Abstract
Objectives
Non-syndromic orofacial clefts, i.e. cleft lip (CL) and cleft palate (CP), are among the most common birth defects. The goal of this study was to identify genomic regions and genes for CL with or without CP (CL/P).
Methods
We performed linkage analyses of a 10 cM genome scan in 820 multiplex CL/P families (6,565 individuals). Significant linkage results were followed by association analyses of 1,476 SNPs in candidate genes and regions, utilizing a weighted false discovery rate (wFDR) approach to control for multiple testing and incorporate the genome scan results.
Results
Significant (multipoint HLOD ≥3.2) or genome-wide-significant (HLOD ≥4.02) linkage results were found for regions 1q32, 2p13, 3q27-28, 9q21, 12p11, 14q21-24 and 16q24. SNPs in IRF6 (1q32) and in or near FOXE1 (9q21) reached formal genome-wide wFDR-adjusted significance. Further, results were phenotype dependent in that the IRF6 region results were most significant for families in which affected individuals have CL alone, and the FOXE1 region results were most significant in families in which some or all of the affected individuals have CL with CP.
Conclusions
These results highlight the importance of careful phenotypic delineation in large samples of families for genetic analyses of complex, heterogeneous traits such as CL/P.
Key Words: Cleft lip, Cleft palate, Linkage, Association, wFDR, IRF6, FOXE1, Genetics
Introduction
Orofacial clefts (OFC) are a major public health problem, affecting one in every 500–1000 births worldwide. It has become increasingly apparent that the genetic contribution to OFC is complex, probably heterogeneous, and likely due to interacting effects of multiple loci coupled to environmental covariates. Over 400 syndromes have been described in which a cleft of the lip and/or palate is a feature [1]. Among these syndromes are numerous genetic examples such as cytogenetic abnormalities (e.g., trisomies 13 and 18, 4p–), and single gene Mendelian disorders (e.g. van der Woude syndrome, Stickler syndrome). Non-syndromic (sometimes termed ‘isolated’) cleft lip with or without cleft palate (NS CL/P) is the most prevalent type of OFC, and is the focus of the current study.
Familial aggregation of NS CL/P has been reported in the scientific literature for 250 years [2,3,4,5,6,7]. Since then, segregation analyses of CL/P have supported models that include genes of major effect [8,9,10], and analyses of recurrence risk patterns have been consistent with estimates ranging from 3 to 14 genes (possibly interacting) for CL/P [11,12,13,14,15].
Mutation screens of more than 20 NS CL/P candidate genes find that 2–6% of the total number of individuals with NS CL/P have mutations in genes such as MSX1, FOXE1, GLI2, JAG2, LHX8, SATB2, RYK1 and others [16,17,18]. That is, the large majority of individuals with NS CL/P (94–98%) do not have mutations in any of a wide range of plausible candidate genes. In parallel, many candidate gene association studies have also been carried out seeking specific polymorphic variants that increase the risk of NS CL/P [1,19,20,21,22].
Most notably, the gene identified in van der Woude syndrome (IRF6 [23]) has been shown by our group [24] and confirmed in multiple other populations (Italy [25]; Belgium [26]; US [27]; Thailand [28]; US/Taiwan/Singapore/Korea [29]; South America [30]; Norway [31]) to show highly significant association with NS CL/P and may explain about 12–18% of NS CL/P [24]. Recently we have identified a specific SNP (rs642961) in IRF6 that disrupts the binding site for the transcription factor AP-2α, and that represents the etiologic locus within IRF6, at least in some populations [32].
In addition to analyses of candidate genes/loci, several genome-wide linkage screens of NS CL/P have been published [33,34,35,36,37,38]. About 8–10 regions have positive, although not highly significant, results in the individual studies. We performed a genome scan meta-analysis of these 5 published genome scans plus data from our additional studies in six countries and found several regions with genome-wide significant results [38], notably a novel region on 9q21. The purpose of this study is to describe a larger genome-wide scan for NS CL/P with twice as many families and individuals as were reported previously [38], to summarize the results of our follow-up fine mapping and candidate gene efforts, and to perform phenotypic-specific analyses. Thus the aim was to identify those genetic regions that appear to contribute to OFC overall, and also those regions that appear to have an effect on specific OFC phenotypes.
Subjects and Methods
Study Populations
This study was conducted in two phases: (1) linkage genome-scan, and (2) fine mapping and candidate gene studies. For the genome-scan, there were 820 families ascertained in six countries (Philippines, Colombia, China, India, Turkey, USA), with 6,565 total individuals (summarized in table 1). Of the total family members, 4,373 were genotyped (1,514 affected and 2,859 unaffected). For fine-mapping and candidate gene studies, there were 861 families, with 7,047 total individuals (summarized in table 2), representing a subset of the 820 genome-scan families plus additional families that were ascertained after the genome-scan was completed. Most of the families were extended multiplex kindreds, i.e. multigenerational families with ≥2 affected individuals. The largest family had 51 individuals; there were 5 pedigrees with 40–50 individuals, 10 with 30–40; 46 with 20–30, many with 10–20 individuals. Each study had approval by the appropriate institutional review boards, and all study subjects provided informed consent to participate. Note that the original 388 genome-scan families [38] were also included in the current study (denoted ‘CIDR-7 families’ in the previous report [38]).
Table 1.
Summary of families and individuals ascertained for genome-scan linkage analyses
| Country | Families | Individuals |
Subsetsa |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| affected |
unaffected |
total |
CL |
CLP |
CL+CLP |
|||||||||||
| genotyped |
genotyped |
genotyped |
n fams | n | n fams | n | n fams | n | ||||||||
| yes | no | total | yes | no | total | yes | no | total | ||||||||
| China | 72 | 98 | 17 | 105 | 246 | 134 | 380 | 344 | 151 | 495 | 19 | 105 | 31 | 189 | 18 | 173 |
| Colombia | 163 | 266 | 15 | 281 | 386 | 369 | 755 | 652 | 384 | 1,036 | 18 | 92 | 112 | 588 | 31 | 342 |
| India | 11 | 18 | 4 | 22 | 63 | 77 | 140 | 81 | 81 | 162 | 5 | 71 | 0 | 0 | 6 | 91 |
| Philippines | 462 | 928 | 44 | 972 | 1,848 | 1,292 | 3,140 | 2,776 | 1,336 | 4,112 | 79 | 362 | 230 | 1,504 | 132 | 1,900 |
| Turkey | 6 | 8 | 0 | 8 | 9 | 32 | 41 | 17 | 32 | 49 | 0 | 0 | 0 | 0 | 2 | 12 |
| USA | 106 | 196 | 24 | 220 | 307 | 184 | 491 | 503 | 208 | 711 | 15 | 70 | 47 | 245 | 33 | 298 |
| Total | 820 | 1,514 | 104 | 1,618 | 2,859 | 2,088 | 4,947 | 4,373 | 2,192 | 6,565 | 136 | 700 | 420 | 2,526 | 222 | 2,816 |
Subsets: CL = those families in which all affected members had CL alone; CLP = those families in which all affected members had CL and CP; CL+CLP = those families in which at least one affected member had CL alone and at least one had CL+CP; n fams = number of families.
Table 2.
Summary of families and individuals ascertained for fine-mapping studies
| Country | Families | Individuals |
Subsetsa |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| affected |
unaffected |
total |
CL | CLP | CL+CLP | |||||||||||
| genotyped |
genotyped |
genotyped |
n fams | n | n fams | n | n fams | n | ||||||||
| yes | no | total | yes | no | total | yes | no | total | ||||||||
| China | 104 | 168 | 33 | 201 | 258 | 461 | 719 | 426 | 494 | 920 | 24 | 174 | 42 | 337 | 33 | 372 |
| Colombia | 232 | 349 | 12 | 361 | 577 | 362 | 939 | 926 | 374 | 1,300 | 23 | 108 | 174 | 826 | 35 | 366 |
| India | 53 | 94 | 42 | 136 | 210 | 380 | 590 | 304 | 422 | 726 | 22 | 253 | 5 | 86 | 24 | 356 |
| Philippines | 221 | 600 | 47 | 647 | 1,090 | 1,108 | 2,198 | 1,690 | 1,155 | 2,845 | 20 | 178 | 82 | 932 | 112 | 1,589 |
| Spain | 36 | 41 | 2 | 43 | 86 | 7 | 93 | 127 | 9 | 136 | 5 | 22 | 25 | 93 | 4 | 14 |
| Turkey | 28 | 32 | 5 | 37 | 53 | 189 | 242 | 85 | 194 | 279 | 6 | 59 | 15 | 158 | 7 | 62 |
| USA | 187 | 231 | 24 | 255 | 413 | 173 | 586 | 644 | 197 | 841 | 42 | 142 | 99 | 335 | 36 | 307 |
| Total | 861 | 1,515 | 165 | 1,680 | 2,687 | 2,680 | 5,367 | 4,202 | 2,845 | 7,047 | 142 | 936 | 442 | 2,767 | 251 | 3,066 |
Subsets: CL = those families in which all affected members had CL alone; CLP = those families in which all affected members had CL and CP; CL+CLP = those families in which at least one affected member had CL alone and at least one had CL+CP; n fams = number of families.
The phenotype was NS CL/P, i.e. for families to be included, it was necessary that the proband have NS CL/P (i.e. no other anomalies), and that no other family member have an indication of an OFC syndrome (e.g. lip pits). Each study included evaluations of family members by clinical geneticists to rule out syndromic forms of CL/P.
For some analyses, the total families were divided into the following non-overlapping subsets based on family members reported to be affected (whether or not all affected family members participated in these studies): CL, those families in which all affected family members had CL only; CLP (all affected family members had CL plus CP); CL+CLP (at least one affected family member had CL only and at least one had CL plus CP). The numbers of families and individuals in each of these subsets are summarized in table 1 (genome-scan dataset) and table 2 (fine-mapping dataset). There was one additional subset that had very few families and was therefore not included in the subset studies reported here, i.e. families in which at least one affected family member had CP only.
Genome Scan Genotyping
The genome scans were conducted in multiple batches (the previous report [38] summarized results from the first two batches of genotyping). Microsatellite genotyping for all batches was performed at the Center for Inherited Disease Research (CIDR) using STRP markers with average spacing of about 9 cM (1–19 cM). Marker set details and methods are available at www.cidr.jhmi.edu.
Trace data was reviewed and genotypes were called using the software package Genotyper from Applied Biosystems. The raw genotype sizes for each genotype were exported from Genotyper and binned using SAS. Study samples were plated with 2 CEPH Utah control samples 1331-1 and 1331-2 repeated on each plate. For each microsatellite marker the CEPH control sample sizes for each plate were compared to the project average and used to adjust the raw sizes for all data for that plate. Data with similar raw sizes were grouped into proposed ‘binned’ alleles. Plots were made for each marker showing the overall as well as by plate allele sizes and proposed bins for both the raw and adjusted size data. Each plot was manually reviewed by at least two data analysts. Trace data for unexpected sizes for CEPH controls and outliers that didn't fall within bins were reviewed in Genotyper. Trace data was also reviewed for markers with high numbers of mendelian errors according to Pedcheck [39]. Binning parameters and plate adjustments were done manually if needed.
After the entire dataset was genotyped, all alleles were re-binned across batches before performing the analyses reported here. The final combined dataset consisted of 375 STRP markers which were genotyped in all three sample submissions. Quality control was monitored by investigator-provided blind duplicate samples. 220 blind duplicate pairs across all three sample submissions resulted in a reproducibility rate of 99.95%.
Candidate Gene SNP Genotyping
After the results of the linkage genome scan, the families were genotyped by CIDR for a custom panel of 1,536 single nucleotide polymorphisms (SNPs), of which 1,476 were analyzed (list available on request, see below for reasons some SNPs were not analyzed).
SNPs were chosen to saturate the 1-LOD regions for each genome-wide significant region found in the linkage scan, targeting all known genes in each region, with 2–6 SNPs per gene and filling intergenic gaps >100 kb with 1 SNP per 20 kb. In addition, SNPs in cleft candidate genes were included in the custom panel, and were chosen from a variety of resources, including published linkage and association studies on clefts, genome-wide scans, gene knockout experiments in mice, studies of chromosomal rearrangements in humans, and gene-expression analyses in human and mouse embryonic tissues [20,40,41,42,43], including the Craniofacial and Oral Gene Expression Network [44] (COGENE). We also searched the Serial Analysis of Gene Expression (SAGE) libraries to see whether a particular gene of interest is expressed in the relevant embryonic tissues at the pertinent developmental stage (weeks 5–6 for fusion of the embryonic lip and weeks 7–10 for fusion of the palatal shelves [45]). Data from two gene expression approaches were also considered, the ENU project [46], and the Developmental Genome Anatomy Project (DGAP [47]). Finally genes involved in Mendelian forms of clefting, particularly those that may manifest as phenocopies of NS CL/P (identified through OMIM) were considered.
To guide the selection of SNPs within the candidate genes and regions, the genome browser of the International HapMap Consortium was used. SNPs were prioritized according to prior evidence of an association with clefting, a preference for coding SNPs and those located in putative regulatory regions, haplotype-tagging properties (htSNPs), and minor allele frequency (MAF) of at least 10%, ideally across the multiple ethnicities represented in this study.
A combination of software, including HAPLOVIEW version 2.05, BEST (Best Enumeration of SNP Tags [48]), and SNP Browser) (Applied Biosystems; Foster City, Calif., USA), was used to evaluate MAF, deviations from Hardy-Weinberg equilibrium (HWE), inter-marker distance, as well as LD patterns and haplotype block structures (for the selection of htSNPs). SNP assays were designed by Illumina (San Diego, Calif., USA) and a ‘design score’ was computed for each SNP using an algorithm that rigorously tests the performance of that SNP on an Illumina GoldenGate platform.
SNP genotyping was performed at CIDR on a BeadLab system using the Illumina GoldenGate technology [49] (Illumina; San Diego, Calif., USA). Genotypic data for 1,489 of 1,536 attempted SNPs were released by CIDR. The overall rate of missing genotypic data was 0.35%. As part of quality control, CIDR genotyped a parent-child trio plus one duplicate child, which yielded a duplicate error rate of 0.009% and an overall parent-child discordance rate of zero. Of the 1,489 SNPs released after CIDR quality control checks, 13 more were not included in our analysis due to poor performance in one or more of the individual study populations (e.g. low MAF or high rates of mendelian inconsistencies). Therefore, results are reported here for the remaining 1,476 SNPs.
Statistical Genetic Analyses
Preliminary Analyses
The inheritance of each marker (genome-wide STRPs and custom SNPs) was assessed with PedCheck [39] to test for inconsistencies due to non-paternity or other errors. Marker allele frequencies are required by linkage analysis approaches and were estimated in the founders of the families, separately by country due to the diverse ethnicities. Other genetic model parameters are summarized in table 3, and were taken from the results of segregation analysis (Philippines – unpublished results; China [10]; India [50]; Colombia [51]; Caucasians [9, 52, 53]).
Table 3.
Summary of genetic model parameters used for parametric linkage analysis in each country, plus weights for meta-analysis
| Recessive |
Dominant |
Weights for meta-analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| FreqA | penAA | penAB | penBB | FreqA | penAA | penAB | penBB | ||
| China | 0.05 | 0.002 | 0.002 | 0.9 | 0.001 | 0.002 | 0.6 | 0.6 | 0.93268 |
| Colombia | 0.05 | 0.002 | 0.002 | 0.9 | 0.001 | 0.002 | 0.6 | 0.6 | 1.28306 |
| India | 0.05 | 0.002 | 0.002 | 0.9 | 0.001 | 0.002 | 0.6 | 0.6 | 0.45258 |
| Philippines | 0.04 | 0.002 | 0.002 | 0.9 | 0.002 | 0.002 | 0.6 | 0.6 | 2.61734 |
| Turkey | 0.05 | 0.002 | 0.002 | 0.9 | 0.001 | 0.002 | 0.6 | 0.6 | 0.12318 |
| USA; Pittsburgh | 0.05 | 0.002 | 0.002 | 0.9 | 0.001 | 0.002 | 0.6 | 0.6 | 0.71294 |
| USA; other than Pittsburgh | 0.05 | 0.002 | 0.002 | 0.9 | 0.001 | 0.002 | 0.6 | 0.6 | 0.87822 |
Parametric Linkage Analyses
Standard multipoint parametric linkage statistics were calculated at each of the 375 genome-scan STRPs, in particular the heterogeneity LOD score or HLOD. HLODs are based on the admixture heterogeneity test [54] where the recombination fraction (θ) and the proportion of linked families (α) are simultaneously estimated. Simulation studies have shown that although the estimate of the proportion of linked families may be not be precise, HLODs are a powerful method for detecting linkage in the presence of heterogeneity [55,56,57]. The descent graph method [58] implemented in computer program SIMWALK2 was used for the multipoint HLOD calculations.
HLOD calculations were done under the best-fitting dominant and recessive models for each study population (estimated from segregation analysis, see preliminary analyses section above and table 3). Maximizing LOD scores over a range of genetic models is valid for simultaneously evaluating linkage and determining the most likely genetic model (without adjustment to significance levels and without need to correct for ascertainment) provided that there is indeed linkage [59]. Furthermore, if an oligogenic model is suspected (as seems likely for CL/P), or if significant heterogeneity exists, some of the causal genes may act in a dominant fashion and others recessive.
To combine the multipoint results across the study populations we summed the multipoint HLODs. Because each study population had different allele frequency estimates and different genetic model parameters it was not optimal to perform a combined linkage analysis pooling all families across all populations. Simulation studies [55] have shown that HLOD analyses that pooled multiple datasets resulted in a loss of power for detecting linkage compared to summing the HLODs from individual studies.
For determining genome-wide significance for the multipoint linkage calculations of the families, standard guidelines [60] were followed. A Bonferroni correction was applied to account for the multiple markers, data subsets, and genetic models tested. To do so, the desired α level of 0.05 was divided by [375 (number of markers) × 4 (number of data subsets) × 2 (number of genetic models tested)] to yield a Bonferroni-adjusted alpha of 0.000017. The χ2 corresponding to the p value (i.e. 18.5) was divided by 2(loge10) = 4.605 to yield 4.02 as the LOD score threshold for genome-wide significance for this study. Values between 3.2 and 4.02 were considered as significant but not genome-wide significant.
Genome Scan Meta-Analysis
As an additional method used primarily to estimate the weighting factors for the weighted FDR approach summarized below, we used the genome scan meta-analysis method (GSMA [38, 61, 62]). The GSMA is a nonparametric rank ordering method that can combine genome scan methods across studies with different markers and different statistical tests. In simulation studies the GSMA detected linkage with power comparable to or greater than that obtained by performing a combined linkage analysis of all data [62, 63].
For the GSMA procedure, the genome was divided into bins with bin-width selected such that there were at least two bins on the smallest chromosome and at least one marker was genotyped within each bin. Therefore, for combining the current 10 cM genome scans a bin width of 30 cM was selected (i.e. 130 bins across the genome). For each of the individual populations, bins were assigned a rank (R, with values from 1 to 130) according to the maximum linkage statistic of markers in each bin (multipoint HLOD scores). Any tied bins were assigned equal R's based on the mean of the sequential ranks for those bins.
Because the study populations covered a wide range of sample sizes (see table 1), we weighted the R statistics based on sample size. Various weighting strategies have been proposed for the GSMA [61, 62], and simulation studies showed that weighting increased the power of the GSMA to detect linked loci [62]. We used a weighting factor based on the total number of genotyped individuals (N-genotyped) – the ranks within each population were multiplied by the square root of N-genotyped divided by the mean of N-genotyped over all studies. The weighting factors calculated for each population are listed in table 3.
The GSMA identified 30 cM bins that are best supported across the studies. In order to narrow the regions of positive findings, we used an extension of the GSMA that involves repeating the GSMA with different bin starting points and then determining the Minimum Region of Maximum Significance (MRMS [64]). In brief, we repeated the GSMA twice more shifting the starting point for the binning procedure to first 10 cM and then to 20 cM. This determined the 10 cM MRMS for each positive finding. Given that the genome scan STRP panel averaged 10 cM between markers, 10 cM was the limit of resolution for the meta-analyses.
Association Analysis Methods (Fine-Mapping and Candidate Genes)
The transmission disequilibrium test (TDT) was used to assess association in the presence of linkage disequilibrium between the 1,476 fine-mapping markers and CL/P. The Family Based Association Test extension of the TDT (FBAT [65,66,67]) was used to assess association between each SNP and CL/P. To control for multiple testing and to include information from the prior linkage genome scan we performed a weighted approach to false discovery rate (wFDR [68]). In this approach, per marker FBAT p values were adjusted using weights derived from the GSMA/MRMS results, and allowing for a maximum 10% FDR.
Results
Multipoint Linkage Results (HLOD and GSMA)
Figure 1 shows a summary of the multipoint HLOD results for the total dataset, and for the CL, CLP and CL+CLP subsets. Depicted are the largest multipoint HLODs on each chromosome, under dominant and recessive models. The HLOD genome scan revealed genome-wide significant linkage results (i.e. multipoint HLOD ≥4.02) in the regions 3q27–28 (under a dominant model for CL/P), 9q21 (dominant model), and 14q21–24 (recessive model). Three additional regions reached nominal significance (i.e. multipoint HLOD ≥3.2): 1q32 (under a dominant model for CL/P), 2p13 (dominant model), and 16q24 (recessive model). Of those regions, 1q32, 9q21, 12p11, 14q,21–24 and 16q24 were also statistically significant in the GSMA analysis. Figure 2A 2B 2C 2D 2E 2F–G shows the detailed HLOD and GSMA results for each of the chromosomal regions with significant results.
Fig. 1.
Summary of the 10 cM genome scan of CL/P. Each graph depicts the maximum summed multipoint HLOD on each chromosome, under both dominant (black) and recessive (grey) genetic models assumed. A Summary for TOTAL, the entire dataset. B Summary for the CL subset, i.e. those families in which all affecteds have CL alone. C Summary for the CL+CLP subset, i.e. those families in which at least one affected has CL alone and at least one has CL+CP. D Summary for the CLP subset, i.e. those families in which all affected members have CL+CP.
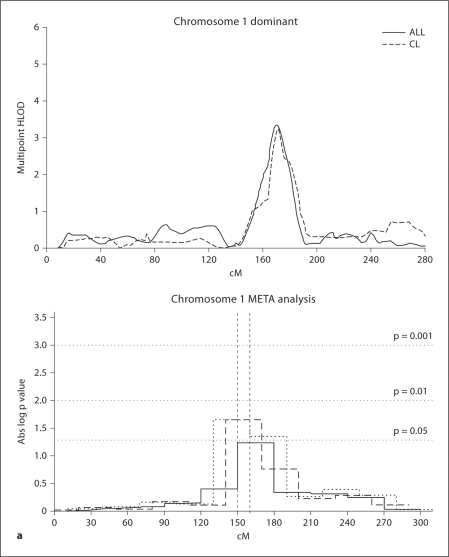
Fig. 2a.
Summed multipoint HLOD plots for each chromosome that had a maximum summed HLOD ≥3.2 (under the best genetic model for each chromosome), in the entire dataset (TOTAL), and the subsets - see definitions in figure 1. For each of those chromosomes, also shown are the graphs of the Minimum Regions of Maximal Significance (MRMS). These graphs summarize the process of repeating the Genome Scan Meta Analyses (GSMA), shifting the bins in order to narrow the region of potential involvement with CL/P. For those chromosomes with statistically significant GSMA/MRMS results (i.e. p values ≤0.05), dashed vertical lines indicate the 10 cM MRMS. a Chromosome 1 under a dominant model for TOTAL and for the CL subset.
Fig. 2b.
Summed multipoint HLOD plots for each chromosome that had a maximum summed HLOD ≥3.2 (under the best genetic model for each chromosome), in the entire dataset (TOTAL), and the subsets - see definitions in figure 1. For each of those chromosomes, also shown are the graphs of the Minimum Regions of Maximal Significance (MRMS). These graphs summarize the process of repeating the Genome Scan Meta Analyses (GSMA), shifting the bins in order to narrow the region of potential involvement with CL/P. For those chromosomes with statistically significant GSMA/MRMS results (i.e. p values ≤0.05), dashed vertical lines indicate the 10 cM MRMS. b Chromosome 2 (dominant).
Fig. 2c.
Summed multipoint HLOD plots for each chromosome that had a maximum summed HLOD ≥3.2 (under the best genetic model for each chromosome), in the entire dataset (TOTAL), and the subsets - see definitions in figure 1. For each of those chromosomes, also shown are the graphs of the Minimum Regions of Maximal Significance (MRMS). These graphs summarize the process of repeating the Genome Scan Meta Analyses (GSMA), shifting the bins in order to narrow the region of potential involvement with CL/P. For those chromosomes with statistically significant GSMA/MRMS results (i.e. p values ≤0.05), dashed vertical lines indicate the 10 cM MRMS. c Chromosome 3 (dominant).
Fig. 2d.
Summed multipoint HLOD plots for each chromosome that had a maximum summed HLOD ≥3.2 (under the best genetic model for each chromosome), in the entire dataset (TOTAL), and the subsets - see definitions in figure 1. For each of those chromosomes, also shown are the graphs of the Minimum Regions of Maximal Significance (MRMS). These graphs summarize the process of repeating the Genome Scan Meta Analyses (GSMA), shifting the bins in order to narrow the region of potential involvement with CL/P. For those chromosomes with statistically significant GSMA/MRMS results (i.e. p values ≤0.05), dashed vertical lines indicate the 10 cM MRMS. d Chromosome 9 (dominant, TOTAL and CL+ CLP).
Fig. 2e.
Summed multipoint HLOD plots for each chromosome that had a maximum summed HLOD ≥3.2 (under the best genetic model for each chromosome), in the entire dataset (TOTAL), and the subsets - see definitions in figure 1. For each of those chromosomes, also shown are the graphs of the Minimum Regions of Maximal Significance (MRMS). These graphs summarize the process of repeating the Genome Scan Meta Analyses (GSMA), shifting the bins in order to narrow the region of potential involvement with CL/P. For those chromosomes with statistically significant GSMA/MRMS results (i.e. p values ≤0.05), dashed vertical lines indicate the 10 cM MRMS. e Chromosome 12 (dominant, TOTAL and CLP subgroup).
Fig. 2f.
Summed multipoint HLOD plots for each chromosome that had a maximum summed HLOD ≥3.2 (under the best genetic model for each chromosome), in the entire dataset (TOTAL), and the subsets - see definitions in figure 1. For each of those chromosomes, also shown are the graphs of the Minimum Regions of Maximal Significance (MRMS). These graphs summarize the process of repeating the Genome Scan Meta Analyses (GSMA), shifting the bins in order to narrow the region of potential involvement with CL/P. For those chromosomes with statistically significant GSMA/MRMS results (i.e. p values ≤0.05), dashed vertical lines indicate the 10 cM MRMS. f Chromosome 14 (recessive).
Fig. 2g.
Summed multipoint HLOD plots for each chromosome that had a maximum summed HLOD ≥3.2 (under the best genetic model for each chromosome), in the entire dataset (TOTAL), and the subsets - see definitions in figure 1. For each of those chromosomes, also shown are the graphs of the Minimum Regions of Maximal Significance (MRMS). These graphs summarize the process of repeating the Genome Scan Meta Analyses (GSMA), shifting the bins in order to narrow the region of potential involvement with CL/P. For those chromosomes with statistically significant GSMA/MRMS results (i.e. p values ≤0.05), dashed vertical lines indicate the 10 cM MRMS. g Chromosome 16 (recessive in TOTAL and CL+ CLP).
As can be seen in figure 1, the phenotypic subsets have differing results. In the CL subset, the region on chromosome 1q32 was significant under a dominant model (see fig. 2A). In the CL+CLP subset, regions on chromosome 9q21 (dominant) and 16q24 (recessive) were significant (see fig. 2D and G for details). In the CLP subset a region on chromosome 12p11 was significant under a dominant model (see fig. 2E). Table 4 summarizes the chromosomal regions with significant linkage results in the TOTAL dataset and the phenotypic subsets, plus potential CL/P candidate genes in those significant regions.
Table 4.
Significant linkage and meta-analysis results
| Chromosomal region | Phenotypic subset | Maximum summed multipoint HLODa | Linkage peak: cM location | Model | Average α | GSMA/MRMS peak (p value)b | Candidate genes in region |
|---|---|---|---|---|---|---|---|
| 1q32 | TOTAL | 3.35 | 170.3 | dom | 0.29 | 150–160 (0.02) | IRF6 |
| CL | 3.24 | 171.0 | dom | 0.47 | – | ||
| 2p13 | TOTAL | 3.25 | 69.0 | dom | 0.31 | – | TGFA |
| 3q27–28 | TOTAL | 4.13 | 204.2 | dom | 0.34 | – | TP63 |
| 9q21 | TOTAL | 5.37 | 118.2 | dom | 0.41 | 100–110 (0.0007) | FOXE1, PTCH, ROR2, TGFBR1, ZNF189 |
| CL+CLP | 5.44 | 116.4 | dom | 0.46 | – | ||
| 12p11 | CLP | 3.22 | 83.0 | dom | 0.25 | 40–50 (0.02) | |
| 14q21–24 | TOTAL | 4.18 | 75.1 | rec | 0.31 | 60–70 (0.01) | PAX9, TGFB3, BMP4 |
| 16q24 | TOTAL | 3.56 | 84.5 | rec | 0.16 | 100–110 (0.01) | CRSPLD2, FOXC2 |
| CL+CLP | 3.42 | 81.0 | rec | 0.24 | – |
Maximum summed HLOD = maximum multipoint HLOD summed over the individual studies (see table 1 for list; see fig. 1 and 2 for full HLOD plots), presented are significant regions,. i.e. with HLOD =3.2). Genome-wide significant results (i.e. HLOD =4.02) are in bold. Peak = cM location of the maximum summed multipoint HLOD; model = genetic model for the maximum summed HLOD; dom = dominant; rec = recessive); Average _ = estimated proportion of linked families at the linkage peak, averaged over the studies.
GSMA/MRSA peak (p value) = significant results from Genome Scan Meta-Analysis over the individual studies; presented is the 10 cM interval best supported by the GSMA/MRMS with the corresponding p value (see fig. 2 for MRMS graphs).
Fine-Mapping and Candidate Gene Analyses
Figure 3 shows the weighted p values and weighted FDR alpha levels for the total dataset, and the CLP and CL+CLP datasets. One SNP in IRF6 and 3 SNPs in or near FOXE1 were the only ones reaching formal weighted-FDR-adjusted significance (p < 10–7, and p < 10–6, respectively) in the total dataset. Although not reaching formal genome-wide significance, additional SNPs on 1q, 6q and 9q were near significant (p < 0.001, results not shown in detail).
Fig. 3.
Summaries of the weighted False Discovery Rate (wFDR) results for 1,476 SNPs selected within candidate genes or to fine-map the linkage peaks. Shown are graphs for the TOTAL dataset, and the CLP and CL+CLP subsets, i.e. those subsets in which there were genome-significant wFDR results. Shown are the −log10 (wFDR p values), with the SNPs arrayed across the genome from chromosome 1 (leftsideof X axis) to chromosome 22.
Of the phenotypic subsets, only CLP had SNPs reaching genome-wide significance: i.e., 5 SNPs in or near FOXE1 on 9q. Although not reaching genome-wide significance, the most significant SNP in both the CL and CL+CLP phenotypic subsets was in IRF6 (p < 0.001 and p < 0.002, respectively), and was the same SNP significant in the TOTAL dataset. Table 5 summarizes the genome-wide significant SNPs in the total dataset and in the CLP subset.
Table 5.
Genome-wide significant SNP results (from weighted FDR analyses of FBAT results) in the TOTAL dataset and CLP pheno-typic subset
| Dataset | Chromosome | SNP name | SNP | Associated allele (freq) | Gene | Chromosome location | Weighted p value |
|---|---|---|---|---|---|---|---|
| TOTAL | 1q32.3-q41 | rs2013162 | A/C | C (62%) | IRF6 | 208035307 | 1.0!10−7 |
| 9q22 | rs1443434 | G/T | T (75%) | FOXE1 | 99657300 | 7.0!10−7 | |
| 9q22 | rs993501 | C/T | T (81%) | 99663198 | 2.1!10−6 | ||
| 9q22.33 | rs6586 | C/T | T (63%) | C9orf156 | 99706752 | 5.8!10−5 | |
| CLP | 9q22 | rs1443434 | G/T | T (72%) | FOXE1 | 99657300 | 4.0!10−7 |
| 9q22.33 | rs6586 | C/T | T (59%) | C9orf156 | 99706752 | 7.5!10−6 | |
| 9q22 | rs993501 | C/T | T (77%) | 99663198 | 1.4!10−5 | ||
| 9q22 | rs1877431 | A/G | G (66%) | 99573968 | 3.3!10−4 | ||
| 9q22.3 | rs4743106 | C/T | T (78%) | TMOD | 99353614 | 3.3!10−4 | |
Discussion
The genome scan revealed multiple significant linkage results (i.e. multipoint HLOD ≥3.2) in the regions 1q32, 2p13, 3q27–28, 9q21, 14q21–24 and 16q24 for the TOTAL dataset, with the 3q, 9q and 14q regions also genome-wide significant (HLOD ≥4.02). The 1q32 region result was also significant in the CL subset but not the others, implying that the significant linkage was due to the CL families. Similarly, the 9q21 and 16q24 results were also genome-wide significant in the CL+CLP subset. In the CLP subset, an additional region of significance was found for the 12p11 region. The remaining two regions (2p11, 3q27–28) were not significant in any individual subset, implying that these regions may be involved in OFC overall, rather than any specific phenotype. Also, note that in each case where there were significant findings in one of the phenotypic subgroups, the estimated proportion of linked families (α) was larger in the subgroup than in the total dataset (see table 4), further supporting the notion that phenotypic sub-grouping may be a useful approach to reduce heterogeneity across cleft families.
Follow-up fine-mapping association studies found SNPs in IRF6 (chromosome 1q) and in or near FOXE1 (chromosome 9q) that reached formal FDR-adjusted significance (see table 5), and SNPs in 6q were near significant. Consistent with the linkage results, the fine-mapping results were also phenotype dependent. The IRF6 SNP rs2013162 (significant in the TOTAL dataset) was only positive in the CL and CL+CLP subsets (although not reaching formal genome-wide significance in those subsets). Similarly, FOXE1 SNPs (significant in the TOTAL dataset) were genome-wide significant only in the CLP subset.
Following is a brief discussion of the most notable results by chromosome. Although not summarized in detail, several of these regions also have chromosomal rearrangements reported in CL/P [47, 69, 70].
Chromosome 1q
The 1q32 region with significant linkage and association results is the location for the IRF6 gene that was identified as the etiologic locus for van der Woude syndrome (VDWS, MIM# 119300 [23]) and also significantly associated with non-syndromic CL/P (SNP rs2235371 [22,24,25,26,28,29,30, 71]).
Recently we have identified a specific IRF6 SNP (rs642961) where the ancestral allele is highly evolutionarily conserved and where functional assays show disruption of an AP-2α binding site that likely represents the major NS CL/P etiologic locus within IRF6 [32]. Notably the results with SNP rs642961 were significant only in families with one or more CL-alone affected members [32], consistent with the linkage and association results reported here. In the current study, IRF6 SNP rs2013162 reached genome-wide significant association with CL/P in the TOTAL dataset; further the association results were positive in only the CL and CL+CLP subsets (although not reaching formal genome-wide significance in those subsets). Based on Haploview version 4.1 [72], the current study SNP (rs2013162) lies between the other two SNPs, but is not in LD with them.
The current results demonstrating significant linkage to the IRF6 region provide powerful support for the IRF6 association findings. Interestingly, until the current study we only found weak linkage signals to the 1q32 region [38] and/or the IRF6 locus itself [24] (LODs <1.0); the current study thus demonstrates the utility of large sample sizes with careful sub-phenotyping in detecting subtle effects that otherwise are only detectable with association methods.
Chromosome 2p and 2q
Note that in our previous linkage report [38], a 2q32–35 region had highly significant GSMA results. In the current study with double the sample size there are no longer genome-wide significant results with this region. The 2q32–35 region contains the gene for DNA-binding protein SATB2 (a.k.a. KIAA1034) that has been identified through translocation breakpoint analysis as a gene involved in cleft palate [73], and that also shows site- and stage-specific expression in murine palate development. It may be that some population or phenotypic subsets led to the previous positive results and additional analyses are on-going to continue to investigate SATB2.
The 2p13 region with significant linkage results contains TGFA, the gene with the first reported association with CL/P [74] and numerous confirmatory reports [19, 75]. There were no genome-wide significant association results with TGFA fine-mapping SNPs (rs3732253, rs1807968, rs374640, rs377122), but the positive linkage results warrant continued study of this region.
Chromosome 3q
The 3q27–28 region had genome-wide significant linkage results, consistent with our previously reported result near this region in Chinese families [76], but there have been no other published reports of either positive linkage or positive association of non-syndromic CL/P to this region. A potential candidate gene in this region is TP63.
TP63 encodes a member of the p53 family of transcription factors. An animal model, p63–/– mice, has been useful in defining the role this protein plays in the development and maintenance of stratified epithelial tissues. p63–/– mice have several developmental defects which include the lack of limbs and other tissues, such as teeth and mammary glands, which develop as a result of interactions between mesenchyme and epithelium. Furthermore, recently completed detailed expression studies [77] during murine craniofacial development to dissect the molecular pathogenesis of the bilateral cleft lip and cleft palate seen in Tp63-deficient mice. Analysis of key signaling molecules revealed that the craniofacial defects resulted from increased Bmp4 signaling acting antagonistically on Fgf8 and Shh [77].
In humans, mutations in the TP63 gene are associated with ectodermal dysplasia and cleft lip/palate syndrome 3 (EEC3 [78, 79]); split-hand/foot malformation 4 (SHFM4); Hay Wells syndrome [80] (ankyloblepharon-ectodermal defects-cleft lip/palate); ADULT syndrome (acro-dermato-ungual-lacrimal-tooth); limb-mammary syndrome; Rap-Hodgkin syndrome (RHS [81, 82]) and NS OFC [83]. Both alternative splicing and the use of alternative promoters results in multiple transcript variants encoding different proteins, which may underlie the wide phenotypic spectrum associated with mutations in TP63 [84, 85].
Therefore, the custom SNP panel included 10 SNPs within TP63: rs4396880, rs1920266, rs4575879, rs7616178, rs4607088, rs7619526, rs4686529, rs7619549, rs9810322, and rs1515490. None of these SNPs reached genome-wide significance in the wFDR analyses, but given the strong linkage signal and the biological plausibility of this gene, our group is continuing analyses in this region. Furthermore, given the interacting nature of the involvement of Tp63 in the murine model [77] we are simultaneously exploring TP63, BMP4, FGF8 and SHH in our multiplex families. Notably, as described below, we also saw genome-wide significant linkage to region 14q which is near the location for BMP4.
Chromosome 9q
Region 9q21 had the most significant linkage result in our previous report [35]. This region also reached genome-wide significance in the current study, and showed the most positive HLOD and GSMA results in the TOTAL and CL+CLP datasets. There are a number of possible candidate genes in this region including the human homolog of patched (PTCH, 9q22.3), receptor tyrosine kinase-like orphan receptor 2 (ROR2, 9q22), transforming growth factor beta receptor type 1 (TGFBR1, 9q33-q34), zinc finger protein 189 (ZNF189, 9q22-q31) and FOXE1 (a.k.a. TTF2, TITF2; 9q22). Therefore multiple SNPs from the 1-LOD and 2-LOD intervals around the 9q linkage peak were genotyped, as well as SNPs in each of these candidate genes.
The only SNPs genotyped in the 9q region to reach genome-wide significant association were in or near FOXE1 (see table 5), and were significantly associated with CL/P in the TOTAL and CLP datasets. FOXE1 is a forkhead domain-containing transcription factor (a.k.a. TTF2, TITF2; 9q22); mutations in FOXE1 are associated with congenital hypothyroidism, thyroid agenesis and cleft palate in humans (Bamford-Lazarus syndrome, MIM 241850) and mice [86,87,88]. The forkhead gene family (Fox), originally identified in Drosophila, encodes transcription factors with a conserved 100-amino acid DNA-binding motif called the ‘forkhead domain’ [89] and regulates diverse developmental processes in eukaryotes. Rare missense mutations in FOXE1 have been associated with isolated clefting [17, 90].
FOXE1 is a major current focus of our research group, including studies of multi-species conservation, structure, function, and etiology. Genotyping the FOXE1 locus at a greater SNP density has excluded adjacent genes and narrowed the mutation to a 43 kb region including and upstream of FOXE1 (Moreno et al., unpublished results). Figure 4 shows the relative positions of the 9q21 SNPs that were genome-wide significant in the current study (see table 5 for list), as well as three additional fine-mapping SNPs recently found to be significantly associated with CL/P (rs894673; rs3758249; rs1867278; Moreno et al. unpublished results), along with the LD patterns estimated in the HapMap CEU population (Data Release 23a/phaseII Mar08, on NCBI B36 assembly, dbSNP b126) utilizing Haploview version 4.1 [72]. Of the recently studied SNPs, rs3758249 is the one most strongly associated with CL/P in multiple populations (Moreno et al., unpublished results) and the associated SNPs from the current study are in a haplotype block with rs3758249 (see fig. 4). Notably, in both the current study and our recent FOXE1 studies, the strongest associations are seen with families in which one or more affected family members have CL plus CP, and little or no evidence of association in families with CL alone or CP alone. Further, the involvement of FOXE1 during primary palatogenesis is supported by the previously uncharacterized epithelial expression in the medial nasal and maxillary processes that will undergo fusion (Moreno et al., unpublished results).
Fig. 4.
Depicted are the LD patterns estimated in the HapMap CEU population for multiple SNPs in FOXE1. The SNPs marked with asterisks (*) are those that were found to be genome-wide significantly associated with CL/P in the current study (TOTAL and CL+CLP subset). The other SNPs have recently been found by our group to be strongly associated with CL/P (Moreno et al., unpublished results).
Chromosome 12p11
This region did not achieve significant linkage results in the TOTAL dataset, however, it did reach significance in the CLP subset. Chromosomal rearrangements involving the 12p region result in phenotypes including CL/P or CP [69, 70], including Pallister-Killian syndrome (mosaic isochromosome 12p) [91] and others (del 12p [92]; Fryns syndrome [93]). However this region has not been previously investigated in non-syndromic CL/P and warrants further investigation.
Chromosome 14q
The 14q21–24 region had genome-wide significant results in our previous report, and also reached genome-wide significant linkage results in the current study. This region contains PAX9, BMP4 and transforming growth factor beta 3 (TGFB3). Both Pax9 and Tgfb3 when inactivated in mice results in clefts of the secondary palate [94]. Positive results have been seen in association studies of CL/P and TGFB3, and one missense mutation has been reported [19, 95]. However, none of the one PAX9 or five TGFB3 SNPs tested in the current study reached genome-wide significant association in any of the datasets.
BMP4 (14q22–23) is another plausible candidate gene in this region. Multiple murine model studies have demonstrated a role for the Bmp4 pathway in lip and palate fusion [77]. In a Bmp4 conditional knockout mouse model [96], all embryos had bilateral cleft lip at 12 days post-conception but by 14.5 days only 22% still exhibited cleft lip, suggesting in utero healing. Our group has found BMP4 nonsense and missense mutations in CL/P, microform, and subclinical cleft cases that are absent in controls [97] and a recent report showed association of a common variant missense change with isolated cleft lip and palate in a Chinese population [98]. Only one BMP4 SNP was included in the custom SNP panel (rs2147105), and was not significantly associated with CL/P in this study.
Given the strong linkage finding, and the biological plausibility of the possible candidates in this region, we are continuing to investigate BMP4 and other candidates in this region, and will incorporate GxG analyses as well (given the interacting patterns in the murine model recently reported [77]).
Chromosome 16q
Chromosome 16q24.1 was first identified in a genome scan of Caucasian NSCLP sib pairs [33]. Two candidate genes in this region were subsequently investigated: IRF8 and CRISPLD2[99]. No association was seen with IRF8, but CRISPLD2 showed association in Caucasian and Hispanic families. Although the function of CRISPLD2 is unknown, its structure featuring the presence of a LCCL (Limulus factor C, Coch-5b2 and Lgl1) domain has been suggested to play a structural or immunologic role, or even be involved in cell motility which is required for effective cell migration. In situ hybridization studies [99] showed that CRISPLD2 is expressed in the mandible, palate, and nasopharynx regions during craniofacial development [99].
FOXC2 is another plausible candidate in this region since FOXC2 mutations cause lymphodema-distichiasis which has incomplete penetrance of cleft palate [100,101,102], the knockout has cleft palate, and decreased Foxc2 expression in the lateral nasal processes occurs in mice with SHH signaling inactivated in the cranial neural crest cells [103]. We are currently testing FOXC2 and CRISPLD2 SNPs in our study populations, as these were not included in the fine-mapping SNPs genotyped in the current study.
Summary and Future Plans
The current study utilized linkage approaches in a large sample of multiplex CL/P families to identify six genomic regions with genome-wide significant findings, plus one additional region significant in one phenotypic subset. A follow-up fine-mapping SNP panel identified two genome-wide significant associations with SNPs in or near candidate genes (IRF6 and FOXE1), which have led to identification of the likely etiologic variants (IRF6 [71]; FOXE1 – Moreno et al., unpublished results). Note that the fine-mapping approach utilized here would only detect relatively common variants associated with CL/P. Therefore, sequencing of candidate genes is now required to find those variants that may have resulted in linkage signals but with family-specific etiologic variants that could not be detected by LD approaches such as the wFDR utilized here. We are also examining the genotyped SNPs for evidence of micordeletions that are causal and that may be flagged from apparent non-mendelian inheritance. As described above, we are also continuing to pursue those regions with significant linkage results with further fine-mapping efforts.
A striking result from these studies is that some of the significant findings could be attributed to specific phenotypic subsets. Indeed, some findings were significant only in specific phenotypic subgroups. Non-syndromic CL/P is a complex trait, with etiologic heterogeneity, so these results highlight the importance of careful phenotypic delineation in large samples of families as one method to begin to understand the observed etiologic heterogeneity in CL/P families, and as a method to implement in other human disorders with similar levels of etiologic heterogeneity.
There are a number of additional sub-clinical phenotypes now under analysis in our group that suggest increased risk for CL/P: dental anomalies [104, 105], deficiencies of the upper lip orbicularis oris muscle [106, 107], asymmetry [108, 109], and specific craniofacial measurements [110] in the non-overtly-affected relatives of individuals affected with CL/P. Such sub-clinical phenotyping holds great promise for addressing etiologic heterogeneity in CL/P families by allowing a subdivision of families into potentially more homogeneous subsets based on both the overt and sub-clinical phenotypes present, thus markedly increasing the power and specificity of our genetic studies.
Finally, identification of specific genes such as IRF6 and FOXE1 as well as the association with specific cleft phenotypes leads to a realistic expectation that improved recurrence risk and OFC prognoses may soon be possible using a combination of molecular testing and improved phenotyping in families.
Electronic Database Information
URL's for this article are: CIDR: Center for Inherited Disease Research (http://www.cidr.jhmi.edu/); OMIM: Online Mendelian Inheritance in Man (http://www.ncbi.nlm.nih.gov/); Haploview: http://www.broad.mit.edu/haploview/haploview; HapMap: http://www.hapmap.org/; FBAT: http://www.biostat.harvard.edu/∼fbat/default.html.
Acknowledgements
Many thanks to the families in our world-wide study sites who graciously agreed to participate, and without whom these studies would not be possible. The staff and collaborators at each study site were critical for successful completion of these studies: Dr. You-e Liu and Dr. Dan-ning Hu (China); Dr. Ajit Ray (India); Dr. Tuncbilek (Turkey); Carla Brandon, Kathy Bardi, Judith Resick, Dr. Katherine Neiswanger, Dr. Joseph Losee (USA, Pittsburgh); Katy Krahn, Amy Mach and Tamara Busch (USA, Iowa), Dora Rivera and Dr. Mauricio Camargo (Colombia). In the Philippines, Operation Smile International, Operation Smile Philippines, the HOPE Foundation, Bill and Kathy Magee, Edith Villanueva, Buena Nepomuceno, Henrietta Gamboa, Salie Onggada, Rachel Lim, and Gloria Melocoton were all critical to sample collection. Many thanks to Dr. Peter Chines of NHGRI for SNP mapping and annotation.
These studies were supported by NIH grants R37-DE08559, R01-DE09886, R01-DE012472, R01-DE014677, R01-DE016148, P50-DE016215, R03-DE016632, KO2-DE015291, M01-RR00084; and a March of Dimes grant #6-FY01-616. Genotyping services were provided by the Center for Inherited Disease Research (CIDR) which is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number N01-HG-65403. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental and Craniofacial Research or the National Institutes of Health.
References
- 1.Murray JC, Schutte BC. Cleft palate: Players, pathways, and pursuits. J Clin Invest. 2004;113:1676–1678. doi: 10.1172/JCI22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trew CJ. Sistens plura exempla palati deficientis. Nova Acta Physio-Medica Academiae Caesareae Leopoldion-Carolinae. 1757;1:445–447. [Google Scholar]
- 3.Darwin C. The Variation of Plants and Animals under Domestication. New York: Appleton; 1875. [Google Scholar]
- 4.Sproule J. Hereditary nature of hare-lip. Br Med J. 1863;1:412. [Google Scholar]
- 5.Rischbieth H. Hare-lip and cleft palate. In: Pearson K, editor. Treasure of Human Inheritance, part iv. London: Dulau; 1910. pp. 79–123. [Google Scholar]
- 6.Fogh-Andersen P. The Inheritance of Hare Lip and Cleft Palate; Contributions to the Elucidation of the Congenital Clefts of the Face. Copenhagen: Nyt Nordisk Forlag Arnold Busck; 1942. [Google Scholar]
- 7.Melnick M. Cleft lip and palate etiology and its meaning in early 20th century England: Galton/Pearson vs. Bateson; polygenically poor protoplasm vs. Mendelism. J Craniofac Genet Dev Biol. 1997;17:65–79. [PubMed] [Google Scholar]
- 8.Melnick M, Marazita ML, Hu DN. Genetic analysis of cleft lip with or without cleft palate in Chinese kindreds. Am J Med Genet Suppl. 1986;2:183–190. doi: 10.1002/ajmg.1320250622. [DOI] [PubMed] [Google Scholar]
- 9.Hecht JT, Yang P, Michels VV, Buetow KH. Complex segregation analysis of nonsyndromic cleft lip and palate. Am J Hum Genet. 1991;49:674–681. [PMC free article] [PubMed] [Google Scholar]
- 10.Marazita ML, Hu DN, Spence MA, Liu YE, Melnick M. Cleft lip with or without cleft palate in Shanghai, China: Evidence for an autosomal major locus. Am J Hum Genet. 1992;51:648–653. [PMC free article] [PubMed] [Google Scholar]
- 11.Farrall M, Holder S. Familial recurrence-pattern analysis of cleft lip with or without cleft palate. Am J Hum Genet. 1992;50:270–277. [PMC free article] [PubMed] [Google Scholar]
- 12.FitzPatrick D, Farrall M. An estimation of the number of susceptibility loci for isolated cleft palate. J Craniofac Genet Dev Biol. 1993;13:230–235. [PubMed] [Google Scholar]
- 13.Schliekelman P, Slatkin M. Multiplex relative risk and estimation of the number of loci underlying an inherited disease. Am J Hum Genet. 2002;71:1369–1385. doi: 10.1086/344779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen K, Mitchell LE. Familial recurrence-pattern analysis of nonsyndromic isolated cleft palate – a Danish registry study. Am J Hum Genet. 1996;58:182–190. [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell LE, Christensen K. Analysis of the recurrence patterns for nonsyndromic cleft lip with or without cleft palate in the families of 3,073 Danish probands. Am J Med Genet. 1996;61:371–376. doi: 10.1002/(SICI)1096-8628(19960202)61:4<371::AID-AJMG12>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 16.Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O'Brien SE, Daack-Hirsch S, Schultz RE, Weber A, Nepomucena B, Romitti PA, Christensen K, Orioli IM, Castilla EE, Machida J, Natsume N, Murray JC. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet. 2003;40:399–407. doi: 10.1136/jmg.40.6.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vieira AR, Avila JR, Daack-Hirsch S, Dragan E, Felix TM, Rahimov F, Harrington J, Schultz RR, Watanabe Y, Johnson M, Fang J, O'Brien SE, Orioli IM, Castilla EE, Fitzpatrick DR, Jiang R, Marazita ML, Murray JC. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 2005;1:e64. doi: 10.1371/journal.pgen.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe A, Akita S, Tin NT, Natsume N, Nakano Y, Niikawa N, Uchiyama T, Yoshiura K. A mutation in RYK is a genetic factor for nonsyndromic cleft lip and palate. Cleft Palate Craniofac J. 2006;43:310–316. doi: 10.1597/04-145.1. [DOI] [PubMed] [Google Scholar]
- 19.Marazita ML, Neiswanger K. Association analysis. In: Wysznyski D, editor. Cleft Lip and Palate: From Origin to Treatment. Oxford: Oxford University Press; 2002. pp. 240–254. [Google Scholar]
- 20.Lidral AC, Moreno LM. Progress toward discerning the genetics of cleft lip. Curr Opin Pediatr. 2005;17:731–739. doi: 10.1097/01.mop.0000185138.65820.7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carinci F, Scapoli L, Palmieri A, Zollino I, Pezzetti F. Human genetic factors in nonsyndromic cleft lip and palate: An update. Int J Pediatr Otorhinolaryngol. 2007;71:1509–1519. doi: 10.1016/j.ijporl.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Vieira AR. Unraveling human cleft lip and palate research. J Dent Res. 2008;87:119–125. doi: 10.1177/154405910808700202. [DOI] [PubMed] [Google Scholar]
- 23.Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti-Ferreira D, Richieri-Costa A, Dixon MJ, Murray JC. Mutations in IRF6 cause van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira AR, Orioli IM, Castilla EE, Moreno L, Arcos-Burgos M, Lidral AC, Field LL, Liu YE, Ray A, Goldstein TH, Schultz RE, Shi M, Johnson MK, Kondo S, Schutte BC, Marazita ML, Murray JC. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
- 25.Scapoli L, Palmieri A, Martinelli M, Pezzetti F, Carinci P, Tognon M, Carinci F. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am J Hum Genet. 2005;76:180–183. doi: 10.1086/427344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghassibe M, Bayet B, Revencu N, Verellen-Dumoulin C, Gillerot Y, Vanwijck R, Vikkula M. Interferon regulatory factor-6: A gene predisposing to isolated cleft lip with or without cleft palate in the Belgian population. Eur J Hum Genet. 2005;13:1239–1242. doi: 10.1038/sj.ejhg.5201486. [DOI] [PubMed] [Google Scholar]
- 27.Blanton SH, Cortez A, Stal S, Mulliken JB, Finnell RH, Hecht JT. Variation in IRF6 contributes to nonsyndromic cleft lip and palate. Am J Med Genet A. 2005;137:259–262. doi: 10.1002/ajmg.a.30887. [DOI] [PubMed] [Google Scholar]
- 28.Srichomthong C, Siriwan P, Shotelersuk V. Significant association between IRF6 820 G->A and non-syndromic cleft lip with or without cleft palate in the Thai population. J Med Genet. 2005;42:e46. doi: 10.1136/jmg.2005.032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JW, McIntosh I, Hetmanski JB, Jabs EW, Vander Kolk CA, Wu-Chou YH, Chen PK, Chong SS, Yeow V, Jee SH, Park BY, Fallin MD, Ingersoll R, Scott AF, Beaty TH. Association between IRF6 and nonsyndromic cleft lip with or without cleft palate in four populations. Genet Med. 2007;9:219–227. doi: 10.1097/GIM.0b013e3180423cca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieira AR, Cooper ME, Marazita ML, Orioli IM, Castilla EE. Interferon regulatory factor 6 (IRF6) is associated with oral-facial cleft in individuals that originate in South America. Am J Med Genet A. 2007;143A:2075–2078. doi: 10.1002/ajmg.a.31884. [DOI] [PubMed] [Google Scholar]
- 31.Jugessur A, Rahimov F, Lie RT, Wilcox AJ, Gjessing HK, Nilsen RM, Nguyen TT, Murray JC. Genetic variants in IRF6 and the risk of facial clefts: Single-marker and haplotype-based analyses in a population-based case-control study of facial clefts in Norway. Genet Epidemiol. 2008;32:413–424. doi: 10.1002/gepi.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jugessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA, Schutte BC, Murray JC. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prescott NJ, Lees MM, Winter RM, Malcolm S. Identification of susceptibility loci for nonsyndromic cleft lip with or without cleft palate in a two stage genome scan of affected sib-pairs. Hum Genet. 2000;106:345–350. doi: 10.1007/s004390051048. [DOI] [PubMed] [Google Scholar]
- 34.Marazita ML, Field LL, Cooper ME, Tobias R, Maher BS, Peanchitlertkajorn S, Liu YE. Genome scan for loci involved in cleft lip with or without cleft palate, in Chinese multiplex families. Am J Hum Genet. 2002;71:349–364. doi: 10.1086/341944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marazita ML, Field LL, Tuncbilek G, Cooper ME, Goldstein T, Gursu KG. Genome-scan for loci involved in cleft lip with or without cleft palate in consanguineous families from Turkey. Am J Med Genet A. 2004;126:111–122. doi: 10.1002/ajmg.a.20564. [DOI] [PubMed] [Google Scholar]
- 36.Wyszynski DF, Albacha-Hejazi H, Aldirani M, Hammod M, Shkair H, Karam A, Alashkar J, Holmes TN, Pugh EW, Doheny KF, McIntosh I, Beaty TH, Bailey-Wilson JE. A genome-wide scan for loci predisposing to non-syndromic cleft lip with or without cleft palate in two large Syrian families. Am J Med Genet A. 2003;123:140–147. doi: 10.1002/ajmg.a.20283. [DOI] [PubMed] [Google Scholar]
- 37.Zeiger JS, Hetmanski JB, Beaty TH, VanderKolk CA, Wyszynski DF, Bailey-Wilson JE, de Luna RO, Perandones C, Tolarova MM, Mosby T, Bennun R, Segovia M, Calda P, Pugh EW, Doheny K, McIntosh I. Evidence for linkage of nonsyndromic cleft lip with or without cleft palate to a region on chromosome 2. Eur J Hum Genet. 2003;11:835–839. doi: 10.1038/sj.ejhg.5201052. [DOI] [PubMed] [Google Scholar]
- 38.Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, Goldstein T, Maher BS, Daack-Hirsch S, Schultz R, Mansilla MA, Field LL, Liu YE, Prescott N, Malcolm S, Winter R, Ray A, Moreno L, Valencia C, Neiswanger K, Wyszynski DF, Bailey-Wilson JE, Albacha-Hejazi H, Beaty TH, McIntosh I, Hetmanski JB, Tuncbilek G, Edwards M, Harkin L, Scott R, Roddick LG. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32–35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connell JR, Weeks DE. Pedcheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown NL, Knott L, Halligan E, Yarram SJ, Mansell JP, Sandy JR. Microarray analysis of murine palatogenesis: Temporal expression of genes during normal palate development. Dev Growth Differ. 2003;45:153–165. doi: 10.1034/j.1600-0854.2004.00686.x. [DOI] [PubMed] [Google Scholar]
- 41.Gong SG, Gong TW, Shum L. Identification of markers of the midface. J Dent Res. 2005;84:69–72. doi: 10.1177/154405910508400112. [DOI] [PubMed] [Google Scholar]
- 42.Mukhopadhyay P, Greene RM, Zacharias W, Weinrich MC, Singh S, Young WW, Jr, Pisano MM. Developmental gene expression profiling of mammalian, fetal orofacial tissue. Birth Defects Res A Clin Mol Teratol. 2004;70:912–926. doi: 10.1002/bdra.20095. [DOI] [PubMed] [Google Scholar]
- 43.Jugessur A, Murray JC. Orofacial clefting: Recent insights into a complex trait. Curr Opin Genet Dev. 2005;15:270–278. doi: 10.1016/j.gde.2005.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai J, Ash D, Kotch LE, Jabs EW, Attie-Bitach T, Auge J, Mattei G, Etchevers H, Vekemans M, Korshunova Y, Tidwell R, Messina DN, Winston JB, Lovett M. Gene expression in pharyngeal arch 1 during human embryonic development. Hum Mol Genet. 2005;14:903–912. doi: 10.1093/hmg/ddi083. [DOI] [PubMed] [Google Scholar]
- 45.Diewert VM. Development of human craniofacial morphology during the late embryonic and early fetal periods. Am J Orthod. 1985;88:64–76. doi: 10.1016/0002-9416(85)90107-1. [DOI] [PubMed] [Google Scholar]
- 46.Beier DR, Herron BJ. Genetic mapping and ENU mutagenesis. Genetica. 2004;122:65–69. doi: 10.1007/s10709-004-1437-5. [DOI] [PubMed] [Google Scholar]
- 47.Higgins AW, Alkuraya FS, Bosco AF, Brown KK, Bruns GA, Donovan DJ, Eisenman R, Fan Y, Farra CG, Ferguson HL, Gusella JF, Harris DJ, Herrick SR, Kelly C, Kim HG, Kishikawa S, Korf BR, Kulkarni S, Lally E, Leach NT, Lemyre E, Lewis J, Ligon AH, Lu W, Maas RL, MacDonald ME, Moore SD, Peters RE, Quade BJ, Quintero-Rivera F, Saadi I, Shen Y, Shendure J, Williamson RE, Morton CC. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet. 2008;82:712–722. doi: 10.1016/j.ajhg.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sebastiani P, Lazarus R, Weiss ST, Kunkel LM, Kohane IS, Ramoni MF. Minimal haplotype tagging. Proc Natl Acad Sci USA. 2003;100:9900–9905. doi: 10.1073/pnas.1633613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, Galver L, Hunt S, McBride C, Bibikova M, Rubano T, Chen J, Wickham E, Doucet D, Chang W, Campbell D, Zhang B, Kruglyak S, Bentley D, Haas J, Rigault P, Zhou L, Stuelpnagel J, Chee MS. Highly parallel snp genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 50.Ray AK, Field LL, Marazita ML. Nonsyndromic cleft lip with or without cleft palate in West Bengal, India: Evidence for an autosomal major locus. Am J Hum Genet. 1993;52:1006–1011. [PMC free article] [PubMed] [Google Scholar]
- 51.Moreno L, Lidral A, Fonseca I, Valencia C, Arcos-Burgos M. Complex segregation analysis of nonsyndromic cleft lip/palate in Antioquia, Colombia. Actualidades Biologica. 2004;23:49–56. [Google Scholar]
- 52.Marazita ML, Spence MA, Melnick M. Genetic analysis of cleft lip with or without cleft palate in Danish kindreds. Am J Med Genet. 1984;19:9–18. doi: 10.1002/ajmg.1320190104. [DOI] [PubMed] [Google Scholar]
- 53.Marazita ML, Goldstein AM, Smalley SL, Spence MA. Cleft lip with or without cleft palate: Reanalysis of a three-generation family study from England. Genet Epidemiol. 1986;3:335–342. doi: 10.1002/gepi.1370030506. [DOI] [PubMed] [Google Scholar]
- 54.Smith CAB. Testing for heterogeneity of recombination fraction values in human genetics. Ann Hum Genet. 1963;17:175–182. doi: 10.1111/j.1469-1809.1963.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 55.Vieland VJ, Wang K, Huang J. Power to detect linkage based on multiple sets of data in the presence of locus heterogeneity: Comparative evaluation of model-based linkage methods for affected sib pair data. Hum Hered. 2001;51:199–208. doi: 10.1159/000053343. [DOI] [PubMed] [Google Scholar]
- 56.Greenberg DA, Abreu PC. Determining trait locus position from multipoint analysis: Accuracy and power of three different statistics. Genet Epidemiol. 2001;21:299–314. doi: 10.1002/gepi.1036. [DOI] [PubMed] [Google Scholar]
- 57.Hodge SE, Vieland VJ, Greenberg DA. Hlods remain powerful tools for detection of linkage in the presence of genetic heterogeneity. Am J Hum Genet. 2002;70:556–559. doi: 10.1086/338923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sobel E, Lange K. Descent graphs in pedigree analysis: Applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 59.Hodge SE, Elston RC. Lods, wrods, and mods: The interpretation of lod scores calculated under different models. Genet Epidemiol. 1994;11:329–342. doi: 10.1002/gepi.1370110403. [DOI] [PubMed] [Google Scholar]
- 60.Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 61.Wise LH, Lanchbury JS, Lewis CM. Meta-analysis of genome searches. Ann Hum Genet. 1999;63:263–272. doi: 10.1046/j.1469-1809.1999.6330263.x. [DOI] [PubMed] [Google Scholar]
- 62.Levinson DF, Levinson MD, Segurado R, Lewis CM. Genome scan meta-analysis of schizophrenia and bipolar disorder, part I: Methods and power analysis. Am J Hum Genet. 2003;73:17–33. doi: 10.1086/376548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O'Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lonnqvist J, Peltonen L, O'Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoega T, Helgason T. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cooper ME, Goldstein TH, Maher BS, Marazita ML. Identifying genomic regions for fine-mapping using genome scan meta-analysis (GSMA) to identify the minimum regions of maximum significance (MRMS) across populations. BMC Genet. 2005;6(suppl 1):S42. doi: 10.1186/1471-2156-6-S1-S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- 66.Horvath S, Xu X, Laird NM. The family based association test method: Strategies for studying general genotype–phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 67.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 68.Roeder K, Bacanu SA, Wasserman L, Devlin B. Using linkage genome scans to improve power of association in genome scans. Am J Hum Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D. A chromosomal duplication map of malformations: Regions of suspected haplo- and triplolethality – and tolerance of segmental aneuploidy – in humans. Am J Hum Genet. 1999;64:1702–1708. doi: 10.1086/302410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D. A chromosomal deletion map of human malformations. Am J Hum Genet. 1998;63:1153–1159. doi: 10.1086/302041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jugessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA, Schutte BC, Murray JC. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 73.FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, Gautier P, McGill N, Hayward C, Firth H, Markham AF, Fantes JA, Bonthron DT. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum Mol Genet. 2003;12:2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- 74.Ardinger HH, Buetow KH, Bell GI, Bardach J, VanDemark DR, Murray JC. Association of genetic variation of the transforming growth factor-alpha gene with cleft lip and palate. Am J Hum Genet. 1989;45:348–353. [PMC free article] [PubMed] [Google Scholar]
- 75.Mitchell LE. Transforming growth factor alpha locus and nonsyndromic cleft lip with or without cleft palate: A reappraisal. Genet Epidemiol. 1997;14:231–240. doi: 10.1002/(SICI)1098-2272(1997)14:3<231::AID-GEPI2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 76.Marazita ML, Field LL, Cooper ME, Tobias R, Maher BS, Peanchitlertkajorn S, Liu YE. Nonsyndromic cleft lip with or without cleft palate in China: Assessment of candidate regions. Cleft Palate Craniofac J. 2002;39:149–156. doi: 10.1597/1545-1569_2002_039_0149_nclwow_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- 77.Thomason HA, Dixon MJ, Dixon J. Facial clefting in Tp63 deficient mice results from altered Bmp4, Fgf8 and Shh signaling. Dev Biol. 2008;321:273–282. doi: 10.1016/j.ydbio.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 78.Ray AK, Marazita ML, Pathak R, Beever CL, Cooper ME, Goldstein T, Shaw DF, Field LL. TP63 mutation and clefting modifier genes in an EEC syndrome family. Clin Genet. 2004;66:217–222. doi: 10.1111/j.1399-0004.2004.00287.x. [DOI] [PubMed] [Google Scholar]
- 79.Barrow LL, van Bokhoven H, Daack-Hirsch S, Andersen T, van Beersum SE, Gorlin R, Murray JC. Analysis of the p63 gene in classical EEC syndrome, related syndromes, and non-syndromic orofacial clefts. J Med Genet. 2002;39:559–566. doi: 10.1136/jmg.39.8.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR, Wessagowit V, Kelly A, Atherton DJ, Griffiths WA, Orlow SJ, van Haeringen A, Ausems MG, Yang A, McKeon F, Bamshad MA, Brunner HG, Hamel BC, van Bokhoven H. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet. 2001;10:221–229. doi: 10.1093/hmg/10.3.221. [DOI] [PubMed] [Google Scholar]
- 81.Bougeard G, Hadj-Rabia S, Faivre L, Sarafan-Vasseur N, Frebourg T. The Rapp-Hodgkin syndrome results from mutations of the TP63 gene. Eur J Hum Genet. 2003;11:700–704. doi: 10.1038/sj.ejhg.5201004. [DOI] [PubMed] [Google Scholar]
- 82.Shotelersuk V, Janklat S, Siriwan P, Tongkobpetch S. De novo missense mutation, s541y, in the p63 gene underlying Rapp-Hodgkin ectodermal dysplasia syndrome. Clin Exp Dermatol. 2005;30:282–285. doi: 10.1111/j.1365-2230.2005.01722.x. [DOI] [PubMed] [Google Scholar]
- 83.Leoyklang P, Siriwan P, Shotelersuk V. A mutation of the p63 gene in non-syndromic cleft lip. J Med Genet. 2006;43:e28. doi: 10.1136/jmg.2005.036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Bokhoven H, Hamel BC, Bamshad M, Sangiorgi E, Gurrieri F, Duijf PH, Vanmolkot KR, van Beusekom E, van Beersum SE, Celli J, Merkx GF, Tenconi R, Fryns JP, Verloes A, Newbury-Ecob RA, Raas-Rotschild A, Majewski F, Beemer FA, Janecke A, Chitayat D, Crisponi G, Kayserili H, Yates JR, Neri G, Brunner HG. P63 gene mutations in EEC syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet. 2001;69:481–492. doi: 10.1086/323123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Bokhoven H, Brunner HG. Splitting p63. Am J Hum Genet. 2002;71:1–13. doi: 10.1086/341450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De Felice M, Ovitt C, Biffali E, Rodriguez-Mallon A, Arra C, Anastassiadis K, Macchia PE, Mattei MG, Mariano A, Scholer H, Macchia V, Di Lauro R. A mouse model for hereditary thyroid dysgenesis and cleft palate. Nat Genet. 1998;19:395–398. doi: 10.1038/1289. [DOI] [PubMed] [Google Scholar]
- 87.Dathan N, Parlato R, Rosica A, De Felice M, Di Lauro R. Distribution of the titf2/foxe1 gene product is consistent with an important role in the development of foregut endoderm, palate, and hair. Dev Dyn. 2002;224:450–456. doi: 10.1002/dvdy.10118. [DOI] [PubMed] [Google Scholar]
- 88.Castanet M, Park SM, Smith A, Bost M, Leger J, Lyonnet S, Pelet A, Czernichow P, Chatterjee K, Polak M. A novel loss-of-function mutation in TTF-2 is associated with congenital hypothyroidism, thyroid agenesis and cleft palate. Hum Mol Genet. 2002;11:2051–2059. doi: 10.1093/hmg/11.17.2051. [DOI] [PubMed] [Google Scholar]
- 89.Lehman JA, Jr, Artz JS. The minimal cleft lip. Plast Reconstr Surg. 1976;58:306–309. doi: 10.1097/00006534-197609000-00008. [DOI] [PubMed] [Google Scholar]
- 90.Venza M, Visalli M, Venza I, Torino C, Saladino R, Teti D. FOXEL gene mutation screening by multiplex PCR/DHPLC in CHARGE syndrome and syndromic and non-syndromic cleft palate. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;836:39–46. doi: 10.1016/j.jchromb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 91.McPherson EW, Ketterer DM, Salsburey DJ. Pallister-Killian and Fryns syndromes: Nosology. Am J Med Genet. 1993;47:241–245. doi: 10.1002/ajmg.1320470219. [DOI] [PubMed] [Google Scholar]
- 92.Glaser B, Rossier E, Barbi G, Chiaie LD, Blank C, Vogel W, Kehrer-Sawatzki H. Molecular cytogenetic analysis of a constitutional de novo interstitial deletion of chromosome 12p in a boy with developmental delay and congenital anomalies. Am J Med Genet A. 2003;116A:66–70. doi: 10.1002/ajmg.a.10878. [DOI] [PubMed] [Google Scholar]
- 93.Alessandri JL, Attali T, Brayer C, Dupuy L, Pilorget H, Ramful D, Samperiz S, Tiran-Rajaofera I, Robin S. [Fryns syndrome. Report on [3] new cases] Arch Pediatr. 2007;14:903–907. doi: 10.1016/j.arcped.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 94.Sasaki Y, O'Kane S, Dixon J, Dixon MJ, Ferguson MW. Temporal and spatial expression of Pax9 and Sonic hedgehog during development of normal mouse palates and cleft palates in TGF-beta3 null embryos. Arch Oral Biol. 2007;52:260–267. doi: 10.1016/j.archoralbio.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 95.Lidral AC, Romitti PA, Basart AM, Doetschman T, Leysens NJ, Daack-Hirsch S, Semina EV, Johnson LR, Machida J, Burds A, Parnell TJ, Rubenstein JL, Murray JC. Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. Am J Hum Genet. 1998;63:557–568. doi: 10.1086/301956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- 97.Suzuki S, Marazita ML, Cooper ME, Miwa N, Hing A, Jugessur A, Natsume N, Shimozato K, Ohbayashi N, Suzuki Y, Niimi T, Minami K, Yamamoto M, Altannamar TJ, Erkhembaatar T, Furukawa H, Daack-Hirsch S, L'Heureux J, Brandon CA, Weinberg SM, Neiswanger K, Deleyiannis FWB, de Salamanca JE, Vieira AR, Lidral AC, Martin JF, Murray JC. Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. Am J Hum Genet. 2009;84:406–411. doi: 10.1016/j.ajhg.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lin JY, Chen YJ, Huang YL, Tang GP, Zhang L, Deng B, Li M, Ma H, Luan RS. Association of bone morphogenetic protein 4 gene polymorphisms with nonsyndromic cleft lip with or without cleft palate in Chinese children. DNA Cell Biol. 2008;27:601–605. doi: 10.1089/dna.2008.0777. [DOI] [PubMed] [Google Scholar]
- 99.Chiquet BT, Lidral AC, Stal S, Mulliken JB, Moreno LM, Arcos-Burgos M, Valencia-Ramirez C, Blanton SH, Hecht JT. CRISPLD2:A novel NSCLP candidate gene. Hum Mol Genet. 2007;16:2241–2248. doi: 10.1093/hmg/ddm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sholto-Douglas-Vernon C, Bell R, Brice G, Mansour S, Sarfarazi M, Child AH, Smith A, Mellor R, Burnand K, Mortimer P, Jeffery S. Lymphoedema-distichiasis and FOXC2: Unreported mutations, de novo mutation estimate, families without coding mutations. Hum Genet. 2005;117:238–242. doi: 10.1007/s00439-005-1275-2. [DOI] [PubMed] [Google Scholar]
- 101.Bahuau M, Houdayer C, Tredano M, Soupre V, Couderc R, Vazquez MP. FOXC2 truncating mutation in distichiasis, lymphedema, and cleft palate. Clin Genet. 2002;62:470–473. doi: 10.1034/j.1399-0004.2002.620608.x. [DOI] [PubMed] [Google Scholar]
- 102.Traboulsi EI, Al-Khayer K, Matsumoto M, Kimak MA, Crowe S, Wilson SE, Finegold DN, Ferrell RE, Meisler DM. Lymphedema-distichiasis syndrome and FOXC2 gene mutation. Am J Ophthalmol. 2002;134:592–596. doi: 10.1016/s0002-9394(02)01642-2. [DOI] [PubMed] [Google Scholar]
- 103.Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vieira AR, McHenry TG, Daack-Hirsch S, Murray JC, Marazita ML. A genome wide linkage scan for cleft lip and palate and dental anomalies. Am J Med Genet A. 2008;146A:1406–1413. doi: 10.1002/ajmg.a.32295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Menezes R, Vieira AR. Dental anomalies as part of the cleft spectrum. Cleft Palate Craniofac J. 2008;45:414–419. doi: 10.1597/07-064.1. [DOI] [PubMed] [Google Scholar]
- 106.Neiswanger K, Weinberg SM, Rogers CR, Brandon CA, Cooper ME, Bardi KM, Deleyiannis FW, Resick JM, Bowen A, Mooney MP, de Salamanca JE, Gonzalez B, Maher BS, Martin RA, Marazita ML. Orbicularis oris muscle defects as an expanded phenotypic feature in nonsyndromic cleft lip with or without cleft palate. Am J Med Genet A. 2007;143:1143–1149. doi: 10.1002/ajmg.a.31760. [DOI] [PubMed] [Google Scholar]
- 107.Weinberg SM, Brandon CA, McHenry TH, Neiswanger K, Deleyiannis FW, de Salamanca JE, Castilla EE, Czeizel AE, Vieira AR, Marazita ML. Rethinking isolated cleft palate: Evidence of occult lip defects in a subset of cases. Am J Med Genet A. 2008;146A:1670–1675. doi: 10.1002/ajmg.a.32291. [DOI] [PubMed] [Google Scholar]
- 108.Neiswanger K, Cooper ME, Weinberg SM, Flodman P, Keglovits AB, Liu Y, Hu DN, Melnick M, Spence MA, Marazita ML. Cleft lip with or without cleft palate and dermatoglyphic asymmetry: Evaluation of a Chinese population. Orthod Craniofac Res. 2002;5:140–146. doi: 10.1034/j.1600-0544.2002.02196.x. [DOI] [PubMed] [Google Scholar]
- 109.Neiswanger K, Cooper ME, Liu YE, Hu DN, Melnick M, Weinberg SM, Marazita ML. Bilateral asymmetry in Chinese families with cleft lip with or without cleft palate. Cleft Palate Craniofac J. 2005;42:192–196. doi: 10.1597/03-032.1. [DOI] [PubMed] [Google Scholar]
- 110.Weinberg SM, Neiswanger K, Richtsmeier JT, Maher BS, Mooney MP, Siegel MI, Marazita ML. Three-dimensional morphometric analysis of craniofacial shape in the unaffected relatives of individuals with nonsyndromic orofacial clefts: A possible marker for genetic susceptibility. Am J Med Genet A. 2008;146A:409–420. doi: 10.1002/ajmg.a.32177. [DOI] [PubMed] [Google Scholar]