Abstract
Context: Kruppel-like factor 15 (KLF15) is a newly discovered transcription factor that plays an important role in glucose homeostasis and lipid accumulation in cells. We present evidence for KLF15 as a transcriptional regulator of the human 17β-hydroxysteroid dehydrogenase type 5 gene (HSD17B5) and its potential role in the pathogenesis of hyperandrogenism.
Objective: The aim was to investigate the molecular mechanism of HSD17B5 regulation.
Methods: Diverse molecular biology techniques were used.
Design and Results: We identified a KLF15 binding site in the HSD17B5 promoter by using luciferase promoter constructs, EMSA, and chromatin immunoprecipitation assays. Overexpression of KLF15 increased HSD17B5 promoter activity and testosterone formation at least 3-fold in cultured H295R cells. Insulin increased KLF15 mRNA expression according to real-time RT-PCR and increased HSD17B5 promoter activity according to luciferase assays. KLF15 overexpression in combination with insulin, glucocorticoid, and cAMP stimulated adipogenesis in H295R cells. In silico and RT-PCR analyses showed that the KLF15 gene promoter undergoes alternative splicing in a tissue-specific manner. Comparison of the HSD17B5 promoter in seven different species revealed that the KLF15 binding site has no human homolog in species other than orangutans.
Conclusions: KLF15 is potentially a novel link between the regulation of testosterone production and fat stores by insulin in humans.
KLF15, a recently discovered multifunctional transcription factor, is a potential novel link between the regulation of human female testosterone production and fat stores by insulin.
Polycystic ovary syndrome (PCOS), a poorly understood chronic hyperandrogenism disorder, is the most common cause of anovulatory infertility, affecting about 5% of reproductive-age women (1,2,3). It is associated with a metabolic syndrome of central obesity, hyperglycemia, dyslipidemia, and hypertension (3), and is a risk factor for type 2 diabetes mellitus and cardiovascular diseases (4). Dysregulation of steroidogenesis seems to be responsible for PCOS. However, the molecular relationships among PCOS, insulin resistance (IR), and obesity are unclear.
Testosterone (T) is the major circulating androgen in all hyperandrogenic disorders (5), and its biosynthesis from androstenedione (AD) requires androgenic 17β-hydroxysteroid dehydrogenase (17β-HSD) activity. T arises by direct secretion from the ovaries and adrenal glands and by conversion from circulating AD in many peripheral tissues. 17β-HSD type 5 appears to account for this in human females (6,7). The 17β-HSD type 5 gene (HSD17B5) is expressed in liver, fat, skin, sebocytes, and hair follicles, as well as other tissues (8,9,10,11,12,13,14,15). This expression pattern suggests that HSD17B5 is differentially regulated through endocrine, paracrine, and/or autocrine mechanisms by both common and tissue-specific transcription factors.
We previously reported that a promoter binding site (CCTCCTCCT) for the ubiquitous transcription factors stimulatory protein 1/3 (Sp1/Sp3) is necessary for human HSD17B5 regulation (16), and a single nucleotide polymorphism in this region is associated with a subtype of PCOS (17). Although this single nucleotide polymorphism has not proven to be associated with PCOS in general (18,19), it is associated with the severity of hyperandrogenemia (19).
A G-rich element (GGGGTGGGGGGGAGGGG) in the human HSD17B5 promoter is equally necessary, but its cognate transcription factor has been unknown (16). In the present study, we found that Kruppel-like factor 15 (KLF15) binds to the G-rich element and, along with Sp1/Sp3, up-regulates the promoter activity. KLF15 is a newly discovered transcription factor that belongs to the Sp1-like/KLF family. KLF15 is a multifunctional transcriptional regulator that is highly expressed in the liver, kidneys, heart, skin, fat, skeletal muscle, brain, and bone (20,21,22,23).
A targeted hepatic deletion of KLF15−/− in mice increased hepatic insulin sensitivity and caused severe hypoglycemia after an overnight fast (24). The main mechanism of this effect was reduced hepatic glucose production due to decreased gluconeogenic substrate availability (24). Thus, KLF15 is the first member of the KLF family shown to be important for glucose homeostasis in vivo.
In vitro studies also indicate that KLF15 plays an important role in the transcriptional regulation of genes during adipogenesis, potently inducing lipid accumulation independently from the peroxisome proliferator activated receptor γ 2 pathway (25). KLF15 directly up-regulates some glucose transport and metabolic genes, such as glucose transporter 4 (GLUT4), acetyl-CoA synthetase 2 (AceCS2), and phosphoenolpyruvate carboxykinase (PEPCK) (21,26).
Insulin together with dexamethasone (Dex) and a cAMP generator (a standard adipogenic induction cocktail) stimulates KLF15 mRNA expression as it induces preadipocyte differentiation to adipocytes (25). However, in hepatocytes, insulin inhibits KLF15 mRNA expression and blocks the stimulating effects of Dex and cAMP on KLF15 mRNA expression (27). These data strongly suggest that insulin differentially regulates KLF15 expression in tissue-specific fashion, up-regulating fat and muscle GLUT4 and AceCS2 gene expression and attenuating hepatic PEPCK gene expression to maintain glucose homeostasis.
Therefore, we propose that KLF15 is a potential novel link between the regulation of female T production and fat stores in humans and that it possibly plays a central role in the pathogenesis of PCOS.
Materials and Methods
Cell culture, nuclear protein extraction, and EMSA
Human adrenal carcinoma cells (H295R) were cultured, nuclear extracts were prepared, and EMSA was performed (16,17). Human and rat primary theca cells (6,16), human ovarian theca-like tumor cells (28), and human sebocytes (SZ95) (29) were also cultured.
Luciferase constructs and promoter analysis of human HSD17B5 promoter
Human HSD17B5 promoter (−82 bp to +68 bp) and its mutant constructs were generated; transfection and luciferase assays were performed (16,17).
Generation of mammalian expression plasmids
We used specific human KLF15 primers to amplify full-length KLF15 coding sequences from H295R cell total RNA. The products were subcloned into pGEM-T easy plasmid. To create KLF15 expression plasmids, the KLF15 was digested from the pGEM-T-KLF15 and subcloned into pIRES2-EGFP and pCMV-Myc vectors (Clontech, Palo Alto, CA). The constructs were designated as pIRES2-EGFP-KLF15 and pCMV-Myc-KLF15. The pCMV-Myc-KLF15 produces a full-length KLF15 bearing an N-terminal Myc tag for EMSA studies. A dominant-negative mutant of KLF15 was generated by cloning truncated KLF15(Δ318) PCR fragment into pIRES2-EGFP vector; the pIRES2-EGFP-KLF15(Δ318) lacks the NH2-terminal 318 amino acids of the transactivation domain, leaving the DNA binding domain intact (25).
Chromatin immunoprecipitation (ChIP) assays
Full-length KLF15 was cloned into a HaloTag vector, and ChIP assays were performed according to manufacturer’s recommendations (Promega, Madison, WI). Control samples were incubated with blocking ligands that bind to the HaloTag protein and prevent interaction with the HaloLink Resin. The purified DNAs were analyzed by PCR using HSD17B5 specific primers (5′-CATTGGTTAACCATCAGTCAG-3′ and 5′-TTCCCTGTCACTTGTCTGACT-3′). The primers are designed to amplify120 bp of the gene promoter region containing only the KLF15 binding site.
Stable transfection cell line
H295R cells stably expressing KLF15 were generated by transfection with pIRES2-EGFP-KLF15. Cells expressing KLF15 were selected on the basis of their expression of EGFP and resistance to G418 (500 μg/ml).
Testosterone formation by cultured cells
H295R cells with or without KLF15 overexpression were grown until subconfluent and then transferred to serum free medium (SFM) with or without 200 ng/ml AD. After 48 h, the media were collected and analyzed for T concentration by RIA (Coat-A-Count; Siemens, Los Angeles, CA).
Real-time RT-PCR
For insulin effects, subconfluent H295R cells were incubated for 24 h in SFM, followed by treatment with or without insulin (5 μg/ml) for 4 h. Total RNA was isolated, and real-time RT-PCR was performed using ribosomal 18S RNA as a reference marker (16) and human KLF15-specific primers (5′-ATGCACAAATGTACTTTCCCT-3′ and 5′-TCAGTTCACGGAGCGCACGGA-3′).
Phylogenetic comparisons
The 500-bp 5′-flanking sequences of hsd17b5 from multiple species were retrieved from the GenBank database (http://www.ncbi.nlm.nih.gov/nuccore) and the Ensembl genome browser (http://www.ensembl.org). Each species’ 5′-flanking sequence was compared with human HSD17B5 promoter sequence (http://www.ncbi.nlm.nih.gov/nuccore) and aligned manually. The accuracy of the monkey hsd17b5 5′-flanking sequence was verified by direct PCR sequence of monkey genomic DNA (data not shown).
Identification of KLF15 genomic organization from expressed sequence tag (EST) databases
Human KLF15 mRNA (GenBank accession no. NM_014079) was used to search the human EST database at the National Center for Biotechnology Information (NCBI) using a BLAST search engine. The ESTs were sorted into 5′ and 3′ sequences, and each was aligned against the KLF15 genomic sequences (GenBank accession nos. NW_921807 and NT_005612), which map to chromosome 3q13-q21. Intron-exon boundaries were identified from the EST maps.
Determination of alternatively spliced products
H295R and human theca cell total RNAs were prepared (6,16), and human tissue RNAs were purchased from Clontech. We designed primers complementary to human KLF15 gene (primer sequences are available upon request) to determine exon 1C, exon 1A, and exon 2 expression. Agarose gel electrophoresis and sequencing were used to analyze and confirm the amplified cDNA products.
Oil red O staining
Adipogenesis in H295R cells stably expressing KLF15 was induced by treatment for 6 d with a standard adipogenic induction cocktail, consisting of 5 μg/ml insulin, 0.25 μm Dex, and 0.5 mm isobutyl methylxanthine. The cells were then returned to the culture medium, which was changed every other day. The cells were stained with oil red O on the 10th day (30,31).
Statistical analysis
Differences between experimental groups were compared by Student’s t test or ANOVA when multiple comparisons were made. Values are expressed as mean ± sd for three independent experiments.
Results
Human HSD17B5 promoter G-rich sequence is a cis-active element
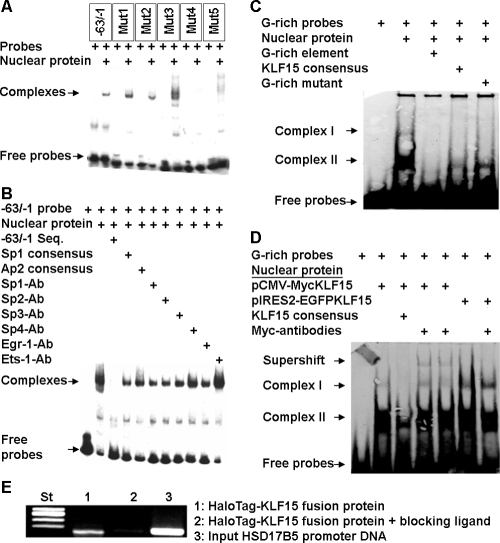
Our previous study suggested that a cis-active element other than the Sp1/Sp3 binding site (−66 bp and −58 bp) also contributes to transcriptional activity of this gene (16). Basal HSD17B5 promoter activity is similar in human and rat primary theca cells, human ovarian theca-like tumor cells, human sebocytes, and H295R cells (data not shown). Therefore, we performed EMSA with a series of 3-bp mutations between −63 bp and −1 bp. Figure 1A demonstrated that the GGGGTGGGGGGGAGGGG sequence at −24 bp and −9 bp is the core sequence of this cis-active element that is involved in HSD17B5 promoter regulation. Figure 1B suggests that Sp1, Sp2, Sp4, AP2 (activator protein 2), Egr-1 (early growth response protein 1), and Ets-1 (v-ets erythroblastosis virus E26 oncogene homolog 1) are not the major transcription factors that participate in the formation of protein-DNA complexes of this G-rich element. Subsequently, we also showed that Myc-associated zinc finger protein and two (BTEB2/KLF5) transcription factors do not bind to this G-rich element either (data not shown). The results were further confirmed using the −32/−1 fragment (data not shown). Therefore, we have ruled out the binding of multiple classic transcription factors to the G-rich element.
Figure 1.
HSD17B5 proximal promoter G-rich sequence is a cis-active element that binds the transcription factor KLF15. A, Double-stranded, 32P end-labeled probes corresponding to the wild-type −63/−1 fragment, mut1, mut2, mut3, mut4, and mut5 were incubated with H295R cell nuclear extract. The wild-type −63/−1 fragment, mut1, and mut2 probes form one major complex, whereas mut3 and mut5 probes form more than one complex. Mut4 does not form any complexes. The study defined the core sequences of binding site as GGGGTGGGGGGAGGGG (G-rich element). B, The DNA-protein complexes were not abolished by Sp1 and AP2 consensus sequences or abolished or supershifted by antibodies to Sp1, Sp2, Sp3, Sp4, Erg-1, or Ets-1; this indicates that this G-rich region directly binds different factors. C and D, Double-stranded, biotin-labeled G-rich element probe was incubated with H295R nuclear extracts (C) and nuclear extracts from overexpression of KLF15 (pCMV-Myc-KLF15 and pIRES2-EGFP-KLF15 constructs) (D), with or without excess unlabeled KLF15 consensus sequence (50-fold molar excess). Two major complexes (complex I and complex II) are seen in the absence of competitor; these complexes are displaced by excess unlabeled G-rich sequence and KLF15 consensus sequence (C and D). The supershifted bands are only seen in pCMV-Myc-KLF15 nuclear extracts treated with Myc antibodies (D, lanes 4 and 5), but not in other lanes of pCMV-Myc-KLF15 nuclear extracts or pIRES2-EGFP KLF15 nuclear extracts. E, ChIP assay was performed using H295R cells overexpressing HaloTag-KLF15 fusion protein according to the manufacturer’s instruction. KLF15 remains bound to the G-rich element of HSD17B5 promoter.
HSD17B5 G-rich element binds the transcription factor KLF15
We investigated the possibility that the complex involving the G-rich element of the HSD17B5 promoter was formed with transcription factor KLF15, which is capable of binding to the AGCCGGGGAGGGGGAGGGGAGGGTGTTG sequence (20). The G-rich element probe was incubated with H295R cell nuclear extracts, with or without excess unlabeled competitor, and submitted to EMSA. The two protein complexes with the G-rich element were abolished by excess unlabeled self-competitor, indicating that the bands were specific (Fig. 1C). A KLF15 consensus sequence and mutant G-rich element reduced the formation of the complex greatly and partially, respectively (Fig. 1C). The data strongly suggest that KLF15 is involved in the formation of the protein-DNA complexes. Next, we extracted nuclear proteins from pCMV-Myc-KLF15 and pIRES2-EGFP-KLF15 transfected cells and performed EMSA. We were indeed able to show that Myc antibodies generated a supershifted complex with nuclear extracts from transfected pCMV-Myc-KLF15 cells, but not with nuclear extracts from pIRES2-EGFP-KLF15 or without adding Myc antibodies (Fig. 1D). We have therefore demonstrated that KLF15 binds to the G-rich element in the HSD17B5 promoter.
KLF15 binds to the G-rich region of HSD17B5 in H295R cells
To investigate KLF15 binding to the HSD17B5 promoter in vivo, we performed ChIP in H295R cells using a HaloCHIP system. We demonstrated that the KLF15 binding site was clearly recruited by HaloTag-KLF15 fusion protein, and this recruitment was markedly decreased by the blocking ligands (Fig. 1E). Therefore, we were able to confirm that KLF15 binds to the G-rich element in the HSD17B5 promoter in vivo.
Binding sites of KLF15 and Sp1/Sp3 are essential for basal and insulin-mediated HSD17B5 promoter activities
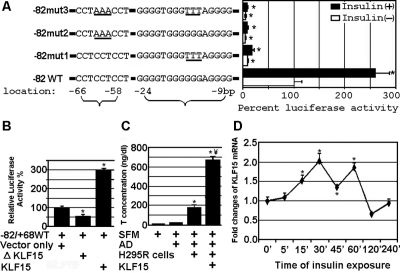
Our data demonstrated that the region between −82 bp and −1 bp contains two elements, a KLF15 binding site and a Sp1/Sp3 binding site. To address the roles of these two elements in gene expression, we individually and combinatorially mutated the two sites within the −82luc reporter constructs. Mutations of 3 bp were created in constructs −82mut1 and −82mut2, between −63 and −61 bp within the Sp1/Sp3 binding site, and between −16 and −14 bp within the KLF15 binding site, respectively. Construct −82mut3 contained double mutant elements. The constructs −82mut1 and −82mut2 and the combinatory mutant construct −82mut3 had significantly reduced luciferase activities both basally and in response to insulin compared with the wild-type construct −82luc (Fig. 2A). These results suggest that both intact KLF15 and Sp1/Sp3 cis-acting elements are required for a high level of basal and insulin-mediated HSD17B5 expression, but neither the individual nor the combined mutations totally abolished the basal or insulin-mediated promoter activities.
Figure 2.
The Sp1/Sp3 and KLF15 elements are necessary for insulin mediation of the HSD17B5 proximal promoter activity. A, Insulin significantly stimulates the HSD17B5 promoter activity in the presence of intact Sp1/Sp3 and KLF15 binding sites. *, P < 0.001 vs. untreated wild-type construct −82luc. B, Expression of a KLF15 dominant-negative mutant decreases HSD17B5 promoter activity by 45%, and overexpression of KLF15 increases HSD17B5 promoter activity 300%. *, P < 0.05 vs. vector transfected cells. C, Overexpression of KLF15 increases T formation from AD 350%. *, P < 0.001 vs. AD; ¥, < 0.01 vs. AD plus H295R cells, and T was undetectable in SFM. D, Effects of insulin on KLF15 mRNA levels in H295R cells. H295R cells were cultured in SFM for 24 h, then treated with insulin for the indicated times, after which KLF15 mRNA abundance was determined by real time RT-PCR. *, P < 0.001 vs. wells not treated with insulin at each time point.
KLF15 regulates human HSD17B5 promoter activity and testosterone formation
To determine whether KLF15 is required for HSD17B5 promoter activity, we constructed an expression vector for a dominant negative KLF15 mutant (Δ318) (25). H295R cells were cotransfected with −82luc constructs and expression plasmids for full-length KLF15 or for the Δ318 mutant. The activity of the HSD17B5 promoter was reduced to 45% by expression of the Δ318 mutant and was increased 300% by overexpression of wild-type KLF15 (Fig. 2B). The conversion of AD to T was also increased 350% by overexpression of wild-type KLF15 (Fig. 2C).
Insulin induces KLF15 mRNA expression in H295R cells
Insulin has been reported to stimulate KLF15 mRNA expression during preadipocyte differentiation (25) but to inhibit KLF15 mRNA expression in hepatocytes (27). We measured KLF15 mRNA levels in H295R cells with or without insulin treatment. Real-time PCR analysis revealed that insulin stimulates KLF15 mRNA expression in H295R cells (Fig. 2D).
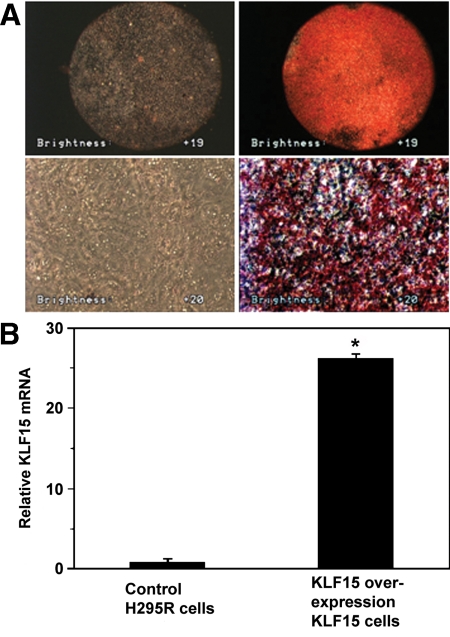
Adipogenesis is inducible by KLF15 overexpression in H295R cells
Ectopic expression of the KLF15 protein successfully induced adipocyte differentiation of 3T3-L1 cells (preadipocyte), NIH 3T3 cells, and C2C12 cells (muscle cells) (25). To investigate the action of KLF15 in H295R cells, we overexpressed this protein in H295R cells. Stable expression of KLF15 protein in H295R cells induced marked lipid accumulation after exposure to a standard adipogenic induction cocktail (Fig. 3); note that the adipocyte induction cocktail alone does not induce lipid adipogenesis in the cells. These results are consistent with the previous observation that KLF15 protein possesses the ability to promote adipogenesis in various other cell lines (25).
Figure 3.
Overexpression of KLF15 induces adipogenesis in H295R cells. A, H295R cells without (left panels) or with (right panels) stable expression of KLF15 were stained with oil red O at 10 d after exposure to inducers of adipocyte differentiation. Macroscopic (upper panels) and microscopic (lower panels; magnification, ×100) views are shown. B, Total RNAs from H295R cells without (left) or with (right) stable expression of KLF15 were subjected to real-time RT-PCR for KLF15 mRNA levels. *, P < 0.001 compared with control cells.
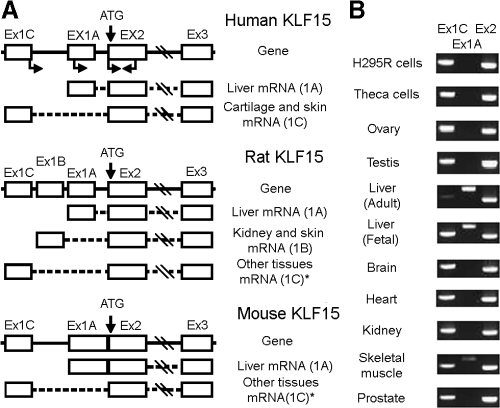
KLF15 uses alternative promoters in a tissue-specific manner
The combination of insulin, Dex, and isobutyl methylxanthine (adipogenic induction cocktail) is known to stimulate KLF15 expression during differentiation from preadipocytes to adipocytes (25). However, in hepatocytes insulin alone inhibits KLF15 expression and also blocks the stimulating effects of Dex and cAMP on KLF15 mRNA expression (27). These data indicate that insulin differentially regulates KLF15 expression in preadipocytes, steroidogenic cells, and hepatocytes. Therefore, we searched ESTs to define the KLF15 expression pattern and gene structure. The search identified 35 human ESTs having sequence identity with the KLF15 mRNA, but two groups of 5′-untranslated sequences (5′-UTR), one from liver and another from skin and cartilage. By comparing the KLF15 ESTs and genomic sequences, we identified four separate exons (Fig. 4A). The coding sequence consists of two exons separated by an 8.5-kb intron sequence and another two exons located upstream of the coding sequence (Fig. 4A). The two groups of ESTs make it possible to predict the existence of two transcripts with alternative exons: exon 1A (272 bp long) is present in liver EST and is located 1.4 kb upstream of exon 2; exon 1C (362 bp long) is present in the skin and cartilage EST and is located 4.8 kb upstream of exon 2 (Fig. 4A). This profile of KLF15 mRNA also exists in rats and mice, although each has a slightly different 5′-UTR from the human (Fig. 4A). To confirm this alternatively spliced phenomenon in humans, we performed RT-PCR using a variety of human tissues. Spliced exon 1C and exon 2 were amplified from all the tissues that were examined, whereas exon 1A was amplified only from human liver and muscle (Fig. 4B).
Figure 4.
Structures of the human, rat, and mouse KLF15 genes. A, The human KLF15 gene sequence was obtained from NW_921807 and NT_005612. Coding exon 2 is preceded by two untranslated exons, 1A and 1C, which are expressed in a tissue-specific manner as transcripts 1A and 1C. Exons 1A and 1C are located 1.4 and 4.8 kb from coding exon 2, respectively. The positions of the EST sequences on the gene (DA_637726, BM_452280, and BG_898891 from liver, cartilage, and skin, respectively) are shown by open boxes. Dashed lines indicate the introns that separated the transcribed regions. The structure of rat and mouse KLF15 genes was obtained by comparison of the rat and mouse ESTs and genomic sequences NM_047694 (rat) and NT_039353 (mouse), respectively. Other tissues include rat brain and bone; mouse brain, bone, heart, thymus, retina, mammary gland, and pineal gland. B, RT-PCR results from human cells and tissues. Spliced product of exon 1A was only amplified from liver and muscle; the steroidogenic cells and tissues have an identical pattern.
The KLF15 binding site of HSD17B5 is unique in humans and orangutans
The sequence of human HSD17B5 KLF15 binding site and its vicinity was compared with that of seven different species. 5′-flanking sequences of hsd17b5 in these species could be classified into three groups based on homologies: human, orangutan, and rhesus monkey were defined as group I; rat and mouse as group II; and cattle, dog, and pig as group III. The 5′-flanking sequences are remarkably different from group to group, although groups I and II have a highly homologous sequence of CATTGGTTAACCATCA (Table 1). The Sp1/Sp3 binding site is almost identical among humans, orangutans, rhesus monkeys, and cattle, but the KLF15 binding site has no human homolog in species other than orangutans and half of the binding site in rhesus monkeys (Table 1).
Table 1.
Alignments of nucleotide sequences of the HSD17B5 gene 5′-flanking regions corresponding to the HSD17B5 gene proximal promoter from seven species
| Species | bp | Sequences |
|---|---|---|
| A | ||
| Human (I) | −147 | agactgcctatatacctcctcctacatgccattggttaaccatcagtcagttt |
| Orangutan (I) | −147 | ………..g………………………………….. |
| Rhesus monkey (I) | −141 | ………..g…………….t…………..t….c…. |
| Rat (II) | −135 | .ag….t.t.gctt..ttc.tc.a.ctt….a….ct…t.caag.a.a |
| Mouse (II) | −135 | .ag….t.tca.ctt.ttcaac.a.ctg………ct…t.caag.aca |
| Cattle (III) | −136 | .ct.a.tta..cctt………..gtgtg.ct.c….at.ggacagtcag |
| Dog (III) | −147 | ttgtata..tgta..aa.gc.g.ga..atac.aaaaactgttgact.gg..aa |
| Pig (III) | −144 | ..cagt..atggat..c.aaa.c.gtgt.tggagct.a.tg..ggtcacc.ca |
| B | ||
| Human (I) | −94 | gcaggggtggggggaggggtttcctgcccattgtttttgtaatctctgaggag |
| Orangutan (I) | −94 | ………c……………………………………. |
| Rhesus monkey (I) | −88 | ……..c..--------…………..c……………..g. |
| Rat (II) | −82 | a.t.t..caa..tc…ca…at.tg.tg..c.g.gaa.g.aga.a…c.. |
| Mouse (II) | −82 | t.ta..a.aa..cc.a.ca…at.tg.tg.cc.g.gaaag.agc.a..at.. |
| Cattle (III) | −81 | ctg…cgttaacttcct…att.ataa.c.c.gaag.c..aagaaaaaa.t |
| Dog (III) | −94 | ttt.aa.g…t.atttta.gg.a..tgac..ac.a.gc..t.t…cccctc |
| Pig (III) | −91 | ttcct.taccat.atttatcac.t.tgtacc..aagag..ttctctcaaca.a |
| C | ||
| Human (I) | −41 | aagcagcagcaaacatttgctagtcagacaagtgacagggaatg |
| Orangutan (I) | −41 | …a……………………………….atg |
| Rhesus monkey (I) | −41 | …a……………………………….atg |
| Rat (II) | −29 | t.t.tt.tcagttggaaga.cgaga..c.atg |
| Mouse (II) | −29 | t.t.tt.tcagttgg.gg…gaga..c.atg |
| Cattle (III) | −30 | tgcagatgtatt.t.a..aga..c….ag….caaa.agatg |
| Dog (III) | −41 | tgcagatgtatt.t.a..aga..c….ag….caaa.agatg |
| Pig (III) | −38 | g..g.aaattggtgt..gtt..cc….ag….---a.agatg |
The first in-frame ATG codons in exon 1 are indicated in bold. A period indicates that the same nucleotide is present in the homologous sequence, and letters refer to the variant nucleotide. Gaps introduced for alignment are denoted by dashes. Sp1/Sp3 (cctcctcct) and KLF15 (ggggtggggggagggg) binding sites are underlined. Group numbers are indicated in parentheses. The sequences were aligned by manual inspection. Access nos. are: human AL391427, orangutan CR860189, rhesus monkey XM001104543, rat NW047492, mouse AF110408, cattle NW974248, dog NW876291, and pig DQ474067.
Discussion
This study provides potentially novel molecular evidence for the relationship of dysregulation of steroidogenesis, fat excess, and insulin excess in the pathogenesis of PCOS. KLF15, a newly discovered transcription factor, appears to mediate both T production and adipogenesis in response to insulin, in addition to playing an important role in glucose homeostasis. Our results clearly demonstrate that human HSD17B5 promoter contains a KLF15 binding site in a human adrenal steroidogenic cell line, and KLF15 binds to this cis-active element endogenously and exogenously (Fig. 1). We also show that overexpression of functional KLF15 protein stimulates HSD17B5 promoter activities and increases T formation from AD; overexpression of dominant negative KLF15 protein inhibits the gene promoter activities (Fig. 2). Together with our previous studies (16,17), we have demonstrated that KLF15 and Sp1/Sp3 directly, specifically, and separately bind to the HSD17B5 promoter. These two separate cis-active elements are essential for the basal and insulin-stimulated promoter activity of HSD17B5 (Fig. 2). Sp1 and Sp3 are ubiquitously expressed transcription factors that play a key role in maintaining basal transcription of many genes. Sp1 and Sp3 mediation of insulin actions have been demonstrated in several genes (26,32,33,34,35,36,37,38,39,40,41), for example, apolipoprotein AI (37), fatty acid synthase (32), and leptin (34). Insulin has been shown to regulate Sp1 and Sp3 gene expression (37), alter transcriptional binding affinities (36,38), and control posttranslational modifications, such as O-glycosylation and phosphorylation (40). The mechanisms of Sp1 mediation of insulin action have been proposed as: 1) Sp1 acts alone in mediating the effects of insulin; 2) Sp1 acts with cooperative interaction with other insulin-responsive transcription factors; and 3) Sp1 binds to an insulin-responsive promoter leading to basal activity, but Sp1 dissociation from this site permits the actions of another factor or factors to modulate gene activity in response to insulin (36). Our data demonstrate that insulin up-regulates KLF15 mRNA levels in H295R cells (Fig. 2). To the best of our knowledge, this is the first molecular observation indicating that insulin action on the regulation of HSD17B5 is mediated by cooperative interaction between KLF15 and Sp1/Sp3. This is in line with the view of insulin actions on the regulation of AceCS2 gene (26).
Expression of HSD17B5 mRNA and androgenic 17β-HSD activity were significantly increased after adipocyte differentiation in response to an adipogenic induction (42). We show that overexpression of KLF15 mediates fat accumulation in H295R cells in response to an adipogenic induction cocktail (Fig. 3), in agreement with previous studies in other cell lines (25). Thus, our results provide the first cogent molecular evidence implicating a transcription factor (KLF15) as a novel link between the regulation of steroidogenesis (HSD17B5) and fat stores in humans. However, future studies must determine whether KLF15 mediates fat accumulation and T production in normal human cells under normal and insulin-resistant conditions.
Alignment of KLF15 and Sp1/Sp3 binding sites of the human HSD17B5 to seven other species revealed that the KLF15 binding site has no human homolog in species other than orangutans and half of a binding site in rhesus monkeys (Table 1). Sp1/Sp3 binding site sequence homology, on the other hand, is highly conserved among humans, orangutans, rhesus monkeys, and cattle. The lack of identity of the KLF15 binding site is a surprising result because of highly conserved coding sequences in this gene family. However, the strong evolutionary conservation of coding sequences for developmentally important genes does not necessarily mean that their expression patterns are as highly conserved. By analogy, mouse hsd17b5 is expressed mainly in the liver, ovary, adrenal, and kidney (43), and rat hsd17b5 is expressed in the liver and kidney (44), contrary to the human enzyme, which is ubiquitously expressed. These observations further support the concept that HSD17B5 genes might have evolved by multiplication from an ancestral gene but eventually formed a gene that is regulated in a unique species-specific manner because of the accumulation of mutations during evolutionary selection (16). This selection may be of physiological relevance, assuming that it is advantageous for T anabolic effects; however, these evolutionarily adaptive changes may be inappropriate and predispose to hyperandrogenism in the current environment of nutritional excess. Our data suggest that humans and orangutans, but not other species, are vulnerable to KLF-mediated hyperandrogenism during IR. However, the covalent modifications of transcription factors and cofactors involved in the transcriptional activation of the human HSD17B5 remain to be investigated before we can unravel the mechanisms underlying these specific patterns of expression and their functional significance.
By in silico and RT-PCR analyses, we show that KLF15 is regulated in a tissue-specific manner by using splice variants that alter the 5′-UTR of the message, but not the protein-coding region (Fig. 4). The human, rat, and mouse KLF15 genes code for at least two transcripts, arising from heterogeneous 5′-UTRs; each of the exons is transcribed independently in transcript 1A, 1B, or 1C, with exons 2 and 3 being common to all transcripts. In addition, the transcripts are expressed in a tissue-specific manner. These molecular data may help to explain the previous findings that differential regulation of KLF15 by insulin among cells is mediated by alternative promoters of the KLF15 gene, which leads to an up-regulation of fat and muscle GLUT4 and AceCS2 gene expression, and an attenuation of hepatic PEPCK gene expression, maintaining glucose homeostasis (21,25,26,27).
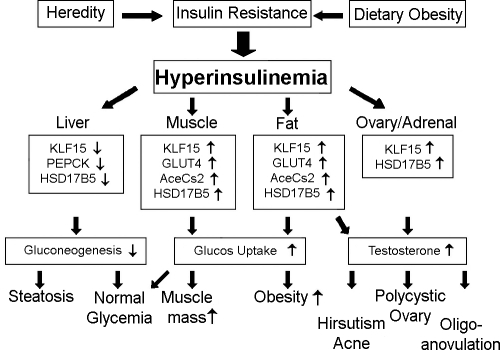
Our results, together with other studies (21,24,25,26,27,42), are compatible with the postulate that KLF15 protein potentially is central in restoring glucose homeostasis to compensate for IR in liver, fat, and muscle. Increased KLF15 expression in response to the compensatory hyperinsulinemia of IR may mediate obesity, steatosis, and T overproduction (Fig. 5). In fat and muscle, KLF15 expression is stimulated by insulin to up-regulate GLUT4 and AceCs2 genes, which increases glucose uptake, causing obesity and muscularity. In liver, KLF15 expression is inhibited by insulin to down-regulate the PEPCK gene, which decreases hepatic gluconeogenesis, predisposing to fatty liver. In steroidogenic cells, KLF15 expression is also stimulated by insulin to up-regulate HSD17B5 expression and to increase T biosynthesis.
Figure 5.
Proposed model of KLF15 mediation of insulin-induced cellular dysfunction of PCOS. KLF15 is postulated to mediate the effects of compensatory insulin secretion on glucose homeostasis, with the side effect of androgen excess. Through KLF15, insulin decreases hepatic gluconeogenesis, increases muscle and fat glucose uptake, and increases T production. Thus, KLF15 is postulated to be a mediator of most hyperinsulinemia-related features of PCOS.
In summary, the current study provides evidence for a potential novel mechanism of insulin-mediated hyperandrogenism in humans that may predispose to PCOS. This aspect of the PCOS phenotype is due to unique species-specific HSD17B5 promoter profiles.
Acknowledgments
We thank Drs. Samuel Refetoff, Roy Weiss, and Christine Yu for helpful discussions. We thank Dr. William Rainey for the human ovarian theca-like cells and Dr. Christos Zouboulis for the sebocytes. We also thank James Helke for skillful testosterone measurement.
Footnotes
This work was supported by National Institutes of Health Grants RO1-HD39267 (to R.L.R. and K.Q.), U54–04185 (to R.L.R.), and K08-HD043279 (to K.Q.).
Disclosure Summary: The authors have nothing to declare.
First Published Online April 14, 2009
Abbreviations: AceCS2, Acetyl-CoA synthetase 2; AD, androstenedione; ChIP, chromatin immunoprecipitation; Dex, dexamethasone; EST, expressed sequence tag; GLUT4, glucose transporter 4; 17β-HSD, 17β-hydroxysteroid dehydrogenase; HSD17B5, 17β-hydroxysteroid dehydrogenase type 5 gene; IR, insulin resistance; KLF15, Kruppel-like factor 15; PCOS, polycystic ovary syndrome; PEPCK, phosphoenolpyruvate carboxykinase; SFM, serum free medium; Sp1/Sp3, stimulatory protein 1/3; T, testosterone; 5′-UTR, 5′-untranslated sequences.
References
- Ehrmann DA, Barnes RB, Rosenfield RL 1995 Polycystic ovary syndrome as a form of functional ovarian hyperandrogenism due to dysregulation of androgen secretion. Endocr Rev 16:322–353 [DOI] [PubMed] [Google Scholar]
- Zawadzki J, Dunaif A 1992 Diagnostic criteria for polycystic ovary syndrome: towards a rational approach. In: Dunaif A, Givens J, Haseltine F, Merriam G, eds. Polycystic ovary syndrome. Cambridge, MA: Blackwell Scientific Publications; 377–384 [Google Scholar]
- Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group 2004 Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril 81:19–25 [DOI] [PubMed] [Google Scholar]
- Ehrmann DA 2005 Polycystic ovary syndrome. N Engl J Med 352:1223–1236 [DOI] [PubMed] [Google Scholar]
- Rosenfield RL 2005 Clinical practice. Hirsutism. N Engl J Med 353:2578–2588 [DOI] [PubMed] [Google Scholar]
- Nelson VL, Qin KN, Rosenfield RL, Wood JR, Penning TM, Legro RS, Strauss 3rd JF, McAllister JM 2001 The biochemical basis for increased testosterone production in theca cells propagated from patients with polycystic ovary syndrome. J Clin Endocrinol Metab 86:5925–5933 [DOI] [PubMed] [Google Scholar]
- Luu-The V, Dufort I, Pelletier G, Labrie F 2001 Type 5 17β-hydroxysteroid dehydrogenase: its role in the formation of androgens in women. Mol Cell Endocrinol 171:77–82 [DOI] [PubMed] [Google Scholar]
- Dufort I, Rheault P, Huang XF, Soucy P, Luu-The V 1999 Characteristics of a highly labile human type 5 17 β-hydroxysteroid dehydrogenase. Endocrinology 140:568–574 [DOI] [PubMed] [Google Scholar]
- Khanna M, Qin KN, Klisak I, Belkin S, Sparkes RS, Cheng KC 1995 Localization of multiple human dihydrodiol dehydrogenase (DDH1 and DDH2) and chlordecone reductase (CHDR) genes in chromosome 10 by the polymerase chain reaction and fluorescence in situ hybridization. Genomics 25:588–590 [DOI] [PubMed] [Google Scholar]
- Lin HK, Jez JM, Schlegel BP, Peehl DM, Pachter JA, Penning TM 1997 Expression and characterization of recombinant type 2 3α-hydroxysteroid dehydrogenase (HSD) from human prostate: demonstration of bifunctional 3 α/17β-HSD activity and cellular distribution. Mol Endocrinol 11:1971–1984 [DOI] [PubMed] [Google Scholar]
- Qin KN, New MI, Cheng KC 1993 Molecular cloning of multiple cDNAs encoding human enzymes structurally related to 3 α-hydroxysteroid dehydrogenase. J Steroid Biochem Mol Biol 46:673–679 [DOI] [PubMed] [Google Scholar]
- Steckelbroeck S, Watzka M, Stoffel-Wagner B, Hans VH, Redel L, Clusmann H, Elger CE, Bidlingmaier F, Klingmüller D 2001 Expression of the 17β-hydroxysteroid dehydrogenase type 5 mRNA in the human brain. Mol Cell Endocrinol 171:165–168 [DOI] [PubMed] [Google Scholar]
- Quinkler M, Sinha B, Tomlinson JW, Bujalska IJ, Stewart PM, Arlt W 2004 Androgen generation in adipose tissue in women with simple obesity—a site-specific role for 17β-hydroxysteroid dehydrogenase type 5. J Endocrinol 183:331–342 [DOI] [PubMed] [Google Scholar]
- Quinkler M, Bujalska IJ, Tomlinson JW, Smith DM, Stewart PM 2006 Depot-specific prostaglandin synthesis in human adipose tissue: a novel possible mechanism of adipogenesis. Gene 380:137–143 [DOI] [PubMed] [Google Scholar]
- Fung KM, Samara EN, Wong C, Metwalli A, Krlin R, Bane B, Liu CZ, Yang JT, Pitha JV, Culkin DJ, Kropp BP, Penning TM, Lin HK 2006 Increased expression of type 2 3α-hydroxysteroid dehydrogenase/type 5 17β-hydroxysteroid dehydrogenase (AKR1C3) and its relationship with androgen receptor in prostate carcinoma. Endocr Relat Cancer 13:169–180 [DOI] [PubMed] [Google Scholar]
- Qin K, Rosenfield RL 2005 Characterization of the basal promoter element of the human type 5 17β-hydroxysteroid dehydrogenase gene. Biochim Biophys Acta 1728:115–125 [DOI] [PubMed] [Google Scholar]
- Qin K, Ehrmann DA, Cox N, Refetoff S, Rosenfield RL 2006 Identification of a functional polymorphism of the human type 5 17β-hydroxysteroid dehydrogenase gene associated with polycystic ovary syndrome. J Clin Endocrinol Metab 91:270–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi MO, Jones MR, Antoine HJ, Pall M, Chen YD, Azziz R 2008 Nonreplication of the type 5 17β-hydroxysteroid dehydrogenase gene association with polycystic ovary syndrome. J Clin Endocrinol Metab 93:300–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioli DJ, Saltamavros AD, Vervita V, Koika V, Adonakis G, Decavalas G, Markou KB, Georgopoulos NA 11 August 2008 Association of the 17-hydroxysteroid dehydrogenase type 5 gene polymorphism (-71A/G HSD17B5 SNP) with hyperandrogenemia in polycystic ovary syndrome (PCOS). Fertil Steril 10.1016/j.fertnstert.2008.06.016 [DOI] [PubMed] [Google Scholar]
- Uchida S, Tanaka Y, Ito H, Saitoh-Ohara F, Inazawa J, Yokoyama KK, Sasaki S, Marumo F 2000 Transcriptional regulation of the CLC-K1 promoter by myc-associated zinc finger protein and kidney-enriched Kruppel-like factor, a novel zinc finger repressor. Mol Cell Biol 20:7319–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Sen-Banerjee S, DePina A, Haspel R, Jain MK 2002 The Kruppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem 277:34322–34328 [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R 2003 Sp1- and Kruppel-like transcription factors. Genome Biol 4:206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, Das H, Petersen CA, Mende U, Burleigh BA, Zhu Y, Pinto YM, Pinto Y, Liao R, Jain MK 2007 Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc Natl Acad Sci USA 104:7074–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Wang B, Orihuela Y, Hong EG, Fisch S, Haldar S, Cline GW, Kim JK, Peroni OD, Kahn BB, Jain MK 2007 Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab 5:305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, Kadowaki T, Matsuki Y, Ogawa W, Hiramatsu R, Kasuga M 2005 Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem 280:12867–12875 [DOI] [PubMed] [Google Scholar]
- Yamamoto J, Ikeda Y, Iguchi H, Fujino T, Tanaka T, Asaba H, Iwasaki S, Ioka RX, Kaneko IW, Magoori K, Takahashi S, Mori T, Sakaue H, Kodama T, Yanagisawa M, Yamamoto TT, Ito S, Sakai J 2004 A Kruppel-like factor KLF15 contributes fasting-induced transcriptional activation of mitochondrial acetyl-CoA synthetase gene AceCS2. J Biol Chem 279:16954–16962 [DOI] [PubMed] [Google Scholar]
- Teshigawara K, Ogawa W, Mori T, Matsuki Y, Watanabe E, Hiramatsu R, Inoue H, Miyake K, Sakaue H, Kasuga M 2005 Role of Kruppel-like factor 15 in PEPCK gene expression in the liver. Biochem Biophys Res Commun 327:920–926 [DOI] [PubMed] [Google Scholar]
- Rainey WE, Sawetawan C, McCarthy JL, McGee EA, Bird IM, Word RA, Carr BR 1996 Human ovarian tumor cells: a potential model for thecal cell steroidogenesis. J Clin Endocrinol Metab 81:257–263 [DOI] [PubMed] [Google Scholar]
- Wróbel A, Seltmann H, Fimmel S, Müller-Decker K, Tsukada M, Bogdanoff B, Mandt N, Blume-Peytavi U, Orfanos CE, Zouboulis CC 2003 Differentiation and apoptosis in human immortalized sebocytes. J Invest Dermatol 120:175–181 [DOI] [PubMed] [Google Scholar]
- Rosenfield RL 1989 Relationship of sebaceous cell stage to growth in culture. J Invest Dermatol 92:751–754 [DOI] [PubMed] [Google Scholar]
- Yu C, Markan K, Temple KA, Deplewski D, Brady MJ, Cohen RN 2005 The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor γ transcriptional activity and repress 3T3-L1 adipogenesis. J Biol Chem 280:13600–13605 [DOI] [PubMed] [Google Scholar]
- Fukuda H, Iritani N, Noguchi T 1997 Transcriptional regulatory regions for expression of the rat fatty acid synthase. FEBS Lett 406:243–248 [DOI] [PubMed] [Google Scholar]
- Fandos C, Sánchez-Feutrie M, Santalucía T, Viñals F, Cadefau J, Gumà A, Cussó R, Kaliman P, Canicio J, Palacín M, Zorzano A 1999 GLUT1 glucose transporter gene transcription is repressed by Sp3. Evidence for a regulatory role of Sp3 during myogenesis. J Mol Biol 294:103–119 [DOI] [PubMed] [Google Scholar]
- Fukuda H, Iritani N 1999 Transcriptional regulation of leptin gene promoter in rat. FEBS Lett 455:165–169 [DOI] [PubMed] [Google Scholar]
- Barth N, Langmann T, Schölmerich J, Schmitz G, Schäffler A 2002 Identification of regulatory elements in the human adipose most abundant gene transcript-1 (apM-1) promoter: role of SP1/SP3 and TNF-α as regulatory pathways. Diabetologia 45:1425–1433 [DOI] [PubMed] [Google Scholar]
- Samson SL, Wong NC 2002 Role of Sp1 in insulin regulation of gene expression. J Mol Endocrinol 29:265–279 [DOI] [PubMed] [Google Scholar]
- Lam JK, Matsubara S, Mihara K, Zheng XL, Mooradian AD, Wong NC 2003 Insulin induction of apolipoprotein AI, role of Sp1. Biochemistry 42:2680–2690 [DOI] [PubMed] [Google Scholar]
- Li T, Chen YH, Liu TJ, Jia J, Hampson S, Shan YX, Kibler D, Wang PH 2003 Using DNA microarray to identify Sp1 as a transcriptional regulatory element of insulin-like growth factor 1 in cardiac muscle cells. Circ Res 93:1202–1209 [DOI] [PubMed] [Google Scholar]
- Wang G, Leiter AB, Englander EW, Greeley Jr GH 2004 Insulin-like growth factor I increases rat peptide YY promoter activity through Sp1 binding sites. Endocrinology 145:659–666 [DOI] [PubMed] [Google Scholar]
- Majumdar G, Harrington A, Hungerford J, Martinez-Hernandez A, Gerling IC, Raghow R, Solomon S 2006 Insulin dynamically regulates calmodulin gene expression by sequential o-glycosylation and phosphorylation of Sp1 and its subcellular compartmentalization in liver cells. J Biol Chem 281:3642–3650 [DOI] [PubMed] [Google Scholar]
- Deng X, Yellaturu C, Cagen L, Wilcox HG, Park EA, Raghow R, Elam MB 2007 Expression of the rat sterol regulatory element-binding protein-1c gene in response to insulin is mediated by increased transactivating capacity of specificity protein 1 (Sp1). J Biol Chem 282:17517–17529 [DOI] [PubMed] [Google Scholar]
- Blouin K, Nadeau M, Mailloux J, Daris M, Lebel S, Luu-The V, Tchernof A 2009 Pathways of adipose tissue androgen metabolism in women: depot differences and modulation by adipogenesis. Am J Physiol Endocrinol Metab 296:E244–E255 [DOI] [PubMed] [Google Scholar]
- Pelletier G, Luu-The V, Li S, Labrie F 2005 Localization of type 5 17β-hydroxysteroid dehydrogenase mRNA in mouse tissues as studied by in situ hybridization. Cell Tissue Res 320:393–398 [DOI] [PubMed] [Google Scholar]
- Ishikura S, Matsumoto K, Sanai M, Horie K, Matsunaga T, Tajima K, El-Kabbani O, Hara A 2006 Molecular cloning of a novel type of rat cytoplasmic 17β-hydroxysteroid dehydrogenase distinct from the type 5 isozyme. J Biochem 139:1053–1063 [DOI] [PubMed] [Google Scholar]