Summary
Previous studies have identified roles of the modulation of Notch activation by Fringe homologues in boundary formation and in regulating the differentiation of vertebrate thymocytes and Drosophila glial cells. We have investigated the role of Lunatic fringe (Lfng) expression during neurogenesis in the vertebrate neural tube. We find that in the zebrafish hindbrain, Lfng is expressed by progenitors in neurogenic regions and downregulated in cells that have initiated neuronal differentiation. Lfng is required cell autonomously in neural epithelial cells to limit the amount of neurogenesis and to maintain progenitors. By contrast, Lfng is not required for the role of Notch in interneuronal fate choice, which we show is mediated by Notch1a. The expression of Lfng does not require Notch activity, but rather is regulated downstream of proneural genes that are widely expressed by neural progenitors. These findings suggest that Lfng acts in a feedback loop downstream of proneural genes, which, by promoting Notch activation, maintains the sensitivity of progenitors to lateral inhibition and thus limits further proneural upregulation.
Keywords: Lateral inhibition, Neurogenesis, Neural progenitors, Notch, Fringe, Zebrafish
INTRODUCTION
Intercellular signalling mediated by the Notch receptor has diverse roles in the regulation of cell differentiation, proliferation and migration during development (Louvi and Artavanis-Tsakonas, 2006). The response of cells to Notch activation is context dependent and can change within a tissue at different stages of development. This is illustrated by roles of Notch in cell differentiation in the vertebrate central nervous system, in which it mediates the lateral inhibition of neurogenesis (Lewis, 1998), promotes the formation of radial glial cells (Gaiano et al., 2000), which can act as neural progenitors (Malatesta et al., 2000; Malatesta et al., 2003), and regulates the choice to differentiate into specific neuronal cell types (Peng et al., 2007). A general feature of these functions is that Notch signalling diversifies adjacent cells by lateral inhibition or induction, in which cells expressing Notch ligands change the differentiation of their neighbours.
During the lateral inhibition of neurogenesis in the vertebrate nervous system, proneural transcription factors that drive the initial steps of neuronal differentiation upregulate expression of the Notch ligands, Delta or Serrate/Jagged. These ligands activate Notch in adjacent cells by promoting a proteolytic cleavage that releases the intracellular domain of Notch (Mumm et al., 2000), which, upon binding to the transcription factor CSL, switches it from a repressor to an activator (Fryer et al., 2002). The activated CSL complex upregulates expression of members of the Hes/Her transcriptional repressor family that inhibit neurogenesis (Kageyama et al., 2007). Consequently, a pool of undifferentiated cells is maintained adjacent to differentiating neurons, within which neurogenesis can be initiated once lateral inhibition is relieved as the forming neuron migrates away from the neural epithelium.
In addition to roles in controlling cell differentiation, in some tissues Notch activation contributes to the inhibition of cell intermingling across boundaries (Dominguez and de Celis, 1998; Micchelli and Blair, 1999; Papayannopoulos et al., 1998). An example is the dorsoventral boundary in the Drosophila wing imaginal disc, in which the role of Notch depends upon the glycosyltransferase Fringe (Kim et al., 1995; Rauskolb et al., 1999). Fringe glycosylates specific sites of the Notch extracellular domain during its intracellular processing, and this glycosylation alters the affinity of Notch binding to its ligands: Delta binds more strongly to Fringe-modified Notch, whereas the binding of Serrate is decreased (Moloney et al., 2000; Panin et al., 1997). Consequently, the dorsal expression of Fringe and Serrate and the ventral expression of Delta leads to a stripe of Notch activation at the dorsoventral interface, which is required to form the compartment boundary (de Celis et al., 1996). Studies of vertebrate homologues of Fringe have suggested analogous roles in boundary formation for lunatic fringe (Lfng) in the chick forebrain (Zeltser et al., 2001), and for radical fringe in the chick limb (Laufer et al., 1997; Rodriguez-Esteban et al., 1997). Similarly, the expression of Radical fringe by zebrafish hindbrain boundary cells may modulate Notch activation that then regulates cell segregation (Cheng et al., 2004).
Some contexts have been found in which Fringe homologues are involved in the regulation of cell differentiation. During lymphopoiesis, Lfng expression in thymocyte progenitors promotes their differentiation by increasing the activation of Notch by limiting amounts of Delta expressed in thymic epithelial cells (Visan et al., 2006). Similarly, in the Drosophila CNS, Fringe is upregulated in specific glial cells and promotes Notch activation required for subtype-specific gene expression (Thomas and van Meyel, 2007). It is therefore intriguing that specific Fringe homologues are expressed in the vertebrate CNS; for example, Lfng expression occurs in dorsoventral stripes in the chick and mouse neural tube (Johnston et al., 1997; Laufer et al., 1997) that could correlate with neurogenesis, and Fringe homologues are expressed in progenitors and differentiating cells in the cerebral cortex in mouse (Ishii et al., 2000). Furthermore, overexpression of Lfng in the chick neural tube was found to increase the number of neurons (de Bellard et al., 2007). These findings raise the possibility that modulation of Notch activity by Fringe homologues regulates neurogenesis in vertebrates.
We set out to investigate the role of Lfng in the zebrafish nervous system, in which gene expression studies have suggested potential roles in boundary formation and/or neurogenesis. At early stages, lfng expression occurs at high levels in alternating segments in the hindbrain (Leve et al., 2001; Prince et al., 2001; Qiu et al., 2004), which by analogy with roles in other tissues could underlie boundary formation. In addition, lfng is expressed in dorsoventrally restricted domains in the neural tube (Prince et al., 2001) that could be associated with zones of neurogenesis. We show that Lfng limits neuronal differentiation and is required to maintain progenitor cells. Lfng acts cell autonomously in progenitors to inhibit their differentiation but, surprisingly, is upregulated downstream of proneural genes. We propose that Lfng acts in a feedback loop that maintains the competence of progenitor cells to receive lateral inhibition from differentiating neurons.
MATERIALS AND METHODS
Zebrafish lines
Wild-type, mibta52b (Jiang et al., 1996; Schier et al., 1996), notch1atp37 (Gray et al., 2001; Holley et al., 2002; van Eeden et al., 1996) and Tg(r3/r5-Gal4::UAS-RFP) embryos were produced and staged according to hours post fertilisation (hpf) and morphological criteria (Kimmel et al., 1995).
Microinjection
Blastomeres (1- to 4-cell) were microinjected with 0.45-1.8 pmol morpholino oligonucleotide (MO; Gene Tools). The lfng splice-blocking MO (ACCGTGTATACCTGTCGCATGTTTC) corresponds to the boundary between exon 1 and intron 1, and a 5 bp mismatch MO (ACCCTCTATAGCTGTGGCATCTTTC) was used as control. To test the effect on splicing, RT-PCR was performed with ∼20 24 hpf embryos using the primers P1, 5′-GGTTTCTGTTGTTTCTCTCGCAG-3′; P2, 5′-CTCGCCGTCTGTGAAGATGTA-3′; and P3, 5′-CTTTAATGGGTTTGTGGTACAGC-3′. For proneural knockdown, ascl1a, ascl1b and ngn1 MOs (Amoyel et al., 2005) were used. When used, 0.45 pmol of p53 MO (Robu et al., 2007) was coinjected. Capped RNA encoding ngn1-myc, dominant-negative CSL [dn-CSL; DBM in Wettstein et al. (Wettstein et al., 1997)], H2B-Citrine (gift from S. Megason and S. Fraser, Caltech, Pasadena, CA, USA) and Tol2 transposase (Balciunas et al., 2006; Kawakami et al., 2000) was prepared and 25-200 pg was injected at the 1-cell stage.
To generate UAS-Lfng::UAS-H2B-Citrine, a plasmid containing two UAS activator sequences followed by the E1b minimal promoter and flanked by miniTol2 sequences (Balciunas et al., 2006) was used to subclone H2B-Citrine downstream of the first UAS and the lfng coding region downstream of the second UAS. The plasmid was injected at 20 ng/μl with 25 pg Tol2 transposase RNA.
In situ hybridisation and immunohistochemistry
RNA probes used are as follows: lfng (Leve et al., 2001; Prince et al., 2001); deltaA, deltaB (Haddon et al., 1998); ngn1 (Blader et al., 1997; Kim et al., 1997; Korzh et al., 1998); ascl1a, ascl1b (Allende and Weinberg, 1994); neurod4 (Wang et al., 2003); isl1 (Chandrasekhar et al., 1997); tbx20 (Ahn et al., 2000); pax2.1 (pax2a - ZFIN) (Mikkola et al., 1992); gad67 (gad1 - ZFIN) (Martin et al., 1998); evx1 (Thaeron et al., 2000); lhx2, lhx9 (Ando et al., 2005; Seth et al., 2006); her4 (Takke et al., 1999); her12 (Bae et al., 2005; Sieger et al., 2004); sox3, sox19a (Okuda et al., 2006); rfng (Cheng et al., 2004; Qiu et al., 2004); foxb1.2 (Moens et al., 1996); krox20 (egr2a - ZFIN) (Oxtoby and Jowett, 1993); ephrin B3 (Chan et al., 2001); notch1a (Bierkamp and Campos-Ortega, 1993); notch1b, notch3 (Westin and Lardelli, 1997); vsx1 (Passini et al., 1998); gata3 (Neave et al., 1995). Digoxigenin- or fluorescein-labelled probes were synthesized and in situ hybridisation performed as described (Thisse et al., 1993). Fluorescent in situ hybridisation was performed as described (Julich et al., 2005), except that detection was with Alexa Fluor 488- or 594-labelled tyramide (Molecular Probes).
For immunohistochemistry, we used: rabbit anti-N-Cadherin [1:400 (Liu et al., 2001)], rabbit anti-EphA4 [1:450 (Irving et al., 1996)], mouse anti-HuC/D (1:100, Molecular Probes), rabbit anti-RFP (1:500, Chemicon International), sheep anti-GFP (1:500, AbD Serotec), mouse anti-neurofilament (RMO-44, 1:25, Zymed) and mouse anti-Zn-5 [1:200 (Trevarrow et al., 1990)]. Detection of primary antibodies was carried out using Alexa Fluor 488, 594 or 647 goat anti-rabbit or anti-mouse, or donkey anti-sheep, conjugates (1:500, Molecular Probes).
Mosaic analysis
Embryos were injected with control MO, lfngE1I1 MO or dn-CSL RNA together with 50 pg H2B-Citrine RNA at the 1-cell stage. Cells (20-30) were transplanted into uninjected or lfngE1I1 MO-injected embryos at 4 hpf, and embryos were allowed to develop until 24 hpf. Embryos were fixed and processed to detect GFP and HuC/D.
DAPT treatment
DAPT treatment was performed as described (Geling et al., 2002). DAPT (46 mM), dissolved in dimethyl sulphoxide (DMSO), was diluted to 0.2 mM in Danieau solution (Shih and Fraser, 1995) and applied to dechorionated embryos from 4 to 16 hpf. Control embryos were treated with Danieau solution containing the same concentration of DMSO.
Data analysis
To quantify neuronal differentiation, HuC/D-labelled cells were counted in stacked images of r3, r4 and r5, visualised by r3/r5-Gal4::UAS-RFP expression. For mosaic analysis, 1 μm optical sections were acquired every 10 μm by confocal microscopy, and the number of transplanted cells that had differentiated into HuC/D-expressing neurons was counted. The significance of results was analysed using Student's t-test.
RESULTS
The expression of lfng is associated with segmentation and neurogenesis
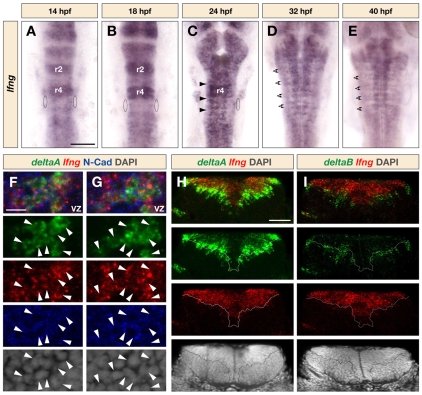
Previous studies have reported that at 12.5-14 hpf, lfng is expressed at higher levels in rhombomeres (r) 2 and 4 in the hindbrain, and by 20 hpf has been upregulated in other segments (Leve et al., 2001; Prince et al., 2001; Qiu et al., 2004). In a more extensive analysis, we observed that lfng expression is at higher levels in r2 and r4 at 14-18 hpf (Fig. 1A,B), by 24 hpf occurs in a punctate pattern (Fig. 1C), and then becomes restricted to zones adjacent to hindbrain boundaries (Fig. 1D,E). This later phase of expression is reminiscent of the pattern of neurogenesis in the zebrafish hindbrain (Amoyel et al., 2005). We therefore investigated whether Lfng has roles in hindbrain boundary formation and/or in neurogenesis.
Fig. 1.
Expression pattern of lfng in the zebrafish hindbrain. (A-E) Dorsal views of wild-type embryos following in situ hybridisation to detect lfng transcripts in the hindbrain between 14 and 40 hpf. Ovals indicate the otocyst, arrowheads in C indicate the location of hindbrain boundaries and double arrows in D and E indicate the neurogenic zones that form adjacent to boundaries. (F,G) Confocal images within the plane of the ventricular zone showing dorsal views of a 40 hpf embryo following detection of lfng (red) and deltaA (green) mRNA combined with anti-N-Cadherin (blue) and DAPI (grey) staining to reveal the cell surface and nucleus, respectively. Arrowheads indicate cells in which lfng and deltaA are coexpressed. VZ, ventricular zone. (H,I) Transverse sections of 40 hpf embryos following fluorescent in situ hybridisation to detect expression of lfng (red) in comparison to deltaA (H, green) and deltaB (I, green) combined with DAPI staining (grey). Dotted line indicates the boundary between the ventricular and mantle zones. Scale bars: 100 μm in A for A-E; 10 μm in F for F,G; 50 μm in H for H,I.
Knockdown of lfng does not affect segmentation or boundary formation
We carried out loss-of-function experiments by using a morpholino oligonucleotide (MO) complementary to the splice donor site of intron 1 of lfng (lfngE1I1 MO) (Fig. S1A in the supplementary material); this is predicted to block splicing of intron 1 and terminate the open reading frame upstream of the region conserved between Fringe proteins (Johnston et al., 1997; Leve et al., 2001; Prince et al., 2001; Qiu et al., 2004). Microinjection of lfngE1I1 MO completely blocked splicing of intron 1, whereas a control MO with five changes in nucleotide sequence was ineffective at blocking intron 1 splicing (Fig. S1B in the supplementary material). We first assessed whether Lfng is involved in boundary formation or segmentation in the hindbrain. We detected no change in the expression of hindbrain boundary cell markers or in the formation of sharp borders of segmental markers following lfng knockdown (see Fig. S2 in the supplementary material). Lfng therefore does not appear to be required for these processes.
Lfng limits the amount of neurogenesis
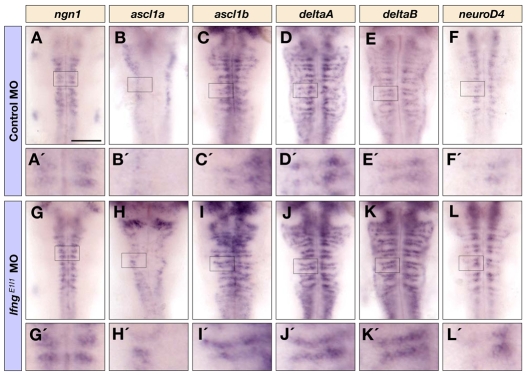
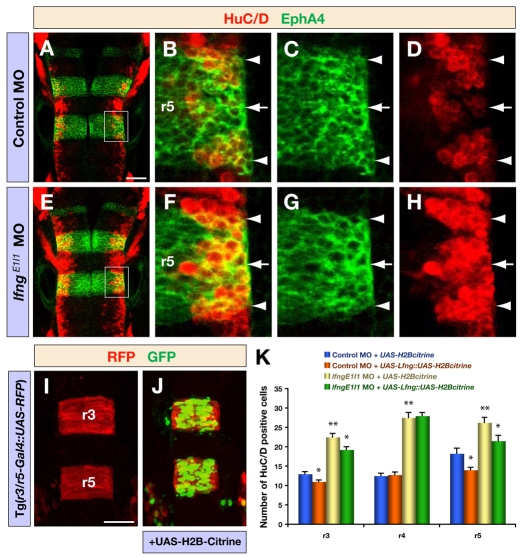
To address whether Lfng has a role in the regulation of neurogenesis, we analysed the effect of lfng knockdown on the expression of proneural and Delta genes that are upregulated at the onset of neuronal differentiation (Allende and Weinberg, 1994; Haddon et al., 1998; Korzh et al., 1998), on neurod4, which is upregulated downstream of proneural genes (Park et al., 2003; Wang et al., 2003), and on HuC/D, which marks differentiating neurons in the mantle zone (Park et al., 2000). We found that lfng knockdown leads to increased expression of proneural and Delta genes, including ngn1 (neurog1 - ZFIN), ascl1a, ascl1b, deltaA and deltaB, and of neurod4 at all stages analysed, for example at 18 and 28 hpf (see Fig. S3 in the supplementary material) and 36 hpf (Fig. 2A-L). This increase appeared to be due in part to a higher level of gene expression per cell, as more intense signals are observed at single cell resolution following lfng knockdown (see, for example, Fig. 2A′-L′). In addition, there was an increased number of cells expressing high levels of proneural and Delta genes, seen for example at 36 hpf, when neurogenesis is confined to narrow neurogenic zones: in control embryos there is a mixture of low- and high-expressing cells within these zones, whereas following lfng knockdown there are more cells with high level expression (Fig. 2B′-D′,H′-J′). Consistent with an increased number of cells initiating neurogenesis, we observed more differentiating neurons in the mantle zone marked by HuC/D (Fig. 3A-H). In order to quantify the effect on neuronal differentiation, we compared the number of HuC/D-positive cells in control and lfng MO embryos. We found that lfng knockdown leads to a 1.7-fold increase in the number of differentiating neurons, both at 18 and 30 hpf (Fig. 3K and data not shown).
Fig. 2.
Knockdown of lfng increases the initiation of neuronal differentiation. Dorsal views of control MO (A-F′) and lfngE1I1 MO (G-L′) injected embryos at 36 hpf showing ngn1 (A,A′,G,G′), ascl1a (B,B′,H,H′), ascl1b (C,C′,I,I′), deltaA (D,D′,J,J′), deltaB (E,E′,K,K′), and neurod4 (F,F′,L,L′) expression in the hindbrain. A′-L′ are higher magnifications of the indicated areas in A-L. Scale bar: 100 μm.
Fig. 3.
Knockdown of lfng produces a neurogenic phenotype. (A-H) Confocal images showing dorsal views of control MO (A-D) and lfngE1I1 MO (E-H) injected embryos at 30 hpf, immunostained for HuC/D (red) and EphA4 (green) to mark postmitotic neurons and r3/r5, respectively. B-D and F-H are higher magnifications of the areas indicated in A and E, respectively. Arrowheads and arrows indicate rhombomere boundaries and centres, respectively. (I,J) Confocal images of the expression of RFP (red) and H2B-Citrine (green) in r3 and r5 in Tg(r3/r5-Gal4::UAS-RFP) embryos at 18 hpf, either noninjected (I) or injected with UAS-H2B-Citrine plasmid (J). Tg(r3/r5-Gal4::UAS-RFP) is an enhancer trap that drives Gal4 and mCherry expression in r3/r5; the expression of UAS-citrine in r3/r5 illustrates the targeting of transgene expression to these rhombomeres. (K) Quantification of the number of differentiating neurons (HuC/D expression) in Tg(r3/r5-Gal4::UAS-RFP) 18 hpf embryos injected with control MO or lfngE1I1 MO together with either a control vector (UAS-H2B-Citrine) or a vector to overexpress Lfng (UAS-Lfng::UAS-H2B-Citrine). Transgenic expression of Lfng in r3/r5 leads to a decreased number of differentiated neurons and partly rescues the effect of lfng knockdown, whereas neurogenesis in r4 is not affected. Values are mean ± standard error, neurons in four embryos were counted for each experimental group; *, P<0.05, **, P<0.01. Scale bars: 50 μm in A for A-H; 50 μm in I for I,J.
These findings raised the question of whether Lfng limits the production of all types of neurons in the hindbrain, or acts in a specific subset. To examine this, we analysed markers of different neuronal types, including interneurons, and reticulospinal, branchiomotor, somatic motor and commissural neurons. We found that there was an increase in the number of all neuronal types examined, apart from reticulospinal neurons (see Fig. S4 in the supplementary material). These findings reveal that Lfng has a widespread role in limiting neurogenesis, with the exception of reticulospinal neurons.
A potential difficulty with MOs is that they can have off-target effects leading to p53-mediated cell death, although this can be suppressed by coinjection with p53 MO (Robu et al., 2007). Since such apoptosis leads to the loss of cells, it seemed unlikely that this underlies the observed increase in neurogenesis. Nevertheless, we tested the effect of coinjecting p53 MO and found that this does not alter the increase in neurogenesis following injection of lfng MO (data not shown). In addition, we assessed the specificity of the phenotypic effect of lfng MO in a rescue experiment in which Lfng is transgenically overexpressed in r3/r5 using a Gal4 enhancer-trap line (Fig. 3I,J). We found that transgenic overexpression of Lfng decreased the amount of neurogenesis in control embryos and partly suppressed the lfng MO-induced increase in neurogenesis (Fig. 3K); both of these effects specifically occurred in r3/r5, where Lfng is ectopically expressed, and not in r4.
Lfng is required to maintain progenitors
A potential explanation for our findings is that lfng knockdown leads to a deficiency in the lateral inhibition of neurogenesis. We therefore analysed expression of Hes/Her family genes that are targets of the Notch pathway during lateral inhibition. We observed no change in the expression of her4 (her4.1 - ZFIN) at 28 hpf (see Fig. S3 in the supplementary material), whereas by 40 hpf the expression of her4 and her12 was decreased following lfng knockdown (Fig. 4A,B,E,F). This appeared to be due to a decrease in the levels of expression, as well as in the number of cells expressing the Notch effectors (Fig. 4A′,B′,E′,F′). However, her4 expression may not provide a sensitive read-out of Notch modulation since, as occurs for the homologous genes in Drosophila (Castro et al., 2005), its expression may also be upregulated by proneural genes (Yeo et al., 2007); the upregulation of proneural genes following lfng knockdown could thus mask any decrease in Notch activation. A potential explanation for the late decrease in her4 and her12 gene expression is that the reduced lateral inhibition of neurogenesis led to a depletion of progenitors, so we analysed the expression of sox3 and sox19a, which mark neural progenitor cells. We found that, although no change was detected at 28 hpf (see Fig. S3 in the supplementary material), by 40 hpf there was lower expression of these markers, which is suggestive of fewer neural progenitors in the neural tube (Fig. 4C′,D′,G′,H′). Taken together, these results suggest that knockdown of lfng eventually leads to a depletion of progenitor cells, consistent with a role in Notch-mediated lateral inhibition of neurogenesis.
Fig. 4.
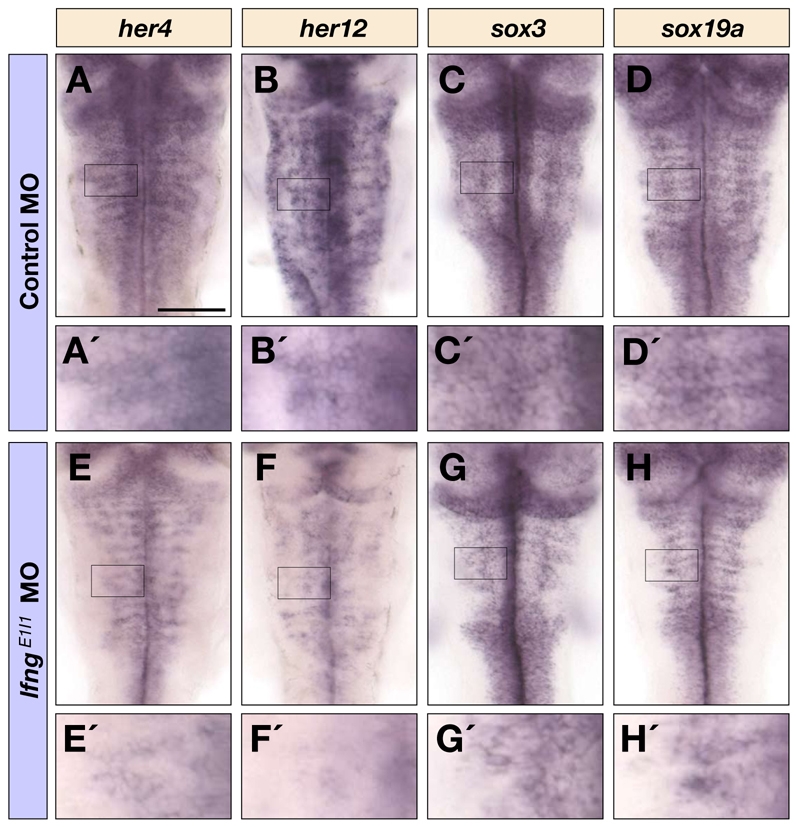
Notch target gene and progenitor marker expression following lfng knockdown. Dorsal views of control MO (A-D′) and lfngE1I1 MO (E-H′) injected embryos at 40 hpf following detection of her4 (A,A′,E,E′), her12 (B,B′,F,F′), sox3 (C,C′,G,G′) and sox19a (D,D′,H,H′) mRNA in the hindbrain. A′-H′ are higher magnifications of the indicated areas in A-H. Scale bar: 100 μm.
Lfng is not required for neuronal subtype specification
Recent work has shown that, in addition to being required in progenitors during the lateral inhibition of differentiation, Notch1 activation regulates the choice to form V2b rather than V2a interneurons in the spinal cord (Batista et al., 2008; Del Barrio et al., 2007; Peng et al., 2007). This raises the possibility that, in addition to a role in the inhibition of neurogenesis, Lfng enables the activation of Notch required for this fate choice. We therefore analysed whether Notch1 is required for interneuron fate choice in the zebrafish hindbrain. We found that, whereas notch1b and notch3 are widely expressed in the nervous system, notch1a expression occurs initially at higher levels in r2 and r4 and subsequently in neurogenic zones (see Fig. S5 in the supplementary material). Furthermore, unlike other Notch receptors (data not shown), notch1a is coexpressed with Delta genes (see Fig. S6 in the supplementary material) and upregulated in neurogenic mib mutants (Fig. 5A-D). We therefore analysed whether interneuron subtype choice regulated by Notch signalling is altered in the notch1atp37 mutant. Indeed, we found that there is a loss of V2b and an increase in V2a interneurons in this mutant (Fig. 5E,F,I,J), as occurs following global Notch inhibition (Batista et al., 2008; Peng et al., 2007).
Fig. 5.
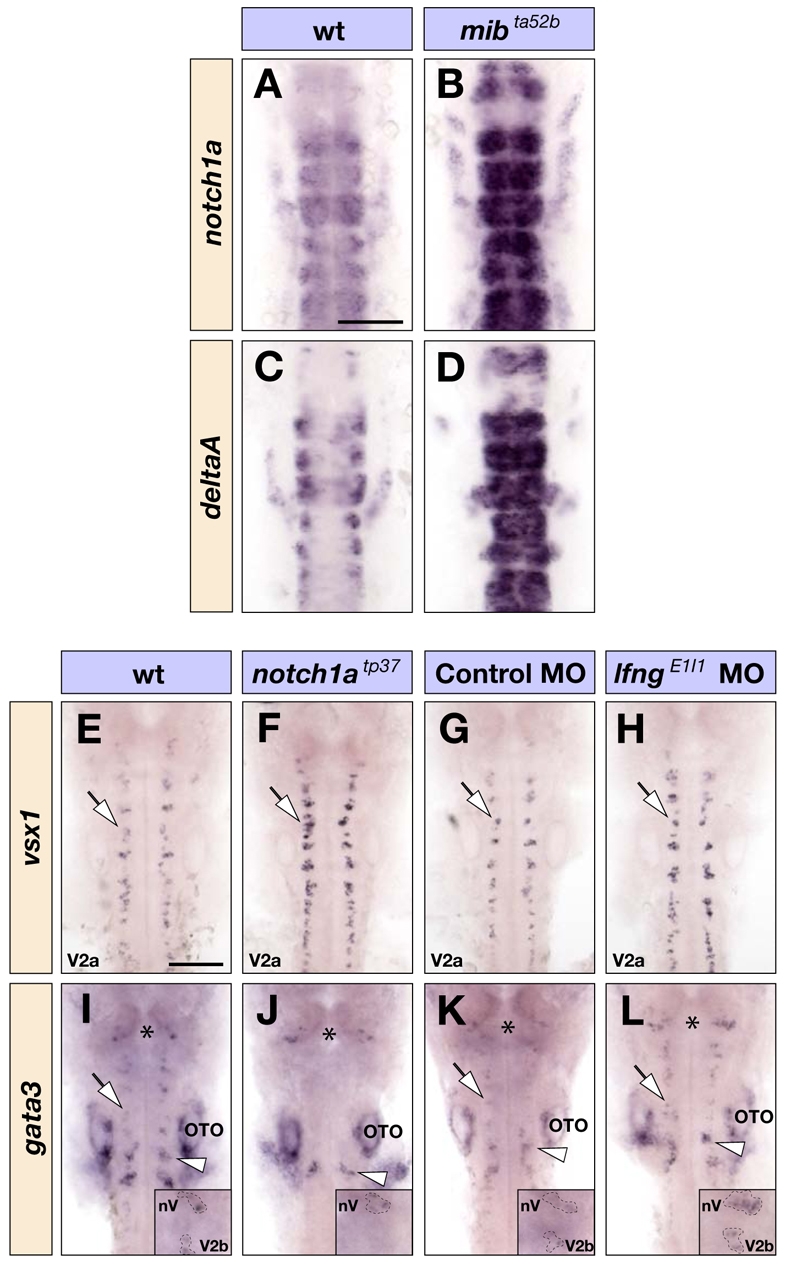
Lfng and Notch1a in neuronal subtype choice. (A-D) Dorsal views of wild-type (A,C) and mibta52b homozygous mutant (B,D) embryos at 18 hpf showing notch1a (A,B) and deltaA (C,D) expression in the hindbrain. (E-L) Dorsal views of wild-type (E,I), notch1atp37 homozygous mutant (F,J) embryos, or control MO (G,K) and lfngE1I1 MO (H,L) injected embryos at 26 hpf showing vsx1 (E-H) and gata3 (I-L) expression in the hindbrain that mark V2a and V2b interneurons (indicated by arrows), respectively. Expression of gata3 in nV and nVII branchiomotor neurons, indicated by the asterisks and arrowheads (I-L), respectively, is not affected in notch1atp37 embryos but increased following knockdown of lfng (insets in I-L). OTO, otocyst. Scale bars: 100 μm
If Lfng is required for Notch1a function in differentiating neurons, knockdown of lfng would have a similar effect on neuronal subtype specification as occurs in the notch1atp37 mutant. However, we found that lfng knockdown does not lead to a switch in the fate of interneuron subtypes, but rather increases the numbers of both interneuron populations (Fig. 5G,H,K,L). We therefore conclude that Lfng is required to promote the lateral inhibition of neurogenesis but not for interneuron subtype specification.
The expression of lfng is regulated by proneural genes
In order to understand how Lfng contributes to the inhibition of neurogenesis, it is essential to determine in which cells lfng is expressed and how its expression is regulated. For example, lfng could be upregulated by Notch activation in progenitors and/or by proneural genes that are widely expressed at low levels and upregulated in differentiating neurons. We therefore compared neuronal marker and lfng expression at single cell resolution. deltaA is widely expressed at low levels in progenitors, and upregulated in cells selected to differentiate during lateral inhibition, whereas deltaB is only expressed in cells that have initiated differentiation (Haddon et al., 1998). In confocal sections in a superficial plane of the neural epithelium we found that lfng overlaps with deltaA expression (Fig. 1F,G). To visualise how lfng expression relates to the transition from progenitors to differentiating neurons, we analysed transverse hindbrain sections and found that lfng expression is confined to the ventricular zone, where it overlaps with the low- and high-level expression of deltaA (Fig. 1H). By contrast, lfng expression occurs complementary to the expression of deltaB in differentiating neurons migrating to the mantle zone (Fig. 1I). We conclude that lfng is coexpressed with Delta genes in neural progenitors and is downregulated in cells that have embarked upon neurogenesis.
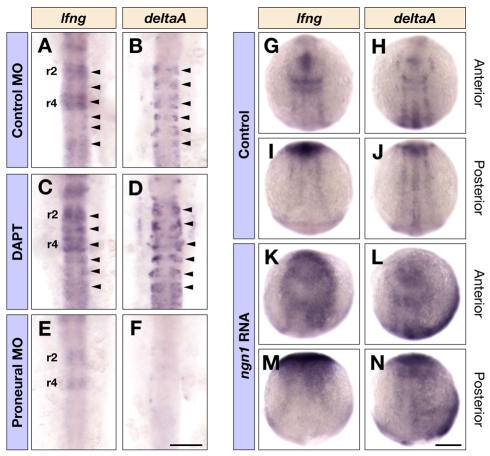
To test whether lfng is regulated by proneural genes and/or by Notch activation, we first carried out knockdown and gain-of-function experiments with proneural genes. Knockdown of ngn1 or ascl1b alone had only a moderate effect on lfng expression, as was also seen for deltaA expression (see Fig. S6 in the supplementary material). By contrast, triple knockdown of ngn1, ascl1b and ascl1a led to a major decrease in lfng expression (Fig. 6A,E and see Fig. S6 in the supplementary material). In gain-of-function experiments, we found that misexpression of ngn1 (Fig. 6G,I,K,M), but not of ascl1b (data not shown), led to upregulation of lfng expression. Taken together, these observations suggest that lfng is a downstream target of multiple proneural genes that can compensate for each other following knockdown, but there are differences between proneural genes in their ability to upregulate lfng.
Fig. 6.
Regulation of lfng expression. (A-F) Dorsal views of control MO (A,B) and triple ascl1a, ascl1b and ngn1 MO (E,F) injected embryos, or DAPT (C,D) treated embryos at 16 hpf showing lfng (A,C,E) and deltaA (B,D,F) expression in the hindbrain. Arrowheads in A-D indicate the expression of lfng and deltaA in the middle of each rhombomere. (G-N) Dorsal views of control (G-J) and ngn1 RNA (K-N) injected embryos at ∼3-somite stage showing lfng (G,I,K,M) and deltaA (H,J,L,N) expression in the anterior and posterior neural plate. Scale bars: 100 μm in F for A-F; 200 μm in N for G-N.
An alternative explanation for the effects of proneural knockdown or misexpression on lfng expression is that they are secondary to the regulation of Delta/Serrate genes that activate Notch. If proneural genes (or downstream transcription factors) regulate lfng directly, blocking Notch activation will lead to more cells expressing lfng due to the consequent loss of lateral inhibition and increase in neurogenesis. By contrast, if Notch activation regulates lfng, blocking Notch activation will decrease lfng expression. We therefore tested the effect of inhibiting Notch activation with DAPT (Geling et al., 2002), and found that this leads to increased lfng expression (Fig. 6A,C), concurrent with more neurogenesis marked by strong deltaA expression (Fig. 6B,D). A further possibility is that proneural genes and Notch activation synergise to upregulate lfng. However, we found no further change in lfng expression by combining triple proneural gene knockdown with the blocking of Notch activation with DAPT, or with the overexpression of ngn1 together with dominant active Su(H) to activate the Notch pathway (data not shown). These data suggest that lfng is upregulated downstream of proneural genes, and not indirectly via Notch activation by Delta ligands.
Lfng acts cell autonomously to inhibit neurogenesis
The finding that lfng expression is upregulated downstream of proneural genes raises the question, which cells is Lfng required in? Lfng could act cell autonomously in progenitors to promote Notch activation and thus inhibit their differentiation. Alternatively, lfng expression in cells selected to differentiate could act non-autonomously to increase Notch activation in adjacent progenitors, as Lfng promotes the translocation of Delta to the cell surface (Sakamoto et al., 2002). These possibilities lead to different predictions for the effect of mosaic knockdown. In the former model, lfng knockdown cells will have increased differentiation, similar to the effect of inhibiting Notch pathway activation with dominant-negative CSL [DBM in Wettstein et al. (Wettstein et al., 1997)]. In the latter model, there would be no increase in the differentiation of lfng knockdown cells, as the increase in neurogenesis occurs in adjacent cells.
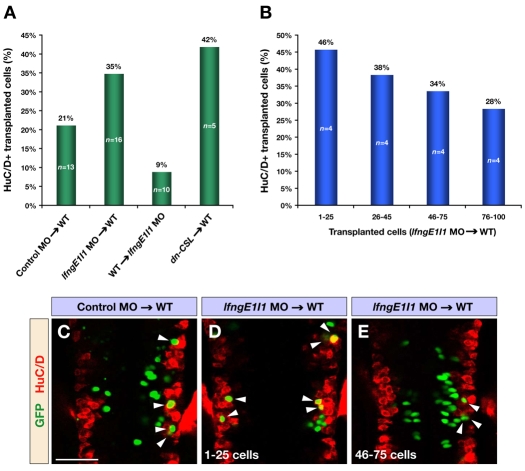
To test these models, we carried out transplantations to generate embryos mosaic for lfng MO plus GFP expression, and determined the relative number of Lfng-deficient cells that had differentiated or remained as progenitors in comparison with mosaic GFP expression alone. We found that there was increased differentiation of lfng MO cells transplanted into a noninjected host, as also occurred for cells overexpressing dn-CSL (Fig. 7A). Furthermore, in the reciprocal experiment there was decreased neuronal differentiation of wild-type cells that had been transplanted into a lfng MO-injected host (Fig. 7A). We observed that, when a large number of transplanted lfng MO cells were present, a lower proportion of these cells differentiated compared with embryos in which there were a low number of Lfng-deficient cells (Fig. 7B). These data are consistent with competition whereby lfng MO cells preferentially undergo neurogenesis at the expense of cells in which Lfng function is not inhibited. In mosaics with a low number of Lfng-deficient cells (Fig. 7D), most are competing with cells expressing lfng, and thus a high proportion of Lfng-deficient cells differentiate. By contrast, in mosaics with a large number of Lfng-deficient cells (Fig. 7E), many will be competing with each other and not be biased to differentiate preferentially at the expense of their neighbours.
Fig. 7.
Mosaic analysis of Lfng function. (A) Transplantations of cells from control or lfngE1I1 MO- and dn-CSL RNA-injected donor embryos into wild-type or lfngE1I1 MO-injected hosts. The bar graph shows the percentage of transplanted cells that have differentiated into neurons marked by HuC/D. For lfngE1I1 MO and dn-CSL cells transplanted into wild-type hosts, only embryos with <100 transplanted cells are included. All changes were significant compared with control MO transplantations (P<0.03). (B) Analysis of lfngE1I1 MO cells transplanted into wild-type embryos. The bar graph shows the percentage of Lfng-deficient cells that have differentiated into neurons marked by HuC/D. Note the linear relationship between the increasing number of transplanted cells and decreasing number of transplanted cells differentiating into neurons. (C-E) Confocal images of dorsal views of wild-type embryos transplanted with control MO (C) and lfngE1I1 MO (D,E) injected cells at 24 hpf immunostained for GFP (green) and HuC/D (red) to mark transplanted cells and postmitotic neurons, respectively. The embryo in E contains a higher number of transplanted cells compared with the one in D. Arrowheads indicate transplanted cells that have differentiated into neurons. Scale bar: 50 μm.
DISCUSSION
The expression of Lfng initially in a segmental pattern and subsequently in neurogenic regions raised the possibility that this Notch modulator may have roles in the regulation of segmentation and/or neurogenesis. We find that lfng is expressed in neurogenic domains of the zebrafish hindbrain, where its expression occurs in progenitors and is rapidly downregulated in differentiating neurons. The results of loss-of-function studies reveal that Lfng is required to limit the amount of neurogenesis in the hindbrain and to maintain neural progenitors at late stages. Analysis of mosaic embryos reveals that Lfng acts cell autonomously in progenitors to inhibit their differentiation. Furthermore, the bias of lfng knockdown cells to preferentially differentiate depends upon the degree of mosaicism, consistent with the competition of cells that occurs during the lateral inhibition of neurogenesis. Taken together, these findings suggest that Lfng acts within progenitor cells to promote the lateral inhibition of neurogenesis.
Significance of segmental expression of lfng
In some tissues, Fringe homologues regulate boundary formation by modulating Notch activity at the interface of expressing and nonexpressing cells (Dominguez and de Celis, 1998; Laufer et al., 1997; Panin et al., 1997; Rodriguez-Esteban et al., 1997). It was therefore possible that the elevated expression of lfng in r2 and r4 reflected a role in hindbrain boundary formation. However, we found no effect of lfng knockdown on segmentation or boundary marker expression. Furthermore, hindbrain boundary cells initially form in mib mutants in which there is a major decrease in Notch activation, but are not maintained, as decreased lateral inhibition of neurogenesis leads to the loss of neural epithelial cells (Cheng et al., 2004). There is thus no evidence to support a role for Notch in hindbrain boundary formation. An alternative explanation for segmental lfng expression is suggested by the observation that the proneural gene ascl1b is initially expressed at higher levels throughout r2 and r4 (Amoyel et al., 2005), correlating with neurogenesis occurring in even-before odd-numbered segments (Bally-Cuif et al., 1998; Maves et al., 2002). The early segmental phase of lfng expression may therefore reflect the fact that neurogenesis is segmentally regulated, rather than suggest a role in segmentation.
Notch in the lateral inhibition of neurogenesis
There are similarities and differences between the effect of lfng knockdown and the major deficiency of Notch activation in mib mutants (Itoh et al., 2003; Jiang et al., 1996). In mib mutants, there is a 2- to 4-fold increase in early differentiating neurons, such as reticulospinal neurons, and the consequent depletion of progenitors leads to a decrease or loss of later-generated branchiomotor and commissural neurons, with neurogenesis almost absent by 24 hpf (Bingham et al., 2003; Jiang et al., 1996). lfng knockdown leads to a 1.7-fold increase in overall neurogenesis and increased production of many neuronal cell types, including branchiomotor neurons, somatic motor neurons, interneurons and commissural neurons. This is a milder neurogenic phenotype than mib mutants, consistent with Lfng increasing, rather than being essential for, Notch activation. As would be anticipated, lfng knockdown leads to later depletion of neural progenitors than in mib mutants, with normal expression of progenitor markers at 24 hpf and a decrease by 40 hpf. However, there is a distinct effect of the mib mutation compared with lfng knockdown on reticulospinal neurons, which are born during gastrulation and are the first to differentiate in the hindbrain (Hanneman et al., 1988; Mendelson, 1986). Whereas in mib mutants the single Mauthner reticulospinal neuron increases to 3-4 neurons (Jiang et al., 1996), there is no increase following lfng knockdown. One possibility is that the milder neurogenic effect of lfng knockdown cannot be detected for reticulospinal neurons due to their low number, or there may be differences in the regulation of their differentiation compared with subsequent neurogenesis.
Our findings appear contrary to a study in which retroviral-mediated overexpression of Lfng in the chick neural tube increases the number of neurons (de Bellard et al., 2007), as we found that transgenic expression of Lfng in r3/r5 decreases neurogenesis. One contributory factor is suggested by our observation that widespread knockdown of lfng has less of an effect on neurogenesis than occurs cell autonomously for mosaic knockdown, consistent with the competition to differentiate during the lateral inhibition of neurogenesis. This can explain our finding that widespread transgenic overexpression of Lfng within r3/r5 inhibits neurogenesis only to a modest extent. A potential explanation for increased neurogenesis in the chick neural tube is suggested by the major increase in cell proliferation that occurs following Lfng expression (de Bellard et al., 2007). Since effects on neuronal differentiation were analysed 24-48 hours after retroviral infection, expansion of the progenitor pool may underlie the increased number of neurons and outweigh a modest inhibition of differentiation by widespread Lfng expression. There would be less of an impact of such effects in our study, in which the number of neurons was analysed 6 hours after the onset of Gal4 driver expression.
Notch1a and neuronal specification
In addition to its role in mediating the lateral inhibition of neurogenesis, Notch mediates inhibitory or inductive signalling required for the specification of interneuronal subtype (Batista et al., 2008; Del Barrio et al., 2007; Peng et al., 2007). We find that in zebrafish, Notch1a is upregulated in differentiating neurons, suggesting that it is expressed downstream of proneural genes, whereas other Notch genes, including notch1b, are expressed predominantly in progenitors. These data suggest that there has been subfunctionalisation, as found for other genes duplicated in the zebrafish genome (Postlethwait et al., 2004), with expression of Notch1 divided between orthologues expressed in progenitors and neurons. We show Notch1a is required for interneuron subtype fate choice, but lfng knockdown leads to an increase of both interneuron subtypes, rather than a decrease in the population that requires Notch1a. We therefore conclude that Lfng acts to limit the amount of neuronal differentiation, but is not required for subtype specification regulated by Notch1a.
Regulation of lfng expression during neurogenesis
We find that lfng expression is decreased following knockdown of proneural genes, and increased following DAPT treatment that inhibits Notch activation, leading to increased proneural gene expression. Furthermore, the effect of proneural knockdown or overexpression on lfng expression was not exacerbated by the alteration of Notch activation, arguing against a synergistic input of proneural and Notch activity. These findings suggest that, like Delta genes, lfng is a target of proneural proteins or downstream transcription factors and does not require Notch activity. This is consistent with studies in mouse suggesting that Lfng is a direct target of proneural transcription factors (Castro et al., 2006), and furthermore, the proneural factor binding motif is present in the zebrafish lfng gene (D. S. Castro, personal communication). However, whereas lfng expression overlaps with deltaA that is widely expressed in progenitors, it is not coexpressed with deltaB in cells committed to neuronal differentiation and initiating migration from the neural epithelial layer. The rapid downregulation of lfng upon the onset of neuronal differentiation suggests that there are regulatory inputs in addition to proneural genes, such as a transcription factor(s) restricted to progenitors.
Analysis of the effects of knockdown or overexpression of single proneural genes suggests that they have overlapping and specific roles in the regulation of lfng. The finding that lfng is upregulated by ngn1 overexpression but little affected by ngn1 knockdown suggests that other proneural genes also regulate lfng. However, ascl1b overexpression did not upregulate lfng, and ascl1a expression overlaps with lfng only in dorsal regions. Further work is required to determine whether this complexity is due, for example, to other factors required for specific proneural genes to regulate lfng.
Lfng in the regulatory logic of the lateral inhibition of neurogenesis
The initiation of neuronal differentiation requires an increase in proneural gene expression from the low level that occurs widely in the neural epithelium. This increase is enabled by positive feedback on proneural expression within cells (Bae et al., 2000; Culi and Modolell, 1998; Dubois et al., 1998; Koyano-Nakagawa et al., 2000), which is countered by inhibitory loops that occur within and between cells (Bai et al., 2007; Heitzler et al., 1996; Kunisch et al., 1994; Ohtsuka et al., 1999). The process of lateral inhibition requires that there is sufficient Notch activation in progenitors to inhibit proneural expression and thus prevent differentiation.
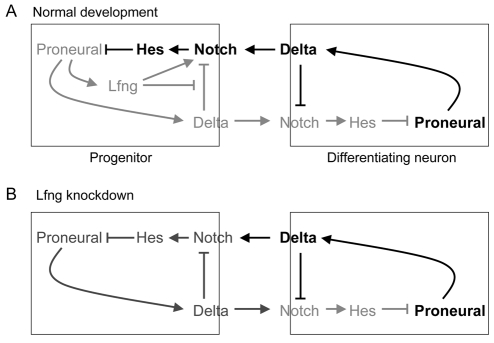
Our findings that lfng is upregulated by proneural genes raises the question of where Lfng acts in the regulatory logic of lateral inhibition. A potential clue is the observation that the level of proneural and Delta gene expression per cell appears to be higher following lfng knockdown. This finding suggests that Lfng acts in a feedback loop downstream of proneural genes, in which the promotion of Notch activation by Lfng limits further upregulation of proneural and Delta genes in progenitors (Fig. 8). As found in other tissues, Lfng may increase the binding of Notch to Delta expressed on adjacent cells, but it is not obvious in this model why lfng would be upregulated in progenitors downstream of proneural genes rather than by Notch activation. Previous studies have found another functional relationship between Lfng and Delta genes that provides a potential explanation for this. Studies in the Drosophila wing margin (Glittenberg et al., 2006; Micchelli et al., 1997) and peripheral nervous system (Jacobsen et al., 1998) have shown that, whereas Delta proteins activate Notch on adjacent cells, Delta also has cis-interactions with Notch that cell autonomously inhibits its activation. Similarly, in vertebrate cell culture, cis-interactions with Delta decrease the level of cell surface Notch, thus decreasing its availability for activation by Delta presented by adjacent cells (Sakamoto et al., 2002). Lfng blocks such cis-interactions, and could thus lead to increased Notch at the cell surface (Sakamoto et al., 2002). Recent work showing that cis-interactions with Notch promote the endocytosis of DeltaD in the zebrafish hindbrain suggests that inhibition occurs by removal of the Notch-Delta complex from the cell surface (Matsuda and Chitnis, 2009).
Fig. 8.
Model of role and regulation of Lfng in the lateral inhibition of neurogenesis. Depiction of regulatory relationships between Lfng and components of the lateral inhibition network. Strongly expressed or activated components are in black and less strongly expressed or activated components in grey. (A) In progenitor cells, expression of proneural genes upregulates lfng, which by increasing Notch activation ensures that progenitors are sensitive to lateral inhibition of their differentiation. Lfng may inhibit further upregulation of proneural genes by promoting trans-activation of Notch and/or by blocking cis-inhibition of Notch that would otherwise occur due to the expression of Delta downstream of proneural genes. Lfng is downregulated in differentiating neurons. (B) Following knockdown of lfng, proneural gene expression that drives neuronal differentiation is elevated in progenitors due to decreased lateral inhibition, possibly as a consequence of cis-inhibition of Notch by Delta that leads to positive feedback on proneural upregulation.
Based upon these findings, we propose the following model (Fig. 8). Progenitor cells have widespread expression of proneural and downstream Delta genes, and compete to further upregulate proneural genes and laterally inhibit their neighbours. Due to cis-inhibition, there will inevitably be decreased activation of Notch in Delta-expressing cells. If cis-inhibition is too strong in progenitor cells, it will decrease their sensitivity to lateral inhibition, leading to positive feedback in which increased expression of proneural and Delta genes inhibits Notch activation that would normally inhibit further upregulation of proneural genes (Fig. 8). The coexpression of lfng with Delta downstream of proneural genes prevents this loop by blocking cis-inhibition, thus preventing the inappropriate upregulation of proneural genes that would lead to neurogenesis. By contrast, in cells that have initiated differentiation, lfng is downregulated and no longer coupled to Delta expression. Consequently, cis-inhibition of Notch by Delta will facilitate the further upregulation of proneural genes. This proposed change in cis-inhibition is consistent with recent work showing that in some progenitors DeltaD endocytosis is mainly due to trans-interactions, whereas in others cis-interactions are more important (Matsuda and Chitnis, 2009).
An alternative view of how Lfng may regulate neurogenesis is suggested by evidence that oscillations of proneural gene expression maintain progenitor cells and sustained high levels of expression promote neurogenesis (Kageyama et al., 2008). Such oscillations require Notch activation in order to downregulate proneural expression from its peak level (Shimojo et al., 2008). A desensitisation of Notch due to cis-inhibition by Delta would lead to decreased signalling, such that there is sustained proneural gene expression and consequently neurogenesis. Further insights into the role of Lfng may therefore be obtained by real-time visualisation of its expression, and by analysing whether lfng knockdown affects oscillations of proneural gene expression.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/15/2523/DC1
Supplementary Material
We thank Francois Guillemot, Qiling Xu and Diogo Castro for discussions; and Julian Lewis, Scott Fraser, Sean Megason, Kamal Sharma and Caroline Erter Burns for constructs and reagents. This work was supported by the Medical Research Council (N.N., T.W.-A., S.G. and D.G.W.), by a BioFuture Grant (BMBF 0311889) of the German Ministry of Education and Research (R.W.K.) and by the Studienstiftung des deutschen Volkes (M.D.). Deposited in PMC for release after 6 months.
References
- Ahn, D. G., Ruvinsky, I., Oates, A. C., Silver, L. M. and Ho, R. K. (2000). tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech. Dev. 95, 253-258. [DOI] [PubMed] [Google Scholar]
- Allende, M. L. and Weinberg, E. S. (1994). The expression pattern of two zebrafish achaete-scute homolog (ash) genes is altered in the embryonic brain of the cyclops mutant. Dev. Biol. 166, 509-530. [DOI] [PubMed] [Google Scholar]
- Amoyel, M., Cheng, Y. C., Jiang, Y. J. and Wilkinson, D. G. (2005). Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development 132, 775-785. [DOI] [PubMed] [Google Scholar]
- Ando, H., Kobayashi, M., Tsubokawa, T., Uyemura, K., Furuta, T. and Okamoto, H. (2005). Lhx2 mediates the activity of Six3 in zebrafish forebrain growth. Dev. Biol. 287, 456-468. [DOI] [PubMed] [Google Scholar]
- Bae, S., Bessho, Y., Hojo, M. and Kageyama, R. (2000). The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development 127, 2933-2943. [DOI] [PubMed] [Google Scholar]
- Bae, Y. K., Shimizu, T. and Hibi, M. (2005). Patterning of proneuronal and inter-proneuronal domains by hairy- and enhancer of split-related genes in zebrafish neuroectoderm. Development 132, 1375-1385. [DOI] [PubMed] [Google Scholar]
- Bai, G., Sheng, N., Xie, Z., Bian, W., Yokota, Y., Benezra, R., Kageyama, R., Guillemot, F. and Jing, N. (2007). Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev. Cell 13, 283-297. [DOI] [PubMed] [Google Scholar]
- Balciunas, D., Wangensteen, K. J., Wilber, A., Bell, J., Geurts, A., Sivasubbu, S., Wang, X., Hackett, P. B., Largaespada, D. A., McIvor, R. S. et al. (2006). Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2, e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally-Cuif, L., Dubois, L. and Vincent, A. (1998). Molecular cloning of Zcoe2, the zebrafish homolog of Xenopus Xcoe2 and mouse EBF-2, and its expression during primary neurogenesis. Mech. Dev. 77, 85-90. [DOI] [PubMed] [Google Scholar]
- Batista, M. F., Jacobstein, J. and Lewis, K. E. (2008). Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons. Dev. Biol. 322, 263-275. [DOI] [PubMed] [Google Scholar]
- Bierkamp, C. and Campos-Ortega, J. A. (1993). A zebrafish homologue of the Drosophila neurogenic gene Notch and its pattern of transcription during early embryogenesis. Mech. Dev. 43, 87-100. [DOI] [PubMed] [Google Scholar]
- Bingham, S., Chaudhari, S., Vanderlaan, G., Itoh, M., Chitnis, A. and Chandrasekhar, A. (2003). Neurogenic phenotype of mind bomb mutants leads to severe patterning defects in the zebrafish hindbrain. Dev. Dyn. 228, 451-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blader, P., Fischer, N., Gradwohl, G., Guillemot, F. and Strahle, U. (1997). The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development 124, 4557-4569. [DOI] [PubMed] [Google Scholar]
- Castro, B., Barolo, S., Bailey, A. M. and Posakony, J. W. (2005). Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development 132, 3333-3344. [DOI] [PubMed] [Google Scholar]
- Castro, D. S., Skowronska-Krawczyk, D., Armant, O., Donaldson, I. J., Parras, C., Hunt, C., Critchley, J. A., Nguyen, L., Gossler, A., Gottgens, B. et al. (2006). Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev. Cell 11, 831-844. [DOI] [PubMed] [Google Scholar]
- Chan, J., Mably, J. D., Serluca, F. C., Chen, J. N., Goldstein, N. B., Thomas, M. C., Cleary, J. A., Brennan, C., Fishman, M. C. and Roberts, T. M. (2001). Morphogenesis of prechordal plate and notochord requires intact Eph/ephrin B signaling. Dev. Biol. 234, 470-482. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar, A., Moens, C. B., Warren, J. T., Jr, Kimmel, C. B. and Kuwada, J. Y. (1997). Development of branchiomotor neurons in zebrafish. Development 124, 2633-2644. [DOI] [PubMed] [Google Scholar]
- Cheng, Y. C., Amoyel, M., Qiu, X., Jiang, Y. J., Xu, Q. and Wilkinson, D. G. (2004). Notch activation regulates the segregation and differentiation of rhombomere boundary cells in the zebrafish hindbrain. Dev. Cell 6, 539-550. [DOI] [PubMed] [Google Scholar]
- Culi, J. and Modolell, J. (1998). Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 12, 2036-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bellard, M., Barembaum, M., Arman, O. and Bronner-Fraser, M. (2007). Lunatic fringe causes expansion and increased neurogenesis of trunk neural tube and neural crest populations. Neuron Glia Biol. 3, 93-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis, J. F., Garcia-Bellido, A. and Bray, S. J. (1996). Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development 122, 359-369. [DOI] [PubMed] [Google Scholar]
- Del Barrio, M. G., Taveira-Marques, R., Muroyama, Y., Yuk, D. I., Li, S., Wines-Samuelson, M., Shen, J., Smith, H. K., Xiang, M., Rowitch, D. et al. (2007). A regulatory network involving Foxn4, Mash1 and delta-like 4/Notch1 generates V2a and V2b spinal interneurons from a common progenitor pool. Development 134, 3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, M. and de Celis, J. F. (1998). A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature 396, 276-278. [DOI] [PubMed] [Google Scholar]
- Dubois, L., Bally-Cuif, L., Crozatier, M., Moreau, J., Paquereau, L. and Vincent, A. (1998). XCoe2, a transcription factor of the Col/Olf-1/EBF family involved in the specification of primary neurons in Xenopus. Curr. Biol. 8, 199-209. [DOI] [PubMed] [Google Scholar]
- Fryer, C. J., Lamar, E., Turbachova, I., Kintner, C. and Jones, K. A. (2002). Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16, 1397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiano, N., Nye, J. S. and Fishell, G. (2000). Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron 26, 395-404. [DOI] [PubMed] [Google Scholar]
- Geling, A., Steiner, H., Willem, M., Bally-Cuif, L. and Haass, C. (2002). A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 3, 688-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glittenberg, M., Pitsouli, C., Garvey, C., Delidakis, C. and Bray, S. (2006). Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. EMBO J. 25, 4697-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, M., Moens, C. B., Amacher, S. L., Eisen, J. S. and Beattie, C. E. (2001). Zebrafish deadly seven functions in neurogenesis. Dev. Biol. 237, 306-323. [DOI] [PubMed] [Google Scholar]
- Haddon, C., Smithers, L., Schneider-Maunoury, S., Coche, T., Henrique, D. and Lewis, J. (1998). Multiple delta genes and lateral inhibition in zebrafish primary neurogenesis. Development 125, 359-370. [DOI] [PubMed] [Google Scholar]
- Hanneman, E., Trevarrow, B., Metcalfe, W. K., Kimmel, C. B. and Westerfield, M. (1988). Segmental pattern of development of the hindbrain and spinal cord of the zebrafish embryo. Development 103, 49-58. [DOI] [PubMed] [Google Scholar]
- Heitzler, P., Bourouis, M., Ruel, L., Carteret, C. and Simpson, P. (1996). Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development 122, 161-171. [DOI] [PubMed] [Google Scholar]
- Holley, S. A., Julich, D., Rauch, G. J., Geisler, R. and Nusslein-Volhard, C. (2002). her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development 129, 1175-1183. [DOI] [PubMed] [Google Scholar]
- Irving, C., Nieto, M. A., DasGupta, R., Charnay, P. and Wilkinson, D. G. (1996). Progressive spatial restriction of Sek-1 and Krox-20 gene expression during hindbrain segmentation. Dev. Biol. 173, 26-38. [DOI] [PubMed] [Google Scholar]
- Ishii, Y., Nakamura, S. and Osumi, N. (2000). Demarcation of early mammalian cortical development by differential expression of fringe genes. Brain Res. Dev. Brain Res. 119, 307-320. [DOI] [PubMed] [Google Scholar]
- Itoh, M., Kim, C. H., Palardy, G., Oda, T., Jiang, Y. J., Maust, D., Yeo, S. Y., Lorick, K., Wright, G. J., Ariza-McNaughton, L. et al. (2003). Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell 4, 67-82. [DOI] [PubMed] [Google Scholar]
- Jacobsen, T. L., Brennan, K., Arias, A. M. and Muskavitch, M. A. (1998). Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development 125, 4531-4540. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. J., Brand, M., Heisenberg, C. P., Beuchle, D., Furutani-Seiki, M., Kelsh, R. N., Warga, R. M., Granato, M., Haffter, P., Hammerschmidt, M. et al. (1996). Mutations affecting neurogenesis and brain morphology in the zebrafish, Danio rerio. Development 123, 205-216. [DOI] [PubMed] [Google Scholar]
- Johnston, S. H., Rauskolb, C., Wilson, R., Prabhakaran, B., Irvine, K. D. and Vogt, T. F. (1997). A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development 124, 2245-2254. [DOI] [PubMed] [Google Scholar]
- Julich, D., Hwee Lim, C., Round, J., Nicolaije, C., Schroeder, J., Davies, A., Geisler, R., Lewis, J., Jiang, Y. J. and Holley, S. A. (2005). beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev. Biol. 286, 391-404. [DOI] [PubMed] [Google Scholar]
- Kageyama, R., Ohtsuka, T. and Kobayashi, T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243-1251. [DOI] [PubMed] [Google Scholar]
- Kageyama, R., Ohtsuka, T., Shimojo, H. and Imayoshi, I. (2008). Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 11, 1247-1251. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., Shima, A. and Kawakami, N. (2000). Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 97, 11403-11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C. H., Bae, Y. K., Yamanaka, Y., Yamashita, S., Shimizu, T., Fujii, R., Park, H. C., Yeo, S. Y., Huh, T. L., Hibi, M. et al. (1997). Overexpression of neurogenin induces ectopic expression of HuC in zebrafish. Neurosci. Lett. 239, 113-116. [DOI] [PubMed] [Google Scholar]
- Kim, J., Irvine, K. D. and Carroll, S. B. (1995). Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosophila wing. Cell 82, 795-802. [DOI] [PubMed] [Google Scholar]
- Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. [DOI] [PubMed] [Google Scholar]
- Korzh, V., Sleptsova, I., Liao, J., He, J. and Gong, Z. (1998). Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev. Dyn. 213, 92-104. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa, N., Kim, J., Anderson, D. and Kintner, C. (2000). Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation. Development 127, 4203-4216. [DOI] [PubMed] [Google Scholar]
- Kunisch, M., Haenlin, M. and Campos-Ortega, J. A. (1994). Lateral inhibition mediated by the Drosophila neurogenic gene delta is enhanced by proneural proteins. Proc. Natl. Acad. Sci. USA 91, 10139-10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer, E., Dahn, R., Orozco, O. E., Yeo, C. Y., Pisenti, J., Henrique, D., Abbott, U. K., Fallon, J. F. and Tabin, C. (1997). Expression of Radical fringe in limb-bud ectoderm regulates apical ectodermal ridge formation. Nature 386, 366-373. [DOI] [PubMed] [Google Scholar]
- Leve, C., Gajewski, M., Rohr, K. B. and Tautz, D. (2001). Homologues of c-hairy1 (her9) and lunatic fringe in zebrafish are expressed in the developing central nervous system, but not in the presomitic mesoderm. Dev. Genes Evol. 211, 493-500. [DOI] [PubMed] [Google Scholar]
- Lewis, J. (1998). Notch signalling and the control of cell fate choices in vertebrates. Semin. Cell Dev. Biol. 9, 583-589. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Babb, S. G., Novince, Z. M., Doedens, A. L., Marrs, J. and Raymond, P. A. (2001). Differential expression of cadherin-2 and cadherin-4 in the developing and adult zebrafish visual system. Vis. Neurosci. 18, 923-933. [PubMed] [Google Scholar]
- Louvi, A. and Artavanis-Tsakonas, S. (2006). Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 93-102. [DOI] [PubMed] [Google Scholar]
- Malatesta, P., Hartfuss, E. and Gotz, M. (2000). Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development 127, 5253-5263. [DOI] [PubMed] [Google Scholar]
- Malatesta, P., Hack, M. A., Hartfuss, E., Kettenmann, H., Klinkert, W., Kirchhoff, F. and Gotz, M. (2003). Neuronal or glial progeny: regional differences in radial glia fate. Neuron 37, 751-764. [DOI] [PubMed] [Google Scholar]
- Martin, S. C., Heinrich, G. and Sandell, J. H. (1998). Sequence and expression of glutamic acid decarboxylase isoforms in the developing zebrafish. J. Comp. Neurol. 396, 253-266. [PubMed] [Google Scholar]
- Matsuda, M. and Chitnis, A. B. (2009). Interaction with Notch determines endocytosis of specific Delta ligands in zebrafish neural tissue. Development 136, 197-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves, L., Jackman, W. and Kimmel, C. B. (2002). FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development 129, 3825-3837. [DOI] [PubMed] [Google Scholar]
- Mendelson, B. (1986). Development of reticulospinal neurons of the zebrafish. I. Time of origin. J. Comp. Neurol. 251, 160-171. [DOI] [PubMed] [Google Scholar]
- Micchelli, C. A. and Blair, S. S. (1999). Dorsoventral lineage restriction in wing imaginal discs requires Notch. Nature 401, 473-476. [DOI] [PubMed] [Google Scholar]
- Micchelli, C. A., Rulifson, E. J. and Blair, S. S. (1997). The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development 124, 1485-1495. [DOI] [PubMed] [Google Scholar]
- Mikkola, I., Fjose, A., Kuwada, J. Y., Wilson, S., Guddal, P. H. and Krauss, S. (1992). The paired domain-containing nuclear factor pax[b] is expressed in specific commissural interneurons in zebrafish embryos. J. Neurobiol. 23, 933-946. [DOI] [PubMed] [Google Scholar]
- Moens, C. B., Yan, Y. L., Appel, B., Force, A. G. and Kimmel, C. B. (1996). valentino: a zebrafish gene required for normal hindbrain segmentation. Development 122, 3981-3990. [DOI] [PubMed] [Google Scholar]
- Moloney, D. J., Panin, V. M., Johnston, S. H., Chen, J., Shao, L., Wilson, R., Wang, Y., Stanley, P., Irvine, K. D., Haltiwanger, R. S. et al. (2000). Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369-375. [DOI] [PubMed] [Google Scholar]
- Mumm, J. S., Schroeter, E. H., Saxena, M. T., Griesemer, A., Tian, X., Pan, D. J., Ray, W. J. and Kopan, R. (2000). A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197-206. [DOI] [PubMed] [Google Scholar]
- Neave, B., Rodaway, A., Wilson, S. W., Patient, R. and Holder, N. (1995). Expression of zebrafish GATA 3 (gta3) during gastrulation and neurulation suggests a role in the specification of cell fate. Mech. Dev. 51, 169-182. [DOI] [PubMed] [Google Scholar]
- Ohtsuka, T., Ishibashi, M., Gradwohl, G., Nakanishi, S., Guillemot, F. and Kageyama, R. (1999). Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda, Y., Yoda, H., Uchikawa, M., Furutani-Seiki, M., Takeda, H., Kondoh, H. and Kamachi, Y. (2006). Comparative genomic and expression analysis of group B1 sox genes in zebrafish indicates their diversification during vertebrate evolution. Dev. Dyn. 235, 811-825. [DOI] [PubMed] [Google Scholar]
- Oxtoby, E. and Jowett, T. (1993). Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 21, 1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panin, V. M., Papayannopoulos, V., Wilson, R. and Irvine, K. D. (1997). Fringe modulates Notch-ligand interactions. Nature 387, 908-912. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos, V., Tomlinson, A., Panin, V. M., Rauskolb, C. and Irvine, K. D. (1998). Dorsal-ventral signaling in the Drosophila eye. Science 281, 2031-2034. [DOI] [PubMed] [Google Scholar]
- Park, H. C., Hong, S. K., Kim, H. S., Kim, S. H., Yoon, E. J., Kim, C. H., Miki, N. and Huh, T. L. (2000). Structural comparison of zebrafish Elav/Hu and their differential expressions during neurogenesis. Neurosci. Lett. 279, 81-84. [DOI] [PubMed] [Google Scholar]
- Park, S. H., Yeo, S. Y., Yoo, K. W., Hong, S. K., Lee, S., Rhee, M., Chitnis, A. B. and Kim, C. H. (2003). Zath3, a neural basic helix-loop-helix gene, regulates early neurogenesis in the zebrafish. Biochem. Biophys. Res. Commun. 308, 184-190. [DOI] [PubMed] [Google Scholar]
- Passini, M. A., Kurtzman, A. L., Canger, A. K., Asch, W. S., Wray, G. A., Raymond, P. A. and Schechter, N. (1998). Cloning of zebrafish vsx1: expression of a paired-like homeobox gene during CNS development. Dev. Genet. 23, 128-141. [DOI] [PubMed] [Google Scholar]
- Peng, C. Y., Yajima, H., Burns, C. E., Zon, L. I., Sisodia, S. S., Pfaff, S. L. and Sharma, K. (2007). Notch and MAML signaling drives Scl-dependent interneuron diversity in the spinal cord. Neuron 53, 813-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait, J., Amores, A., Cresko, W., Singer, A. and Yan, Y. L. (2004). Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 20, 481-490. [DOI] [PubMed] [Google Scholar]
- Prince, V. E., Holley, S. A., Bally-Cuif, L., Prabhakaran, B., Oates, A. C., Ho, R. K. and Vogt, T. F. (2001). Zebrafish lunatic fringe demarcates segmental boundaries. Mech. Dev. 105, 175-180. [DOI] [PubMed] [Google Scholar]
- Qiu, X., Xu, H., Haddon, C., Lewis, J. and Jiang, Y. J. (2004). Sequence and embryonic expression of three zebrafish fringe genes: lunatic fringe, radical fringe, and manic fringe. Dev. Dyn. 231, 621-630. [DOI] [PubMed] [Google Scholar]
- Rauskolb, C., Correia, T. and Irvine, K. D. (1999). Fringe-dependent separation of dorsal and ventral cells in the Drosophila wing. Nature 401, 476-480. [DOI] [PubMed] [Google Scholar]
- Robu, M. E., Larson, J. D., Nasevicius, A., Beiraghi, S., Brenner, C., Farber, S. A. and Ekker, S. C. (2007). p53 activation by knockdown technologies. PLoS Genet. 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Esteban, C., Schwabe, J. W., De La Pena, J., Foys, B., Eshelman, B. and Belmonte, J. C. (1997). Radical fringe positions the apical ectodermal ridge at the dorsoventral boundary of the vertebrate limb. Nature 386, 360-366. [DOI] [PubMed] [Google Scholar]
- Sakamoto, K., Ohara, O., Takagi, M., Takeda, S. and Katsube, K. (2002). Intracellular cell-autonomous association of Notch and its ligands: a novel mechanism of Notch signal modification. Dev. Biol. 241, 313-326. [DOI] [PubMed] [Google Scholar]
- Schier, A. F., Neuhauss, S. C., Harvey, M., Malicki, J., Solnica-Krezel, L., Stainier, D. Y., Zwartkruis, F., Abdelilah, S., Stemple, D. L., Rangini, Z. et al. (1996). Mutations affecting the development of the embryonic zebrafish brain. Development 123, 165-178. [DOI] [PubMed] [Google Scholar]
- Seth, A., Culverwell, J., Walkowicz, M., Toro, S., Rick, J. M., Neuhauss, S. C., Varga, Z. M. and Karlstrom, R. O. (2006). belladonna/(Ihx2) is required for neural patterning and midline axon guidance in the zebrafish forebrain. Development 133, 725-735. [DOI] [PubMed] [Google Scholar]
- Shih, J. and Fraser, S. E. (1995). Distribution of tissue progenitors within the shield region of the zebrafish gastrula. Development 121, 2755-2765. [DOI] [PubMed] [Google Scholar]
- Shimojo, H., Ohtsuka, T. and Kageyama, R. (2008). Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52-64. [DOI] [PubMed] [Google Scholar]
- Sieger, D., Tautz, D. and Gajewski, M. (2004). her11 is involved in the somitogenesis clock in zebrafish. Dev. Genes Evol. 214, 393-406. [DOI] [PubMed] [Google Scholar]
- Takke, C., Dornseifer, P. v. Weizsacker, E. and Campos-Ortega, J. A. (1999). her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development 126, 1811-1821. [DOI] [PubMed] [Google Scholar]
- Thaeron, C., Avaron, F., Casane, D., Borday, V., Thisse, B., Thisse, C., Boulekbache, H. and Laurenti, P. (2000). Zebrafish evx1 is dynamically expressed during embryogenesis in subsets of interneurones, posterior gut and urogenital system. Mech. Dev. 99, 167-172. [DOI] [PubMed] [Google Scholar]
- Thisse, C., Thisse, B., Schilling, T. F. and Postlethwait, J. H. (1993). Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119, 1203-1215. [DOI] [PubMed] [Google Scholar]
- Thomas, G. B. and van Meyel, D. J. (2007). The glycosyltransferase Fringe promotes Delta-Notch signaling between neurons and glia, and is required for subtype-specific glial gene expression. Development 134, 591-600. [DOI] [PubMed] [Google Scholar]
- Trevarrow, B., Marks, D. L. and Kimmel, C. B. (1990). Organization of hindbrain segments in the zebrafish embryo. Neuron 4, 669-679. [DOI] [PubMed] [Google Scholar]
- van Eeden, F. J., Granato, M., Schach, U., Brand, M., Furutani-Seiki, M., Haffter, P., Hammerschmidt, M., Heisenberg, C. P., Jiang, Y. J., Kane, D. A. et al. (1996). Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development 123, 153-164. [DOI] [PubMed] [Google Scholar]
- Visan, I., Tan, J. B., Yuan, J. S., Harper, J. A., Koch, U. and Guidos, C. J. (2006). Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat. Immunol. 7, 634-643. [DOI] [PubMed] [Google Scholar]
- Wang, X., Emelyanov, A., Korzh, V. and Gong, Z. (2003). Zebrafish atonal homologue zath3 is expressed during neurogenesis in embryonic development. Dev. Dyn. 227, 587-592. [DOI] [PubMed] [Google Scholar]
- Westin, L. and Lardelli, M. (1997). Three novel Notch genes in zebrafish: implications for vertebrate Notch gene evolution and function. Dev. Genes Evol. 207, 51-63. [DOI] [PubMed] [Google Scholar]
- Wettstein, D. A., Turner, D. L. and Kintner, C. (1997). The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development 124, 693-702. [DOI] [PubMed] [Google Scholar]
- Yeo, S. Y., Kim, M., Kim, H. S., Huh, T. L. and Chitnis, A. B. (2007). Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev. Biol. 301, 555-567. [DOI] [PubMed] [Google Scholar]
- Zeltser, L. M., Larsen, C. W. and Lumsden, A. (2001). A new developmental compartment in the forebrain regulated by Lunatic fringe. Nat. Neurosci. 4, 683-684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.