Summary
There is a growing appreciation that in addition to well-documented intraspecies quorum sensing systems [1], small molecules act as signals between microbes of different species [2]. This review will focus on how bacterial small molecules modulate these interspecies interactions. We will particularly emphasize complex relationships such as those between microbes and insects, interactions resulting in non-antagonistic outcomes (i.e. developmental and morphological processes), how co-culture can lead to the discovery of new small molecules, and the use of known compounds to evoke unexpected responses and mediate crosstalk between microbes.
Introduction
Historically, interspecies interactions have focused on growth-inhibitory interactions, yet a variety of phenotypic outcomes other than antibiosis are possible, including alterations in developmental processes such as sporulation and biofilm formation or production of secondary metabolites (Fig 1). Over the years, studies of antibiosis have undoubtedly led to a deeper understanding of how microbes relate to a major component of their natural environments—their fellow microbes—as well as to the discovery of clinically-useful compounds. Examining interspecies interactions using a broader framework that encompasses both alternative signals and more diverse responses will accordingly continue to advance these vital fields.
Figure 1. Schematic illustrating potential interspecies interactions.
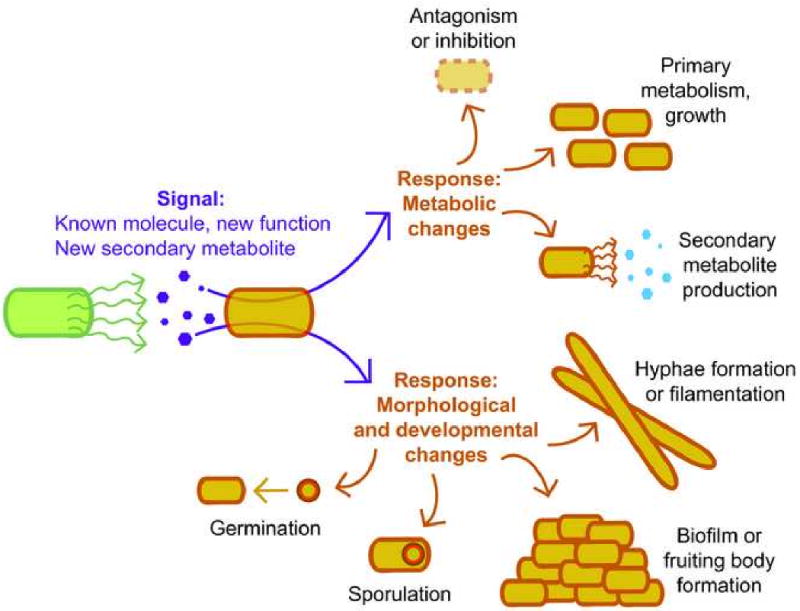
An interaction between two microbes is illustrated on the left of the figure, with the green microbe producing a signal (purple hexagons) that causes the orange microbe to respond in one of the manners illustrated on the right. The signals discussed here fall primarily into two classes: known metabolites (such as peptidoglycan, antibiotics, and intraspecies signals) that cause unexpected responses affecting other microbial speces, and novel secondary metabolites; in some cases the signals are still unknown. Upon detecting the signal, the responding organism may experience changes in metabolism (growth inhibition or stimulation, or production of new small molecules) or morphological and developmental changes (alterations in cell shape or morphology; production of biofilms or fruiting bodies; or specialized processes such as sporulation and germination). More than one response is possible to a single signal.
The last few years have seen a surge of studies [2] covering all aspects of these possible interactions (Fig. 1). Detecting phenotypic or developmental biomodulation between two organisms can indicate when they are communicating via small molecules, and thus can denote the presence of overlooked compounds. In other cases, signaling has been shown to occur via “repurposed” compounds—known molecules that are functioning in an unexpected manner. One exciting potential result of interspecies interactions is the induction of novel secondary metabolite production by the responding organism. Thus, examination of microbial relationships can lead to the discovery of new molecules—in some cases as the small molecule mediating the interaction, and in others as the consequent result of two microbes interacting.
The scope of this article will be limited primarily to microbial interactions, although a few studies are referenced that highlight the complex relationship that microbes have with multicellular eukaryotes, and all demonstrate how little we understand of the complicated interplay occurring between microbes and the potential chemical eavesdropping occurring between them (Table 1).
Table 1. Tabulated interspecies interactions discussed in this paper.
This table lists the recently-described interactions discussed. The first column indicates the review section they are included in. (F) indicates a fungal participant; N/A indicates not applicable; N/D indicates not determined.
| Signal-producing organism | Small molecule signal | Responding organism | Response | Ref. | |
|---|---|---|---|---|---|
| IA | Alliances and Antagonisms: Microbial-Eukaryotic | ||||
| Attine ant food cultivar (F) | N/D | Escovopsis spp. (F) | Growth inhibition or growth attraction | [4] | |
| Streptomyces thermosacchari | Mycangimycin | Entomocorticium sp. (F) | No effect | [5] | |
| Streptomyces thermosacchari | Mycangimycin | Ophiostoma minus (F) | Growth inhibition | [5] | |
| Enterobacter sp. | N/D | Bacillus thuringiensis | Insecticidal toxin activation | [6] | |
| Bacillus thuringiensis AND Enterobacter sp. | Active inseticidal toxin | Insect larvae | Death | [6] | |
| Endosymbiotic bacteria | N/D | Fusarium sp. (F) | Virulence inhibition | [7] | |
| Fusarium sp. AND endosymbiotic bacteria | Volatile sesquiterpenes | Fusarium sp. (F) | Virulence inhibition | [8] | |
| IB | Alliances and Antagonisms: Microbial-Microbial | ||||
| Pseudomonas aeruginosa | HQNO | Staphylococcus aureus | Small colony variant formation | [9] | |
| Streptococcus sanguinis | H2O2 | Streptococcus mutans | Growth inhibition | [10] | |
| Streptococcus mutans | Mutacins (bacteriocins) | Streptococcus sanguinis | Growth inhibition | [10] | |
| Streptococcus oligofermentas | H2O2 | Streptococcus mutans | Growth inhibition | [11] | |
| IIA | New roles for known molecules: Peptidoglycan | ||||
| Bacillus cereus | Peptidoglycan: from vegetative cells | Cytophaga-Flavobacterium rhizosphere bacteria | Growth stimulation | [14] | |
| Unknown gut microbiota | Peptidoglycan: muramyl dipeptides | Candida albicans (F) | Hyphae stimulation | [15] | |
| Diaminopimelic acid-peptidoglycan producing bacteria | Peptidoglycan: disaccharide tripeptides | Bacillus subtilis | Germination stimulation | [16] | |
| Streptomyces spp. | Staurosporine | Bacillus subtilis | Germination stimulation | [16] | |
| Bacillus subtilis, Escherichia coli, Saccharomyces cerevisiae (F) | Peptidoglycan; DNA; insoluble cell fractions | Myxococcus xanthus | Rippling | [17] | |
| Availability of prey | N/D; not nutrients | Myxococcus xanthus | Fruiting body formation | [19] | |
| Unknown | N-acetylglucosamine | Streptomyces coelicolor | Antibiotic production altered | [20] | |
| Unknown | N-acetylglucosamine | Streptomyces coelicolor | Secondary metabolite induction | [20] | |
| IIB | New roles for known molecules: Antibiotics | ||||
| N/A | Fluoroquinolones (synthetic) | Staphylococcus aureus | SOS-response and methyl-mismatch repair upregulation | [22] | |
| N/A | Azithromycin | Haemophilus influenzae | Biofilm inhibition | [23] | |
| N/A | Azithromycin, ceftazidime, ciprofloxacin | Pseudomonas aeruginosa | Virulence inhibition | [24] | |
| N/A | Tobramycin and tetracycline | Pseudomonas aeruginosa | Biofilm stimulation | [25] | |
| Pseudomonas aeruginosa | Phenazines | Pseudomonas aeruginosa | Complex colony morphology repression | [26] | |
| IIC | New roles for known molecules: Signaling system cross-talk | ||||
| Streptococcus oralis | Autoinducer-2 | Actinomyces naeslunii | Biofilm stimulation | [27] | |
| Candida albicans (F) | Farnesol | Candida albicans (F) | Hyphae inhibition | [28] | |
| Xanthomonas campestris | DSF | Xanthomonas campestris | Biofilm alteration; Virulence stimulation | [29,30] | |
| Xanthomonas campestris | DSF | Candida albicans (F) | Hyphae inhibition | [32] | |
| Burkholderia cenocepacia | BDSF | Xanthomonas campestris | Virulence stimulation | [33] | |
| Burkholderia cenocepacia | BDSF | Candida albcans (F) | Hyphae inhibition | [33] | |
| Candida albicans (F) | Farnesol | Pseudomonas aeruginosa | Secondary metabolite and swarming inhibition | [34] [35] | |
| Stenotrophomonas maltophilia | DSF | Pseudomonas aeruginosa | Biofilm filamentation stimulation | [37] | |
| Escherichia coli Bacillus subtilis Candida albicans (F), other organisms | cis-2-decenoic acid | Pseudomonas aeruginosa | Biofilm dispersal | [40] | |
| IIIA | Identifying new compounds using co-culture: Eliciting directly | ||||
| Marine α-proteobacterium | N/D | Libertella sp. (F) | Secondary metabolite induction | [42] | |
| Salinispora arenicola | N/D | Emericella sp. (F) | Secondary metabolite induction | [43] | |
| Streptomyces padanus | Horizontal gene transfer | Rhodococcus fascians | Secondary metabolite induction | [44] | |
| IIIB | Identifying new compounds using co-culture: Using development as read-out | ||||
| Bacillus subtilis | Surfactin | Streptomyces coelicolor | Sporulation and aerial hyphae inhibition | [45] | |
| Bacillus subtilis | Bacillaene | Streptomyces coelicolor | Secondary metabolite inhibition | [46] | |
| IVB | Missing pieces: Biofilm formation | ||||
| Fusobacterium nucleatum | N/D | Staphylococcus epidermidis | Biofilm stimulation | [50] | |
| Fusobacterium nucleatum | N/D | Porphyromonas gingivalis | Biofilm stimulation | [50] | |
| Saccharomyces cerevisae (F) | N/D | Lactobacillus casei | Biofilm stimulation | [51] | |
| N/D | Norspermine | Vibrio cholerae | Biofilm stimulation | [54] | |
| N/D | Glycine betaine | Vibrio cholerae | Biofilm stimulation | [55] | |
| Human respiratory tracts | Mucin | Pseudomonas aeruginosa | Biofilm alteration | [56] | |
| N/D | Nitric oxide | Pseudomonas aeruginosa | Biofilm dispersal | [57] | |
| IVC | Missing pieces: New small molecules | ||||
| Unknown plant? | p-coumaryl-homoserine lactone | Rhodopseudomonas palustris | 17-gene regulon | [58] | |
I. Alliances and Antagonisms
A. Microbial-Eukaryotic
Here we focus on insects that have evolved specific associations with fungi and bacteria, a biological context that has selected the evolution of myriad antagonistic and beneficial interactions that highlight microorganisms' ability to exert exquisite biological specificity in mediating their interactions.
Attine ants grow fungal cultivars as food, and have been shown to have co-evolved with both their food cultivar and actinomycetes that help protect their food from being infected by a parasitic Escovopsis fungus [3]. There is specificity in both the attraction and repulsion between these two sets of fungi and these conflicting forces explain why in natural environments particular Escovopsis fungi infect only a restricted set of the food fungi [4]. Although the active compounds driving these responses are not yet known, Escovopsis spp. are attracted to and grow especially on cultivars that are hosts to that parasite, and the food cultivars produce compounds that actively inhibit the growth of other Escovopsis strains [4].
The southern pine beetle, Dendeoctonus frontalis, exemplifies another example of the intriguing symbioses between the insect, fungal, and bacterial worlds. The beetle is symbiotically associated with an Entomocorticium sp. fungus that helps nourish the beetles' larvae, but an antagonistic fungus, Ophiostoma minus, can outcompete this beneficial symbiont to the detriment of the beetle larvae [5]. An actinomycete bacterium mediates the retention of the beneficial fungus by producing mycangimycin, a novel linear polyene peroxide antifungal that selectively inhibits only the antagonistic fungus and not the symbiotic one [5]. This discovery shows that examining insect symbioses can reveal not only new biology, but also interspecies signaling molecules, some of which will be chemically novel.
The importance of how the larger microbial context can influence biological activity was highlighted in a study that overturned a long-asserted understanding regarding the mechanism of the anti-insecticidal activity of Bacillus thuringiensis. The presence of the insect mid-gut microbiota (in particular an Enterobacter sp.) was shown to be required for anti-insecticidal activity, and the B. thuringiensis toxin alone—in the absence of enteric bacteria—was insufficient to kill insects [6].
The biology of the fungus Fusarium also underscores the significance of microbial context. Some strains act as plant pathogens, while others are protective agents against pathogenic Fusarium strains [7]. The non-pathogenic Fusarium are associated with a consortium of endosymbiotic bacteria that alter fungal gene expression and eliminate their ability to invade plants [7]. The protective capacity of these non-pathogenic strains is explained because Fusarium associated with its endosymbionts—but not the endosymbionts alone—produce volatile sesquiterpenes that repress virulence genes in pathogenic Fusarium strains [8].
B. Microbial-Microbial
Two recent studies follow-up on phenomena described years ago. Pseudomonas aeruginosa and Staphylococcus aureus, two human-associated bacteria, have been known since the 1950s to have a paradoxical relationship in which 4-hydroxy-2-heptylquinoline-N-oxide (HQNO) produced by P. aeruginosa suppresses the respiration of S. aureus, but also increases its resistance to antibiotics in co-culture. It was shown that HQNO selects for small colony variants—a form of S. aureus that conveys antibiotic resistance [9]. HQNO was only detected in the sputum of cystic fibrosis patients infected with P. aeruginosa, highlighting the potential relevance of the interaction of these pathogens in clinical cases [9].
Since the 1970s, Streptococcus sanguinis, a commensal oral bacterium, and Streptococcus mutans, a common resident of the mouth that contributes to dental caries, have been known to be antagonistic towards one another, but no chemical mediators were known. It was demonstrated that S. sanguinis makes hydrogen peroxide (H2O2) and S. mutans produces mutacins (bacteriocins), each at levels sufficient to effectively inhibit each other [10]. Interestingly, the production of these inhibitory compounds is decreased in co-culture versus mono-culture via an unknown mechanism [10].
Streptococcus oligofermentas is another oral bacterium that inhibits S. mutans by producing H2O2 either from peptone or lactic acid [11,12]. In this case there is an additional twist: S. mutans produces the lactic acid substrate, which typically acts as an inhibitory compound towards other microbes [11]. This illustrates how a microbial competitor may turn a potentially antagonistic signal produced by S. mutans against itself in its environmental setting.
II. New Roles for Known Molecules
A. Peptidoglycan
Peptidoglycan and cell wall fragments are increasingly being recognized as important signaling molecules [13], and recent studies continue to elucidate additional roles for these microbial products.
1. Growth
In a microbial interaction of unknown specificity, both Bacillus cereus and its purified peptidoglycan stimulated the growth of rhizosphere bacteria from the Cytophaga-Flavobacterium group in a medium containing root exudates in a manner that did not depend on either B. cereus sporulation or growth inhibition [14]. The rhizosphere bacteria produce a cell-wall degrading enzyme that presumably permits them to mobilize B. cereus peptidoglycan fragments as a carbon source for their growth, although the details of what the degradative enzyme is, or whether it is preferentially expressed in the presence of root exudate as the data suggests are not clear [14].
2. Development
Recent studies examine how peptidoglycan acts as a signal influencing microbial development. A beautiful combination of chemistry and biochemistry was used to identify peptidoglycan fragments in human serum that are powerful inducers of hyphae in Candida albicans [15]. In a splendid example delineating how a responding cell detects and transduces a signal, purified muramyl dipeptides were shown to enter fungal cells and bind to a specific adenylate cyclase that induced cAMP production and hyphal growth [15]. While initially this study seems to involve an exclusively eukaryotic interaction, an intriguing aspect is that mammals cannot produce muramic acid, suggesting that the high quantities of these compounds found in human serum are originally produced by bacterial intestinal microbiota [15].
Another fascinating study investigated how disaccharide tripeptides stimulated germination in Bacillus subtilis at concentrations equivalent to that available from a single cell [16]. A peptidoglycan-specific receptor was identified that was a eukaryotic-like serine/threonine membrane kinase and was localized in the inner membrane of the spore [16]. In addition to phosphorylating a ribosomal GTPase potentially responsible for the downstream effects of peptidoglycan-fragment binding, this receptor kinase is specific for disaccharide tripeptides containing diaminopimelic acid and does not bind to those containing lysine [16]. This is germane because the composition of different species' peptidoglycan differs and thus only disaccharide tripeptides from certain bacteria stimulate B. subtilis to germinate [16]. Finally, natural products such as staurosporine (produced by Streptomyces spp.) were shown to control germination even in the absence of a peptidoglycan cue by directly interacting with this receptor kinase, hinting at potentially relevant interspecies signaling [16].
The predator Myxococcus xanthus responds to prey signals by altering both its chemotactic and developmental patterns. Groups of M. xanthus cells form ripples as a predatory behavior in response to direct contact with B. subtilis, Escherichia coli, or Saccharomyces cerevisiae prey [17]. Peptidoglycan and some of its components were previously known as rippling signals [18], but other cell components such as DNA and insoluble cell fractions from organisms that do not contain peptidoglycan also stimulate rippling behavior [17]. These macromolecules only stimulated rippling upon physical contact with M. xanthus, and their breakdown into their component parts diminished their activity [17]. The availability of prey was also shown to spatially direct M. xanthus' development into fruiting bodies in a manner that is not simply due to changes in nutrient levels [19]. Thus, the presence of prey governs both chemotaxis and development in this organism.
3. Secondary Metabolite Production
Finally, the peptidoglycan component N-acetylglucosamine blocks development and antibiotic production of Streptomyces coelicolor on nutrient-rich media through the transcriptional regulator DasR, while both processes are stimulated on nutrient-poor media [20]. An intriguing aspect of this work is that transcription stimulation of an otherwise cryptic biosynthetic gene cluster was observed [20].
B. Antibiotics
1. At Subinhibitory Concentrations
Antibiotics are likely the best known bacterial secondary metabolites. Now there is a growing appreciation that they can act as signaling molecules in their own right and mediate outcomes other than death [21]. Unfortunately, many recent papers exploring this hypothesis have used derivatives of natural antibiotics or synthetic ones, making it difficult to argue that the observed effects are representative of natural microbial interactions. That said, subinhibitory levels of various antibiotics were seen to variously upregulate expression of SOS-response and methyl-mismatch repair genes [22], decrease biofilm mass [23], and alter virulence factor expression in different bacteria [24,25]. It is challenging to consolidate these disparate results into a straightforward model of how antibiotics at subinhibitory concentrations affect bacteria, but they encourage us to consider the potentially pleiotropic and complicated responses of bacteria to small molecule secondary metabolites.
2. Altering Morphology
Phenazines—redox-active, pigmented antibiotics—have recently been shown to have a role controlling colony morphology in P. aeruginosa, and a similar phenotype was seen in S. coelicolor in response to its own pigmented antibiotics [26]. This effect was mediated in both organisms through a related transcriptional regulator, SoxR, and demonstrates how secondary metabolites can alter development in unexpected ways. In the future, it will be exciting to determine whether there is any crosstalk between these different signaling systems in these organisms.
C. Signaling System Crosstalk
1. Autoinducer-2
The well-characterized quorum-sensing molecule autoinducer-2 (AI-2) dictates the ability of the oral microbes Actinomyces naeslundii and Streptococcus oralis to form dual-species biofilms in flowing saliva under experimental conditions analogous to their natural setting [27]. Only very low concentrations of AI-2 were optimal for biofilm formation, reminding us of the precision with which organisms sense signals and that in the natural environment the concentrations of small molecules mediating interactions may be well below those observed in typical laboratory growth conditions [27].
2. Fatty Alcohols and Acids
Exploration of related signaling systems in different organisms has revealed biologically-relevant crosstalk (Fig. 2). In particular, intraspecies small molecules signals may modulate the microbial development of different species. The sesquiterpene farnesol is an intraspecies signal for C. albicans that inhibits filamentation [28] and the diffusible signal factor (DSF) from Xanthomonas campestris—cis-11-methyl-2-dodecenoic acid—controls its biofilm formation and virulence capacities [29-31]. DSF can act as an interspecies signal as well, mimicking the effect of farnesol on C. albicans [32]. A novel signaling molecule from Burkholderia cenocepacia, BDSF—cis-2-dodecenoic acid—has been identified that restores biofilm production in X. campestris DSF-deficient mutants and inhibits C. albicans hyphal growth either as a pure compound or in co-culture [33]. Thus BDSF functions similarly to DSF with regards to X. campestris, and similarly to both DSF and farnesol by inhibiting hyphae formation in C. albicans; however, farnesol does not appear to be effective at stimulating DSF-controlled genes in X. campestris [32].
Figure 2. Schematic illustrating signal cross-talk between organisms.
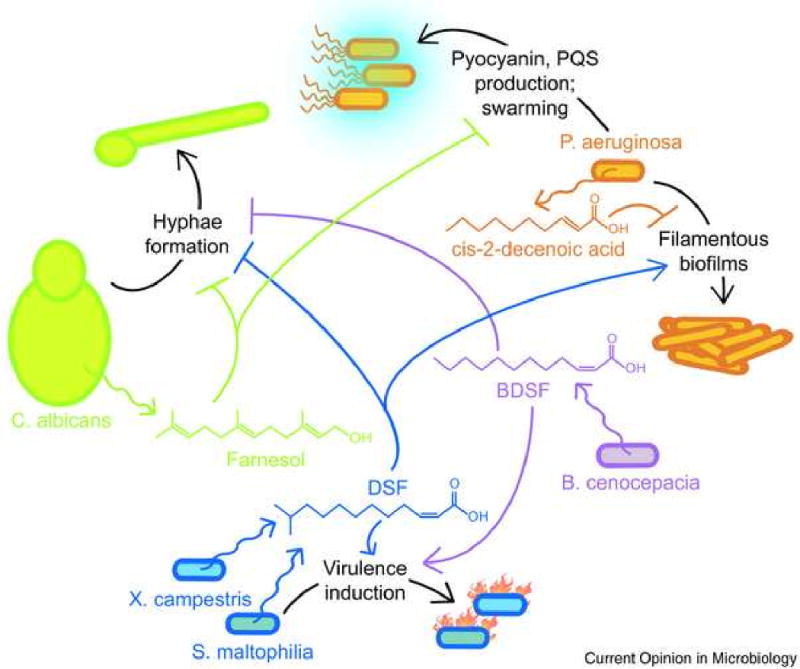
Some fatty acids and alcohols produced by fungi and bacteria have roles as interspecies signals. The wavy arrows indicate the production of small molecules by particular microbes; if known, the effects of these compounds on the behavior of other organisms is indicated by smooth arrow- or bar-headed lines. See text in section IIC for more details.
Farnesol does affect P. aeruginosa, however [34]. Farnesol alters the activity of a transcriptional regulator in P. aeruginosa when applied as a purified compound or during C. albicans co-culture, decreasing production of the secondary metabolites pyocyanin and PQS [34] and decreasing swarming [35]. Considering the ability of farnesol and DSF to mediate crosstalk between fungi and bacteria, one wonders whether DSF also has an effect on P. aeruginosa [34]. DSF from Stenotrophomonas maltophilia [36] indeed affects P. aeruginosa, stimulating the development of filamentous biofilms and increasing its resistance to the antibiotic polymyxin [37]. The sensor kinase in P. aeruginosa responsible for mediating this response was identified, and has homologs in many pseudomonads, suggesting that interspecies communication between the pseudomonads and xanthomonads may be common [37]. The sensor kinase receptor was also shown to be specific to DSF, not responding to either dodecanoic acid or farnesoic acid [37]. Thus, although both DSF and farnesol affect C. albicans in similar ways, they may instead function in parallel signaling pathways in P. aeruginosa. Indeed, this is one of the more intriguing aspects of interspecies signaling crosstalk—how related signals are recognized by different organisms, and whether their downstream signaling systems are analogous or different. DSF triggering different downstream effects in two related Xanthomonas spp. [38] and homologous two-component systems in X. campestris and Xylella fastidiosa resulting in different virulence regulation [39] are examples that illustrate this diversity of effects.
Finally, exciting work shows that P. aeruginosa produces a newly identified fatty acid, cis-2-decenoic acid, that is structurally related to DSF and BDSF (Fig. 2) [40]. Cis-2-decenoic acid is capable of completely dispersing biofilms from a diverse range of microorganisms (not only P. aeruginosa, but also E. coli, Streptococcus pyogenes, B. subtilis, and C. albicans among others) as well as preventing the biofilms from forming initially [40]. Although the structural similarity of these two compounds leads one to speculate that DSF is hijacking the native cis-2-decenoic acid receptor, the disparate resulting phenotypes observed suggest potentially distinct receptors with different downstream effects. These findings demonstrate that crosstalk between related signaling systems can occur (Fig. 2), and imply that we have only begun to understand how related signals and receptors are involved in interspecies communication.
III. Identifying New Compounds Using Co-culture
A. Eliciting Directly
Many bacteria have the genetic capacity to produce numerous and chemically diverse secondary metabolites but do not do so under common laboratory conditions [41]. Provoking production of such compounds is a particularly exciting potential outcome of interspecies interactions. Two recently published studies describe the stimulation of new compound production in co-cultures of marine bacteria and fungi, in striking examples that biological interactions can elicit the production of novel compounds [42,43]. The mechanisms of induction are still unclear, although in one case it is mediated by cell-cell contact [42].
The induction of a novel aminoglycoside with a new ring structure resulted from the competitive co-culture of Rhodococcus fascians with Streptomyces padanus [44]. Surprisingly, this phenomenon appears to be the result not of a diffusible small molecule, but due to the horizontal gene transfer of DNA from the actinomycete [44]. This unexpected result underscores the possible novel interaction mechanisms and phenotypic consequences that might result from interspecies interactions in the wild.
B. Using Development as Readout
Antibiosis is certainly not the only consequence of interspecies interactions, although it is often the easiest to observe. However, by looking for other types of responses in co-culture (Fig. 1) it is possible to identify small molecule signals, some of which may be structurally and biologically interesting.
This approach was validated by characterizing the development and secondary metabolite production of B. subtilis and S. coelicolor in co-culture. When grown together, S. coelicolor's development of aerial hyphae and sporulation were inhibited; a similar effect was observed between a range of Bacillus and Streptomycete spp. [45]. The active compound modulating these processes was the secondary metabolite surfactin, produced by B. subtilis. While this compound was previously known, this result is noteworthy because it clearly demonstrated that developmental phenotypes observed in co-culture can lead to the identification of the small molecules mediating the effect and revealed a new biological role for this secondary metabolite [45].
Having characterized the co-culture phenotype of these two organisms, a B. subtilis transposon mutant library was used to identify developmental deviations [46]. In this way the compound bacillaene, produced by a gene cluster previously believed to be cryptic in B. subtilis, was shown to delay the production of a pigmented antibiotic in S. coelicolor and also inhibit the growth of S. avermitilis [46,47]. Thus, by examining the interaction of two species and observing alterations in secondary metabolite production, a cryptic signaling molecule with a new biological role was identified.
IV. Missing Pieces…
A number of studies have observed co-culture phenotypes mediated by unknown signals, while others have identified new metabolites that could act as signals. These tantalizing studies point to potential interactions that may provide a starting point for future work.
A. Growth Enhancement
It has been suggested that one reason environmental bacteria do not grow under laboratory conditions is because of a lack of signals from their microbial neighbors [48], an idea supported by the finding that a higher diversity of organisms were obtained by incubating sediment samples in quasi-natural environments than with traditional laboratory culturing techniques [49]. The signals involved in facilitating this growth are undefined, however, and may well be non-species-specific.
B. Biofilm Formation
Biofilms are relevant to many human diseases and natural biofilms are frequently composed of multiple species. These features have inspired interest in interspecies interactions in biofilm formation. Many microbes form better biofilms in combination with other organisms or in the presence of their partners' diffusible compounds, as has been demonstrated for both the oral microbes Fusobacterium nucleatum with either Staphylococcus epidermidis or Porphyromonas gingivalis [50] as well as S. cerevisiae and Lactobacillus casei [51]. The results were less clear-cut in work that examined biofilm formation of dual-species mixes of bacteria isolated from drinking water, in which some co-cultures exhibited enhancement and others antagonism via unknown mechanisms [52]. In work that lays the groundwork for studying a naturally-occurring, simple gut symbiosis, the spatial arrangement, dynamics, and synergistic interactions of the two organisms found in the medicinal leech gut—a Rikenella and Aeromonas sp.—were determined, and may provide insight into other more complex digestive-tract communities [53].
Many naturally-produced compounds have been shown to affect biofilm development in individual microbes, but have not been shown to exert their effect when produced by a signaling partner in a co-culture context. The polyamine norspermine and glycine betaine both increase biofilm cell density in Vibrio cholerae and may be produced by organisms in this bacterium's natural marine environment [54,55]. P. aeruginosa had an altered biofilm morphology when grown on mucin surfaces compared to other polymeric substrates, a phenotype particularly prominent on mucin from human respiratory tracts [56]. Meanwhile, nitric oxide, a known signal for microbial and eukaryotic cells, was shown to disperse cells from P. aeruginosa biofilms, although it is unclear which organism may exert such an influence in natural environments [57].
C. New Small Molecules
A thought-provoking paper describes a new intraspecies communication molecule in Rhodopseudomonas palustris, p-coumaryl-homoserine lactone, produced by a LuxI homolog [58]. This molecule is related to the well-known acyl-homoserine lactones but is produced from p-coumaric acid rather than a fatty acid. Interestingly, the p-coumaric acid must be obtained from the environment or an as-yet-unidentified plant partner [58]. This result raises the exciting possibility that other bacterial LuxI homologs produce—not canonical acyl homoserine lactones—but potentially a whole range of new signaling molecules [58]. It remains to be seen whether particular p-coumarate-producing partners have a specific interaction with R. palustris.
Conclusions
The last years have brought an explosion of work investigating many aspects of interspecies interactions, and revealed myriad new signal molecules, communicating partners, and phenotypic responses. In most cases we still have much to learn about some aspect of these systems, chiefly how these signals are detected and transduced within the responding cell. This area deserves attention, particularly in those systems in which crosstalk between organisms seems likely, and considering the indications that many intraspecies signaling molecules may also have roles affecting other microbes. A number of exciting new intraspecies signal systems have been uncovered in the last few years [59-62]; an exciting possibility is that these small molecule signals may also modulate the physiology of other microbes in as-yet-undiscovered interspecies interactions.
Finally, the genome sequences of many bacteria have revealed a huge biosynthetic capacity for producing small molecules [41]. These gene clusters frequently encode compounds that are not produced under laboratory conditions and thus have no known chemical structure or biological role. This mysterious biosynthetic potential begs the question of what the ecological function of such compounds might be—recent work indicates the possibility that they function in interspecies interactions.
Acknowledgments
E.A.S. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship. Work related to interspecies signaling in our laboratory is supported by a grant from the National Institutes of Health (GM82137). We thank Erin Gontang for useful suggestions and comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 2.Ryan RP, Dow JM. Diffusible signals and interspecies communication in bacteria. Microbiology. 2008;154:1845–1858. doi: 10.1099/mic.0.2008/017871-0. [DOI] [PubMed] [Google Scholar]
- 3.Currie CR. A community of ants, fungi, and bacteria: a multilateral approach to studying symbiosis. Annu Rev Microbiol. 2001;55:357–380. doi: 10.1146/annurev.micro.55.1.357. [DOI] [PubMed] [Google Scholar]
- 4.Gerardo NM, Jacobs SR, Currie CR, Mueller UG. Ancient host-pathogen associations maintained by specificity of chemotaxis and antibiosis. PLoS Biol. 2006;4:e235. doi: 10.1371/journal.pbio.0040235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott JJ, Oh DC, Yuceer MC, Klepzig KD, Clardy J, Currie CR. Bacterial protection of beetle-fungus mutualism. Science. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• By studying the interplay between a beetle and its microbial partners, this group discovered a new secondary metabolite: mycangimycin. This exciting work shows both how small molecules are involved in maintaining specific associations between symbionts and their hosts, as well as how investigating these interactions can lead to the identification of novel small molecules.
- 6.Broderick NA, Raffa KF, Handelsman J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc Natl Acad Sci U S A. 2006;103:15196–15199. doi: 10.1073/pnas.0604865103. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This interesting work shows that the B. cereus insecticidal activity is reliant on the midgut microbiota of the insect, demonstrating how unexpected bacterial interactions may be necessary for activties long-thought to be understood.
- 7.Minerdi D, Moretti M, Gilardi G, Barberio C, Gullino ML, Garibaldi A. Bacterial ectosymbionts and virulence silencing in a Fusarium oxysporum strain. Environ Microbiol. 2008;10:1725–1741. doi: 10.1111/j.1462-2920.2008.01594.x. [DOI] [PubMed] [Google Scholar]
- 8.Minerdi D, Bossi S, Gullino ML, Garibaldi A. Volatile organic compounds: a potential direct long-distance mechanism for antagonistic action of Fusarium oxysporum strain MSA 35. Environ Microbiol. 2008 doi: 10.1111/j.1462-2920.2008.01805.x. Early view. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman LR, Deziel E, D'Argenio DA, Lepine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2006;103:19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. 2005;187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong H, Chen W, Merritt J, Qi F, Shi W, Dong X. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol Microbiol. 2007;63:872–880. doi: 10.1111/j.1365-2958.2006.05546.x. [DOI] [PubMed] [Google Scholar]
- 12.Tong H, Chen W, Shi W, Qi F, Dong X. SO-LAAO, a novel L-amino acid oxidase that enables Streptococcus oligofermentans to outcompete Streptococcus mutans by generating H2O2 from peptone. J Bacteriol. 2008;190:4716–4721. doi: 10.1128/JB.00363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cloud-Hansen KA, Peterson SB, Stabb EV, Goldman WE, McFall-Ngai MJ, Handelsman J. Breaching the great wall: peptidoglycan and microbial interactions. Nat Rev Microbiol. 2006;4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- 14.Peterson SB, Dunn AK, Klimowicz AK, Handelsman J. Peptidoglycan from Bacillus cereus mediates commensalism with rhizosphere bacteria from the Cytophaga-Flavobacterium group. Appl Environ Microbiol. 2006;72:5421–5427. doi: 10.1128/AEM.02928-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu XL, Lee RT, Fang HM, Wang YM, Li R, Zou H, Zhu Y, Wang Y. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe. 2008;4:28–39. doi: 10.1016/j.chom.2008.05.014. [DOI] [PubMed] [Google Scholar]; •• Bacterial muramyl dipeptides were shown to enter fungal cells, bind to the adenylate cyclase Cyr1p, and cause cAMP production, explaining the powerful ability of human serum to induce hyphae in C. albicans. This study beautifully identified a signal compound and its mechanism of detection and signal transduction.
- 16.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Disaccharide tripeptides stimulated germination of B. subtilis, but only when they contained a particular diaminopimelic acid moiety, and thus only peptidogylcan from particular organisms was active. The serine/threonine membrane kinase PrkC was identified as the receptor transducing this peptidoglycan-specific germination signal.
- 17.Berleman JE, Chumley T, Cheung P, Kirby JR. Rippling is a predatory behavior in Myxococcus xanthus. J Bacteriol. 2006;188:5888–5895. doi: 10.1128/JB.00559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimkets LJ, Kaiser D. Induction of coordinated movement of Myxococcus xanthus cells. J Bacteriol. 1982;152:451–461. doi: 10.1128/jb.152.1.451-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berleman JE, Kirby JR. Multicellular development in Myxococcus xanthus is stimulated by predator-prey interactions. J Bacteriol. 2007;189:5675–5682. doi: 10.1128/JB.00544-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper shows that prey availability—not exclusively nutrient levels—governs the spatial structure of fruiting body formation in M xanthus
- 20.Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fajardo A, Martinez JL. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008;11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Mesak LR, Miao V, Davies J. Effects of subinhibitory concentrations of antibiotics on SOS and DNA repair gene expression in Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:3394–3397. doi: 10.1128/AAC.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starner TD, Shrout JD, Parsek MR, Appelbaum PC, Kim G. Subinhibitory concentrations of azithromycin decrease nontypeable Haemophilus influenzae biofilm formation and diminish established biofilms. Antimicrob Agents Chemother. 2008;52:137–145. doi: 10.1128/AAC.00607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skindersoe ME, Alhede M, Phipps R, Yang L, Jensen PO, Rasmussen TB, Bjarnsholt T, Tolker-Nielsen T, Hoiby N, Givskov M. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52:3648–3663. doi: 10.1128/AAC.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linares JF, Gustafsson I, Baquero F, Martinez JL. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A. 2006;103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietrich LE, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rickard AH, Palmer RJ, Jr, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, Bassler BL, Kolenbrander PE. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60:1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- 28.Hogan DA. Talking to themselves: autoregulation and quorum sensing in fungi. Eukaryot Cell. 2006;5:613–619. doi: 10.1128/EC.5.4.613-619.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torres PS, Malamud F, Rigano LA, Russo DM, Marano MR, Castagnaro AP, Zorreguieta A, Bouarab K, Dow JM, Vojnov AA. Controlled synthesis of the DSF cell-cell signal is required for biofilm formation and virulence in Xanthomonas campestris. Environ Microbiol. 2007;9:2101–2109. doi: 10.1111/j.1462-2920.2007.01332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dow JM, Crossman L, Findlay K, He YQ, Feng JX, Tang JL. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci U S A. 2003;100:10995–11000. doi: 10.1073/pnas.1833360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He YW, Zhang LH. Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol Rev. 2008;32:842–857. doi: 10.1111/j.1574-6976.2008.00120.x. [DOI] [PubMed] [Google Scholar]
- 32.Wang LH, He Y, Gao Y, Wu JE, Dong YH, He C, Wang SX, Weng LX, Xu JL, Tay L, et al. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol. 2004;51:903–912. doi: 10.1046/j.1365-2958.2003.03883.x. [DOI] [PubMed] [Google Scholar]
- 33.Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. Isme J. 2008;2:27–36. doi: 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]; • This paper identified a new B. cenocepacia signaling compound and demonstrated it could complement DSF-deficient X. campestris as well as inhibit C. albicans filamentation, supporting its potential role as an interspecies signal.
- 34.Cugini C, Calfee MW, Farrow JM, 3rd, Morales DK, Pesci EC, Hogan DA. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:896–906. doi: 10.1111/j.1365-2958.2007.05840.x. [DOI] [PubMed] [Google Scholar]
- 35.McAlester G, O'Gara F, Morrissey JP. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J Med Microbiol. 2008;57:563–569. doi: 10.1099/jmm.0.47705-0. [DOI] [PubMed] [Google Scholar]
- 36.Fouhy Y, Scanlon K, Schouest K, Spillane C, Crossman L, Avison MB, Ryan RP, Dow JM. Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J Bacteriol. 2007;189:4964–4968. doi: 10.1128/JB.00310-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, Tolker-Nielsen T, Dow JM. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol. 2008;68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee S, Almeida RP, Lindow S. Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu Rev Phytopathol. 2008;46:243–271. doi: 10.1146/annurev.phyto.45.062806.094342. [DOI] [PubMed] [Google Scholar]
- 39.Dow M. Diversification of the function of cell-to-cell signaling in regulation of virulence within plant pathogenic xanthomonads. Sci Signal. 2008;1:pe23. doi: 10.1126/stke.121pe23. [DOI] [PubMed] [Google Scholar]
- 40.Davies DG, Marques CN. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol. 2008 doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gross H. Strategies to unravel the function of orphan biosynthesis pathways: recent examples and future prospects. Appl Microbiol Biotechnol. 2007;75:267–277. doi: 10.1007/s00253-007-0900-5. [DOI] [PubMed] [Google Scholar]
- 42.Oh DC, Jensen PR, Kauffman CA, Fenical W. Libertellenones A-D: induction of cytotoxic diterpenoid biosynthesis by marine microbial competition. Bioorg Med Chem. 2005;13:5267–5273. doi: 10.1016/j.bmc.2005.05.068. [DOI] [PubMed] [Google Scholar]
- 43.Oh DC, Kauffman CA, Jensen PR, Fenical W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp in competing co-culture. J Nat Prod. 2007;70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 44.Kurosawa K, Ghiviriga I, Sambandan TG, Lessard PA, Barbara JE, Rha C, Sinskey AJ. Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians. J Am Chem Soc. 2008;130:1126–1127. doi: 10.1021/ja077821p. [DOI] [PubMed] [Google Scholar]
- 45.Straight PD, Willey JM, Kolter R. Interactions between Streptomyces coelicolor and Bacillus subtilis: Role of surfactants in raising aerial structures. J Bacteriol. 2006;188:4918–4925. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Straight PD, Fischbach MA, Walsh CT, Rudner DZ, Kolter R. A singular enzymatic megacomplex from Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:305–310. doi: 10.1073/pnas.0609073103. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Bacillaene produced by B. subtilis affected the development of S. coelicolor, inhibiting its production of a pigmented antibiotic. The enzymes required to produce this secondary metabolite are shown to associate into a large megacomplex. This work is notable because it demonstrates the ability of using developmental phenotypes to identify new small molecules.
- 47.Butcher RA, Schroeder FC, Fischbach MA, Straight PD, Kolter R, Walsh CT, Clardy J. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 49.Bollmann A, Lewis K, Epstein SS. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl Environ Microbiol. 2007;73:6386–6390. doi: 10.1128/AEM.01309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito Y, Fujii R, Nakagawa KI, Kuramitsu HK, Okuda K, Ishihara K. Stimulation of Fusobacterium nucleatum biofilm formation by Porphyromonas gingivalis. Oral Microbiol Immunol. 2008;23:1–6. doi: 10.1111/j.1399-302X.2007.00380.x. [DOI] [PubMed] [Google Scholar]
- 51.Kawarai T, Furukawa S, Ogihara H, Yamasaki M. Mixed-species biofilm formation by lactic acid bacteria and rice wine yeasts. Appl Environ Microbiol. 2007;73:4673–4676. doi: 10.1128/AEM.02891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simoes LC, Simoes M, Vieira MJ. Biofilm interactions between distinct bacterial genera isolated from drinking water. Appl Environ Microbiol. 2007;73:6192–6200. doi: 10.1128/AEM.00837-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kikuchi Y, Graf J. Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl Environ Microbiol. 2007;73:1984–1991. doi: 10.1128/AEM.01833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karatan E, Duncan TR, Watnick PI. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J Bacteriol. 2005;187:7434–7443. doi: 10.1128/JB.187.21.7434-7443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapfhammer D, Karatan E, Pflughoeft KJ, Watnick PI. Role for glycine betaine transport in Vibrio cholerae osmoadaptation and biofilm formation within microbial communities. Appl Environ Microbiol. 2005;71:3840–3847. doi: 10.1128/AEM.71.7.3840-3847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landry RM, An D, Hupp JT, Singh PK, Parsek MR. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol Microbiol. 2006;59:142–151. doi: 10.1111/j.1365-2958.2005.04941.x. [DOI] [PubMed] [Google Scholar]
- 57.Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, et al. A new class of homoserine lactone quorum-sensing signals. Nature. 2008;454:595–599. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]; •• This paper demonstrates that a luxI homolog in R. palustris produces a novel signaling compound using a substrate moiety obtained from an as-yet-unidentified partner, possibly a plant.
- 59.Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science. 2007;318:652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- 60.Ibrahim M, Guillot A, Wessner F, Algaron F, Besset C, Courtin P, Gardan R, Monnet V. Control of the transcription of a short gene encoding a cyclic peptide in Streptococcus thermophilus: a new quorum-sensing system? J Bacteriol. 2007;189:8844–8854. doi: 10.1128/JB.01057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nichols D, Lewis K, Orjala J, Mo S, Ortenberg R, O'Connor P, Zhao C, Vouros P, Kaeberlein T, Epstein SS. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl Environ Microbiol. 2008;74:4889–4897. doi: 10.1128/AEM.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen H, Fink GR. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 2006;20:1150–1161. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]


