Summary
This work reports the characterization and functional analysis of disrupted in schizophrenia 1 (disc1), a well-documented schizophrenia-susceptibility gene, in zebrafish cranial neural crest (CNC). Our data demonstrated that disc1 was expressed in zebrafish CNC cells. Loss of Disc1 resulted in persistent CNC cell medial migration, dorsal to the developing neural epithelium, and hindered migration away from the region dorsal to the neural rod. General CNC cell motility was not affected by Disc1 knockdown, however, as the speed of CNC cells was indistinguishable from that of wild-type counterparts. We determined that the failure of CNC cells to migrate away from the neural rod correlated with the enhanced expression of two transcription factors, foxd3 and sox10. These transcription factors have many functions in CNC cells, including the maintenance of precursor pools, timing of migration onset, and the induction of cell differentiation. Our work, in conjunction with previous studies, suggests that the perpetuation of expression of these factors affects several aspects of CNC cell development, leading to a loss of craniofacial cartilage and an expansion of peripheral cranial glia. Based on our data, we propose a model in which Disc1 functions in the transcriptional repression of foxd3 and sox10, thus mediating CNC cell migration and differentiation.
Keywords: foxd3, sox10, disc1, Disrupted In Schizophrenia 1, Zebrafish, Cranial neural crest, Neural crest migration, Schizophrenia
INTRODUCTION
Schizophrenia is a debilitating psychiatric illness. It is hypothesized that its origin lies, in part, in defective neurodevelopment (Lewis and Levitt, 2002). One of the most highly associated susceptibility genes in humans is disrupted in schizophrenia 1 (DISC1) (reviewed by Chubb et al., 2008). Although this protein does not belong to a known protein family, it is expressed in a developmentally regulated pattern in the brain (Austin et al., 2003; Ma et al., 2002; Meyer and Morris, 2008; Schurov et al., 2004). In addition, yeast two-hybrid studies have identified DISC1-interacting proteins with crucial developmental functions (Millar et al., 2003; Morris et al., 2003).
The developmental regulation, cellular localization and protein interactors identified implicate Disc1 in brain development. Both in vivo and in vitro studies demonstrate that this protein functions in neurite outgrowth and neuronal migration (Duan et al., 2007; Kamiya et al., 2005), in part through its interaction with Ndel1, a centrosome-associated protein (Shu et al., 2004). However, Disc1 interacts with a plethora of proteins in addition to Ndel1 (Camargo et al., 2007), has multiple isoforms (Porteous and Millar, 2006) and localizes to several cellular compartments (Brandon et al., 2005; James et al., 2004; Morris et al., 2003), indicating it might be involved in numerous molecular mechanisms impacting cell motility. To explore the evolutionarily conserved roles of this protein in development, we chose to examine its function in the zebrafish, a vertebrate system amenable to developmental and cell biology studies.
The studies presented here identified a previously uncharacterized function of Disc1, which affects the migration and differentiation of cranial neural crest (CNC) cells. We determined that disc1 has a dynamic expression pattern in CNC cells. disc1 is expressed in premigratory neural crest and in the anterior streams of migrating CNC cells. During development, CNC cells migrate long distances over stereotyped paths from their place of induction to give rise to several tissues, including the peripheral nervous system, craniofacial cartilage and pigment (Knight and Schilling, 2006). Cell intrinsic and extrinsic cues govern the survival, proliferation and migration of CNC cells (Morales et al., 2005; Sauka-Spengler and Bronner-Fraser, 2008; Steventon et al., 2005). Proper timing and direction of migration onset is crucial for CNC cells to encounter cell extrinsic cues necessary for their survival and differentiation (Knight and Schilling, 2006; Tapadia et al., 2005).
Important for our studies, CNC cells share many characteristics with cells of the developing brain, including a common neuroectodermal origin, long-range migratory ability, the derivation of multiple cell types, reliance on common subset of transcription factors regulating cellular potency/differentiation, and responsiveness to the same extracellular signals (Lefcort et al., 2007). Furthermore, their location near the surface of the embryo allows for unobstructed monitoring of cellular motility in vivo. Using live-embryo imaging, we determined that wild-type CNC cells migrate medially over the neural keel, preceding their migration away from the neural rod. Loss of Disc1 in these cells does not affect general cell motility but does result in the perpetuation of medial CNC cell movement and hinders migration into the periphery. We determined that this aberrant migration is likely to be due to the enhanced expression of foxd3 and sox10, transcription factors implicated in neural crest migration and differentiation. The perpetuation of expression of these factors in our studies resulted in a loss of craniofacial cartilage and an expansion of peripheral cranial glia. Based on our data, we propose that Disc1 functions in CNC cell migration and differentiation through the transcriptional repression of foxd3 and sox10.
MATERIALS AND METHODS
Zebrafish husbandry
Wild-type and sox10m241 mutant (Malicki et al., 1996) zebrafish were maintained according to Institutional Animal Care and Use approved protocols on a 14:10 light:dark cycle. Embryos were collected, maintained and staged as previously described (Kimmel et al., 1995; Westerfield, 1995).
In situ hybridization and immunohistochemistry
Chromatogenic in situ hybridizations were performed as described (Thisse et al., 1993). Probes used were dlx2a (Akimenko et al., 1994), hand2 (Angelo et al., 2000), myod1 (Weinberg et al., 1996), sox10 (Dutton et al., 2001b), crestin (Luo et al., 2001), snai1b (Thisse et al., 1995) and foxd3 (Odenthal and Nusslein-Volhard, 1998). A disc1 probe was generated by cloning the open-reading frame into the pCR4-Topo vector (Invitrogen). Fluorescent in situ hybridization was done as described (Julich et al., 2005). For all experiments imaging fixed Tg(-4.9sox10:egfp)ba2 embryos, EGFP immunohistochemistry was performed as described (Drerup et al., 2007) with an anti-GFP antibody (Invitrogen).
Mitotic and cell death index
Phosphohistone-H3 immunohistochemistry (Upstate) was done on heterozygous Tg(-4.9sox10:egfp)ba2 embryos. TUNEL staining was performed using the In Situ Cell Death Kit (Roche). Counts of total EGFP- and TUNEL/EGFP-positive cells were carried out in z-stacks projected onto a single plane.
Morpholino and mRNA microinjection
Morpholino antisense oligonucleotides were obtained from Gene Tools (MOE3, GGACTCGATCTAATGAGAGAAATCA; MOE8, TGTTTCTGTACCTGATACGGCTGTG; SMO2, ACATGCTGTGCTCCTCCGCCGACAT; Mismatch, CTGCTAAATGCCCCAAAGTACTTCG) and Open Biosystems (SMO1, ATGCTGTGCTCCTCCGCCGACATCG) (Dutton et al., 2001a). The open-reading frame of human DISC1 and zebrafish foxd3 were cloned into the pCS2+ vector and mRNA synthesized according to manufacturer's instructions (Ambion).
RT-PCR analysis
RT-PCR analysis was performed as described (Drerup et al., 2007) with disc1 and β-actin primers.
Alcian Blue analysis
Larvae at 5 days post-fertilization (dpf) were stained with Alcian Blue as described (Kimmel et al., 1998).
Time-lapse imaging
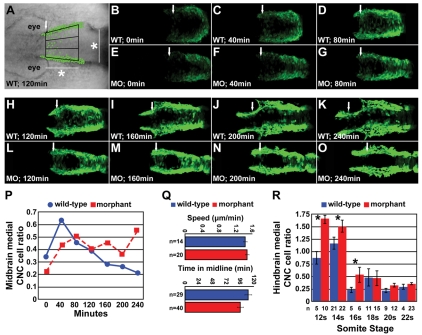
Tg(-4.9sox10:egfp)ba2 heterozygous embryos were dechorionated at 6 somites (s) and embedded in 1.5% low-melt agarose. Time-lapse imaging was done with a 510 META Zeiss confocal microscope. Every 8 minutes, over a 4-hour period, a z-stack of approximately 130 μm was taken with 5 μm between each z-slice. For movies, each z-stack was projected onto a single plane. For cell speed analysis, the distance traveled and the time elapsed for a single cell was tracked manually. Total time spent in the midline was determined in a different subset of cells. To determine the proportion of the CNC cell population occupying the midline at each point of the time-lapse, the area dorsal to the neural tube was divided into thirds (see Fig. 4A) and the number of cells in each region was counted. The rostral boundary was defined by the most anterior medially migrating cells (arrows in Fig. 4). The distance to the caudal edge of the region of interest was set as the width of the neural tube at the midbrain-hindbrain boundary at 10s.
Fig. 4.
Time-lapse imaging of medial CNC cell movement. (A) Time-lapse, live embryo imaging in Disc1 morphant and control Tg(sox10:egfp) zebrafish was performed to analyze the movement of CNC cells in the area depicted, dorsal to the developing midbrain (white arrow, anterior border; *, the width of the neural rod at 10s, see Materials and methods for details). (B-O) Representative images from the time-lapse of Disc1 morphants and controls at seven evenly spaced time-points (from minute 0 to 240) starting at 6s. The posterior border of the developing eye was aligned at each time-point. (P) A high proportion of CNC cells occupy the midline in Disc1 morphants throughout the time-lapse, whereas this phenotype is short lived in controls. (Q) Neither the speed of CNC cell medial migration (top) nor the time spent in the midline (bottom) were altered in Disc1 morphants (n, number of cells). (R) In a separate experiment, we determined that CNC cells in the hindbrain region behaved similarly; the ratio of medially lying CNC dorsal to the hindbrain (arches 2 and 3 in Fig. 5A) was elevated in Disc1 morphants until the 16s stage. *, P<0.05.
Migratory distance and hindbrain CNC cell counts
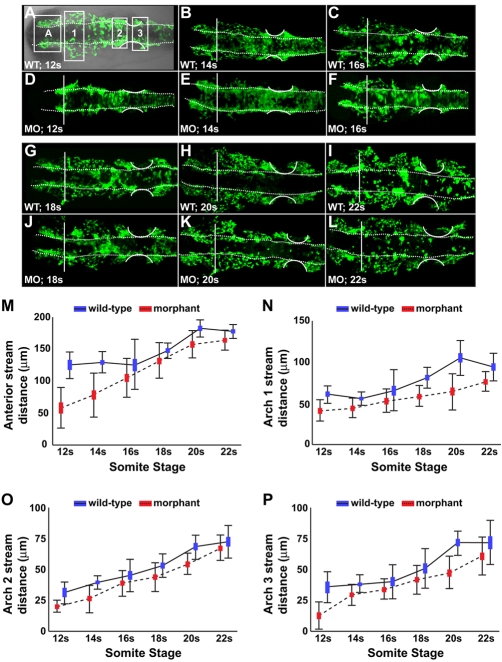
Fixed Tg(-4.9sox10:egfp)ba2 embryos were flat mounted and imaged with a 510 META Zeiss confocal microscope. Migratory distance (see Fig. 5A) was measured using LSM image browser software. Hindbrain CNC cell counts were done in z-stacks projected onto a single plane in regions defined in Fig. 5A.
Fig. 5.
CNC migration from the neural rod was hindered in Disc1 morphants. (A) DIC overlay showing a flat-mounted Tg(sox10:egfp) zebrafish (dorsal view, anterior to the left), demonstrating the four streams analyzed. (A-L) Flat-mounted control and Disc1 morphant Tg(sox10:egfp) zebrafish (dorsal view, anterior to the left) were fixed at evenly spaced intervals from 12s to 22s. CNC migratory distance was measured in the anterior (A), arch 1 (1), arch 2 (2) and arch 3 (3) streams of the neural crest (as indicated by the boxed regions in A). Vertical line marks the posterior eye; dotted lines outline the neural rod; curved lines outline the developing ear. (M-P) Graphical representation of the maximum migratory distance achieved by neural crest cells in each stream. Disc1 morphant CNC cells are hindered in their migration away from the dorsolateral midbrain and hindbrain. Height of the box is the mean±s.e.m. Error bars represent s.d.
Statistical analysis
Data sets suitable for parametric analysis were analyzed using one-way analysis of variance with a Tukey HSD test of pairwise comparisons or a multiple analysis of variance with post-hoc contrasts. Some data sets were log transformed prior to analysis for normalization. Non-parametric data was analyzed using the Wilcoxon/Kruskal-Wallis test for χ2 approximation. Significance was set at P<0.05.
RESULTS
Expression of the zebrafish disc1 ortholog
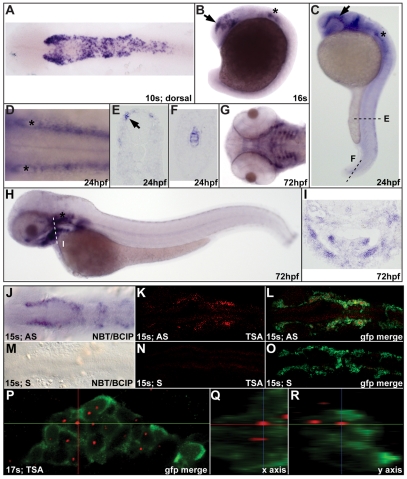
The full-length zebrafish (Danio rerio) disc1 ortholog was cloned using 5′RACE based on a partial sequence identified previously (Taylor et al., 2003). Bioinformatic analyses indicate that this is probably the only disc1 ortholog present (C.M.D. and J.A.M., unpublished). Semi-quantitative RT-PCR analysis demonstrated that disc1 expression is developmentally regulated, with a peak in expression at cleavage and early blastula stages, and downregulation during gastrulation (see Fig. S1A in the supplementary material). Specific expression was not detectable by in situ hybridization until the 10-somite stage (s), after which it is expressed in numerous tissues in the embryonic and larval zebrafish, including the notochord, ear, pectoral fin, trunk neural crest, CNC, and the developing craniofacial skeleton (a CNC derivative; Fig. 1). The CNC expression of disc1 was particularly intriguing as it is expressed throughout the premigratory neural crest but expression in the migratory neural crest is limited to CNC around the eye (Fig. 1B-D). disc1 is not expressed in CNC cells migrating ventrolaterally into the posterior pharyngeal arches, but expression is detectable in the developing craniofacial skeleton by 3 dpf (Fig. 1G-I). These data might indicate separate functions of Disc1 in the premigratory neural crest and the developing craniofacial skeleton.
Fig. 1.
disc1 expression in zebrafish CNC. (A) Dorsal view of a 10s (14 hpf) embryo showing disc1 expression in the CNC. (B,C) Lateral views with anterior to the left show continued disc1 expression in CNC migrating around the eye (arrow) and the developing ear (*) at 16s and 24 hpf. (D) Dorsal view of the hindbrain at 24 hpf shows disc1 expression in the neural crest on either side of the neural tube. (E,F) Sections through the trunk (E) and tail (F) show disc1 expression in trunk neural crest and notochord at 24 hpf. (G-I) Ventral and lateral views show disc1 expression in the developing craniofacial skeleton at 72 hpf. This was corroborated by a coronal section through this region (I). (J-L) Fluorescent in situ hybridization for disc1 replicated the staining pattern observed using standard, chromatogenic methods (J) at 15s. (K,L) disc1 in situ hybridization in Tg(sox10:egfp) zebrafish illustrated specific expression in EGFP-positive CNC cells. (M-O) Sense probes showed no specific staining. (P-R) Orthogonal views of the third stream of migrating CNC at 17s confirmed disc1 expression. AS, antisense probe; S, sense probe; NBT/BCIP, Nitro blue tetrazolium chloride/5-Bromo-4-chloro-3-indolyl phosphate, toluidine salt; TSA, Tyramide Signal Amplification. Dorsal views with anterior to the left.
To confirm disc1 expression specifically in CNC cells, fluorescent disc1 in situ hybridization was performed in Tg(-4.9sox10:egfp)ba2 zebrafish (hereafter referred to as Tg(sox10:egfp); gift of Dr T. Schilling, University of California, Irvine, CA, USA), which express enhanced green fluorescent protein (EGFP) under the control of the sox10 promoter in neural crest cells (Carney et al., 2006; Wada et al., 2005). Confocal analysis of 15s [16.5 hours post-fertilization (hpf)] embryos illustrated specific expression of disc1 (red) in sox10-expressing CNC cells (Fig. 1J-O). Orthogonal views of CNC cells anterior to the otic vesicle at 17s (17.5 hpf) confirmed strong expression of disc1 in the nucleus of EGFP-positive cells (Fig. 1P-R). As the fluorescent substrate used in this study precipitates in a very localized manner, the nuclear disc1 expression indicates that this cellular compartment has the highest level of disc1 mRNA at this time of high disc1 transcription. disc1 is also probably co-expressed in a subset of sox10- and foxd3-expressing CNC cells (see Fig. S1B-G in the supplementary material). The temporal overlap of expression of disc1 and foxd3 is very short, however, as strong disc1 expression begins at 10s when foxd3 expression in this cell population is being rapidly downregulated.
Disc1 knockdown results in abnormal craniofacial development
We next examined the function of Disc1 in CNC. A morpholino antisense oligonucleotide (MOE3) was designed for the intron 2/exon 3 splice-acceptor site to alter splicing of the disc1 pre-mRNA, deleting exon 3, which introduces a premature stop site and a severely truncated protein. Injection of MOE3 at the one-cell stage effectively altered disc1 transcript size (Fig. 2A).
Fig. 2.
Two splice-site targeted Disc1 morpholinos altered disc1 transcript size and craniofacial development. (A) RT-PCR analysis of MOE3-injected embryos demonstrated alternative splicing of disc1 pre-mRNA resulting in a smaller transcript. At 6 hpf, maternally expressed disc1 could not be modified by the splice site-targeted morpholino, which resulted in the wild-type transcript present. (B) MOE8 injection also effectively altered transcript size. (C-H) Morpholino-mediated Disc1 knockdown resulted in three classes of craniofacial cartilage defects. (C,F) Class 1 (normal); (D,G) class 2; (E,H) class 3. (C-E) Lateral views; (F-H) ventral views with anterior to the left. (I) The ratio of MOE3 and MOE8 morphant embryos in each class was dose dependent. (I, right) Although half-effective doses of MOE3 or MOE8 did not affect craniofacial development, they acted synergistically to produce a high ratio of larvae with abnormal craniofacial cartilage.
Alcian Blue staining of the cartilaginous craniofacial skeleton, a CNC derivative, revealed abnormal development in a high proportion of Disc1 morphants (embryos with Disc1 knockdown) at 5 dpf (Fig. 2C-H). Disc1 morphants fell into three classes based on their Alcian Blue staining: (1) normal; (2) mild malformations of the ventral craniofacial cartilage; and (3) severe malformation and reductions/loss of the ventral craniofacial cartilage and anterior neurocranium. The ratio of morphants in each class was dose dependent (Fig. 2I). To validate this effect, a second morpholino (MOE8) was designed to target the exon 8/intron 8 splice site of the disc1 pre-mRNA. The protein produced from this altered transcript is truncated at the same point as the mutant form of DISC1 first associated with increased schizophrenia susceptibility (Millar et al., 2000). The analysis of MOE8 morphants confirmed that this morpholino effectively altered disc1 pre-mRNA splicing (Fig. 2B) and caused dose-dependent abnormalities in craniofacial development that were similar to those caused by MOE3 (Fig. 2I). In addition, sub-optimal doses of the MOE3 and MOE8 morpholinos worked synergistically to generate defects in craniofacial development that were identical to those resulting from full doses of either morpholino individually (Fig. 2I). This synergism, in addition to the rescue experiment discussed below, indicates that the reported phenotype is due to the loss of functional Disc1.
The role of Disc1 in craniofacial development was further analyzed by investigating the CNC contribution to the developing pharyngeal arches in Disc1 morphants. Zebrafish CNC migrate a substantial distance from the dorsolateral neural rod into the pharyngeal arches before forming the ventral craniofacial cartilage. In situ hybridization with dlx2a and hand2 showed a reduction in the ectomesenchymal CNC cell population in the pharyngeal arches at 26 hpf, 30 hpf and 32 hpf (see Fig. S2A,B,D,E,G-L in the supplementary material). In addition, lateral views of Tg(sox10:egfp) Disc1 morphants illustrated that CNC cells both in and dorsal to the arches were abnormally arranged (see Fig. S2C,F in the supplementary material). Thus, Disc1 loss reduced the ectomesenchymal CNC cell population in the pharyngeal arches and altered its arrangement, which probably led to the abnormal craniofacial development.
Medial expansion of CNC markers correlates with abnormal craniofacial development in Disc1 morphants
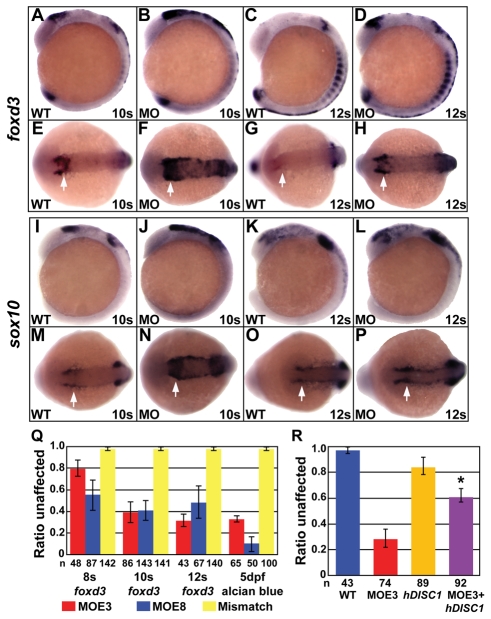
Loss of pharyngeal arch CNC cells could result from the disruption of a number of steps, from CNC induction to migration and differentiation. CNC cell induction was normal in Disc1 morphants (see Fig. S3A-H in the supplementary material). At later stages, however, the expression of the CNC markers foxd3 (Fig. 3A-H) and sox10 (Fig. 3I-P) dorsal to the developing midbrain was enhanced and seemed to be more medially localized to the midline in Disc1 morphants (arrows) than in wild-type embryos. Enhanced expression persisted in morphants around the posterior portion of the developing eye at later stages, when the signal has substantially decreased in controls.
Fig. 3.
Disc1 morphants had a medial expansion of foxd3 and sox10 expression. (A-H) At 10s and 12s, lateral (A-D) and dorsal (E-H) views show the medial expansion and increased level of foxd3 expression in Disc1 morphants (arrows). (I-P) sox10 expression showed similar results. (Q) foxd3 in situ hybridization and Alcian Blue staining of craniofacial cartilage was done on Disc1 morphants and mismatch control-injected embryos from within the same clutch. The ratio of embryos with normal foxd3 expression was significantly reduced in embryos injected with either MOE3 or MOE8. The ratio of larvae from the same clutches with normal Alcian Blue staining (class 1) was similarly reduced. Injection of a mismatch control produced no defect in foxd3 expression or cartilage development. (R) The medial expansion of foxd3 expression can be rescued in Disc1 morphants by co-injection of 100 pg of human DISC1 mRNA (*, P<0.01).
To determine if there was a correlation between the expanded expression of foxd3 and the craniofacial defects in Disc1 morphants, the preponderance of both phenotypes within individual clutches was analyzed. Embryos from the same clutch were injected with the MOE3 or MOE8 morpholino and the resulting morphants were divided into four groups: 8s, 10s, or 12s for foxd3 in situ hybridization or 5 dpf for Alcian Blue. Within clutches, the proportion of embryos with expanded foxd3 expression closely mirrors the proportion of larvae with abnormal craniofacial cartilage (Fig. 3Q). A mismatch Disc1 morpholino had no effect on foxd3 expression or craniofacial development. Finally, the expansion of foxd3 expression at 10s was suppressed by co-injection of 100 pg of human DISC1 mRNA (Fig. 3R; P<0.01), which confirms the dependence of this phenotype on Disc1 loss.
CNC cell proliferation and apoptosis were unaffected in Disc1 morphants
Increased cell proliferation could underlie the expansion of foxd3- and sox10-positive CNC cells. To determine whether proliferation was increased in Disc1 morphants, phosphohistone H3 immunohistochemistry was performed on control and Disc1 morphant Tg(sox10:egfp) zebrafish (see Fig. S4A-D in the supplementary material). We determined that the ratio of actively dividing CNC cells to the total number of CNC cells dorsal to the midbrain (the mitotic index) was not changed in Disc1 morphants (8s, P=0.668; 10s, P=0.492; see Fig. S4E in the supplementary material). Notably, total CNC cell number was unchanged in Disc1 morphants (8s, P=0.601; 10s, P=0.227; 12s, P=0.080; see Fig. S4F in the supplementary material), indicating that a change in CNC cell population size was not the cause of the expansion of CNC markers. Cell death was also analyzed in Disc1 morphant CNC cells. Apoptosis was monitored in this population during its migratory phase. Although the temporal dynamics of cell death were slightly altered in Disc1 morphants (see Fig. S5A in the supplementary material), overall there was no difference in the CNC cell death ratio (P=0.347; see Fig. S5B in supplementary material). Hence, the CNC population was not significantly altered in size as a result of proliferation or cell death.
CNC cell migration into the periphery was hindered in Disc1 morphants
We next hypothesized that abnormal medial CNC cell movement led to the documented expansion phenotype. To test this, the segment of CNC dorsal to the developing midbrain (Fig. 4A) was monitored in Disc1 morphant and control Tg(sox10:egfp) zebrafish by using confocal time-lapse microscopy. In our initial analysis of wild-type CNC cell movement, we observed medial movement of a surprising number of EGFP-positive cells shortly after the start of the time-lapse at 6s (see Movie 1 in the supplementary material). CNC cells moved medially, dorsal to the developing neural keel, and either crossed or returned to their side of origin. To analyze whether CNC cell localization over time was altered in Disc1 morphants, the ratio of CNC cells in the medial third of this region (Fig. 4A) was determined in 40-minute intervals from 6s to 12s (Fig. 4B-O). In controls, the ratio of medial cells increased soon after the start of the time-lapse but decreased consistently thereafter (Fig. 4B-D,H-K,P). In morphants, the ratio of CNC cells lying in the dorsal midline increased similarly to controls but failed to subsequently decrease. Instead, the ratio of CNC cells in the midline remained consistent through the last time-point measured (Fig. 4E-G,L-P). The increased ratio of CNC cells in the dorsal midline was not due to decreased speed of cell migration through this region (P=0.80) or to a failure of these cells in morphants to exit the midline, i.e. the time spent in the midline by single cells was unchanged in Disc1 morphants (P=0.11; Fig. 4Q). Rather, it seems that CNC cells in Disc1 morphants initiate medial migration at the same time as controls but that these cells continue to migrate through the midline dorsal to the neural rod at later stages than in their control counterparts. Whether it is the same cells or different cells that perform this migratory behavior at later stages of development in the morphants is unclear at this time. Single cells were not tracked throughout the course of the time-lapse. As it appears that only a subset of CNC cells normally perform this early, medial migratory behavior, it is likely that they are the ones competent to persist in this phenotype in Disc1 morphants and likely that they repeatedly migrate into/through the midline. This migratory phenotype is an interesting phenomenon in itself and will be the subject of future studies.
To determine if CNC cells originating near the hindbrain, which give rise to the posterior ventral craniofacial skeleton, behaved similarly to those originating near the midbrain, the ratio of CNC cells occupying the midline in this region was analyzed in a separate set of embryos. Disc1 morphant and control Tg(sox10:egfp) embryos from the same clutch were fixed at regularly spaced somitic stages (12s-22s) and the medial to lateral CNC cell ratio was determined dorsal to the hindbrain (streams 2 and 3, Fig. 5A). Similar to CNC originating near the midbrain, the ratio of hindbrain CNC cells residing medially was significantly greater in Disc1 morphants at 12s (P<0.005), 14s (P<0.05) and 16s (P<0.005; Fig. 4R) than in control embryos. At later stages, there was no difference between wild type and morphants, indicating that midline clearance was hindered but not abolished (18s, P=0.823; 20s, P=0.076; 22s, P=0.560). In summary, loss of Disc1 in CNC cells results in a failure of these cells to halt their migration through the midline dorsal to the neural tissue at the time when their wild-type counterparts are beginning their long-range migration from the dorsolateral neural tube.
The impact of this prolonged medial migration phenotype on CNC cell migration away from the neural rod was analyzed by measuring the maximum distance moved by these cells in the same subset of 12s-22s embryos described above. The distance that the CNC cells moved anteriorly between the eyes, as well as away from the neural rod, in the three outlined streams was measured and compared (Fig. 5A). MANOVA analysis of data from each age group demonstrated that, in addition to the prolonged occupation of the midline, CNC cells in Disc1 morphants were inhibited in their migration from their site of origin, lateral to the midbrain and hindbrain (12s, P<0.05; 14s, P<0.0001; 16s, P<0.005; 18s, P<0.005; 20s, P<0.0001; 22s, P<0.05; Fig. 5). This trend was consistent between the individual streams of CNC cells from the midbrain through the hindbrain (Fig. 5M-P).
These data, in combination with the aforementioned persistent medial migration of CNC in Disc1 morphants, implicated Disc1 not in general CNC cell motility but, rather, in the transition from medial CNC cell migration to long-range migration into the periphery. This hindered lateral and anterior migration might directly impact on the development of CNC cell derivatives in Disc1 morphants.
Disc1 repressed expression of foxd3 and sox10 in CNC cells
In addition to the medial expansion of foxd3- and sox10-positive CNC cells, there appeared to be an increase in overall expression of these transcription factors (see Fig. 3). We considered the possibility that the enhanced expression of foxd3 and sox10 could perhaps be a causative factor in delayed CNC migration. To determine whether the expansion of CNC markers observed was specific to foxd3 and sox10, we analyzed the expression of other CNC markers. Expression of crestin and snai1b was relatively normal in Disc1 morphants, indicating a specific effect on the expression of foxd3 and sox10 in this cell population (see Fig. S6A-D in the supplementary material).
We directly tested the ability of Disc1 to repress the expression of these transcription factors by asymmetrically overexpressing human DISC1 in zebrafish. To validate our experimental strategy, 200 pg of gfp mRNA was injected into a single cell of a two- to four-cell-stage embryo. This resulted in asymmetrical expression of GFP at 5s (Fig. 6A). A similar injection of 200 pg of DISC1 resulted in the asymmetrical loss or reduction of expression of foxd3 and sox10 in a proportion of injected embryos at 4s and 6s, respectively (Fig. 6B-E; foxd3, n=42, 48% affected; sox10, n=58, 40% affected). Normal myod1 expression in these embryos indicated a specific effect on transcription factor expression (Fig. 6D,E). These data provide further evidence that Disc1 functions to repress the expression of foxd3 and sox10. The action of Disc1 on sox10 expression might be direct or indirect, as there is evidence that Foxd3 regulates sox10 expression (Stewart et al., 2006). To examine the relationship between Foxd3 function and sox10 expression, Foxd3 was overexpressed by mRNA injection at the one-cell stage and sox10 expression was assayed at 11s. In embryos overexpressing Foxd3, sox10 expression was augmented (arrows, Fig. 6F-I). Interestingly, the expansion of sox10 expression in these embryos and the decreased lateral CNC cell movement were remarkably similar to the phenotypes observed with loss of Disc1 in morphants (see Fig. 3). Overall, these data implicate Disc1 in the repression of foxd3 and sox10 expression, an essential step in the initiation of CNC migration into the periphery.
Fig. 6.
Disc1 repressed foxd3 and sox10 expression. (A) Injection of 200 pg of gfp mRNA into one cell of a two- to four-cell-stage embryo resulted in unilateral expression at 5s. Dotted line outlines the edge of the embryo. (B-E) Asymmetrical overexpression of the human ortholog of disc1 resulted in a unilateral loss of foxd3 (B,C) and sox10 (D,E) expression (arrows). Expression of myod1 was normal. Dorsal views with anterior to the left. (F-I) Overexpression of Foxd3 resulted in an expansion of sox10 expression (arrows in H,I) similar to that seen in Disc1 morphants. F and G are lateral views, H and I, dorsal views, both with anterior to the left.
Defects in Disc1 morphants were suppressed by a partial reduction of Sox10 levels
To test for a genetic interaction between Disc1 and these transcription factors, we attempted to suppress the craniofacial deficits and expansion of CNC cell markers at 10s through a partial reduction of Sox10. We chose to repress Sox10 instead of Foxd3 as we, and others, have demonstrated that sox10 expression is at least partially regulated by Foxd3 (Stewart et al., 2006) and is directly involved in the specification of non-cartilaginous CNC derivatives (Dutton et al., 2001b). To partially repress Sox10 function, Disc1 morphants were co-injected with a previously described sox10-targeted morpholino, SMO1 (Dutton et al., 2001a). Alcian Blue staining at 5 dpf revealed a decrease in the proportion of embryos with abnormal craniofacial cartilage in the co-injected embryos (Fig. 7, left). This result was confirmed in a separate experiment by the co-injection of MOE3 and a second, Sox10 start site-targeted morpholino, SMO2 (Fig. 7, middle). Similarly, co-injection of SMO2 significantly reduced the proportion of embryos with a medial expansion of sox10-expressing CNC cells at the 10s stage (Fig. 7, right; P<0.005). In summary, these data support a model in which Disc1 repression of sox10 expression is necessary for migration of CNC cells into the periphery where they can properly differentiate.
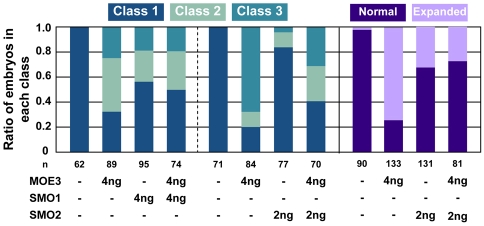
Fig. 7.
Partial reduction of Sox10 in Disc1 morphants suppressed craniofacial defects and CNC expansion. Sox10 levels were reduced in Disc1 morphants by the co-injection of sox10-targeted morpholinos. Co-injection of the 5′UTR-targeted Sox10 morpholino (SMO1) decreased the proportion of Disc1 morphants in class 2 or 3 slightly. As this morpholino has non-specific effects on craniofacial development, a start site-targeted Sox10 morpholino (SMO2) was independently co-injected with MOE3. This morpholino has significantly fewer off-target effects and more convincingly suppressed the craniofacial defects in Disc1 morphants. Additionally, co-injection of SMO2 also suppressed the medial expansion of sox10-positive CNC cells at 10s in Disc1 morphants (purple bars).
Peripheral cranial glial populations were expanded in Disc1 morphants
Together, our data provide evidence for a loss of craniofacial cartilage precursors owing to enhanced transcription factor expression and abnormal CNC migration. However, CNC cells are not lost to apoptosis, so the question of their fate remained. We analyzed other CNC derivatives in Disc1 morphants. Peripheral neuronal and melanocyte development was relatively normal in Disc1 morphants (see Fig. S7A-C in the supplementary material); however, the peripheral glial population was enhanced with loss of Disc1. In situ hybridization for foxd3 clearly showed an expansion of this cell population in Disc1 morphants beginning at 48 hpf (Fig. 8A-F,L). Although foxd3 also marks premigratory neural crest, its expression in CNC cells is downregulated prior to migration. Expression is detectable again during glial differentiation (Kelsh et al., 2000), validating its use as an independent marker for both tissues. Partial knockdown of Sox10 using SMO2 significantly suppressed the expansion of glial precursors in Disc1 morphants at 51 hpf, providing further evidence that this phenotype originates as a result of the enhanced expression of sox10 (Fig. 8C,F-H,L; P<0.0001). Additionally, we determined whether partial depletion of Sox10 in a previously identified sox10 mutant (sox10m241) could suppress the expansion of glia in Disc1 morphants. sox10m241 (Malicki et al., 1996) heterozygotes were crossed to wild-type zebrafish generating clutches in which 50% of embryos were predicted to be heterozygous for this sox10 mutation. When injected with the MOE3 Disc1-targeted morpholino, 49% of these clutches exhibited normal glial foxd3 expression (Fig. 8I-J; n=144). This percentage of embryos with normal peripheral glia is not statistically different from the percentage of embryos expected to be heterozygous for the sox10 mutation (χ2=0.49<χ2[0.05],3=7.83). A second observer corroborated these findings (inter-rater correlation=0.81). It can be inferred from this experiment that partial knockdown of Sox10 in heterozygotes, which make up 50% of these clutches, suppressed the glial phenotype in Disc1 morphants. In summary, these data suggest a possible switch in CNC cell fate, from craniofacial cartilage precursors to peripheral glia, due to loss of Disc1-dependent repression of sox10.
Fig. 8.
Peripheral cranial glia populations were expanded in Disc1 morphants. (A-F) foxd3 expression, which marks the developing glia, was expanded at 48 hpf and 51 hpf, but not 40 hpf, in Disc1 morphants (arrowheads indicate enhanced expression in the trigeminal ganglion and the posterior lateral line ganglion). (G,H) Glial expansion was suppressed by SMO2 co-injection. (I-K) Glial expansion was also suppressed in 49% of Disc1 morphants from a sox10m241/+ by wild-type cross. Uninjected embryos (I; n=47) showed normal foxd3 expression, whereas Disc1 morphants (n=144) showed two distinct phenotypes, expanded (J, arrowheads) and normal (K) foxd3 expression, which probably correlates to their genotype (wild-type or heterozygous). (L) Table showing the percentage of Disc1 morphants with glial expansion (left) and suppression of expansion by SMO2 co-injection (right).
DISCUSSION
The data presented are the first evidence of the role of Disc1 in CNC migration, CNC differentiation, and the repression of foxd3 and sox10, transcription factors implicated in these processes as well as in progenitor maintenance (Kelsh, 2006; Lister et al., 2006; Raible, 2006; Stewart et al., 2006; Teng et al., 2008). Disc1 has previously been implicated in cell migration in the developing brain (Kamiya et al., 2005). In addition, a role for Disc1 in transcriptional regulation is supported by previous data showing Disc1 localization to the nucleus (Pletnikov et al., 2007; Sawamura et al., 2005) and evidence that it can bind to transcription factors (Morris et al., 2003). Our data combine these seemingly disparate functions of Disc1. In CNC, correct timing of foxd3 and sox10 expression is crucial for migration and the development of neural crest-derived structures. In addition, foxd3 and members of the Sox family are expressed in the developing brain (Karoutzou et al., 2008; Lein et al., 2007; Wegner and Stolt, 2005), and we speculate that their regulation by Disc1 might be conserved in this related tissue.
Disc1-dependent transcriptional repression of Foxd3 and Sox10 in CNC cells
Our data implicate Disc1 in the transcriptional repression of foxd3 and sox10, two transcription factors that are crucial for multiple steps of CNC development. Evidence points to a role for Foxd3 as a transcriptional repressor crucial for the maintenance of neural crest progenitor pools, neural crest migration and the differentiation of some neural crest derivatives (Cheung et al., 2005; Lister et al., 2006; Montero-Balaguer et al., 2006; Stewart et al., 2006; Teng et al., 2008). Zebrafish foxd3 mutants have normal numbers of premigratory neural crest, but delayed neural crest migration and depletion of certain neural crest derivatives (Lister et al., 2006; Stewart et al., 2006). Interestingly, the only neural crest derivative with foxd3 expression during and after terminal differentiation is peripheral glia (Kelsh et al., 2000), indicating a possible role for this transcription factor in the differentiation of this derivative. Similar to the Disc1 morphants reported here, colgate (hdac1) mutant zebrafish (Ignatius et al., 2008) have an upregulation of foxd3 expression and an increase in the number of peripheral glia. Craniofacial cartilage is also reduced in this mutant (Nambiar and Henion, 2004). In addition, colgate mutants show delayed neural crest migration, similar to Disc1 morphants, further substantiating the relationship between enhanced foxd3 expression and cell migration in this population. Interestingly, Foxd3 also has a role in progenitor maintenance that is not restricted to the neural crest. First identified as an embryonic stem cell marker (Clevidence et al., 1993; Sutton et al., 1996), Foxd3 is necessary for progenitor maintenance and survival of the early mouse embryo (Hanna et al., 2002). In summary, our data implicate Disc1 in the negative regulation of foxd3 in the neural crest, which subsequently regulates the Foxd3-dependent processes of cell migration and differentiation.
Our research in Disc1 morphants demonstrates a regulatory function for Disc1 in sox10 expression. Loss of Disc1 increased the expression of sox10, whereas overexpression of human DISC1 represses sox10 expression. The functional relevance of this repression is evident by the suppression of craniofacial defects in Disc1 morphants by partial Sox10 reduction. Past research has implicated Sox10 in numerous processes that might underlie the craniofacial and peripheral glial phenotypes in Disc1 morphants, including CNC formation, the maintenance of progenitor pools, and the specification and differentiation of certain derivatives (Kelsh, 2006). In zebrafish, the specification of peripheral neurons relies on Sox10 function (Carney et al., 2006). Additionally, the formation of glial populations, both peripherally and centrally, is regulated by Sox10 across species (Dutton et al., 2001b; Li et al., 2007; Liu et al., 2007; Svaren and Meijer, 2008). Although our Disc1 morphants have fairly normal peripheral neuronal populations, there is a dramatic increase in the number of peripheral glia. This could, in fact, be due to the misregulation of both foxd3 and sox10 expression in Disc1 morphants, which could predispose CNC cells to a glial fate at the expense of craniofacial cartilage.
Increased sox10 expression might occur directly as a result of Disc1 function, or indirectly as a result of the increase in foxd3 expression observed in Disc1 morphants. Studies in Foxd3 mutant zebrafish have illustrated that loss of Foxd3 directly affects the expression of other neural crest-associated genes, including sox10 (Montero-Balaguer et al., 2006; Stewart et al., 2006). Similarly, our studies point to a relationship between foxd3 and sox10 expression. We show that overexpression of Foxd3 did not affect sox10 expression at early somitic stages (data not shown) but led to elevated levels of sox10 at 11s. These data are similar to the results seen in Disc1 morphants, where expression of foxd3 and sox10 are normal at 6s but increased by 10s. Whether the effect on sox10 expression is directly due to the loss of Disc1-dependent transcriptional repression or indirectly due to the upregulation of foxd3 expression is yet to be determined. In addition, the abnormal medial placement of neural crest cells dorsal to the developing midbrain in embryos overexpressing Foxd3 (see Fig. 6F-I) implies migration abnormalities similar to those studied in Disc1 morphants. This further implicates the upregulation of these transcription factors in the perpetuation of medial movement and lack of migration from the region dorsal to the neural rod. Future studies will address the mechanisms of Disc1-dependent repression of foxd3 and sox10 expression, and the impact of enhanced expression of these factors on neural crest migration directly.
The role of Disc1 role in CNC migration and differentiation
Our in vivo analysis of CNC cell movement in wild-type zebrafish revealed a medial movement of CNC cells dorsal to the neural keel prior to their emigration from their rostrocaudal area of origin. This movement begins at approximately the 6s stage, which is earlier than CNC cells have previously been thought to be highly motile. The function of and factors controlling this medial movement are, as of yet, unknown. These studies illustrate that the perpetuation of this medial movement in Disc1 morphants, which is probably due to the enhanced foxd3 and sox10 expression, significantly delays CNC migration from the region dorsal to the neural rod and results in a loss of the ectomesenchymal CNC cell population in the pharyngeal pouches. Concurrently in these morphants, we observed an increase in the number of peripheral glia. There are several possible, but not necessarily exclusive, reasons for these two phenotypes in Disc1 morphants.
First, it is possible that the loss of Disc1, upregulating foxd3 and sox10 expression, results in a switch of CNC cell fate from craniofacial cartilage precursors to glial precursors prior to migration into the periphery. This might affect the temporal dynamics of migration, leading to the documented phenotypes. Second, delayed migration from the region dorsal to the neural rod might itself underlie the abnormal transcription factor expression and CNC differentiation in Disc1 morphants. If the CNC population is instead multipotent in the early migratory phase, a delay in migration towards the periphery might result in them missing the environmental signals necessary for differentiation into the appropriate tissues. For craniofacial cartilage precursors, for example, environmental sources of fibroblast growth factor and sonic hedgehog are necessary for CNC survival and craniofacial cartilage patterning (Crump et al., 2004; Eberhart et al., 2006). As CNC cells in Disc1 morphants appear to survive, at least during their migratory phase, delayed migration might rather result in a change in the tissue to which they give rise.
Finally, migration and differentiation of CNC might be impacted independently by the continued upregulation of foxd3 and sox10 expression. As mentioned above, the expression of both transcription factors has been associated with progenitor cell maintenance (Kim et al., 2003; McKeown et al., 2005; Paratore et al., 2002). Persistent upregulation of foxd3 and sox10 expression might hold CNC cells in a multipotent, progenitor state, which could impact on their ability to migrate normally into the pharyngeal pouches. Independently, the upregulation of these factors could also impact on the differentiation of the CNC. Both foxd3 and sox10 are expressed in differentiating peripheral glia but are not expressed highly in other CNC derivatives (Dutton et al., 2001b; Kelsh et al., 2000). In addition, Sox10 has been shown to regulate the promoter activity of genes necessary for glial differentiation in CNC cells and the brain (Ghislain and Charnay, 2006; Li et al., 2007; Stolt et al., 2002).
Conclusions
We have shown that Disc1 is a potent regulator of foxd3 and sox10 expression, CNC migration and CNC specification in the zebrafish. Although this study highlights the role of Disc1 in the neural crest, it provides insight into a basic cellular function of this protein in transcription factor regulation that might be preserved in the central nervous system. Foxd3 is a well-characterized marker of stem cells expressed in neurogenic regions of the rodent brain (Lein et al., 2007). Sox10 has been repeatedly emphasized as an integral transcription factor necessary for oligodendrocyte differentiation. Abnormal white matter volume and expression of oligodendrocyte-related genes, including SOX10, have consistently been associated with schizophrenia (Karoutzou et al., 2008; Wegner and Stolt, 2005). Additionally, SOX10 has been implicated as a schizophrenia-susceptibility gene itself (Dracheva et al., 2006; Maeno et al., 2007). In conclusion, although the studies documented here were not done in the central nervous system, they provide insight into protein function. Understanding the basic functions of Disc1 in transcriptional regulation, cell migration and cell differentiation in the zebrafish neural crest might give insight into the cellular processes that, when disrupted, predispose individuals to mental illness.
Note added in proof
After submission of this manuscript, work from Wood et al. (Wood et al., 2009) was published describing Disc1 function in oligodendrocyte development in zebrafish. This work also described craniofacial deficits in Disc1 morphants and corroborates our findings.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/15/2623/DC1
Supplementary Material
We thank J. M. Topczewska and K. D. Meyer for their assistance. Funding for this work was provided to J.A.M. by the McKnight Brain Disorders Award, NARSAD (National Alliance for Research on Schizophrenia and Depression) and the Illinois Department of Public Aid, and to J.T. by NIH/NIDCR DE016678. Deposited in PMC for release after 12 months.
References
- Akimenko, M. A., Ekker, M., Wegner, J., Lin, W. and Westerfield, M. (1994). Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 14, 3475-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo, S., Lohr, J., Lee, K. H., Ticho, B. S., Breitbart, R. E., Hill, S., Yost, H. J. and Srivastava, D. (2000). Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech. Dev. 95, 231-237. [DOI] [PubMed] [Google Scholar]
- Austin, C. P., Ma, L., Ky, B., Morris, J. A. and Shughrue, P. J. (2003). DISC1 (Disrupted in Schizophrenia-1) is expressed in limbic regions of the primate brain. Neuroreport 14, 951-954. [DOI] [PubMed] [Google Scholar]
- Brandon, N. J., Schurov, I., Camargo, L. M., Handford, E. J., Duran-Jimeniz, B., Hunt, P., Millar, J. K., Porteous, D. J., Shearman, M. S. and Whiting, P. J. (2005). Subcellular targeting of DISC1 is dependent on a domain independent from the Nudel binding site. Mol. Cell. Neurosci. 28, 613-624. [DOI] [PubMed] [Google Scholar]
- Camargo, L. M., Collura, V., Rain, J. C., Mizuguchi, K., Hermjakob, H., Kerrien, S., Bonnert, T. P., Whiting, P. J. and Brandon, N. J. (2007). Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry 12, 74-86. [DOI] [PubMed] [Google Scholar]
- Carney, T. J., Dutton, K. A., Greenhill, E., Delfino-Machin, M., Dufourcq, P., Blader, P. and Kelsh, R. N. (2006). A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development 133, 4619-4630. [DOI] [PubMed] [Google Scholar]
- Cheung, M., Chaboissier, M. C., Mynett, A., Hirst, E., Schedl, A. and Briscoe, J. (2005). The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell 8, 179-192. [DOI] [PubMed] [Google Scholar]
- Chubb, J. E., Bradshaw, N. J., Soares, D. C., Porteous, D. J. and Millar, J. K. (2008). The DISC locus in psychiatric illness. Mol. Psychiatry 13, 36-64. [DOI] [PubMed] [Google Scholar]
- Clevidence, D. E., Overdier, D. G., Tao, W., Qian, X., Pani, L., Lai, E. and Costa, R. H. (1993). Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc. Natl. Acad. Sci. USA 90, 3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump, J. G., Maves, L., Lawson, N. D., Weinstein, B. M. and Kimmel, C. B. (2004). An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development 131, 5703-5716. [DOI] [PubMed] [Google Scholar]
- Dracheva, S., Davis, K. L., Chin, B., Woo, D. A., Schmeidler, J. and Haroutunian, V. (2006). Myelin-associated mRNA and protein expression deficits in the anterior cingulate cortex and hippocampus in elderly schizophrenia patients. Neurobiol. Dis. 21, 531-540. [DOI] [PubMed] [Google Scholar]
- Drerup, C. M., Ahlgren, S. C. and Morris, J. A. (2007). Expression profiles of ndel1a and ndel1b, two orthologs of the NudE-Like gene, in the zebrafish. Gene Expr. Patterns 7, 672-679. [DOI] [PubMed] [Google Scholar]
- Duan, X., Chang, J. H., Ge, S., Faulkner, R. L., Kim, J. Y., Kitabatake, Y., Liu, X. B., Yang, C. H., Jordan, J. D., Ma, D. K. et al. (2007). Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 130, 1146-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton, K., Dutton, J. R., Pauliny, A. and Kelsh, R. N. (2001a). A morpholino phenocopy of the colourless mutant. Genesis 30, 188-189. [DOI] [PubMed] [Google Scholar]
- Dutton, K. A., Pauliny, A., Lopes, S. S., Elworthy, S., Carney, T. J., Rauch, J., Geisler, R., Haffter, P. and Kelsh, R. N. (2001b). Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 128, 4113-4125. [DOI] [PubMed] [Google Scholar]
- Eberhart, J. K., Swartz, M. E., Crump, J. G. and Kimmel, C. B. (2006). Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development 133, 1069-1077. [DOI] [PubMed] [Google Scholar]
- Ghislain, J. and Charnay, P. (2006). Control of myelination in Schwann cells: a Krox20 cis-regulatory element integrates Oct6, Brn2 and Sox10 activities. EMBO Rep. 7, 52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, L. A., Foreman, R. K., Tarasenko, I. A., Kessler, D. S. and Labosky, P. A. (2002). Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 16, 2650-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius, M. S., Moose, H. E., El-Hodiri, H. M. and Henion, P. D. (2008). colgate/hdac1 Repression of foxd3 expression is required to permit mitfa-dependent melanogenesis. Dev. Biol. 313, 568-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, R., Adams, R. R., Christie, S., Buchanan, S. R., Porteous, D. J. and Millar, J. K. (2004). Disrupted in Schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Mol. Cell. Neurosci. 26, 112-122. [DOI] [PubMed] [Google Scholar]
- Julich, D., Hwee Lim, C., Round, J., Nicolaije, C., Schroeder, J., Davies, A., Geisler, R., Lewis, J., Jiang, Y. J. and Holley, S. A. (2005). beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev. Biol. 286, 391-404. [DOI] [PubMed] [Google Scholar]
- Kamiya, A., Kubo, K., Tomoda, T., Takaki, M., Youn, R., Ozeki, Y., Sawamura, N., Park, U., Kudo, C., Okawa, M. et al. (2005). A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell. Biol. 7, 1167-1178. [DOI] [PubMed] [Google Scholar]
- Karoutzou, G., Emrich, H. M. and Dietrich, D. E. (2008). The myelin-pathogenesis puzzle in schizophrenia: a literature review. Mol. Psychiatry 13, 245-260. [DOI] [PubMed] [Google Scholar]
- Kelsh, R. N. (2006). Sorting out Sox10 functions in neural crest development. BioEssays 28, 788-798. [DOI] [PubMed] [Google Scholar]
- Kelsh, R. N., Dutton, K., Medlin, J. and Eisen, J. S. (2000). Expression of zebrafish fkd6 in neural crest-derived glia. Mech. Dev. 93, 161-164. [DOI] [PubMed] [Google Scholar]
- Kim, J., Lo, L., Dormand, E. and Anderson, D. J. (2003). SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron 38, 17-31. [DOI] [PubMed] [Google Scholar]
- Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. [DOI] [PubMed] [Google Scholar]
- Kimmel, C. B., Miller, C. T., Kruze, G., Ullmann, B., BreMiller, R. A., Larison, K. D. and Snyder, H. C. (1998). The shaping of pharyngeal cartilages during early development of the zebrafish. Dev. Biol. 203, 245-263. [DOI] [PubMed] [Google Scholar]
- Knight, R. D. and Schilling, T. F. (2006). Cranial neural crest and development of the head skeleton. In Neural crest induction and differentiation, Vol. 589 (ed. J. P. Saint-Jeannet), pp. 120-133. Georgetown, Texas: Landes Bioscience. [DOI] [PubMed] [Google Scholar]
- Lefcort, F., O'Connor, T. and Kulesa, P. M. (2007). Comparative Analysis of Neural Crest Cell and Axonal Growth Cone Dynamics and Behavior. In Intracellular Mechanisms for Neuritogenesis (ed. I. de Curtis), pp. 282-301. New York, NY, USA: Springer.
- Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., Boe, A. F., Boguski, M. S., Brockway, K. S., Byrnes, E. J. et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168-176. [DOI] [PubMed] [Google Scholar]
- Lewis, D. A. and Levitt, P. (2002). Schizophrenia as a disorder of neurodevelopment. Annu. Rev. Neurosci. 25, 409-432. [DOI] [PubMed] [Google Scholar]
- Li, H., Lu, Y., Smith, H. K. and Richardson, W. D. (2007). Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J. Neurosci. 27, 14375-14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, J. A., Cooper, C., Nguyen, K., Modrell, M., Grant, K. and Raible, D. W. (2006). Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev. Biol. 290, 92-104. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Hu, X., Cai, J., Liu, B., Peng, X., Wegner, M. and Qiu, M. (2007). Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev. Biol. 302, 683-693. [DOI] [PubMed] [Google Scholar]
- Luo, R., An, M., Arduini, B. L. and Henion, P. D. (2001). Specific pan-neural crest expression of zebrafish Crestin throughout embryonic development. Dev. Dyn. 220, 169-174. [DOI] [PubMed] [Google Scholar]
- Ma, L., Liu, Y., Ky, B., Shughrue, P. J., Austin, C. P. and Morris, J. A. (2002). Cloning and characterization of Disc1, the mouse ortholog of DISC1 (Disrupted-in-Schizophrenia 1). Genomics 80, 662-672. [DOI] [PubMed] [Google Scholar]
- Maeno, N., Takahashi, N., Saito, S., Ji, X., Ishihara, R., Aoyama, N., Branko, A., Miura, H., Ikeda, M., Suzuki, T. et al. (2007). Association of SOX10 with schizophrenia in the Japanese population. Psychiatr. Genet. 17, 227-231. [DOI] [PubMed] [Google Scholar]
- Malicki, J., Schier, A. F., Solnica-Krezel, L., Stemple, D. L., Neuhauss, S. C., Stainier, D. Y., Abdelilah, S., Rangini, Z., Zwartkruis, F. and Driever, W. (1996). Mutations affecting development of the zebrafish ear. Development 123, 275-283. [DOI] [PubMed] [Google Scholar]
- McKeown, S. J., Lee, V. M., Bronner-Fraser, M., Newgreen, D. F. and Farlie, P. G. (2005). Sox10 overexpression induces neural crest-like cells from all dorsoventral levels of the neural tube but inhibits differentiation. Dev. Dyn. 233, 430-444. [DOI] [PubMed] [Google Scholar]
- Meyer, K. D. and Morris, J. A. (2008). Immunohistochemical analysis of Disc1 expression in the developing and adult hippocampus. Gene Expr. Patterns 8, 494-501. [DOI] [PubMed] [Google Scholar]
- Millar, J. K., Wilson-Annan, J. C., Anderson, S., Christie, S., Taylor, M. S., Semple, C. A., Devon, R. S., Clair, D. M., Muir, W. J., Blackwood, D. H. et al. (2000). Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415-1423. [DOI] [PubMed] [Google Scholar]
- Millar, J. K., Christie, S. and Porteous, D. J. (2003). Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem. Biophys. Res. Commun. 311, 1019-1025. [DOI] [PubMed] [Google Scholar]
- Montero-Balaguer, M., Lang, M. R., Sachdev, S. W., Knappmeyer, C., Stewart, R. A., De La Guardia, A., Hatzopoulos, A. K. and Knapik, E. W. (2006). The mother superior mutation ablates foxd3 activity in neural crest progenitor cells and depletes neural crest derivatives in zebrafish. Dev. Dyn. 235, 3199-3212. [DOI] [PubMed] [Google Scholar]
- Morales, A. V., Barbas, J. A. and Nieto, M. A. (2005). How to become neural crest: from segregation to delamination. Semin. Cell Dev. Biol. 16, 655-662. [DOI] [PubMed] [Google Scholar]
- Morris, J. A., Kandpal, G., Ma, L. and Austin, C. P. (2003). DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum. Mol. Genet. 12, 1591-1608. [DOI] [PubMed] [Google Scholar]
- Nambiar, R. M. and Henion, P. D. (2004). Sequential antagonism of early and late Wnt-signaling by zebrafish colgate promotes dorsal and anterior fates. Dev. Biol. 267, 165-180. [DOI] [PubMed] [Google Scholar]
- Odenthal, J. and Nusslein-Volhard, C. (1998). fork head domain genes in zebrafish. Dev. Genes Evol. 208, 245-258. [DOI] [PubMed] [Google Scholar]
- Paratore, C., Eichenberger, C., Suter, U. and Sommer, L. (2002). Sox10 haploinsufficiency affects maintenance of progenitor cells in a mouse model of Hirschsprung disease. Hum. Mol. Genet. 11, 3075-3085. [DOI] [PubMed] [Google Scholar]
- Pletnikov, M. V., Xu, Y., Ovanesov, M. V., Kamiya, A., Sawa, A. and Ross, C. A. (2007). PC12 cell model of inducible expression of mutant DISC1: new evidence for a dominant-negative mechanism of abnormal neuronal differentiation. Neurosci. Res. 58, 234-244. [DOI] [PubMed] [Google Scholar]
- Porteous, D. J. and Millar, J. K. (2006). Disrupted in schizophrenia 1, building brains and memories. Trends Mol. Med. 12, 255-261. [DOI] [PubMed] [Google Scholar]
- Raible, D. W. (2006). Development of the neural crest: achieving specificity in regulatory pathways. Curr. Opin. Cell Biol. 18, 698-703. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler, T. and Bronner-Fraser, M. (2008). A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9, 557-568. [DOI] [PubMed] [Google Scholar]
- Sawamura, N., Sawamura-Yamamoto, T., Ozeki, Y., Ross, C. A. and Sawa, A. (2005). A form of DISC1 enriched in nucleus: altered subcellular distribution in orbitofrontal cortex in psychosis and substance/alcohol abuse. Proc. Natl. Acad. Sci. USA 102, 1187-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurov, I. L., Handford, E. J., Brandon, N. J. and Whiting, P. J. (2004). Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicates its role in neurodevelopment. Mol. Psychiatry 9, 1100-1110. [DOI] [PubMed] [Google Scholar]
- Shu, T., Ayala, R., Nguyen, M. D., Xie, Z., Gleeson, J. G. and Tsai, L. H. (2004). Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263-277. [DOI] [PubMed] [Google Scholar]
- Steventon, B., Carmona-Fontaine, C. and Mayor, R. (2005). Genetic network during neural crest induction: from cell specification to cell survival. Semin. Cell Dev. Biol. 16, 647-654. [DOI] [PubMed] [Google Scholar]
- Stewart, R. A., Arduini, B. L., Berghmans, S., George, R. E., Kanki, J. P., Henion, P. D. and Look, A. T. (2006). Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev. Biol. 292, 174-188. [DOI] [PubMed] [Google Scholar]
- Stolt, C. C., Rehberg, S., Ader, M., Lommes, P., Riethmacher, D., Schachner, M., Bartsch, U. and Wegner, M. (2002). Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 16, 165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, J., Costa, R., Klug, M., Field, L., Xu, D., Largaespada, D. A., Fletcher, C. F., Jenkins, N. A., Copeland, N. G., Klemsz, M. et al. (1996). Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J. Biol. Chem. 271, 23126-23133. [DOI] [PubMed] [Google Scholar]
- Svaren, J. and Meijer, D. (2008). The molecular machinery of myelin gene transcription in Schwann cells. Glia 56, 1541-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapadia, M. D., Cordero, D. R. and Helms, J. A. (2005). It's all in your head: new insights into craniofacial development and deformation. J. Anat. 207, 461-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, M. S., Devon, R. S., Millar, J. K. and Porteous, D. J. (2003). Evolutionary constraints on the Disrupted in Schizophrenia locus. Genomics 81, 67-77. [DOI] [PubMed] [Google Scholar]
- Teng, L., Mundell, N. A., Frist, A. Y., Wang, Q. and Labosky, P. A. (2008). Requirement for Foxd3 in the maintenance of neural crest progenitors. Development 135, 1615-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse, C., Thisse, B., Schilling, T. F. and Postlethwait, J. H. (1993). Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119, 1203-1215. [DOI] [PubMed] [Google Scholar]
- Thisse, C., Thisse, B. and Postlethwait, J. H. (1995). Expression of snail2, a second member of the zebrafish snail family, in cephalic mesendoderm and presumptive neural crest of wild-type and spadetail mutant embryos. Dev. Biol. 172, 86-99. [DOI] [PubMed] [Google Scholar]
- Wada, N., Javidan, Y., Nelson, S., Carney, T. J., Kelsh, R. N. and Schilling, T. F. (2005). Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development 132, 3977-3988. [DOI] [PubMed] [Google Scholar]
- Wegner, M. and Stolt, C. C. (2005). From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci. 28, 583-588. [DOI] [PubMed] [Google Scholar]
- Weinberg, E. S., Allende, M. L., Kelly, C. S., Abdelhamid, A., Murakami, T., Andermann, P., Doerre, O. G., Grunwald, D. J. and Riggleman, B. (1996). Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development 122, 271-280. [DOI] [PubMed] [Google Scholar]
- Westerfield, M. (1995). The Zebrafish Book: Guide for the Laboratory Use of Zebrafish (Danio rerio), 3rd edn. Eugene, Oregon: University of Oregon Press.
- Wood, J. D., Bonath, F., Kumar, S., Ross, C. A. and Cunliffe, V. T. (2009). Disrupted-in-schizophrenia 1 and neuregulin 1 are required for the specification of oligodendrocytes and neurones in the zebrafish brain. Hum. Mol. Genet. 18, 391-404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.