Abstract
Background & Aims
Autoimmune hepatitis (AIH) is a chronic inflammatory liver disease associated with autoantibodies and liver-infiltrating lymphocytes. Whereas autoantibodies are routinely tested to diagnose and classify AIH, liver-infiltrating lymphocytes are regarded as the primary factor for disease pathogenesis. The purpose of this study was to identify and characterize autoantigenic peptides within human AIH-specific soluble liver antigen / liver pancreas antigen (SLA/LP) that are targeted by CD4+ T cells and restricted by the disease susceptibility gene HLA-DRB1*0301.
Methods
HLA- DRB1*0301 transgenic mice were immunized with SLA/LP. Antibody and T cell responses were analyzed with SLA/LP-overlapping peptides in EIA, proliferation and Elispot assays. Minimal optimal T cell epitopes were identified, characterized with cloned T cell hybridomas and confirmed in tetramer and ELISpot assays with AIH patients’ PBMC.
Results
All mice developed SLA/LP-specific IgG1/IgG2a antibodies against the same SLA/LP peptides as humans. T cells targeted several peptides within SLA/LP, two of which were DR3-restricted and one overlapped the sequence recognized by human autoantibodies. Minimal optimal epitopes were mapped, DRB1*0301/epitope-tetramers were generated, and frequency and function of HLA-DRB1*0301-restricted autoantigen-specific T cells in AIH patients were analyzed with tetramer and IFN-γ ELISpot analysis.
Conclusions
This study identifies the first T cell epitopes within SLA/LP, restricted by the disease susceptibility gene DRB1*0301 and in close proximity to the human autoantibody epitope. These results and the generated reagents now provide the opportunity to directly monitor autoreactive T cells in AIH patients in clinical studies.
Introduction
Autoimmune hepatitis (AIH) is a chronic inflammatory liver disease, which has a prevalence of approximately 1/10,000 in the United States and Europe and accounts for 2−6% of all liver transplantations 1. AIH is diagnosed by elevated alanine amino transferase levels, an intrahepatic lymphocytic mononuclear cell infiltrate, hypergammaglobulinemia and autoantibodies, and the absence of liver disease of viral, toxic or metabolic etiology 2, 3.
Autoantibodies target nuclear antigens (antinuclear antibodies, ANA), smooth muscle antigens (SMA), liver-kidney microsomes (LKM-1) and/or soluble liver antigen/liver-pancreas antigen (SLA/LP). Autoantibodies to SLA/LP are detectable in about 20% of AIH cases 4, 5, and are unique in that they are the only autoantibodies specific for AIH 4. The target of SLA/LP autoantibodies has been cloned 6 and named SEPSECS by the Nomenclature Commission of the Human Genome Organization. However, even though autoantibodies are well accepted as serologic markers for diagnosis and classification of AIH, it is still unclear whether they play any role in disease pathogenesis 7.
The degree of the intrahepatic lymphocytic infiltrate, the histologic hallmark of AIH, correlates well with disease progression and is therefore thought to contribute directly to disease pathogenesis. The clonal restriction of the intrahepatic T cell population and the observation that HLA-DRB1*0301 and HLA-DRB1*0401 predispose to AIH and influence disease severity 8, 9, have led to the hypothesis that intrahepatic CD4+ T cells recognize self-antigens in the context of HLA-DRB1*0301 and HLA-DRB1*0401 2. HLA-DRB1*0301, the principal AIH susceptibility allele among white Europeans and Americans 8, 9, and HLA-DRB1*0401 may even present the same autoantigen to CD4+ T cells because both alleles share an amino acid motif in their antigen binding groove (‘shared binding hypothesis’)10.
Despite much effort, it still remains unknown which autoantigens HLA-DRB1*0301 and HLA-DRB1*0401-restricted CD4+ T cells recognize in AIH. This question has been declared a high priority investigational challenge 11, but proven difficult to answer for the following reasons. As in other autoimmune diseases 12, the frequency of autoreactive CD4+ T cells in the peripheral blood is too low to screen a large number of candidate peptides for T cell recognition. Autoreactive CD4+ T cells are presumably more frequent at the site of inflammation, but only a small piece of tissue and thereby only a small number of lymphocytes can be obtained by liver biopsy. It therefore remains necessary to expand and/or clone liver-biopsy-derived T cells in vitro before they can be studied in functional assays 13. This approach renders an ex vivo assessment of the number and function of autoantigen-specific T cells impossible. Furthermore, assays that solely rely on the function of autoantigen-specific T cells are often compromised due to immunosuppressive therapy of AIH.
We therefore immunized ‘humanized’ mice, which are transgenic for HLA-DRB1*0301 and do not express murine MHC class II 14, with the recently cloned SLA/LP autoantigen 6 and established T cell hybridomas to identify the optimal DRB1*0301-restricted sequences (epitopes) within SLA/LP (Fig. 1). These epitopes were then used in Elispot assays and in flow cytometry assays with custom-made fluorescently labeled tetrameric DRB1*0301 complexes to monitor AIH-specific T cell responses in the blood of patients with AIH. The identification of these T cell epitopes may also be a step towards the identification of those immunogenic self- and/or crossreactive foreign antigens that set off the initial autoimmune reaction.
Fig. 1. Experimental strategy.
β-gal, β-galactosidase.
Material and Methods
SLA/LP protein, peptides, tetramer and antibodies
Recombinant human SLA/LP (AAD33963) was synthesized and purified as previously described 6. Twenty-mer peptides overlapping by 12 amino acids (aa) and spanning aa 1−422 of human SLA/LP (Mimotopes, Clayton, Australia), shorter SLA/LP peptides and the DRB1*0301-restricted hepatitis C virus (HCV) peptide HCV261−269 (VLVLNPSVA) 15 (Facility for Biotechnology Resources, CBER, FDA, Bethesda, MD) were resuspended at 20 mg/ml in dimethyl sulfoxide (DMSO) and diluted with phosphate-buffered saline solution (PBS) to 2 mg/ml. Tetramers DRB1*0301-SLA/LP184−198 and DRB1*0301-SLA/LP373−386 were custom-made by Beckman Coulter (Fullerton, CA).
Anti-mouse CD4-PE or -FITC (clone 129.19), CD3e-APC-Cy7 (clone 145−2C11), CD19-PE (clone 1D3), CD11a-PE-Cy7 (clone 2D7), CD62L-APC (clone MEL-14) and anti-human CD3-APC (clone UCHT1), CD4-PE-Cy7 or -APC-Cy7 (clone SK3), CD25-APC-Cy7 (clone MA 251), CD8-PE-Cy5 or -PerCP (clone RPA T-8), CD14-PerCP (clone MΦP9), CD16-PE-Cy5 (clone 3G8), HLA-DR-FITC (clone L243) and CD19-PE-Cy5 or -PerCP (clone HIB 19) as well as purified antibodies to I-A/I-E (clone 2G9), HLA-DR, -DP, -DQ (clone TÜ39) were purchased from BD Pharmingen (San Diego, CA). Anti-human CD14-PE-Cy5 (clone TÜK4) was purchased from Serotec (Raleigh, NC).
Mouse immunization
C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME) and HLA-DRB1*0301-transgenic mice that do not express endogenous class II at detectable level (DR3.AB0) 14 were maintained in a clean conventional facility. Experiments were approved by the Animal Care and Use Committee of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Eight-to-twelve week-old mice were immunized at the base of the tail and both hind foot pads with (i) 40 μg of human recombinant SLA/LP (11 mice) or (ii) 50 μg of peptides SLA/LP81−100, SLA/LP89−108 and SLA/LP97−116 (5 mice) or (iii) 50 μg of peptides SLA/LP177−196 and SLA/LP185−204 (5 mice) or (iv) 50 μg of peptides SLA/LP403−416 and SLA/LP408−418 (5 mice), in 100−200 μl PBS emulsified with an equal amount of AS01B adjuvant (GlaxoSmithKline, Rixensart, Belgium) 16, reimmunized after 3 weeks and killed 10 days after the last immunization. Lymphocytes were isolated by either mechanical disruption or enzymatic digestion (400 ug/ml Liberase CI (Roche Diagnostics, Indianapolis, IN) of draining lymph nodes and spleen, followed by filtration through a 70 μm nylon cell strainer (BD Falcon, Bedford, MA) and red blood cell lysis with ACK lysing buffer (Bio-Whittaker, Walkersville, MD).
Analysis of the humoral immune response
SLA/LP-specific antibodies were detected with the QUANTA Lite™ SLA/LP EIA kit (Inova Diagnostics, San Diego, CA) according to the manufacturer's instructions except for the use of an HRP-labeled polyclonal goat anti-mouse Ig (BD Pharmingen, dilution 1:2000) for detection. Isotyping was performed with a mouse immunoglobulin screening/isotyping kit (Zymed Laboratories, San Francisco, CA). Fine mapping of the antibody specificity was performed by EIA with 1/100 dilutions of sera using microtiter plates (Falcon) coated with SLA/LP-overlapping peptides (10 μg/ml).
Analysis of the cellular immune response of immunized mice
(i) Proliferation assays
Quadruplicate cultures of 5 × 105 mouse spleen cells were stimulated for 3 days with or without human recombinant SLA/LP at the indicated concentration or with 10 ng/ml phorbol myristic acetate (PMA) / 1 μM ionomycin (Sigma-Aldrich) in 96-well round bottom plates in RPMI1640 containing 10% heat-inactivated FBS (Bio-Whittacker, Walkersville, MD), 50 μM β-mercaptoethanol (Gibco), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 10 mM HEPES, and 1 mM sodium pyruvate (Cellgro, Herndon, VA). For the last 16 h, 1 μCi 3H-thymidine (ICN, Costa Mesa, CA) was added per well. Cells were harvested with a Packard Filtermate 96-well-harvester (Perkin Elmer, Wellesley, MA) and incorporated radioactivity was measured with a Packard Topcount (Perkin Elmer).
(ii) Enzyme-linked immunospot (ELISpot) assays
Ex vivo ELISpot assays were performed as described 17 with the following modifications. Triplicate cultures of 5×105 splenocytes were stimulated with or without recombinant SLA/LP (2.5 μg/ml), synthetic peptides (10 μg/ml) or PMA/ionomycin on plates coated with 2 μg/ml anti-mouse interleukin-2 (IL-2) (clone JES6−1A12, BD Pharmingen. Biotin-conjugated anti-mouse IL-2 antibody (2 μg/ml, clone JES6-SH4, BD Pharmingen) and 3-amino-9-ethylcarbazole (AEC) substrate solution (Sigma-Aldrich) were used for detection. Spots were counted with a KS ELISpot Reader (Carl Zeiss Inc., Thornwood, NY).
(iii) Tetramer analysis
CD4+ T cells were negatively isolated from spleen and lymph node cells with magnetic beads and MACS columns (Miltenyi Biotec, Auburn, CA), incubated for 10 min with antiCD16/antiCD32 as Fc-block (clone 2.4G2, BD Pharmingen), washed and stained with 2.5 μg/ml EMA (Sigma-Aldrich) under a fluorescent light source for 10 min. Cells were washed again, and stained with 10 μl of either DRB1*0301-SLA/LP184−198-tetramer or DRB1*0301-SLA/LP373−386-tetramer in the presence of unconjugated anti-mouse TCR-β (clone H57−597, eBiosciences, San Diego, CA) at 37° C for 3h. After addition of antibodies against cell surface molecules for the last 30 min, cells were washed and analyzed on an LSRII flow cytometer (BD Biosciences, San Jose, CA) using FACS Diva (BD Biosciences, San Jose, CA) and FlowJo (TreeStar, San Carlos, CA) software. EMA+ (dead) cells were excluded and CD3+CD4+ T cells were analyzed for DRB1*0301/epitope-tetramer binding and expression of CD62L and CD11a.
(iv) Generation of T cell hybridomas
Sixty million spleen cells of SLA/LP-immunized mice were stimulated with 5 μg/ml SLA/LP or 10 μg/ml of the immunization peptides in 5 ml culture medium. Four days later, cells were fused to BWZ.36 (kindly provided by Nilabh Shastri, University of California, Berkeley, CA), a TCRαβ− cell line transfected with the β-galactosidase (LacZ) reporter gene under the control of the NFAT enhancer element of the IL-2 gene 18. SLA/LP-specific hybridoma were subcloned at 0.3 cells/well.
(v) Hybridoma activation and LacZ assays
Duplicate or triplicate cultures of 0.2 − 1×105 T cell hybridomas were stimulated overnight with 2×105 irradiated (3,000 rad) spleen cells from HLA-DRB1*0301+ transgenic mice with or without SLA/LP peptides at the indicated concentrations and with or without 2.5 or 10 μg anti I-A / anti I-E (clone 2G9, BD Pharmingen), anti-HLA-DR, -DP, -DQ (TÜ39, BD Pharmingen) or isotype control antibodies. TCR activation resulted in activation of the NFAT enhancer element of the IL-2 gene and transcription of the lacZ reporter gene. To measure LacZ activity, cultures were washed extensively and lysed with 100 μl buffer (1 mM MgCl2, 0.125 % NP-40 (Calbiochem, La Jolla, CA) and 0.15 mM chlorophenol red β-galactoside (CPRG, Roche) in PBS). After 8h at 37°C, the reaction was stopped with 100 μl buffer (300 mM glycine and 15 mM Na2-EDTA in water, pH 12). Absorption was read at 575 nm with a 96-well Benchmark Microplate reader (Bio-Rad, Hercules, CA) with 620 nm as reference wavelength.
Detection of SLA/LP-specific CD4+ T cells in humans
PBMC were isolated from 9 patients with anti-SLA/LP+ AIH and, as negative controls, from an HLA-DRB1*0301− patient with anti-SLA/LP+ AIH and two HLA-DRB1*0301+ , anti-SLA/LP− patients with past history of hepatitis B (anti-HBs+ , anti-HBe+ , HB-Ag−, HBeAg−). Patients were followed either at Johannes Gutenberg Universität, Mainz, Germany, Universitätsklinikum Hamburg Eppendorf, Hamburg, Germany, Medizinische Universitätsklinik Freiburg, Germany or the Liver Diseases Branch, NIDDK, NIH, Bethesda, MD, USA and provided informed consent under protocols approved by the Ethics Committee and Institutional Review Board of the respective institutions.
Thawed PBMC were stained with 2.5 μg/ml EMA under a fluorescent light source for 10 min, washed in PBS and stained with 10 μl of either the DRB1*0301-SLA/LP373−386 tetramer or the DRB1*0301-SLA/LP184−196 tetramer (both coupled to PE) in complete RPMI medium for 30 min at 37°C. After washing in PBS, cells were stained with antibodies against CD3, CD8, CD14 and CD19 and CD4 antibodies for 20 min at 4°C, washed and subsequently treated with anti-PE magnetic beads (Miltenyi Biotec, Auburn, CA) for 15 min at 4°C. Ten percent of the cells were analyzed directly by flow cytometry (pre-enrichment sample), the remaining 90% were applied to MS columns (Miltenyi Biotec, Auburn, CA) to enrich PE-conjugated tetramer+ cells (post-enrichment sample). After exclusion of doublets by flow cytometry, forward and side scatter-gating of lymphocytes and exclusion of EMA/CD8/CD14/CD19+ events, the CD3+ T cell population was analyzed for expression of CD4 and for tetramer binding.
For proliferation assays, 107 PBMC were stained with 1 μM carboxyfluorescein diacetate succinimidylester (CFSE, Molecular Probes, Eugene, Oregon) in 2 ml PBS for 5 min at room temperature. After addition of 5 ml FCS and three washes in PBS, cells were stimulated with 10 μg/ml DRB1*0301-restricted SLA/LP373−386 peptide or the DRB1*0301-restricted HCV261−269 control peptide 15, respectively. Forty U/ml recombinant IL2 were added on days 4 and 7. On day 10, cells were stained with EMA, the respective DRB1*0301/epitope-tetramer and antibodies against cell surface proteins and analyzed by flow cytometry. After EMA/CD8/CD14/CD16/CD19 exclusion, CD3+CD4+ lymphocytes were analyzed for tetramer binding and CFSE content.
For IFN-γ ELISpot assays, CD4+ T cells were isolated with the Miltenyi Multisort Kit, released from the beads, stained with the DRB1*0301-SLA/LP373−386 tetramer or the DRB1*0301-SLA/LP184−196 tetramer as described above and sorted manually with anti-PE beads (Miltenyi) following the manufacturer's protocol. Part of the tetramer-enriched cell population was directly used in an Elispot, the other part was expanded with 40 ng/ml anti-CD3 under twice weekly addition of 200 U/ml IL-2, 10 ng/ml IL-7 and 100 ng/ml IL-15 prior to ELISpot analysis. IFN-γ ELISpot assays were performed as described 19 using 60,000 − 300,000 tetramer-enriched cells and 200,000 irradiated (3,000 rad) autologous tetramer-negative PBMC as feeder cells in presence or absence of 10 μg/ml peptide.
Results
Humoral immune response of HLA-DRB1*0301 transgenic mice to human recombinant SLA/LP
Sixteen DRB1*0301-transgenic mice were subcutaneously immunized with human recombinant SLA/LP and AS01B adjuvant 16. As shown in Fig. 2A, SLA/LP-specific antibodies were detectable after a single immunization and increased further after a booster immunization. Isotyping revealed a large IgG1, IgG2b and IgG3 fraction (Fig. 2B) consistent with a strong Th2 CD4+ T cell response in the context of this adjuvant 16. Because the humoral immune response of patients with AIH is targeted against the carboxyterminus of SLA/LP 4, antibodies against this region were mapped with overlapping 20mer peptides. As shown in Fig. 2C, the 20mer peptide spanning aa 403−422 elicited the most vigorous response in HLA-DRB1*0301-transgenic mice. This peptide was significantly less vigorously recognized by nontransgenic C57BL/6 mice suggesting a role of HLA-DR-restricted CD4+ T cell help in the generation of these autoantibodies.
Fig. 2. Humoral immune response of SLA/LP-immunized HLA-DRB1*0301-transgenic mice.
(A, B) HLA-DRB1*0301 transgenic mice (n=16) were immunized with adjuvanted SLA/LP on days 0 and 21 and SLA/LP-specific antibody responses were quantified at the indicated time points (A) and studied for isotype distribution on day 42 (B). The dashed horizontal line indicates the cut-off of positivity. (C) Fine-mapping of the antibody response of immunized HLA-DRB1*0301 transgenic (filled bars) and C57BL/6 mice (open bars). Mean and SD of the results of 3 mice are shown in panels B and C. A p-value of <0.05 (Student t test) was considered significant. Aa, amino acid.
Cellular immune response to SLA/LP in HLA-DRB1*0301 transgenic mice
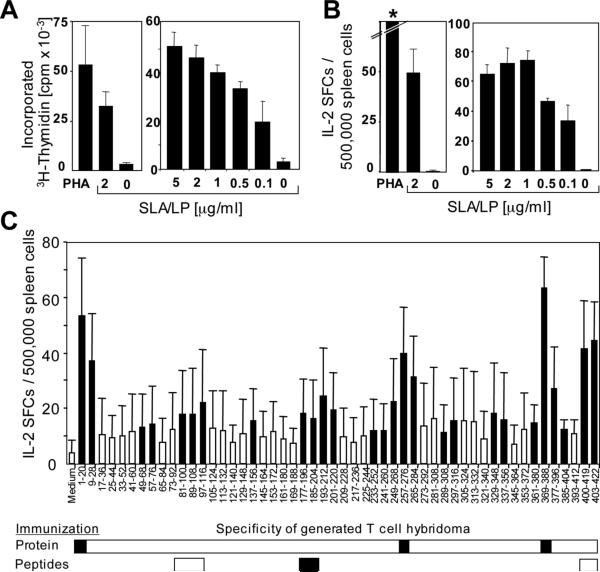
Spleen-derived T cells of SLA/LP-immunized HLA-DRB1*0301-transgenic mice proliferated (Fig. 3A) and produced IL-2 (Fig. 3B) in a dose-dependent manner in response to SLA/LP. When T cell responses of SLA/LP-immunized mice were assessed with a panel of overlapping 20mer peptides, positions 1−28, 49−76, 81−116, 137−156, 177−220, 233−284, 289−316, 329−356, 361−404, and 400−422 were identified as immunogenic regions within the SLA/LP protein (Fig. 3C).
Fig. 3. Cellular immune response of SLA/LP-immunized HLA-DRB1*0301-transgenic mice.
(A, B) SLA/LP dose-dependent proliferation (A) and IL-2 ELISpot (B) using spleen cells from SLA/LP-immunized HLA-DRB1*0301-transgenic mice. The left graphs in panels A and B, respectively, show results (mean of triplicate values) from representative SLA/LP-immunized mice, the right graphs in panels A and B show mean and SD of three mice per group. The asterisk indicates confluent spots too numerous to count. (C) Fine-mapping of the immune response of SLA/LP-immunized mice (n=11) using overlapping peptides. Filled bars indicate responses higher than the mean plus 3 SD of the medium control. Immunogenic regions for which T cell hybrids could be established from either SLA/LP protein-immunized mice (n=11) or peptide-immunized mice (n=5 per peptide) are indicated by filled horizontal bars.
To fine-map the immunodominant regions, T cells of SLA/LP-immunized HLA- DRB1*0301-transgenic mice were fused with the lymphoma cell line BWZ.36. The generated CD4+ T cell hybridomas recognized peptides spanning SLA/LP positions 1−20, 257−276 and 369−388, respectively, indicating that these sequences were immunodominant (Fig. 2C). To increase the efficiency of T cell hybridoma generation against sequences that were less vigorously recognized, additional groups of 5 HLA-DRB1*0301-transgenic mice were subsequently immunized with peptide mix SLA/LP81−100/SLA/LP89−108/SLA/LP97−116, peptide mix SLA/LP177−196 /SLA/LP185−204 and peptide mix SLA/LP403−416/SLA/LP408−418, respectively. This strategy generated CD4+ T cell hybridomas against SLA/LP177−196 /SLA/LP185−204. No T cell hybridomas were generated for SLA/LP81−116 and SLA/LP400−422 indicating that CD4 T cells with these specificities were at low frequency or did not respond well to in vitro stimulation.
Fine-specificity of the SLA/LP-specific T cells
Stimulation of T cell hybridoma with their cognate peptide activated the β-galactosidase reporter gene and resulted in a color reaction. The immunogenic 20mer peptide SLA/LP369−388 was of specific interest (Fig. 4A, top bar), because it overlapped with the SLA/LP sequence that is recognized by antibodies of patients with AIH 4. T cell recognition increased further when three amino acids were removed from the aminoterminus and two or three amino acids were removed from the carboxyterminus (Fig. 4A). T cell recognition was abrogated, however, upon further stepwise truncation of the peptide and peptide SLA/LP373−386 NRLDRCLKAVRKER was identified as the optimal epitope (Fig. 4A, underlined sequence) and confirmed in dose titration experiments (Suppl. Fig. 1A). To analyze MHC restriction, the same T cell hybridoma was then stimulated with the optimal peptide and DRB1*0301+ spleen cells in the context of I-A/I-E antibody, HLA-DR antibody or an isotype control. Recognition of peptide SLA/LP373−386 was inhibited by HLA-DR antibodies, but not by I-A/I-E antibodies and isotype controls (Fig. 4B), indicating that it was restricted by HLA-DR.
Fig. 4. Fine-mapping of epitope SLA/LP373−386 NRLDRCLKAVRKER and SLA/LP186−197 LIQQGARVGRID.
CD4+ T cell hybrids were stimulated with 10 μg/ml indicated truncated peptides to determine the optimal epitopes (underlined) (A, C) which were shown to be HLA-DR-restricted (B, D).
A second DRB1*0301-restricted optimal epitope was mapped in a similar way within the sequence of SLA/LP184−204 (Fig. 4C). Stepwise amino- and carboxyterminal truncation identified peptide SLA/LP186−197 LIQQGARVGRID as the optimal epitope (Fig. 4A, Suppl. Fig. 1B), because deletion of either its aminoterminal lysine or its carboxyterminal aspartic acid reduced recognition. Antibody blocking experiments revealed that this epitope was also restricted by HLA-DR (Fig. 4D).
Next, we mapped the fine specificity of SLA/LP257−276-specific hybridomas (Suppl. Fig. 2A, top bar). Analysis of the two shorter overlapping peptides SLA/LP257−268 and SLA/LP262−276 clearly demonstrated that immunogenicity resided in the carboxyterminal sequence (Suppl. Fig. 2A, third bar). Removal of an arginine and lysine from the aminoterminus and a lysine from the carboxyterminal did not significantly reduce activation of the T cell hybridomas. Recognition was abolished by further removal of glutamic acid from the aminoterminus, revealing SLA/LP264−275 EMFSYLSNQIKK as the optimal epitope (Suppl. Figure 2A,B). In contrast to the two epitopes identified in Fig. 4, this epitope was not restricted by HLA-DR because anti-I-A/anti-I-E rather than anti-HLA-DR antibodies blocked its recognition. Although DR3.AB0 transgenic mice do not express endogenous class II at detectable level, they do express the murine Eb gene 14. It is therefore possible that low level expression of a DRα/I-Eb fusion molecule was responsible for the presentation of this peptide.
Finally, using the same technique, the immunogenic epitope within SLA/LP1−20 was identified as SLA/LP7−18 CFWRRFIHGIGR (not shown). The sequence of this epitope is not present in the major SLA/LP splice variant (NM_153825), but only in the less expressed splice variant derived from a Jurkat-cell cDNA library (NM_016955) 6. Since expression of the minor variant in liver is uncertain and the relevance of its recognition by liver-derived T cells thus questionable, this epitope was not further evaluated.
In summary, the use of a humanized, HLA-DRB1*0301-transgenic mouse model resulted in the identification of two HLA-DR-restricted SLA/LP epitopes that are recognized by CD4+ T cells and one of these epitopes (Fig. 4A,B) overlapped the sequence of the major B cell autoantigen that is recognized by patients with anti-SLA/LP+ AIH 4, 6.
Generation of DRB1*0301-SLA/LP373−386- and DRB1*0301-SLA/LP184−198-tetramers to detect SLA/LP-specific CD4+ T cells
Next, we investigated whether the identified HLA-DR-restricted epitopes could be used to detect SLA/LP-specific CD4+ T cells with fluorescently labeled tetrameric DRB1*0301 complexes. DRB1*0301-tetramers were synthesized for epitope SLA/LP373−386 and attempted for epitope SLA/LP186−197. Since the latter tetramer did not fold with the minimal epitope SLA/LP186−197, we chose a longer precursor peptide, SLA/LP184−198 instead.
Both, the DRB1*0301-SLA/LP373−386-tetramer and the DRB1*0301-SLA/LP184−198-tetramer were first titered and validated with CD4+ T cells of immunized HLA-DRB1*0301-transgenic mice. A large DRB1*0301-SLA/LP373−386-tetramer+ CD4+ T cell population was identified in the spleens of two representative DRB1*0301-transgenic, immunized mice, but not in a DRB1*0301-transgenic, nonimmunized control mouse (Fig. 5A). A CD4+ T cell population of similar size was detected with the second, DRB1*0301-SLA/LP184−198-tetramer (Fig. 5B). Both DRB1*0301-tetramer+ CD4+ T cell populations expressed CD11a and, when compared to DRB1*0301/epitope-tetramer− CD4+ T cells had downregulated CD62L expression indicating loss of their lymph node homing potential as expected for antigen-induced effector T cells. Serum alanine aminotransferase (ALT) levels remained within the upper limit of normal and no significant lymphocyte infiltrate was detected in the liver (not shown).
Fig. 5. Verification of the DRB1*0301-SLA/LP373−386 tetramer and DRB1*0301-SLA/LP184−198 tetramers.
DRB1*0301-SLA/LP373−386 tetramer+ (A) and DRB1*0301-SLA/LP184−198+ (B) CD4+ T cells were detectable in spleens of SLA/LP-immunized, but not nonimmunized DRB1*0301-transgenic mice. CD62L- and CD11a expression were compared on tetramer+ and tetramer− CD4+ T cells. Numbers indicate the percentage of positive cells.
Detection of SLA/LP-specific CD4+ T cells in the blood of AIH patients
As a proof of principle, we also studied PBMC from patients with anti-SLA/LP+ AIH using the DRB1*0301-SLA/LP373−386- and DRB1*0301-SLA/LP184−198-tetramers. As specificity controls, PBMC from an HLA-DRB1*0301− patient with anti-SLA/LP+ AIH and two HLA-DRB1*0301+ , anti-SLA/LP− patients with past history of hepatitis B were studied in parallel. After gating on single cells and lymphocytes, EMA+/CD8+/CD14+/CD19+ cells were excluded and the CD3+ cells were analyzed for CD4 expression and binding of the respective tetramer (Fig. 6A, B). Magnetic bead enrichment concentrated tetramer+ cells by a factor of 23 to 1042 (Table 1). Overall, tetramer+ CD4 T cells were detected in PBMC of 7/9 AIH patients after enrichment with anti-PE-labeled magnetic beads as compared to 0/3 (0%) control patients (p=0.045, Fisher's exact test, Table 1). When CFSE-labeled PBMC were stimulated with the cognate SLA/LP373−386 peptide more tetramer+ cells than tetramer− cells proliferated (47.0% versus 6.2%). In contrast, only 3.7% tetramer+ cells proliferated upon stimulation with the unrelated, DRB1*0301-restricted HCV peptide HCV261−269 15 (Fig. 6E).
Fig. 6. Detection of tetramer+ SLA/LP-specific CD4+ T cells in humans.
(A) Gating strategy. After exclusion of doublets (left graph), gates were set on lymphocytes based on forward and side scatter (second graph from left), and on CD3+ cells after exclusion of CD8+ T cells, CD16+ monocytes and CD19+ B cells (second gate from right). (B-C) Staining of CD4+ tetramer+ T cells ex vivo (B) and after enrichment with anti-PE-coupled magnetic beads (C). (D-E) PBMC were CFSE-labelled and stimulated with either the SLA/LP373−383 peptide (D) or an DRB1*0301-restricted HCV peptide (E). Proliferation of DRB1*0301-SLA/LP373−386+ and DRB1*0301-SLA/LP373−386− cells were compared. Numbers indicate the percentage of events in the respective gate or quadrant.
Table 1.
Patient Characteristics
| Patient | Age | Diagnose | Gender | ALT [U/L] |
AST [U/L] |
IgG | Treatment | Tetramer+ Cells1/CD4+T Cells [%] | Tetramer+ Cells2/CD4+T Cells [%] | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ex vivo | Enriched3 | Factor4 | Ex vivo | Enriched3 | Factor4 | ||||||||
| Patients with anti-SLA+ autoimmune hepatitis | |||||||||||||
| AIH 1 | 23 | anti-SLA/LP+, anti-LKM1+ | f | 18 | 16 | 11.5 | tacrolimus, mycophenolate | 0.01% | 0.40% | 51 | 0.00% | 0.26% | 65 |
| AIH 2 | 56 | anti-SLA/LP+ | f | 23 | 19 | 17.9 | prednisolone | 0.00% | 0.12% | n.a. 5 | n.d. 6 | ||
| AIH 3 | 77 | anti-SLA/LP+ | f | 72 | 32 | 20.3 | prednisone | 0.00% | 0.05% | 53 | 0.01% | 0.16% | 23 |
| AIH 4 | 58 | anti-SLA/LP+ | f | 65 | 41 | 11.7 | prednisone | 0.01% | 1.32% | 152 | 0.01% | 0.27% | 26 |
| AIH 5 | 25 | anti-SLA/LP+ | f | 18 | 24 | 15.1 | azathioprine, prednisone | 0.00% | 1.21% | 1042 | 0.00% | 1.39% | 332 |
| AIH 6 | 63 | anti-SLA/LP+, anti-SMA+ | f | 53 | 38 | 22.3 | none | 0.00% | 0.00% | 0 | 0.00% | 0.00% | 0 |
| AIH 7 | 35 | anti-SLA/LP+, anti-ANA+ | f | 61 | 48 | n.a. | azathioprine, prednisone | 0.00% | 0.55% | 199 | 0.00% | 0.00% | 0 |
| AIH 8 | 45 | anti-SLA/LP+ | f | 20 | 25 | n.a. | azathioprine, budenoside | 0.01% | 0.00% | 0 | 0.00% | 0.00% | 0 |
| AIH 9 | 46 | anti-SLA/LP+ | f | 18 | 22 | n.a. | azathioprine | 0.02% | 0.17%7 | 9 | n.d. 6 | ||
| Controls | |||||||||||||
| Control 1 | 65 | anti-SLA/LP-neg., HBV+, HLA-DR3+ | f | 10 | 17 | n.a. | none | 0.00% | 0.00% | 0 | 0.00% | 0.00% | 0 |
| Control 2 | 62 | anti-SLA/LP-neg., HBV+, HLA-DR3+ | m | 17 | 18 | n.a. | none | 0.00% | 0.00% | 0 | 0.00% | 0.00% | 0 |
| Control 3 | 50 | anti-SLA/LP+, not HLA-DR3+ | m | 40 | 78 | n.a. | none | 0.00% | 0.00% | 0 | 0.00% | 0.00% | 0 |
Refers to the DRB1*0301/SLA/LP 184−196 Tetramer.
Refers to the DRB1*0301/SLA/LP 373−383 Tetramer.
Enrichment was performed with magnetic beads unless otherwise indicated. Positive values are shown in bold and underlined.
Frequency of tetramer+ cells after enrichment divided by ex vivo frequency.
N.a., not applicable.
N.d., not done.
Peptide-stimulated T cell line.
Finally, the newly identified CD4 T cell epitopes were also tested in IFN-γ ELISpot assays. As shown in Fig. 7, tetramer-enriched CD4+ T cells of HLA-DR1B1*0301-positive, recognized both epitopes and produced IFN-γ, but tetramer-enriched CD4+ T cells of HLA-DR1B1*0301-negative AIH patients or DR1B1*0301-positive controls without AIH did not. These cells were tested ex vivo and after several weeks of antigen-nonspecific in vitro expansion with 40 ng/ml anti-CD3, 200 U/ml IL-2, 10 ng/ml IL-7 and 100 ng/ml IL-15. The rapid, antigen-nonspecific expansion protocol was chosen over a peptide-specific expansion protocol because of the very low number (< 105) of tetramer-positive cells with which the expansion was started. ELISpot assays with the nonspecifically expanded T cell lines (filled symbols in panel B) confirmed the ex vivo ELISpot results (open symbols in panel B).
Fig. 7. Detection of IFN-γ+ SLA/LP-specific CD4+ T cells in humans.
(A, B) ELISpot images of tetramer-enriched CD4+ T cells of AIH patients. (B) IFN-γ producing, SLA-LP-specific cells are detected ex vivo (open symbols) or after antigen-nonspecific in vitro expansion (filled symbols) in tetramer-enriched CD4 T cell populations of HLA-DRB1*0301-positive patients with anti-SLA/LP positive AIH but not HLA-DRB1*0301-negative patients with anti-SLA/LP positive AIH and not HLA-DRB1*0301-positive patients without anti-SLA/LP positive AIH. Horizontal lines indicate the mean. Please note that the number of IFN-γ producing cells in this panel is calculated for 106 cells but that the number of cells in the Elispot wells in panel A was lower.
Discussion
We here report that SLA/LP is not only targeted by humoral but also by cellular immune responses and describe two CD4+ T cell epitopes that are recognized in a DRB1*0301-restricted manner. CD4+ T cells specific for these SLA/LP epitopes were detected in HLA-DRB1*0301-transgenic mice and in PBMC of HLA-DRB1*0301-positive patients with AIH. Only HLA-DRB1*0301-transgenic, but not nontransgenic mice mounted SLA/LP-specific autoantibodies of the same specificity as in AIH in humans, which suggests that HLA-DRB1*0301-restricted CD4+ T cells shape the antibody repertoire in the context of the genetic background. Interestingly, one of the HLA-DR-restricted CD4+ T cell epitopes (Fig. 4A,B) was located in the carboxyterminal sequence of SLA/LP overlapping with the sequence that is recognized by antibodies 4, 6. This close proximity of T and B cell epitopes within the SLA/LP autoantigen appears to be a characteristic of autoimmune diseases 12, 20, 21. HLA-DRB1*0301-restricted SLA/LP-specific CD4+ T cells may for example help autoreactive B cells to produce autoantibodies and to secrete proinflammatory cytokines and chemokines that recruit plasma cells and CD8+ T cells into the liver. Vice versa, autoantigen-specific B cells may bind and internalize SLA/LP via specific autoantibodies on their cell surface, process it into short peptides and present these peptides on DRB1*0301 molecules to stimulate CD4+ T cells 22.
The absence of liver disease in the SLA/LP-immunized HLA-DRB1*0301-transgenic mice was not unexpected. Recent studies on autoimmune diabetes suggest that the frequency of autoantigen-specific T cells needs to be much higher (>80% of all CD8+ T cells in that study) and that additional inflammatory cytokine- and TLR-mediated signals are required to initiate autoimmune disease 23. Furthermore, all published T cell-mediated autoimmune models in HLA-transgenic mice involve organs other than the liver 24, 25, which is characterized by a constitutive tolerogenic microenvironment 26. Thus, additional factors, such as tissue destruction by unrelated mechanisms (toxic or infectious) might be necessary to induce intrahepatic inflammation and to trigger AIH. Since SLA/LP 6 and LKM, the second cloned autoantigen in AIH, are cytosolic enzymes 27, it is possible that specific enzyme-substrate interactions initiate hepatocyte injury and death 28, followed by transport of cell-associated autoantigens into the regional lymph nodes and priming of autoantigen-specific T and B cells.
In the analyzed patients with anti-SLA/LP+ AIH, the frequency of autoantigen-specific, CD4+ T cells detectable by Elispot and tetramer analysis was in the same range as frequencies reported for other autoimmune diseases such as type I diabetes 29 and chronic infectious diseases such as mycobacterium tuberculosis 30, borrelia burgdorferi 31 and HCV infection 32, 33. Upon immunosuppressive treatment, as it was the case for the patients in our study, the frequency of autoreactive T cells may decrease further. In this scenario, enrichment of tetramer+ T cells with either magnetic beads 32, 33 may increase the sensitivity of detection and allow their functional characterization.
The tetramer-staining profiles displayed arelatively low mean fluorescence intensity. Similar profiles have been reported for other MHC class II tetramers in the literature 30. In this context, it has recently been shown that CD4+ T cells in blood, spleen and lymph node differ in their MHC class II tetramer binding profiles with tetramer+ T cells of lymph nodes and spleen exhibiting higher mean fluorescence intensities than tetramer+ T cells in the blood 34. The identified SLA/LP-specific T cell epitopes and their corresponding tetramers should therefore be useful to monitor AIH-specific T cells during acute AIH and active disease progression and to study their role in the liver, the site of this disease.
Supplementary Material
Acknowledgement
We thank Dr. Masaaki Shiina for help with multi-color flow cytometry, Dr. Martine Wettendorff, GlaxoSmithKline Biologicals, Rixensart, Belgium for adjuvant AS01B, Dr. Nilab Shastri (University of California, Berkeley, CA) for cell lines and Gary Norman (Inova Diagnostics, San Diego, CA) for SLA/LP EIA kits. This study was supported by the Intramural Research Program of NIDDK, NIH.
This study was supported by the intramural research program of NIDDK, NIH. H.M. was supported by grant MI 677/1-1, G.A. by grant AH 173/1-1 from the Deutsche Forschungsgemeinschaft, Bonn, Germany. J. H. and A. W. L. were supported by SFB 548 from the Deutsche Forschungsgemeinschaft, Bonn, Germany. The authors have no conflict of interest to disclose.
Abbreviations used in this paper
- AIH
autoimmune hepatitis
- ELISpot
enzyme-linked immunospot
- SLA/LP
soluble liver antigen/liver pancreas
References
- 1.Milkiewicz P, Hubscher SG, Skiba G, Hathaway M, Elias E. Recurrence of autoimmune hepatitis after liver transplantation. Transplantation. 1999;68:253–6. doi: 10.1097/00007890-199907270-00016. [DOI] [PubMed] [Google Scholar]
- 2.McFarlane IG. Definition and classification of autoimmune hepatitis. Semin Liver Dis. 2002;22:317–24. doi: 10.1055/s-2002-35702. [DOI] [PubMed] [Google Scholar]
- 3.Czaja AJ. Autoimmune hepatitis--approach to diagnosis. MedGenMed. 2006;8:55. [PMC free article] [PubMed] [Google Scholar]
- 4.Herkel J, Heidrich B, Nieraad N, Wies I, Rother M, Lohse AW. Fine specificity of autoantibodies to soluble liver antigen and liver/pancreas. Hepatology. 2002;35:403–8. doi: 10.1053/jhep.2002.30699. [DOI] [PubMed] [Google Scholar]
- 5.Manns M, Gerken G, Kyriatsoulis A, Staritz M, Meyer zum Buschenfelde KH. Characterisation of a new subgroup of autoimmune chronic active hepatitis by autoantibodies against a soluble liver antigen. Lancet. 1987;1:292–4. doi: 10.1016/s0140-6736(87)92024-1. [DOI] [PubMed] [Google Scholar]
- 6.Wies I, Brunner S, Henninger J, Herkel J, Kanzler S, Meyer zum Buschenfelde KH, Lohse AW. Identification of target antigen for SLA/LP autoantibodies in autoimmune hepatitis. Lancet. 2000;355:1510–5. doi: 10.1016/s0140-6736(00)02166-8. [DOI] [PubMed] [Google Scholar]
- 7.Yamauchi K, Yamaguchi N, Furukawa T, Takatsu K, Nakanishi T, Ishida K, Komatsu T, Tokushige K, Nagahara H, Hashimoto E, Shiratori K. A murine model of acute liver injury induced by human monoclonal autoantibody. Hepatology. 2005;42:149–55. doi: 10.1002/hep.20726. [DOI] [PubMed] [Google Scholar]
- 8.Donaldson PT, Doherty DG, Hayllar KM, McFarlane IG, Johnson PJ, Williams R. Susceptibility to autoimmune chronic active hepatitis: human leukocyte antigens DR4 and A1-B8-DR3 are independent risk factors. Hepatology. 1991;13:701–6. [PubMed] [Google Scholar]
- 9.Czaja AJ, Strettell MD, Thomson LJ, Santrach PJ, Moore SB, Donaldson PT, Williams R. Associations between alleles of the major histocompatibility complex and type 1 autoimmune hepatitis. Hepatology. 1997;25:317–23. doi: 10.1002/hep.510250211. [DOI] [PubMed] [Google Scholar]
- 10.Czaja AJ, Donaldson PT. Genetic susceptibilities for immune expression and liver cell injury in autoimmune hepatitis. Immunol Rev. 2000;174:250–9. doi: 10.1034/j.1600-0528.2002.017401.x. [DOI] [PubMed] [Google Scholar]
- 11.Czaja AJ, Manns MP, McFarlane IG, Hoofnagle JH. Autoimmune hepatitis: the investigational and clinical challenges. Hepatology. 2000;31:1194–200. doi: 10.1053/he.2000.5980. [DOI] [PubMed] [Google Scholar]
- 12.Reijonen H, Mallone R, Heninger AK, Laughlin EM, Kochik SA, Falk B, Kwok WW, Greenbaum C, Nepom GT. GAD65-specific CD4+ T-cells with high antigen avidity are prevalent in peripheral blood of patients with type 1 diabetes. Diabetes. 2004;53:1987–94. doi: 10.2337/diabetes.53.8.1987. [DOI] [PubMed] [Google Scholar]
- 13.Lohr HF, Schlaak JF, Lohse AW, Bocher WO, Arenz M, Gerken G, Meyer Zum Buschenfelde KH. Autoreactive CD4+ LKM-specific and anticlonotypic T-cell responses in LKM-1 antibody-positive autoimmune hepatitis. Hepatology. 1996;24:1416–21. doi: 10.1002/hep.510240619. [DOI] [PubMed] [Google Scholar]
- 14.Kong YC, Lomo LC, Motte RW, Giraldo AA, Baisch J, Strauss G, Hammerling GJ, David CS. HLA-DRB1 polymorphism determines susceptibility to autoimmune thyroiditis in transgenic mice: definitive association with HLA-DRB1*0301 (DR3) gene. J Exp Med. 1996;184:1167–72. doi: 10.1084/jem.184.3.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diepolder HM, Gerlach J- T, Zachoval R, Hoffmann RM, Jung MC, Wierenga EA, Scholz S, Santantonio T, Houghton M, Southwood S, Sette A, Pape GR. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J. Virol. 1997;71:6011–6019. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandepapeliere P, Rehermann B, Koutsoukos M, Moris P, Garcon N, Wettendorff M, Leroux-Roels G. Potent enhancement of cellular and humoral immune responses against recombinant hepatitis B antigens using AS02A adjuvant in healthy adults. Vaccine. 2005;23:2591–601. doi: 10.1016/j.vaccine.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 17.Wedemeyer H, Gagneten S, Davis A, Bartenschlager R, Feinstone S, Rehermann B. Oral immunization with HCV-NS3-transformed salmonella: induction of HCV-specific CTL in a transgenic mouse model. Gastroenterology. 2001;121:1158–66. doi: 10.1053/gast.2001.29311. [DOI] [PubMed] [Google Scholar]
- 18.Malarkannan S, Mendoza LM, Shastri N. Generation of antigen-specific, lacZ-inducible T-cell hybrids. Methods Mol Biol. 2001;156:265–72. doi: 10.1385/1-59259-062-4:265. [DOI] [PubMed] [Google Scholar]
- 19.Manigold T, Shin EC, Mizukoshi E, Mihalik K, Murthy KK, Rice CM, Piccirillo CA, Rehermann B. Foxp3+CD4+CD25+ T cells control virus-specific memory T cells in chimpanzees recovered from Hepatitis C. Blood. 2006;107:4424–4432. doi: 10.1182/blood-2005-09-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galperin C, Gershwin ME. Immunopathology of primary biliary cirrhosis. Baillieres Clin Gastroenterol. 1996;10:461–81. doi: 10.1016/s0950-3528(96)90053-6. [DOI] [PubMed] [Google Scholar]
- 21.Wucherpfennig KW, Catz I, Hausmann S, Strominger JL, Steinman L, Warren KG. Recognition of the immunodominant myelin basic protein peptide by autoantibodies and HLA-DR2-restricted T cell clones from multiple sclerosis patients. Identity of key contact residues in the B-cell and T-cell epitopes. J Clin Invest. 1997;100:1114–22. doi: 10.1172/JCI119622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mamula MJ. Epitope spreading: the role of self peptides and autoantigen processing by B lymphocytes. Immunol Rev. 1998;164:231–9. doi: 10.1111/j.1600-065x.1998.tb01223.x. [DOI] [PubMed] [Google Scholar]
- 23.Lang KS, Recher M, Junt T, Navarini AA, Harris NL, Freigang S, Odermatt B, Conrad C, Ittner LM, Bauer S, Luther SA, Uematsu S, Akira S, Hengartner H, Zinkernagel RM. Toll-like receptor engagement converts T-cell autoreactivity into overt autoimmune disease. Nat Med. 2005;11:138–45. doi: 10.1038/nm1176. [DOI] [PubMed] [Google Scholar]
- 24.Nabozny GH, Baisch JM, Cheng S, Cosgrove D, Griffiths MM, Luthra HS, David CS. HLA-DQ8 transgenic mice are highly susceptible to collagen-induced arthritis: a novel model for human polyarthritis. J Exp Med. 1996;183:27–37. doi: 10.1084/jem.183.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito K, Bian HJ, Molina M, Han J, Magram J, Saar E, Belunis C, Bolin DR, Arceo R, Campbell R, Falcioni F, Vidovic D, Hammer J, Nagy ZA. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. J Exp Med. 1996;183:2635–44. doi: 10.1084/jem.183.6.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crispe IN. Hepatic T cells and liver tolerance. Nat Rev Immunol. 2003;3:51–62. doi: 10.1038/nri981. [DOI] [PubMed] [Google Scholar]
- 27.Manns MP, Johnson EF, Griffin KJ, Tan EM, Sullivan KF. Major antigen of liver kidney microsomal autoantibodies in idiopathic autoimmune hepatitis is cytochrome P450db1. J Clin Invest. 1989;83:1066–72. doi: 10.1172/JCI113949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Lu Y, Cederbaum AI. Induction of cytochrome P450 2E1 increases hepatotoxicity caused by Fas agonistic Jo2 antibody in mice. Hepatology. 2005;42:400–10. doi: 10.1002/hep.20792. [DOI] [PubMed] [Google Scholar]
- 29.Oling V, Marttila J, Ilonen J, Kwok WW, Nepom G, Knip M, Simell O, Reijonen H. GAD65- and proinsulin-specific CD4+ T-cells detected by MHC class II tetramers in peripheral blood of type 1 diabetes patients and at-risk subjects. J Autoimmun. 2005;25:235–43. doi: 10.1016/j.jaut.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 30.Hohn H, Kortsik C, Zehbe I, Hitzler WE, Kayser K, Freitag K, Neukirch C, Andersen P, Doherty TM, Maeurer M. MHC class II tetramer guided detection of Mycobacterium tuberculosis-specific CD4+ T cells in peripheral blood from patients with pulmonary tuberculosis. Scand J Immunol. 2007;65:467–78. doi: 10.1111/j.1365-3083.2007.01924.x. [DOI] [PubMed] [Google Scholar]
- 31.Meyer AL, Trollmo C, Crawford F, Marrack P, Steere AC, Huber BT, Kappler J, Hafler DA. Direct enumeration of Borrelia-reactive CD4 T cells ex vivo by using MHC class II tetramers. Proc Natl Acad Sci U S A. 2000;97:11433–8. doi: 10.1073/pnas.190335897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Day CL, Seth NP, Lucas M, Appel H, Gauthier L, Lauer GM, Robbins GK, Szczepiorkowski ZM, Casson DR, Chung RT, Bell S, Harcourt G, Walker BD, Klenerman P, Wucherpfennig KW. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest. 2003;112:831–42. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulsenheimer A, Lucas M, Seth NP, Tilman Gerlach J, Gruener NH, Loughry A, Pape GR, Wucherpfennig KW, Diepolder HM, Klenerman P. Transient immunological control during acute hepatitis C virus infection: ex vivo analysis of helper T-cell responses. J Viral Hepat. 2006;13:708–14. doi: 10.1111/j.1365-2893.2006.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fazilleau N, Eisenbraun MD, Malherbe L, Ebright JN, Pogue-Caley RR, McHeyzer-Williams LJ, McHeyzer-Williams MG. Lymphoid reservoirs of antigen-specific memory T helper cells. Nat Immunol. 2007;8:753–61. doi: 10.1038/ni1472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.