBO J. 1995;14:2269–2280. doi: 10.1002/j.1460-2075.1995.tb07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlando R. A., Farquhar M. G. Functional domains of the receptor-associated protein (RAP) Proc. Natl. Acad. Sci. U.S.A. 1994;91:3161–3165. doi: 10.1073/pnas.91.8.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersen O. M., Schwartz F. P., Eisenstein E., Jacobsen C., Moestrup S. K., Etzerodt M., Thøgersen H. C. Dominant thermodynamic role of the third independent receptor binding site in the receptor-associated protein. Biochemistry. 2001;40:15408–15417. doi: 10.1021/bi0110692. [DOI] [PubMed] [Google Scholar]
- 15.Jensen G. A., Andersen O. M., Bonvin A. M. J. J., Bjerrum-Bohr I., Etzerodt M., Thøgersen H. C., O'Shea C., Poulsen F. M., Kragelund B. B. Binding site structure of one LRP-RAP complex: implications for a common ligand-receptor binding motif. J. Mol. Biol. 2006;362:700–716. doi: 10.1016/j.jmb.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Dolmer K., Gettins P. G. W. Three complement-like repeats compose the complete α2-macroglobulin binding site in the second ligand binding cluster of the low density lipoprotein receptor-related protein. J. Biol. Chem. 2006;281:34189–34196. doi: 10.1074/jbc.M604389200. [DOI] [PubMed] [Google Scholar]
- 17.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balbo A., Minor K. H., Velikovsky C. A., Mariuzza R. A., Peterson C. B., Schuck P. Studying multiprotein complexes by multisignal sedimentation velocity analytical centrifugation. Proc. Natl. Acad. Sci. U.S.A. 2005;102:81–86. doi: 10.1073/pnas.0408399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen O. M., Christensen L. L., Christensen P. A., Sorensen E. S., Jacobsen C., Moestrup S. K., Etzerodt M., Thøgersen H. C. Identification of the minimal functional unit in the low density lipoprotein receptor-related protein for binding the receptor-associated protein (RAP) J. Biol. Chem. 2000;275:21017–21024. doi: 10.1074/jbc.M000507200. [DOI] [PubMed] [Google Scholar]
- 20.Medved L. V., Migliorini M., Mikhailenko I., Barrientos L. G., Llinás M., Strickland D. K. Domain organization of the 39-kDa receptor-associated protein. J. Biol. Chem. 1999;274:717–727. doi: 10.1074/jbc.274.2.717. [DOI] [PubMed] [Google Scholar]
- 21.Fisher C., Beglova N., Blacklow S. C. Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol. Cell. 2006;22:277–283. doi: 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 22.Lee D., Walsh J. D., Mikhailenko I., Yu P., Migliorini M., Wu Y., Krueger S., Curtis J. E., Harris B., Lockett S., et al. RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi. Mol. Cell. 2006;22:423–430. doi: 10.1016/j.molcel.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Estrada K., Fisher C., Blacklow S. C. Unfolding of the RAP-D3 helical bundle facilitates dissociation of RAP-receptor complexes. Biochemistry. 2008;47:1532–1539. doi: 10.1021/bi702076y. [DOI] [PubMed] [Google Scholar]
- 24.Herz J., Hamann U., Rogne S., Myklebost O., Gausepohl H., Stanley K. K. Surface location and high calcium affinity of a 500kD liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moestrup S. K., Kaltoft K., Sottrup-Jensen L., Gliemann J. The human α2-macroglobulin receptor contains high affinity calcium binding sites important for receptor conformation and ligand recognition. J. Biol. Chem. 1990;265:12623–12628. [PubMed] [Google Scholar]
- 26.Simonovic M., Dolmer K., Huang W., Strickland D. K., Volz K., Gettins P. G. W. Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor. Biochemistry. 2001;40:15127–15134. doi: 10.1021/bi015688m. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q.-Y., Dolmer K., Huang W., Gettins P. G. W., Rong L. Role of calcium in protein folding and function of Tva, the receptor of subgroup A avian sarcoma and leukosis virus. J. Virol. 2001;75:2051–2058. doi: 10.1128/JVI.75.5.2051-2058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazic A., Dolmer K., Strickland D. K., Gettins P. G. W. Dissection of RAP-LRP interactions. Binding of RAP and RAP fragments to complement-like repeats 7 and 8 from cluster II of LRP. Arch. Biochem. Biophys. 2006;450:167–175. doi: 10.1016/j.abb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 28a.Lee D., Walsh J. D., Migliorini M., Yu P., Cai T., Schwieters C. D., Krueger S., Strickland D. K., Wang Y. X. The structure of receptor-associated protein (RAP) Protein Sci. 2007;16:1628–1640. doi: 10.1110/ps.072865407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao J., Lahiri J., Isaacs L., Weis R. M., Whitesides G. M. A trivalent system from vancomycin.D-Ala-D-Ala with higher affinity than avidin.biotin. Science. 1998;280:708–711. doi: 10.1126/science.280.5364.708. [DOI] [PubMed] [Google Scholar]
- 30.Zhou H.-X. Quantitative relation between intermolecular and intramolecular binding of Pro-rich peptides to SH3 domains. Biophys. J. 2006;91:3170–3181. doi: 10.1529/biophysj.106.090258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolkatchev D., Vinogradova A., Ni F. Transforming bivalent ligands into retractable enzyme inhibitors through polypeptide-protein interactions. Bioorg. Med. Chem. Lett. 2005;15:5120–5123. doi: 10.1016/j.bmcl.2005.08.085. [DOI] [PubMed] [Google Scholar]
- 32.Springer T. A. An extracellular β-propellor module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J. Mol. Biol. 1998;283:837–862. doi: 10.1006/jmbi.1998.2115. [DOI] [PubMed] [Google Scholar]
- 33.Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Z., Michaely P. The epidermal growth factor homology domain of the LDL receptor drives lipoprotein release through an allosteric mechanism involving H190, H562, and H586. J. Biol. Chem. 2008;283:26528–26537. doi: 10.1074/jbc.M804624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obermoeller-McCormick L. M., Li Y., Osaka H., FitzGerald D. J., Schwartz A. L., Bu G. Dissection of receptor folding and ligand-binding property with functional minireceptors of LDL receptor-related protein. J. Cell Sci. 2001;114:899–908. doi: 10.1242/jcs.114.5.899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.