Abstract
Objective: We performed this study to test the hypothesis that variation in the lamin a/c gene (LMNA) contributes to milder phenotypes of insulin resistance, hyperandrogenism, and/or metabolic syndrome associated with polycystic ovary syndrome (PCOS).
Research Design and Methods: We resequenced the coding region, flanking intronic, and proximal promoter regions of the lamin a/c gene in 43 women with PCOS with evidence of upper-body obesity (waist circumference >88 cm) and identified 56 variants, two of which were nonsynonymous substitutions (lmna11 exon1 E98D; lmna24 exon 7 R455C). We genotyped 53 single-nucleotide polymorphisms (44 identified through resequencing and nine included to maximize informativeness of the entire gene) in 624 index (PCOS) cases and 544 controls of European ancestry. We tested for association between these variants and PCOS. In a subset of individuals, we also tested for association with metabolic syndrome and quantitative traits (body mass index, waist circumference, total testosterone, dehydroepiandrosterone sulfate, fasting glucose and insulin, low-density lipoprotein, and total triglycerides).
Results: After correction for multiple testing, none of the variants showed significant evidence for association with PCOS, the metabolic syndrome, or any of the quantitative traits tested.
Conclusions: Whereas these studies cannot exclude the role of genetic variation in the lamin a/c gene in isolated cases of PCOS, we can conclude that common variation in the lamin a/c gene does not contribute to the etiology of PCOS in women of European ancestry.
Genetic variants in lamin a/c are not common in Caucasian women with PCOS.
Polycystic ovary syndrome (PCOS) is characterized by hyperandrogenemia and irregular menses and is associated with features of the metabolic syndrome including insulin resistance, obesity, and a 7-fold increased risk of developing type 2 diabetes (1). There is a well-documented familial clustering of PCOS, with about 40% of reproductive-age sisters affected with hyperandrogenemia, and a recent twin study (2) demonstrated a high heritability for PCOS (h2 = 0.70), which taken together provide evidence for a genetic susceptibility to the disorder (3). One approach to identifying susceptibility genes for complex disorders is to characterize genetic variation in genes causing Mendelian disorders with similar phenotypes. For example, variation in peroxisomal proliferator-activated receptor (PPAR)-γ can cause familial partial lipodystrophy (4,5), an autosomal dominant disorder characterized by gradual loss of sc fat from the extremities, whereas the PPARγ Pro12Ala variant is a well-established type 2 diabetes susceptibility locus (6). The cardinal features of PCOS, hyperandrogenemia and polycystic ovaries, are also features of partial lipodystrophy syndromes, which can be due to mutations in the gene for lamin a/c (LMNA) or PPARγ (7). PPARγ (8,9) has been investigated in PCOS but LMNA has not. Variation in LMNA has been shown to be associated with Dunnigan-type familial partial lipodystrophy (FPLD) (reviewed in Ref. 10) in which there is a sc fat loss from the extremities and trunk during adolescence. Lamin a/c mutations have also been implicated in severe metabolic syndrome (7), and type A insulin resistance syndrome (11).
To characterize the role of genetic variation in the gene for lamin a/c (LMNA) in the etiology of PCOS, we sequenced LMNA in 43 PCOS patients and tested for association between the PCOS-specific variants and anonymous single-nucleotide polymorphisms (SNPs) encompassing the LMNA genomic region in a large PCOS cohort.
Subjects and Methods
Subjects
This study was approved by the Institutional Review Boards of the Brigham and Women’s Hospital, Northwestern University Feinberg School of Medicine, Pennsylvania State University College of Medicine, and University of Pennsylvania Medical Center. Written informed consent was obtained from all participants. We studied 624 index cases (probands) with PCOS and 544 control women (72 intensively phenotyped and 472 minimally phenotyped from the NUgene DNA repository, http://www.nugene.org) of European Caucasian ancestry. Phenotypic characteristics of cases and controls are given in Table 1.
Table 1.
Phenotypic characteristics of study participants
| PCOS (n = 624)
|
Minimally phenotyped controls (n = 472)
|
Intensively phenotyped controls (n = 72)
|
||||
|---|---|---|---|---|---|---|
| n | Median (range) | n | Median (range) | n | Median (range) | |
| Age | 624 | 28 (14–48) | 472 | 35 (18–45) | 72 | 29 (18–40) |
| BMI (kg/m2) | 624 | 35.0 (16.5–64.5)a | 463 | 23.2 (16.7–68.1) | 72 | 27.7 (18.0–53.5) |
| Waist circumference (cm) | 408 | 101 (58–170)a | ND | ND | 61 | 85 (63–134) |
| T (ng/dl) [(mmol/liter)] | 624 | 73 (29–337)b [2.5 (1.0–11.7)]b | ND | ND | 72 | 28 (6–49) [1.0 (0.2–1.7)] |
| uT (ng/dl) [(mmol/liter)] | 621 | 24 (1.7–109)b [0.8 (0.06–3.8)]b | ND | ND | 72 | 7.0 (1–16.0) [0.2 (0.03–0.6)] |
| DHEAS (ng/ml) [(μmol/liter)] | 616 | 2091 (50–13,336)b [5.7 (0.1–36.2)]b | ND | ND | 70 | 1363 (390–3484) [3.7 (1.1–9.5)] |
| SHBG (nmol) | 514 | 56 (12–426)b | ND | ND | 30 | 104 (46–331) |
| Fasting insulin (μU/ml) [(pmol/liter)] | 601 | 22 (3–152)b [132 (18–912)] | ND | ND | 65 | 11 (4–29) [66 (24–174)] |
| Fasting glucose (mg/dl) [(mmol/liter)] | 607 | 88 (58–189) [4.9 (3.2–10.5)] | ND | ND | 71 | 89 (72–130) [5.0 (4.0–7.2)] |
| HDL (mg/dl) [(mmol/liter)] | 577 | 40 (15–107)b [1.03 (0.39–2.77)]b | ND | ND | 57 | 49 (27–85) [1.27 (0.70–2.20)] |
| TTG (mg/dl) [(mmol/liter)] | 578 | 137 (35–2427)b [3.54 (0.91–62.76)]b | ND | ND | 57 | 78 (37–306) [0.88 (0.41–3.45)] |
P < 0.0001 vs. intensively phenotyped controls;
P < 0.0001 vs. intensively phenotyped controls after adjusting for BMI and age. ND, Not determined.
Conversion factors: uT and T, nanograms per deciliter to millimoles per liter, multiply by 0.03467; DHEAS, nanograms per milliliter to micromoles per liter, multiply by 0.002714; insulin, microunits per milliliter to picomoles per liter, multiply by 6.0; glucose, milligrams per deciliter to millimoles per liter, multiply by 0.05551; HDL, milligrams per deciliter to millimoles per liter, multiply by 0.02586; TTG, milligrams per deciliter to millimoles per liter, multiply by 0.01129.
PCOS cases
PCOS was defined according to the National Institute of Child Health and Human Development criteria as previously implemented by us (3,12); affected women also fulfilled the Rotterdam and Androgen Excess Society criteria for PCOS (13,14,15). All women with PCOS had hyperandrogenemia [total (T) or bioavailable testosterone (uT) levels 2 sd above the mean established in reproductively normal women] and chronic anovulation (less than six menses per year) with the exclusion of specific disorders of the ovaries, adrenal, or pituitary (3). We have previously reported phenotypic and genetic data on many of the PCOS cases (16).
Controls
Intensively phenotyped reproductively normal control women (n = 72) were of comparable age, weight, and ethnicity to PCOS cases and had normal androgen levels and regular menses (17,18). Minimally phenotyped women (n = 472) were selected from NUgene, a large scale GenBank that combines centralized DNA collection with electronic medical records. We enriched the minimally phenotyped control cohort for reproductively normal women by excluding women with self-reported history of diabetes and by preferentially included women with documented pregnancies (272 women had at least one pregnancy, average 2.2 pregnancies, range one to 11 pregnancies). Nevertheless, our power calculations assumed that the control population would have the general population prevalence of PCOS. The use of unphenotyped or population-based controls in genetic association studies is now a standard approach and may result in a small reduction (5–10%) in the power to detect a positive result (19,20).
Study protocols
Medications known to alter reproductive hormone levels or glucose homeostasis were stopped for 1 month or more before the study. Contraceptive steroids were stopped 3 or more months before the study. Anthropometric measurements (blood pressure, waist circumference, weight, and height) were taken as reported (3). Fasting blood samples were obtained for reproductive and metabolic hormones as reported (3,21).
Biochemical assays
Circulating levels of glucose, insulin, proinsulin, T, uT, dehydroepiandrosterone sulfate (DHEAS), SHBG, high-density lipoprotein (HDL) cholesterol, and triglyceride were determined as previously reported (3).
Genetic analyses
Sequencing
We selected 43 women with PCOS with phenotypic similarities to FPLD based on the presence of central obesity (i.e. increased waist circumference >88 cm according to American Heart Association criteria (22) in the presence of relatively low body mass index (BMI) (<35 kg/m2). We sequenced both strands of about 2 kb putative promoter regions, all exons and 200 bp flanking each exon using an 3100 genetic analyzer (Applied Biosystems, Foster City, CA). Sequencing results were assembled and analyzed using the SeqMan II software program (DNASTAR Inc., Madison, WI) and checked manually.
Genotyping
SNPs were genotyped using Illumina Goldengate array system (Illumina, San Diego, CA) as part of a 384-SNP bundle. The remaining SNPs mapped to other potential PCOS candidate genes. Assays by Design 5′nuclease Taqman technology (Applied Biosystems) was used for SNPs that did not pass Illumina Goldengate quality control specification. Genotyping was performed according to manufacturers’ recommendations.
Data analyses
Secondary phenotypes
Metabolic syndrome.
The metabolic syndrome phenotype was assigned according to the American Heart Association criteria (22). Women were considered affected if they fulfilled three of five criteria: systolic blood pressure 130 mm Hg or greater and/or diastolic blood pressure 85 mm Hg or greater, waist circumference 88 cm or greater, fasting glucose 100 mg/dl or greater, HDL less than 500 mg/dl, and triglycerides 150 mg/dl or greater. Two hundred twenty-five women with PCOS had metabolic syndrome and 232 women with PCOS were unaffected.
Power analysis
We used the Genetic Power Calculator package to calculate the power to detect association (23) with a range of disease allele frequencies (0.05–0.4) and effect sizes [genotype relative risk (GRR) 1.1–2.0] in our cohort at a significance level of P = 0.001 and PCOS population prevalence of 0.05. Under the multiplicative and additive models, we had greater than 80% power for all allele frequencies and GRR 2.0 or greater and greater than 50% power for GRR 1.5 or greater. Similarly, power analyses for quantitative traits were carried out for 624 PCOS probands using the program CaTSQT2 (Skol, A., personal communication, and Ref. 24)] assuming an additive model. We had greater than 75% power to detect a quantitative trait loci (QTL), which explains greater than 3% of the variance for all but waist circumference. For waist circumference, we had 60% power to detect the same QTL.
Genetic analysis
We tested for association with two dichotomous traits: PCOS and metabolic syndrome. We corrected for multiple testing using permutation to generate corrected significance levels. Genetic analyses were implemented and pairwise linkage disequilibrium (LD) plots of D′ were generated using Haploview 3.2 software (haploview@broad.mit.edu) (25).
We tested for association with eight quantitative traits in the PCOS subjects using the PLINK 0.99 software (http://pngu.mgh.harvard.edu/purcell/plink/) (26). BMI was also tested in the complete cohort. The phenotypes tested were BMI (all n = 1166; PCOS n = 624), waist circumference (n = 624), total testosterone (n = 624), DHEAS (n = 616), fasting glucose (n = 607), fasting insulin (n = 601), and total triglycerides (n = 570).
Results
Sequencing
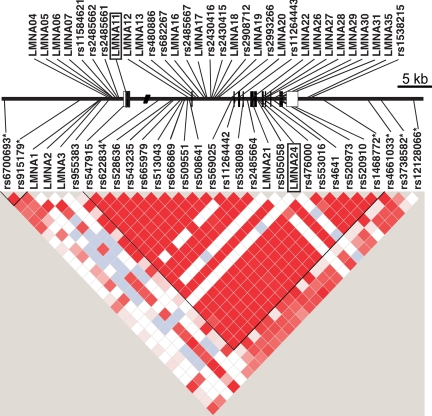
We detected 56 variants in the 43 women with PCOS that were sequenced (Fig. 1 and supplemental Table 1, published as supplemental data on The Endocrine Society’s Journals Online Web site at http://jcem.endojournals.org). Twenty-five variants were unique to this study and were named using the designation lmnaX. Five polymorphisms mapped to coding regions, two were nonsynonymous substitutions (lmna11 exon 1 E98D; lmna24 exon 7 R455C), and three were synonymous substitutions. Carriers of either lmna11 or lmna24 did not differ phenotypically from women with typical PCOS. Of these variants, 31 were successfully genotyped with the Illumina platform. Thirteen were genotyped using Taqman and 12 variants could not be genotyped using either the Taqman or Illumina platforms.
Figure 1.
Schematic of the lamin a/c gene sequencing and genetic analysis. The boxes indicate the location of each exon. The open boxes are nontranslated sequences. The filled boxes are translated exons. The relative location of SNPs is indicated. SNPs identified by sequencing but not included in the association studies are shown above the lamin a/c gene map, and SNPs used for the association studies are shown below the gene map. Asterisks indicate SNPs that were not identified in PCOS subjects but were selected from the database to increase informativeness of the LMNA genomic region. The pair-wise LD (D′) plot was generated with Haploview. Dark red indicates strong LD. White indicates no LD.
Genotyping
We genotyped 53 SNPs: 44 SNPs identified by sequencing and nine anonymous SNPs to complete coverage of the gene including potential regulatory elements mapping within 20 kb of LMNA (supplemental Table 2). We failed to design robust genotyping assays for 12 variants of the 56 variants identified by sequencing. Of the 53 SNPs that were successfully genotyped, 40 were genotyped using the Illumina platform and 13 were genotyped using the Taqman platform. Fortunately, there is sufficient LD within LMNA that there is also sufficient redundancy among the markers to haplotype tag or measure association with all regions of the gene.
Statistical analyses
Association studies were carried out for all markers with minor allele frequencies (MAFs) greater than 1% (n = 30) and lmn24, which was included in the analysis because it is a coding variant (supplemental Table 2). Twenty-two genotyped markers had MAFs less than 1%. The strongest evidence for association with PCOS was observed with the SNP lmna21 (χ2 = 10.55 and corrected P = 0.03). Because this association is due to a reduced frequency of the minor allele in the women with PCOS compared with controls, lmna21 minor allele may be a protective allele in the general population, but due to the low frequency of lmna21, even in the control population (MAF in controls = 1.7%), the relevance of this finding is unclear. No other variants showed significant evidence for association with PCOS or metabolic syndrome.
For the quantitative traits, the strongest evidence for association was observed with waist circumference (rs6700693, P = 0.003) and TGG levels (lmn24, P = 0.007). However, none of these findings remain statistically significant after correction for multiple testing.
Discussion and Conclusions
LMNA is mutated in two syndromes of hyperandrogenism, extreme insulin resistance, and polycystic ovaries, Dunnigan’s FPLD (7), and type A syndrome (11). LMNA is therefore a very plausible PCOS candidate gene. To address the role of genetic variation at LMNA in PCOS, we used two approaches. First, we determined the extent of common genetic variation in the lamin a/c gene in the general PCOS population by sequencing LMNA in 43 women with PCOS. Second, we used the variants thus identified in a case-control study to determine their association with PCOS, metabolic syndrome and associated quantitative traits in a cohort of 624 women with PCOS and 544 control women. This study was designed to study common variation in LMNA: by sequencing about 100 chromosomes, we could identify variants with a MAF greater than 1–2%. Our association study was powered to detect moderate genetic effects. For PCOS we have greater than 80% power for all allele frequencies and GRR 2.0 or greater and greater than 50% power for GRR 1.5 or greater, whereas for most quantitative traits, we had greater than 75% power to detect QTLs, which explain greater than 3% of the variance. For waist circumference we had slightly less power (60% power) to detect a QTL.
We sequenced the genomic region encompassing the coding region of LMNA and 2 kb of putative upstream and 600 bp putative downstream regulatory sequences. The majority (31 of 56) of the variants identified have previously been reported in public databases; however, we observed none of the variants previously found in patients with laminopathies. All 25 novel variants were extremely rare (observed on one or two chromosomes). Two missense mutations were identified (LMNA11 and LMNA24). LMNA24 (R455C) has not been previously observed, and maps to exon 7 of the lamin a/c gene, the exon that harbors the majority of metabolic syndrome and PCOS-associated variants (10). However, in our population this variant was not associated with PCOS or metabolic syndrome. Although LMNA24 was nominally associated with TTG levels, these findings did not remain significant after correction for multiple testing. LMNA11 (E98D) is much more frequent in than LMNA24 with a MAF of 0.059 but shows no evidence for association with any of the traits tested in our cohort. Furthermore, none of the remaining 29 SNPs or eight haplotypes tested showed statistically significant evidence for association with PCOS, metabolic syndrome, or any of the quantitative traits.
Analysis of cohorts of women with laminopathies clearly shows that there is an increased prevalence of PCOS in women with mutations in the lamin a/c gene (reviewed in Refs. 10 and 27,28,29). Therefore, it is of interest to determine whether the converse is also true. In other words, are lamin a/c mutations common in women with PCOS from the general population of European ancestry? Here we report the results of the first study that has examined the role of genetic variation in LMNA in PCOS in a PCOS cohort that was not selected based on the presence of lipodystrophy.
We conclude from this study that genetic variation in LMNA is not a major contributor to the etiology of PCOS. These conclusions are based on three findings. First, although in previous studies LMNA mutations contributing to laminopathies have clustered to specific regions of the gene and the same variants have been found in multiple studies, we did not find any of these variants in our sequence analysis. Therefore, these established variants are not common (frequency <2%) in PCOS. Second, we did not identify any missense mutations in our sequence analysis, and to date all LMNA mutations identified in women with PCOS symptoms have been coding variants. Finally, we did not detect any evidence for association with PCOS among any of the 31 variants tested. However, we cannot exclude the possibility that variation in the gene for lamin a/c contributes to the PCOS phenotype in a small subset of patients (2–5%). Studies with larger cohorts will be needed to test for loci with more modest effect sizes.
Supplementary Material
Acknowledgments
We thank all the women and their families for participating in this study and Dr. Deborah Driscoll for sharing with us DNAs from her PCOS cohort.
Footnotes
This work was supported by National Institutes of Health Grants P50 HD44405 (to M.U. and A.D.), U54 HD34449 (to A.D.), M01 RR00048 (Northwestern University General Clinical Research Center), M01 RR10732 and C06 RR016499 (Pennsylvania State University General Clinical Research Center), and M01 RR02635 (Brigham and Women’s Hospital General Clinical Research Center).
Disclosure Summary: M.U., G.N., J.D., E.S.,C.A., R.S.L., and A.D. have nothing to declare.
First Published Online April 28, 2009
Abbreviations: BMI, Body mass index; DHEAS, dehydroepiandrosterone sulfate; FPLD, familial partial lipodystrophy; GRR, genotype relative risk; HDL, high-density lipoprotein; LD, linkage disequilibrium; MAF, minor allele frequency; PCOS, polycystic ovary syndrome; PPAR, peroxisomal proliferator-activated receptor; QTL, quantitative trait loci; SNP, single-nucleotide polymorphism; T, total testosterone; uT, bioavailable testosterone.
References
- Sam S, Dunaif A 2003 Polycystic ovary syndrome: syndrome XX? Trends Endocrinol Metab 14:365–370 [DOI] [PubMed] [Google Scholar]
- Vink J, Sadrzadeh SM, Lambalk CB, Boomsma DI 2006 Heritability of polycystic ovary syndrome (PCOS) in a Dutch twin-family study. J Clin Endocrinol Metab 91:2100–2104 [DOI] [PubMed] [Google Scholar]
- Legro RS, Driscoll D, Strauss 3rd JF, Fox J, Dunaif A 1998 Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci USA 95:14956–14960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele RA, Cao H, Frankowski C, Mathews ST, Leff T 2002 PPARG F388L, a transactivation-deficient mutant, in familial partial lipodystrophy. Diabetes 51:3586–3590 [DOI] [PubMed] [Google Scholar]
- Agarwal AK, Garg A 2002 A novel heterozygous mutation in peroxisome proliferator-activated receptor-γ gene in a patient with familial partial lipodystrophy. J Clin Endocrinol Metab 87:408–411 [DOI] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES 2000 The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80 [DOI] [PubMed] [Google Scholar]
- Monajemi H, Stroes E, Hegele RA, Fliers E 2007 Inherited lipodystrophies and the metabolic syndrome. Clin Endocrinol (Oxf) 67:479–484 [DOI] [PubMed] [Google Scholar]
- Antoine HJ, Pall M, Trader BC, Chen YD, Azziz R, Goodarzi MO 2007 Genetic variants in peroxisome proliferator-activated receptor γ influence insulin resistance and testosterone levels in normal women, but not those with polycystic ovary syndrome. Fertil Steril 87:862–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S, Fingerhut A, Khomtsiv U, Khomtsiv L, Tan S, Quadbeck B, Herrmann BL, Knebel B, Mèuller-Wieland D, Mann K, Janssen OE 2005 The peroxisome proliferator activated receptor γ Pro12Ala polymorphism is associated with a lower hirsutism score and increased insulin sensitivity in women with polycystic ovary syndrome. Clin Endocrinol (Oxf) 62:573–579 [DOI] [PubMed] [Google Scholar]
- Worman HJ, Bonne G 2007 “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res 313:2121–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Morbois-Trabut L, Couzinet B, Lascols O, Dion E, Bâerâeziat V, Fáeve B, Richard I, Capeau J, Chanson P, Vigouroux C 2005 Type A insulin resistance syndrome revealing a novel lamin A mutation. Diabetes 54:1873–1878 [DOI] [PubMed] [Google Scholar]
- Urbanek M, Legro RS, Driscoll DA, Azziz R, Ehrmann DA, Norman RJ, Strauss 3rd JF, Spielman RS, Dunaif A 1999 Thirty-seven candidate genes for polycystic ovary syndrome: strongest evidence for linkage is with follistatin. Proc Natl Acad Sci USA 96:8573–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadski JK, Dunaif A 1992 Diagnostic criteria for polycystic ovary syndrome. In: Givens J, Haseltine F, Merriman G, eds. The polycystic ovary syndrome. Cambridge, MA: Blackwell Scientific; 377–384 [Google Scholar]
- 2004 Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum Reprod 19:41–47 [DOI] [PubMed] [Google Scholar]
- Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor AE, Witchel SF 2006 Positions statement: criteria for defining polycystic ovary syndrome as a predominantly hyperandrogenic syndrome: an Androgen Excess Society guideline. J Clin Endocrinol Metab 91:4237–4245 [DOI] [PubMed] [Google Scholar]
- Urbanek M, Sam S, Legro RS, Dunaif A 2007 Identification of a polycystic ovary syndrome susceptibility variant in fibrillin-3 and association with a metabolic phenotype. J Clin Endocrinol Metabol 92:4191–4198 [DOI] [PubMed] [Google Scholar]
- Sam S, Legro RS, Essah PA, Apridonidze T, Dunaif A 2006 Evidence for metabolic and reproductive phenotypes in mothers of women with polycystic ovary syndrome. Proc Nat Acad Sci USA 103:7030–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam S, Legro RS, Bentley-Lewis R, Dunaif A 2005 Dyslipidemia and metabolic syndrome in the sisters of women with polycystic ovary syndrome. J Clin Endocrinol Metab 90:4797–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium 2007 Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN 2008 Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9:356–369 [DOI] [PubMed] [Google Scholar]
- Legro RS, Finegood D, Dunaif A 1998 A fasting glucose to insulin ratio is a useful measure of insulin sensitivity in women with polycystic ovary syndrome. J Clin Endocrinol Metab 83:2694–2698 [DOI] [PubMed] [Google Scholar]
- Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, Gordon DJ, Krauss RM, Savage PJ, Smith Jr SC, Spertus JA, Costa F 2005 Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation 112:2735–2752 [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC 2003 Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics (Oxford, England) 19:149–150 [DOI] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M 2007 Optimal designs for two-stage genome-wide association studies. Genetic Epidemiol 31:776–788 [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ 2005 Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC 2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaudain A, Vantyghem MC, Guerci B, Hâecart AC, Auclair M, Reznik Y, Narbonne H, Ducluzeau PH, Donadille B, Lebbâe C, Bâerâeziat V, Capeau J, Lascols O, Vigouroux C 2007 New metabolic phenotypes in laminopathies: LMNA mutations in patients with severe metabolic syndrome. J Clin Endocrinol Metab 92:4835–4844 [DOI] [PubMed] [Google Scholar]
- Agarwal AK, Garg A 2006 Genetic basis of lipodystrophies and management of metabolic complications. Ann Rev Med 57:297–311 [DOI] [PubMed] [Google Scholar]
- Gambineri A, Semple RK, Forlani G, Genghini S, Grassi I, Hyden CS, Pagotto U, O'Rahilly S, Pasquali R 2008 Monogenic polycystic ovary syndrome due to a mutation in the lamin a/c gene is sensitive to thiazolidinediones but not to metformin. Eur J Endocrinol 159:347–353 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.