Abstract
X-chromosome inactivation is an epigenetic process used to regulate gene dosage in mammalian females by silencing genes on one X-chromosome. While the pattern of X-chromosome inactivation is typically random in normal females, abnormalities of the X-chromosome may result in skewing due to disadvantaged cell growth. We describe a female patient with an X;1 translocation [46,X,t(X;1)(q28;q21)] and unusual pattern of X-chromosome inactivation who demonstrates functional disomy of the Xq28 region distal to the translocation breakpoint. There was complete skewing of X-chromosome inactivation in the patient, along with the atypical findings of an active normal X chromosome and an inactive derivative X. Characterization of the translocation revealed that the patient’s Xq28 breakpoint interrupts the DKC1 gene. Molecular analysis of the breakpoint region revealed functional disomy of Xq28 genes distal to DKC1. We propose that atypical X-chromosome inactivation occurred in the patient due to a post-inactivation cell selection mechanism likely initiated by disruption of DKC1. As a result, the pattern of X-chromosome inactivation is opposite that of the expected for an X;autosome translocation. Therefore, we suggest the phenotypic abnormalities found in the patient are a result of functional disomy in the Xq28 region.
Keywords: X-Chromosome Inactivation, X;1 Translocation, Xq28 Functional Disomy, DKC1
INTRODUCTION
X-chromosome inactivation (XCI) is a critical epigenetic process that regulates gene dosage in mammalian females [Lyon, 1961]. Cytogenetic or molecular abnormalities on the X-chromosome may influence XCI, resulting in a non-random or “skewed” pattern due to a survival disadvantage of cells containing an imbalance in genetic material [Disteche et al., 1979; Mattei et al., 1982]. Skewed XCI is a common finding in a balanced X;autosome translocation, with the derivative X chromosome [der(X)] typically remaining active and the normal X inactivated.
In the case of a balanced X;autosome translocation, the potential consequences of atypical XCI include monosomy of autosomal genes (due to the spread of XCI) and functional disomy of chromosome X genes [Schmidt and Du Sart, 1992]. Digression from the expected XCI pattern has been cited as the cause of phenotypic abnormality in a number of X;autosome translocation patients [Wolff et al., 2000].
In this report, we describe a female patient with an X;1 translocation and an unusual pattern of XCI. Interpretation of the patient's cytogenetic Xq28 breakpoint and examination of phenotypic features initially led to a diagnosis of Otopalatodigital syndrome (OPD) type I, known to be caused by mutations in the FLNA gene [Robertson et al., 2003]. Despite a thorough genetic analysis, no FLNA defect was identified suggesting an alternate underlying cause of the patient's phenotype. Based on successive analyses we propose that the patient's abnormal phenotypic features arose due to atypical XCI and subsequent Xq28 functional disomy.
MATERIALS AND METHODS
Subjects and Consent
Informed consent was obtained for the patient and her parents with IRB approval granted by The Research Institute at Nationwide Children's Hospital.
Tissue Sources, Cell Lines, DNA and RNA Preparations
Peripheral blood lymphocytes (PBL) and buccal swabs were obtained from the patient and parents. PBL were used to create immortalized lymphoblastoid cell lines (LCL) by Epstein-Barr Virus (EBV) transformation using the Marmoset cell line GM07404E (Coriell, Camden, NJ). Somatic cell hybrid (SCH) lines were established as previously described [Cirullo et al., 1983]. Separate hybrid cell lines containing a normal X chromosome, a der(X) chromosome, a normal chromosome 1, or a derivative chromosome 1 were isolated.
Molecular and Cytogenetic Breakpoint Mapping
Nick-translated biotin-labeled BAC clones from the RPCI-11 panel (Invitrogen, Carlsbad, CA) were used to map the X-chromosome breakpoint region by fluorescence in-situ hybridization (FISH). Information on mapping and sequence data was obtained from the Ensembl Genome Browser (http://www.ensembl.org), the National Center for Biotechnology Information, (http://www.ncbi.nlm.nih.gov), and the University of California Santa Cruz (http://genome.ucsc.edu). Concurrent with the effort to map the breakpoint by FISH, SCH lines were analyzed for the presence or absence of markers in Xq28 and 1q21 by locus-specific PCR.
X-Inactivation Analysis
To determine XCI pattern, a CAG triplet repeat in the first exon of the androgen receptor (AR) gene (Xq11.2-Xq12) was analyzed by methylation-specific PCR as described by Kubota [1999]. Reaction products were electrophoresed on an AB 310 or 3130 Genetic Analyzer and genotype determined using Genescan or Genemapper software (Applied Biosystems, Foster City, CA).
Expression by RT-PCR
Reverse-transcription PCR (RT-PCR) was used to study the expression of genes near the translocation breakpoints. Total RNA extracted from EBV-LCL and SCH lines (RNeasy Mini Kit, Qiagen, Valencia, CA) was used for first strand cDNA synthesis (Superscript II RT, Invitrogen, Carlsbad, CA).
Sequencing
After PCR analysis to refine the breakpoint region to less than 1 kb, primer pairs were designed to amplify across the breakpoint on each derivative chromosome and the resulting products were analyzed by bi-directional sequencing. BLAST was used to determine the origin of the sequence inserted into the derivative X breakpoint region [McGinnis and Madden, 2004]. Transfac Matrix Database (v7.0), Biobase was used to screen for transcription factor binding sites and sequence conservation was evaluated using the Vertebrate Multiz Alignment & PhastCons Conservation (28 Species) track on the UCSC Genome Bioinformatics website [Karolchik et al., 2003; Matys et al., 2003; Siepel et al., 2005]. Big Dye version 1.1 dye terminator chemistry was used in all sequencing reactions and the products were separated by capillary electrophoresis on an AB 3130 genetic analyzer (Applied Biosystems, Foster City, CA).
Expression Array Analysis
Patient and parental RNA (isolated from PBL) was converted to cDNA for use in microarray analysis (NuGEN Inc, San Carlos, CA). For each array, 2.2 µg of cDNA was hybridized to Human 133 Plus 2.0 GeneChips (Affymetrix Inc., Santa Clara, CA). The probe set signals were generated using the RMA algorithm in ArrayAssist 3.4 (Stratagene, La Jolla, CA) and were used to determine differential gene expression by pair-wise comparisons.
RESULTS
Clinical Report
The patient was born to a 30-year-old, gravida 2, para 2 female. A cleft lip was detected on prenatal ultrasound. Subsequent amniocentesis revealed an apparently balanced X;1 translocation [46,X,t(X;1)(q28;q21)dn] (Fig 1). On neonatal examination a unilateral cleft lip, midline cleft palate, hypertelorism, a broad and flat nasal bridge, small low-set ears, preauricular pits, proximally positioned thumbs, short distal phalanges, fifth digit clinodactyly, camptodactyly, and a hemangioma over the sacrum were documented. A brain MRI revealed an abnormal, hypoplastic corpus collosum.
Figure 1.
Cytogenetic characterization of the patient’s balanced translocation, 46,X,t(X;1)(q28;q21)dn
The patient was followed on a regular basis by a clinical geneticist (AS). Despite receiving growth hormone therapy for several years, the patient continued to be extremely small in size with her height and weight proportionate (Fig 2). At the age of 8 years she was 115.5 cm tall (below the 3rd centile), weighed 22.1 kg (at the 3rd centile), and head circumference measured 50.5 cm (10th centile).
Figure 2.
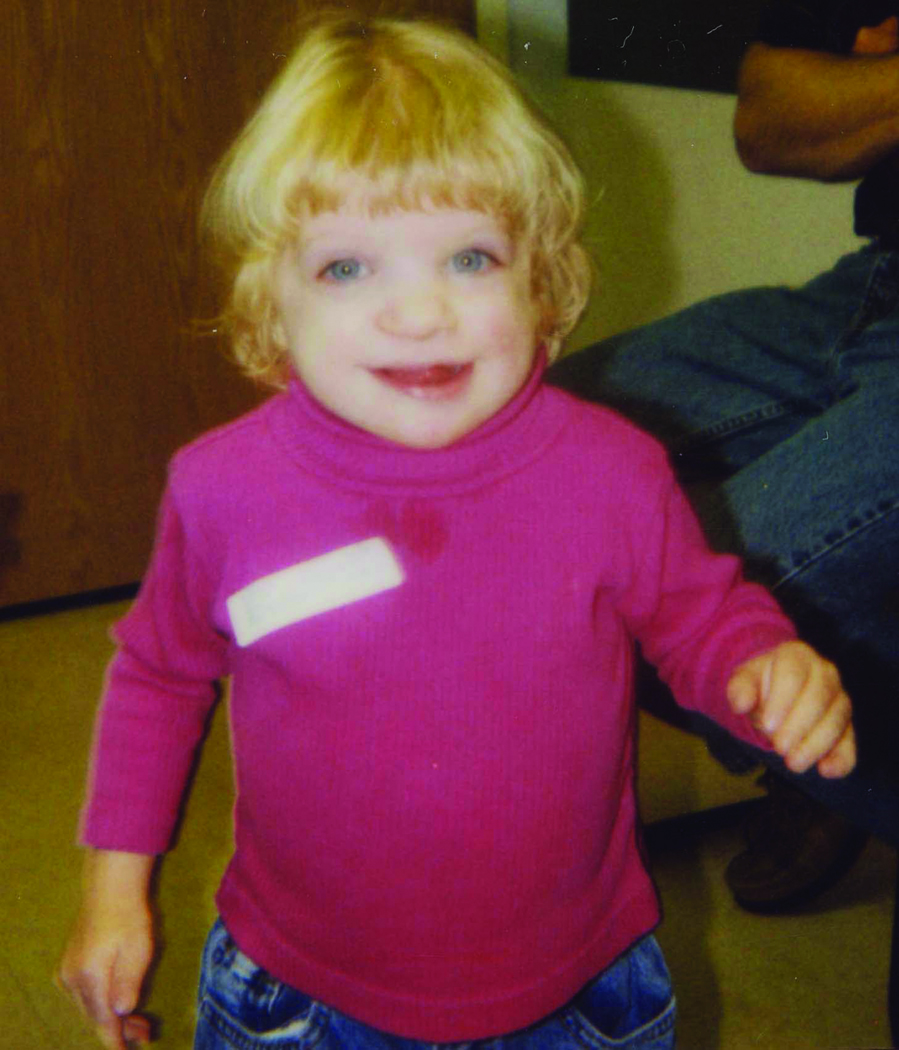
A. The X;1 translocation patient at age 3 years. Note the patient's abnormal facial features including hypertelorism, clefting and a depressed nasal bridge with a prominent tip.
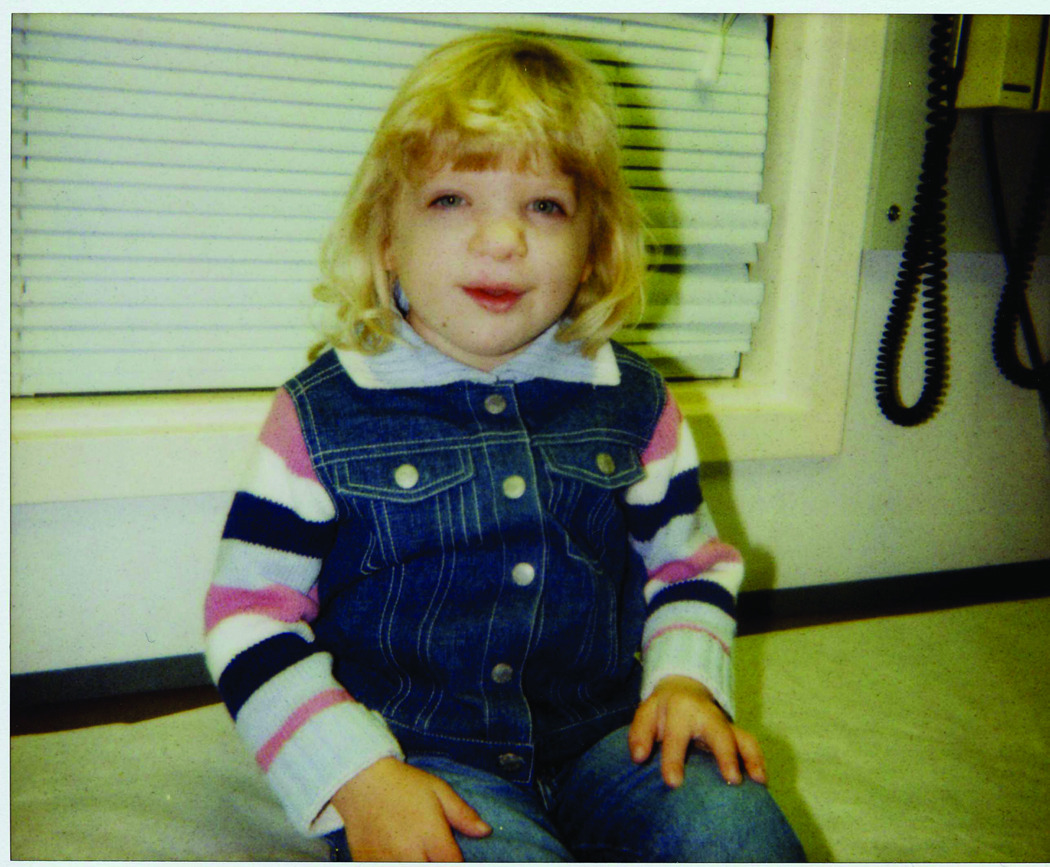
B. The patient at 5 years of age with significant growth and developmental delay.
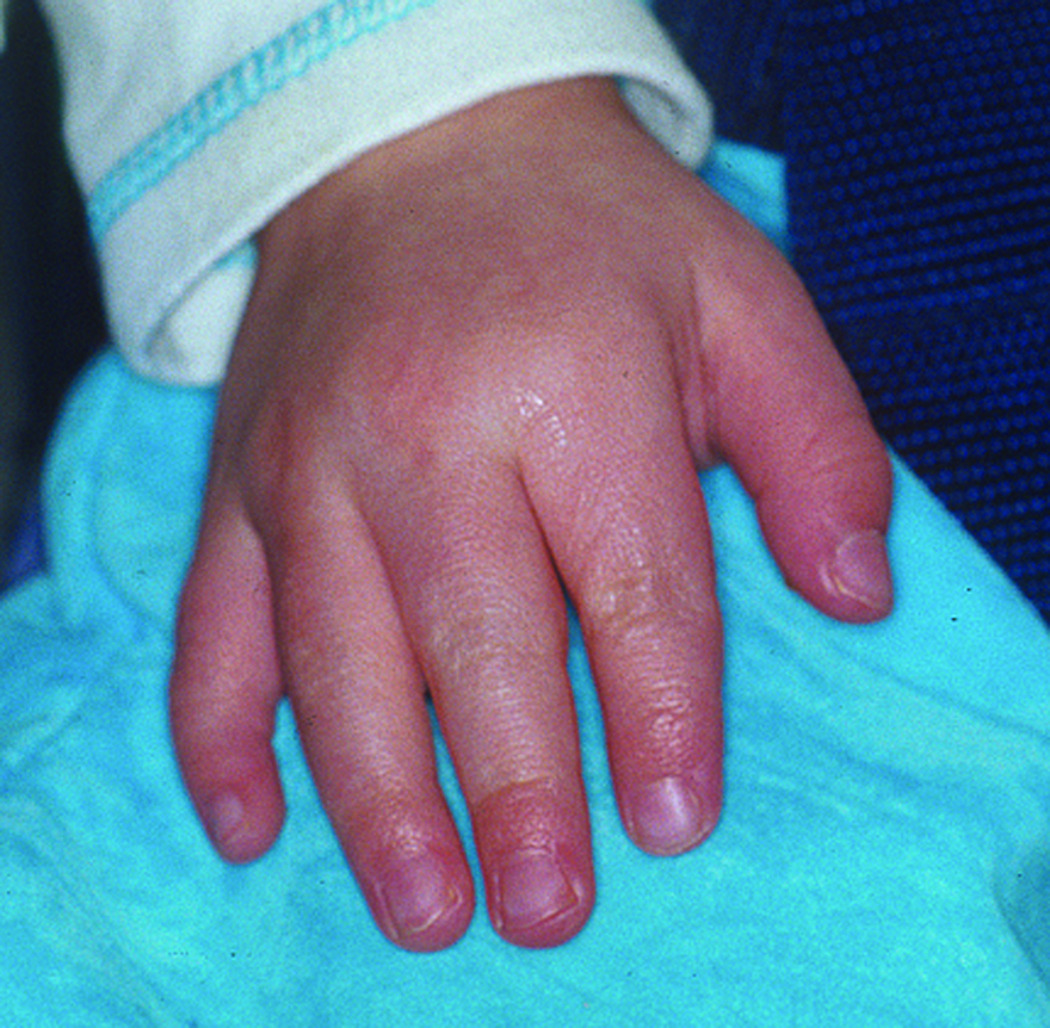
C. Abnormalities of the patient’s hand include distally tapering digits, ulnar deviation of the index finger, and 5th finger clinodactyly.
Facial features at age 8 included persistent hypertelorism (with an inner canthal distance of 3.7 cm, >99th centile, and an outer canthal distance of 9.3 cm, >97th centile, as measured at age 6) and a very broad, depressed nasal bridge with a prominent tip. Her cleft lip and palate were repaired. She had small, cup-shaped auricles and hypermobile small joints. Her extremities were symmetrical, with all digits tapering distally, ulnar deviation of both index fingers, and 5th finger clinodactyly.
Radiological findings included a delayed bone age, bilateral coxa valga and a steep left acetabular angle. An MRI of the brain revealed a very small corpus callosum, with almost no splenuim or posterior elements; the myelination pattern was normal. She had some developmental delay. At age 8 years, she was in 3rd grade in a special needs school and doing well, however, she was still not toilet trained. Her speech had been improving. The family history was negative for birth defects, mental retardation, or genetic disease.
Breakpoint Characterization
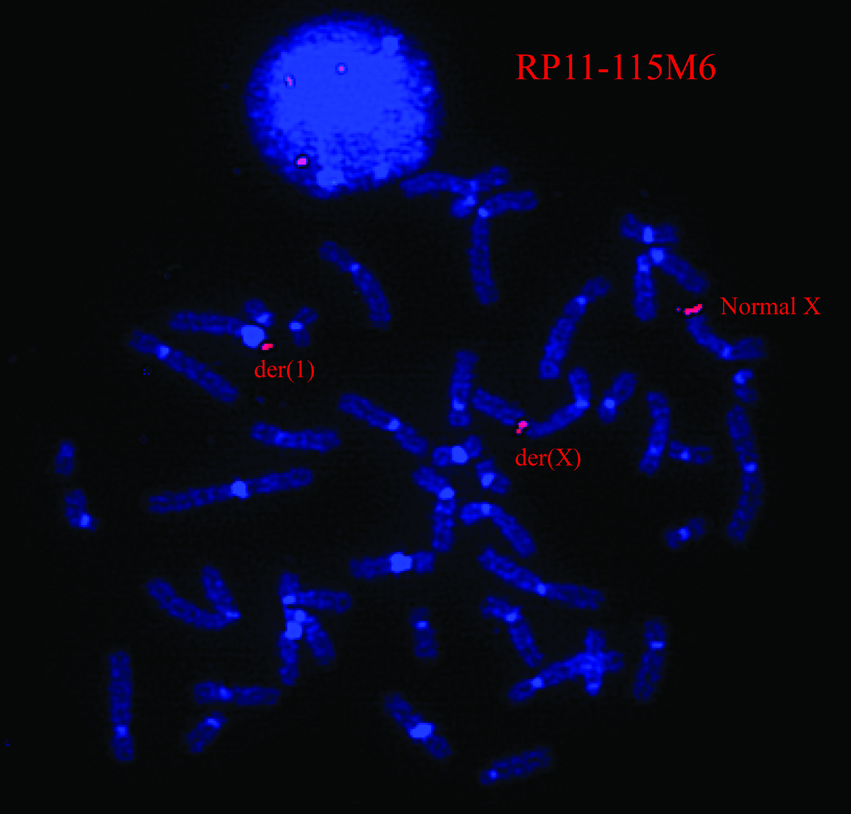
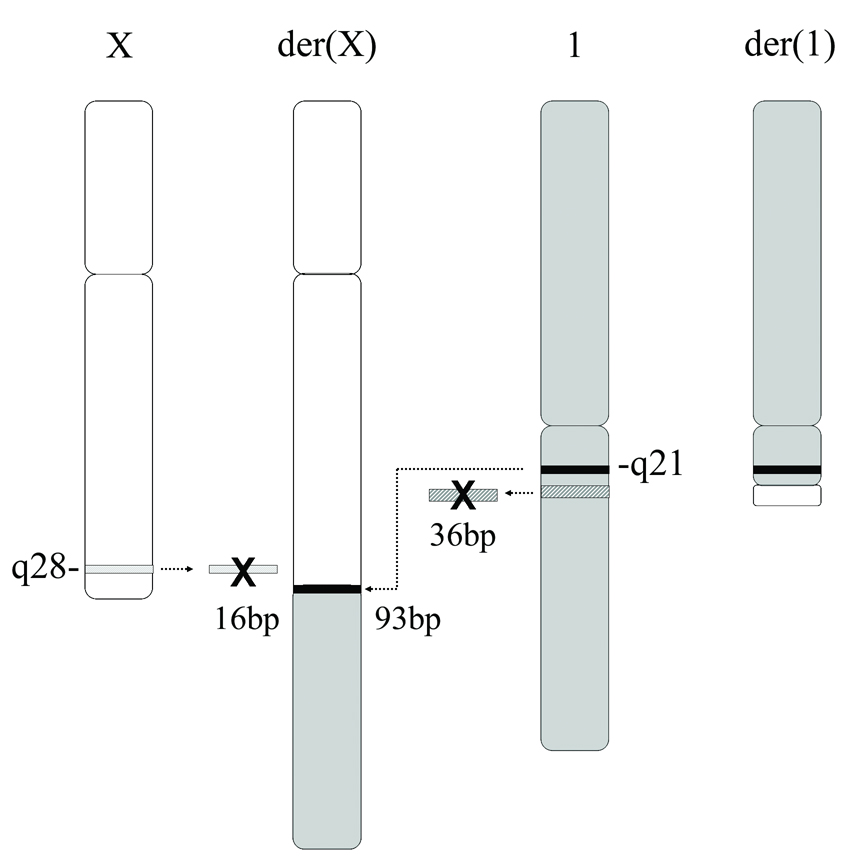
Cytogenetic and molecular techniques were used to define the patient’s translocation breakpoints. Multiple BAC clones in the Xq28 region were used to narrow the critical region by FISH (See Supplemental Table I available online), and BAC RP11–115M6 was found to span the Xq28 translocation breakpoint in the patient (Fig 3). Mapping of the SCH lines by PCR identified the X-chromosome breakpoint to basepair 153,644,715 (Mar 2006, NCBI Bld 36.1) within the first intron of the DKC1 gene on Xq28. The breakpoint at 1q21 was located at basepair 153,386,489 (Mar 2006, NCBI Bld 36.1) within the 35 kb intergenic span between the DPM3 and KRTCAP2 genes. Sequencing across the breakpoints of both derivative chromosomes revealed a deletion of 16 bp of chromosome X involving bps 153,644,716 to 153,644,731 (Mar 2006, NCBI Bld 36.1) and 36 bp from chromosome 1 involving bps 153,386,474–153,386,509 (Mar 2006, NCBI Bld 36.1). In addition, a duplicate copy of 93 bp from 153,384,859–153,384,951 (Mar 2006, NCBI Bld 36.1) of chromosome 1 was inserted at the breakpoint on the derivative X (See Supplemental Figure 1 available online). None of these, nor the surrounding sequences were within highly conserved regions, except the nearby DKC1 exons. The chromosome 1 breakpoint was within a simple (TA)n repeat bounded by two SINE alu repeats, and the chromosome X breakpoint region fell within a region of regulatory potential [King et al., 2005].
Figure 3.
By FISH analysis, BAC RP11–115M6 (containing Xq28 sequence) spanned the translocation breakpoint, as the probe hybridized to the patient’s normal X, der(X) and der(1) chromosomes.
X-Chromosome Inactivation Status
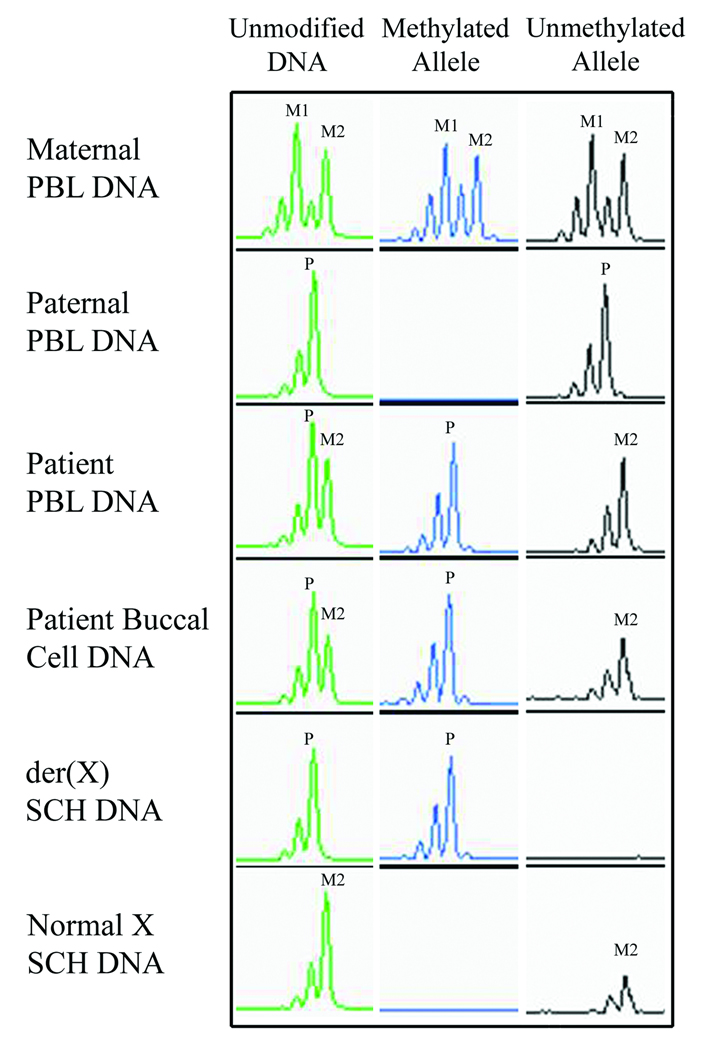
X-chromosome inactivation analysis was performed on patient DNA from buccal cells, PBL, EBV-LCL and SCH lines (Fig 4). The patient displayed a completely non-random XCI pattern with near 100% skewing found in all sources (within a 5% limit of detection). Analysis of SCH lines established that the patient’s normal X was active and maternally inherited, while the der(X) was inactive and of paternal origin. Analysis of maternal PBL DNA revealed a random pattern of XCI.
Figure 4.
XCI analysis at the AR locus established that the patient inherited maternal allele 2 (M2) and the paternal allele (P) as shown using unmodified DNA. Methylation specific PCR utilizing bisulfite modified DNA revealed complete skewing in the patient in both PBL and buccal cells. Analysis of SCH DNA determined that inactivation was biased toward the paternally derived der(X) chromosome (P), with the normal X remaining active (M2).
RT-PCR Results
The expression of critical genes near the breakpoint regions was determined by RT-PCR (Table I). Examination of the SCH containing the normal X indicated it was the active X-chromosome, as transcripts were detected for all X-chromosome genes studied. Analysis of the SCH containing the der(X) revealed a pattern representative of an inactive X-chromosome with a majority of genes not actively producing transcripts.
TABLE I.
A Combined Analysis of Expression Array and RT-PCR Data
| Gene Information | Microarray Data | RT-PCR Results | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Name | Cytogenetic Band |
Location (Mb) Ensembl 1-11-07 |
Undergoes XCI1 | Patient/ Paternal Gene Expression |
Patient/ Maternal Gene Expression |
Patient | SCH Der (1) |
SCH Der (X) |
SCH Normal X |
| X Chromosome | |||||||||
| SLC9A6 | Xq26.3 | 134.90 | Yes | + | 0 | 0 | |||
| FLNA | Xq28 | 153.23 | Yes | 0.89 | 0.86 | + | 0 | + | + |
| EMD | Xq28 | 153.26 | Yes | 0.99 | 0.99 | + | 0 | 0 | + |
| RPL10 | Xq28 | 153.28 | Yes | 1.02 | 1.01 | + | 0 | 0 | |
| DNASE1L1 | Xq28 | 153.28 | Yes | 1.08 | 1.09 | + | 0 | +/- | + |
| TAZ | Xq28 | 153.29 | Yes | 1.00 | 1.03 | + | 0 | 0 | |
| ATP6AP1 | Xq28 | 153.31 | Yes | + | 0 | 0 | + | ||
| GDI1 | Xq28 | 153.32 | Yes | 0.96 | 1.00 | + | 0 | 0 | |
| FAM50A | Xq28 | 153.33 | Yes | + | 0 | 0 | |||
| PLXNA3 | Xq28 | 153.34 | Yes | + | 0 | 0 | + | ||
| LAGE3 | Xq28 | 153.36 | Yes | + | 0 | 0 | |||
| UBL4 | Xq28 | 153.37 | Yes | 1.19 | 1.19 | + | 0 | 0 | |
| FAM3A | Xq28 | 153.39 | Yes | + | 0 | 0 | |||
| IKBKG | Xq28 | 153.42 | 9/9 Xi expressed | 1.02 | 1.02 | + | 0 | + | + |
| GAB3 | Xq28 | 153.56 | 8/9 Xi expressed | 0.98 | 0.89 | + | 0 | 0 | + |
| DKC1 (Breakpoint) | Xq28 | 153.64 | Yes | 0.99 | 0.97 | + | 0 | 0 | + |
| MPP1 | Xq28 | 153.66 | Yes | 1.67 | 1.67 | + | + | 0 | + |
| F8 | Xq28 | 153.72 | Yes | 1.45 | 1.46 | ||||
| FUNDC2 | Xq28 | 153.91 | Yes | 1.64 | 1.70 | ||||
| MTCP1 | Xq28 | 153.94 | Yes | 1.43 | 1.54 | ||||
| BRCC3 | Xq28 | 153.95 | 3/9 Xi expressed | 1.96 | 1.85 | ||||
| VBP1 | Xq28 | 154.10 | 1/9 Xi expressed | 1.33 | 1.38 | + | + | 0 | + |
| RAB39B | Xq28 | 154.14 | Yes | 1.60 | 1.64 | ||||
| F8A1 | Xq28 | 154.34 | Yes | 1.59 | 1.67 | ||||
| CLIC2 | Xq28 | 154.16 | 3/9 Xi expressed | 2.67 | 1.54 | + | + | 0 | + |
| TMHLE | Xq28 | 154.37 | Yes | 2.18 | 2.09 | ||||
| Chromosome 1 | |||||||||
| ARNT | 1q21.2 | 149.05 | + | + | 0 | 0 | |||
| (Genes distal to bkpt) | |||||||||
| RUSC1 | 1q22 | 153.56 | + | 0 | + | ||||
| LMNA | 1q22 | 154.32 | + | 0 | + | 0 | |||
| NUF2 (CDCA1) | 1q23.3 | 161.56 | + | 0 | + | 0 | |||
| SERPINC1 | 1q25.1 | 172.14 | + | 0 | + | 0 | |||
| IRF6 | 1q32.2 | 208.02 | + | 0 | + | ||||
XCI expresssion data from human-mouse somatic cell hybrids (SCH) [Carrel and Willard 2005]
The XCI ratio represents the number of times biallelic expression was observed out of 9 independent SCHs
Xi = inactive X chromosome (from human-mouse SCH lines)
Gene expression distal to each breakpoint (1q21 and Xq28) was studied in the SCH lines containing the derivative chromosomes to determine if partial monosomy or functional disomy was occurring in these regions. The spread of XCI was not detected in RT-PCR analysis of chromosome 1 genes, as genes distal to the 1q21 breakpoint were expressed in the SCH with the isolated der(X). In Xq28, RT-PCR results suggest that genes distal to the breakpoint were expressed from both the normal X-chromosome and from the translocated Xq28 material on the der(1), indicative of functional disomy for this region. The expression of DKC1 occurred solely from the normal active X-chromosome, apparently due to the disruption of the gene on the der(X) by the translocation breakpoint.
Expression Array Interpretation
An Affymetrix microarray chip was used to assess genome-wide expression in our patient compared to parental controls. Gene expression that was increased or decreased by two-fold as compared to parental samples was further analyzed in an attempt to explain the patient’s phenotype. Additionally, genes near the translocation breakpoint regions were examined for possible monosomic or disomic expression. Notably, FLNA and DKC1 gene expression were found to be normal in the patient. Decreased expression of genes distal to the 1q21 breakpoint was not observed. Evidence of functional disomy in Xq28 was apparent from the microarray results, as genes distal to the breakpoint region demonstrated increased levels of expression (Table I).
DISCUSSION
The X;1 translocation patient described in this study displayed an atypical pattern of XCI and abnormal phenotypic features. Possible causative mechanisms for her phenotype included a direct gene disruption due to the translocation breakpoint, a position effect mutation, or altered gene dosage due to the consequences of her translocation and XCI status.
A direct gene disruption was not the apparent cause of the phenotypic features described in our patient, but it likely served as a contributing factor. Based on experimental evidence, it was determined that DKC1 was interrupted in our patient at the Xq28 translocation breakpoint. The DKC1 gene encodes the 58kD dyskerin protein that participates in rRNA processing and functions in telomere maintenance [Marrone et al., 2005]. Mutations in DKC1 are known to cause dyskeratosis congenita (DKC) [OMIM 30500], a disorder characterized by nail dystrophy, hyperpigmentation of the skin, and leukoplakia associated with aplastic anemia. Female carriers of a DKC1 mutation are reported to have a completely skewed X-chromosome lineage, with the X-chromosome bearing the mutant allele inactivated [Ferraris et al., 1997; Vulliamy et al., 1997]. One of the hallmarks of DKC is a defect in cell proliferation due to bone marrow dysfunction. As a result, affected cells have a survival disadvantage over normal cells, especially in rapidly dividing tissues. Therefore, while the patient displayed no phenotypic features associated with DKC, it is probable that the cause of the patient’s skewed XCI pattern in blood was due to the interruption of the DKC1 gene by the translocation breakpoint.
A position effect mutation was also considered as a possible cause of the patient's clinical phenotype. Position effect mutations have been reported to cause a variety of genetic disorders. When originally considering a diagnosis of OPD I it was noted that the translocation breakpoint in DKC1 lay distal to the FLNA gene by approximately 400 kb, a distance well within the reported range of up to 1 Mb for position effect mutations to occur [de Kok et al., 1996; Kleinjan and van Heyningen, 2005]. However, the patient’s X-inactivation status was a confounding variable in the argument for a position effect mutation. In buccal cells and lymphocytes, the patient displayed a completely skewed XCI pattern with her normal X remaining active. This was unusual in a balanced X;autosome translocation, as the der(X) is typically found to remain active, and suggests that a position effect mutation due to the translocation breakpoint on the der(X) was not the root cause of her clinical features. There was not strong evidence of a position effect mutation involving chromosome 1, as all of the tested chromosome 1 genes were expressed from both the normal 1 and der(X) chromosome. Furthermore, the chromosome 1 breakpoint did not interrupt any genes or highly conserved regions. While skewing was observed in two types of tissue (PBL and buccal cells), it is important to note that this pattern may not be representative of all tissues in the patient.
We hypothesize that the most likely cause of the patient’s phenotypic abnormalities is altered gene dosage caused by an X;1 translocation in combination with an atypical pattern of XCI. Consequences of the patient’s XCI pattern, with the XIST locus translocated to the der(1), included the potential for monosomy of 1q21-qter and functional disomy of Xq28. The spread of XCI is known to occur in a discontinuous manner with the inactivation signal capable of crossing over 100 Mb of DNA [White et al., 1998]. However, evidence for this in our patient was not found.
Genes distal to the Xq28 breakpoint were tested for functional disomy in our patient. Findings were compared to published XCI data from human-mouse SCH lines in order to confirm inactivation status [Carrel and Willard, 2005]. Functional disomy of Xq28 was observed in a number of genes by RT-PCR and microarray analysis (Table I). Quantifiable gene expression, as determined by microarray analysis, revealed that a majority of genes distal to the breakpoint demonstrated increased expression in comparison to parental controls.
Functional disomy in the Xq28 region has been reported previously in a number of patients, and is thought to be associated with an abnormal phenotype. Sanlaville et al. [2005], recently compiled the phenotypic findings among patients with functional disomy of the Xq28 region. Common features among 19 patients include growth retardation, microcephaly, hypotonia, feeding difficulties, developmental delay, abnormal palate, hypoplastic genitalia, and increased susceptibility to infections. It is important to consider that these patients represent a heterogenous group which includes members of both sexes and that variable chromosomal mechanisms contributed to Xq28 disomy. Although no balanced X;autosome translocation patients were presented, six patients with an unbalanced form of an X;autosome translocation and Xq28 disomy were described. Clinical features such as growth retardation, developmental delay and abnormal palate are shared in common with our patient. Less frequently reported features also found in common with our patient include agenesis of the corpus callosum, abnormally shaped ears, tapering fingers, and clinodactyly [Akiyama et al., 2001; Sanlaville et al., 2005]. Overall, it appears that our patient displayed a less severe phenotype as compared to other patients described in the literature with Xq28 functional disomy. This is not unexpected as she carried a balanced form of an X;autosome translocation and has a relatively small region of Xq28 that is functionally disomic.
To summarize, our patient carried a balanced X;1 translocation and demonstrated an atypical pattern of XCI which resulted in functional disomy for part of band Xq28. An XCI pattern precisely opposite of that expected for an X;autosome translocation occurred and was likely due to the selective pressure caused by an interruption of the DKC1 gene. As a result, genetic imbalance occurred and was almost certainly the mechanism for the phenotypic abnormalities found in our patient. This suggests that there are discreet loci within the genome which serve such a critical genetic function that imbalance at a chromosomal level is tolerated in order to preserve expression of vital genes. As Schmidt and Du Sart [1992] have previously proposed, the pattern of XCI may not be expected to conform to the model of least genomic imbalance if a metabolically important X-linked gene is disrupted. Such may be the case if the interruption of DKC1 resulted in a post-inactivation growth disadvantage for cells bearing an active der(X) chromosome in our patient. As a consequence, in mature tissues, only those cells bearing an active normal X survived and the result was a non-random pattern of XCI. This situation illustrates the need for XCI studies and extensive genetic counseling in all patients referred for apparently balanced X;autosome translocations. The use of microarray analysis may become an important means of determining gene expression in translocation patients, and may serve as a critical instrument for predicting phenotype.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to the family for their participation in this research project. Support to AL was provided by NIH grants R01 DE014677 and KO2DE015291, and a March of Dimes grant #6-FY01–616.
REFERENCES
- Akiyama M, Kawame H, Ohashi H, Tohma T, Ohta H, Shishikura A, Miyata I, Usui N, Eto Y. Functional disomy for Xq26.3-qter in a boy with an unbalanced t(X;21)(q26.3;p11.2) translocation. Am J Med Genet. 2001;99:111–114. doi: 10.1002/1096-8628(2001)9999:9999<::aid-ajmg1150>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Cirullo RE, Dana S, Wasmuth JJ. Efficient procedure for transferring specific human genes into Chinese hamster cell mutants: interspecific transfer of the human genes encoding leucyl- and asparaginyl-tRNA synthetases. Mol Cell Biol. 1983;3:892–902. doi: 10.1128/mcb.3.5.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok YJ, Vossenaar ER, Cremers CW, Dahl N, Laporte J, Hu LJ, Lacombe D, Fischel-Ghodsian N, Friedman RA, Parnes LS, Thorpe P, Bitner-Glindzicz M, Pander HJ, Heilbronner H, Graveline J, den Dunnen JT, Brunner HG, Ropers HH, Cremers FP. Identification of a hot spot for microdeletions in patients with X-linked deafness type 3 (DFN3) 900 kb proximal to the DFN3 gene POU3F4. Hum Mol Genet. 1996;5:1229–1235. doi: 10.1093/hmg/5.9.1229. [DOI] [PubMed] [Google Scholar]
- Disteche CM, Eicher EM, Latt SA. Late replication in an X-autosome translocation in the mouse: correlation with genetic inactivation and evidence for selective effects during embryogenesis. Proc Natl Acad Sci U S A. 1979;76:5234–5238. doi: 10.1073/pnas.76.10.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris AM, Forni GL, Mangerini R, Gaetani GF. Nonrandom X-chromosome inactivation in hemopoietic cells from carriers of dyskeratosis congenita. Am J Hum Genet. 1997;61:458–461. doi: 10.1016/S0002-9297(07)64075-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Nonoyama S, Tonoki H, Masuno M, Imaizumi K, Kojima M, Wakui K, Shimadzu M, Fukushima Y. A new assay for the analysis of X-chromosome inactivation based on methylation-specific PCR. Hum Genet. 1999;104:49–55. doi: 10.1007/s004390050909. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Marrone A, Walne A, Dokal I. Dyskeratosis congenita: telomerase, telomeres and anticipation. Curr Opin Genet Dev. 2005;15:249–257. doi: 10.1016/j.gde.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Mattei MG, Mattei JF, Ayme S, Giraud F. X-autosome translocations: cytogenetic characteristics and their consequences. Hum Genet. 1982;61:295–309. doi: 10.1007/BF00276593. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R, Orstavik KH, Quarrell OW, Schwartz CE, Shears DJ, Suri M, Kendrick-Jones J, Wilkie AO. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- Sanlaville D, Prieur M, de Blois MC, Genevieve D, Lapierre JM, Ozilou C, Picq M, Gosset P, Morichon-Delvallez N, Munnich A, Cormier-Daire V, Baujat G, Romana S, Vekemans M, Turleau C. Functional disomy of the Xq28 chromosome region. Eur J Hum Genet. 2005;13:579–585. doi: 10.1038/sj.ejhg.5201384. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Du Sart D. Functional disomies of the X chromosome influence the cell selection and hence the X inactivation pattern in females with balanced X-autosome translocations: a review of 122 cases. Am J Med Genet. 1992;42:161–169. doi: 10.1002/ajmg.1320420205. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy TJ, Knight SW, Dokal I, Mason PJ. Skewed X-inactivation in carriers of X-linked dyskeratosis congenita. Blood. 1997;90:2213–2216. [PubMed] [Google Scholar]
- White WM, Willard HF, Van Dyke DL, Wolff DJ. The spreading of X inactivation into autosomal material of an x;autosome translocation: evidence for a difference between autosomal and X-chromosomal DNA. Am J Hum Genet. 1998;63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff DJ, Schwartz S, Carrel L. Molecular determination of X inactivation pattern correlates with phenotype in women with a structurally abnormal X chromosome. Genet Med. 2000;2:136–141. doi: 10.1097/00125817-200003000-00004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.