Abstract
The objectives of this study were to perform a quantitative comparison of proteins released from cartilage explants in response to treatment with IL-1β, TNF-α, or mechanical compression injury in vitro and to interpret this release in the context of anabolic-catabolic shifts known to occur in cartilage in response to these insults in vitro and their implications in vivo. Bovine calf cartilage explants from 6–12 animals were subjected to injurious compression, TNF-α (100 ng/ml), IL-1β (10 ng/ml), or no treatment and cultured for 5 days in equal volumes of medium. The pooled medium from each of these four conditions was labeled with one of four iTRAQ labels and subjected to nano-2D-LC/MS/MS on a quadrupole time-of-flight instrument. Data were analysed by ProQuant for peptide identification and quantitation. k-means clustering and biological pathways analysis were used to identify proteins that may correlate with known cartilage phenotypic responses to such treatments. IL-1β and TNF-α treatment caused a decrease in the synthesis of collagen subunits (p < 0.05) as well as increased release of aggrecan G2 and G3 domains to the medium (p < 0.05). MMP-1, MMP-3, MMP-9, and MMP-13 were significantly increased by all treatments compared with untreated samples (p < 0.10). Increased release of proteins involved in innate immunity and immune cell recruitment were noted following IL-1β and TNF-α treatment, whereas increased release of intracellular proteins was seen most dramatically with mechanical compression injury. Proteins involved in insulin-like growth factor and TGF-β superfamily pathway modulation showed changes in pro-anabolic pathways that may represent early repair signals. At the systems level, two principal components were sufficient to describe 97% of the covariance in the data. A strong correlation was noted between the proteins released in response to IL-1β and TNF-α; in contrast, mechanical injury resulted in both similarities and unique differences in the groups of proteins released compared with cytokine treatment.
Degeneration of cartilage is a primary feature of osteoarthritis (OA)1 (1). Although OA is a disease of the entire joint, chondrocytes are thought to play a primary role in mediating cartilage destruction. Changes in the cell's biochemical, biophysical, and mechanical environment during OA drive changes in normal chondrocyte phenotype, which may further potentiate disease progression (2).
Pro-inflammatory stimuli, particularly the inflammatory cytokines, TNF-α and IL-1β, are commonly present in both rheumatoid and osteoarthritic joints and synovial fluid (3). These cytokines, through their actions on chondrocytes, synoviocytes and macrophages, can promote cartilage degeneration and stimulate local and systemic inflammation (for review see Ref. 2). The importance of these cytokines to rheumatoid arthritis has been illustrated by the effectiveness of anti-TNF-α and anti-IL-1 therapies, which have abrogated disease progression in many patients (4).
Joint injury dramatically increases the risk for developing OA. Human knee injury, such as anterior cruciate ligament (ACL) or meniscal tear, significantly increases the relative risk of developing OA, with mean relative risk or odds ratio values ranging from 3 to 20, and increasing with age and with time following initial injury (for review, see Ref. 5). Acute knee injury is normally accompanied by an increase in synovial fluid levels of MMP-3 (6) and pro-inflammatory cytokines including TNF-α and IL-1β (7), as well as increased release to the synovial fluid of cartilage COMP fragments, collagen II cross-links, and aggrecan fragments (8, 9). In Vitro models of cartilage injury have shown that injurious compression promotes cell death, predominantly by apoptosis (10), decreases matrix synthesis, decreases chondrocyte anabolic biosynthetic response to moderate dynamic compression (11), increases type II collagen degradation (12), increases protease production (13), and alters gene transcription and subsequent protein release from the tissue (14–16). Following injury, chondrocytes make initial, but mostly unfruitful, attempts at repair (17).
Recent proteomics studies have helped to identify proteins relevant to cartilage injury and OA degradation. Gel-based proteomics studies have shown the release of FGF-2 following mechanical injury and the release of MMP-3, MMP-1, ykl40, serum amyloid A (SAA), and TIMP-1 following IL-1β treatment of cartilage explants (18, 19). In addition, Hermansson et al. (20) showed increased expression of inhibin β-A and increased type II collagen synthesis (collagen II C-terminal telopeptide) in human OA cartilage. Proteomics technologies have shown that protein samples can also be compared at the peptide level by mass spectrometry using chemical isotope labels such as iTRAQ (21). The iTRAQ four-plex system is an amine-reactive succinimidyl ester coupled to an isobaric reporter tag that in collision-induced decomposition experiments releases four different isotope-labeled signature ions (m/z 114.1, 115.1, 116.1, 117.1), which then provide relative quantitation at the MS/MS level (21).
The objective of the present study was to use a quantitative mass spectrometry approach to compare proteins released to the medium after treatment of cartilage explants with TNF-α, IL-1β, or injurious mechanical compression. Such results are of importance to understanding the mechanisms by which injury to synovial joints may lead to cartilage degradation in vivo as well as the individual contributions of cartilage mechanical injury and cytokine insult to the initiation of reparative processes versus degradative pathways that may lead to the progression of OA. We used k-means clustering and focused biological pathway analysis to identify groups of released proteins associated with cell death, matrix integrity, catabolic and anabolic behavior, innate immunity and inflammatory responses, probing mechanisms relevant to cartilage damage, inflammation, and repair. Because newly synthesized and/or degraded proteins that are actively or passively released to medium reflect cell and tissue response to treatments, we hoped to understand the consequences of these treatments that may represent shifts in the anabolic-catabolic axes of cartilage.
EXPERIMENTAL PROCEDURES
Cartilage Explantation and Culture
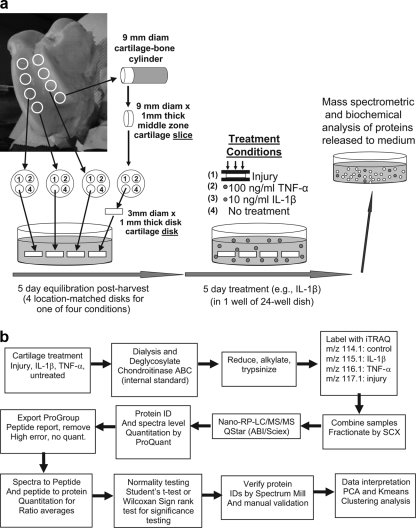
Cartilage was explanted from stifle joints of 2–3-week-old bovine calves obtained from the local abattoir (Research '87, Hopkinton, MA) as described previously (22). Briefly, 9-mm cartilage-bone cylinders were drilled from the patellofemoral groove perpendicular to the joint surface, and one or two sequential 1-mm thick cartilage slices were microtomed from the upper-middle zone (Fig. 1a). Using a dermal punch, four 3-mm diameter disks were punched from each cartilage slice yielding 4 position-matched explant disks, each 1-mm thick. Eight or 12 disks per condition were obtained from each of 6–12 animals. The explants were rested for 5 days in medium containing 1% ITS (high glucose (25 mm), Dulbecco's modified Eagle's medium supplemented with 10 mm HEPES, 0.1 mm non-essential amino acids, 115 μm ascorbic acid, 400 μm l-proline, 100 units/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B, and 1 mm sodium pyruvate).
Fig. 1.
a, schematic of cartilage harvest and explant cultures system in which location-matched cartilage disks from the femoropatellar groove are subjected to treatment with mechanical injury, IL-1β, TNF-α, or left untreated. All disks for each animal were maintained in a 24-well dish. b, schematic of experimental procedures following cartilage treatments, sample processing, iTRAQ-2D/LS/MS/MS, and data analysis.
Mechanical Injury and TNF-α and IL-1β Treatment
Strain and strain rate-controlled unconfined compression was performed in an incubator-housed loading apparatus as described previously (14, 23). Each explant was individually loaded into a polysulfone chamber and subjected to a single uniaxial, radially unconfined compression to 50% strain at a strain rate of 100%/sec (velocity = 1 mm/sec), against a non-porous platen. The compression was then immediately released using the same strain rate (velocity). This compression protocol resulted in a measured peak stress of 18.9 ± 0.3 MPa (mean ± S. E.). Injured explants were then placed in sets of four in 2.0 ml of medium as above but without 1% ITS. Separate sets of four explants were treated with cytokines (R&D systems) IL-1β (10 ng/ml) or TNF-α (100 ng/ml) or no additional treatment, also in sets of four in 2.0 ml of medium as above but without 1% ITS. All explants were then cultured for 5 days with a 10% medium removal and 10% supplementation every 24 h (Fig. 1a). Medium was collected and stored at −80 °C. In each set of four explant disks, sGAG release was measured every 24 h for the duration of the 5-day treatment, and biosynthesis was assessed after the experiment on day 6 (data not shown) to confirm that no obvious outliers existed that might skew the data from approaching a true mean, given the sample size of between 6–12 animals with 8 or 12 position-matched explants per animal. The 5-day treatment was chosen because the total measured sGAG release to the medium had begun to plateau by that time, in a manner similar to that observed previously after mechanical injury (24) or cytokine treatment (25) using the same cartilage explant system. No outliers were identified and the coefficient of variation for sGAG loss on day 5 averaged 25% and ranged from 13 to 40% for the groups with the highest variance found after IL-1β treatment. The effects of injury and cytokine treatment on 35S-sulfate and 3H-proline incorporation (as measures of proteoglycan and protein synthesis respectively) were also similar to those described previously (11, 26, 27), and the coefficient of variation ranged from 17–35% and 15–25%.
Sample Preparation and Trypsinization
See Fig. 1b for complete experimental design. Pooled medium (3 ml) from each treatment group, obtained from 6–12 animals total (sample set 1 and sample set 2), was supplemented with 5 mm EDTA, 100 μg/ml phenylmethanesulphonyl fluoride, 5 mg/ml iodoacetamide and dialyzed in a 7.5 kDa cutoff membrane for 3 h at room temperature against buffer containing 10 mm Tris acetate, pH 8.0, 40 mm NaCl, and 5 mm EDTA. Chondroitinase ABC (Seikagaku, Japan) was added (70 milliunit/sample), and samples were dialyzed overnight at 37 °C to remove chondroitin sulfate. The chondroitinase ABC also served as an ideal protein internal standard for this experiment because it is a non-mammalian protein, and there is no risk that there may be an underlying inadvertent expression of this protein in one or more of the samples. The samples were then dialyzed against 2 mm and then 0.5 mm triethylammonium bicarbonate (TEAB) for 8–12 h each at room temperature. The samples were frozen, concentrated 20-fold, and subjected to protein estimation by micro BCA protein assay kit (Pierce). The samples were then alklyated by sequential addition of 2 mm TCEP and 5 mm iodoacetamide in 100 mm TEAB, precipitated by addition of six volumes of acetone, placed at −20 °C overnight, pelleted by centrifugation at 15,000 × g for 30 min at 4 °C, and finally resuspended in 25 μl of 50 mm TEAB containing 0.1% SDS. Trypsin (0.1 μg/μl in 50 mm TEAB; Promega) was added to each sample at a ratio of 1:37.5, trypsin to protein. To verify complete trypsinization, sample set 2 was subjected to a second trypsinization at 1:75. Sample volume was taken to 40 μl by addition of water and acetonitrile (to 10%) and then incubated at 37 °C overnight and dried.
iTRAQ Labeling
iTRAQ labeling was performed according to manufacturer's instructions. Briefly, 100 μg (sample set 1) and 75 μg (sample set 2) of peptides were suspended in 500 mm TEAB. iTRAQ labels were diluted with 70 μl of ethanol and added to each sample, assigning 114.1 to untreated, 115.1 to IL-1β, 116.1 to TNF-α, 117.1 to injury. The samples were mixed and incubated at room temperature for 55 min, diluted with 200 μl of water to end the reaction, and combined based on volume equivalents of proteins (1.5 ml sample set 1 or 1.0 ml sample set 2). (Because the different conditions resulted in the release of different amounts of protein in the originally identical medium volumes containing equal volumes of cartilage, we labeled equal amounts of protein and then split it back to the original control volumes.) The combined sample mixture was dried down to ∼50 μl and combined with 1.0 ml of strong cation exchange buffer A (10 mm potassium phosphate, pH 2.8, 25% acetonitrile) and 1% phosphoric acid to pH 3.5. Samples were further diluted with 3.0 ml of strong cation exchange buffer.
Strong Cation Exchange Chromatography
The combined sample was injected via a 4.0-ml sample loop onto a 2.1 mm × 100 mm strong cation exchange column (PolyLC) at a flow rate of 250 μl/min of 100% Buffer A (10 mm phosphate, pH 2.8) on an Agilent 1100 HPLC equipped with a UV cell and micro-fraction collector. Peptides were eluted on a gradient from 0–40% buffer B (10 mm phosphate buffer pH 2.8, 400 mm KCl, 25% acetonitrile) over a 40 min period and then from 40–95% buffer B over a 10 min period, holding at 95% buffer B for 5 min before returning to the starting conditions (total run time was 80 min). One minute fractions (2-adjacent 0.5 min fractions) were collected over the peptide eluting region (λ = 214 nm). Fractions were concentrated by SpeedVac and desalted by Ziptip™ (Millipore) to obtain ∼45 fractions for LC/MS/MS analysis.
LC/MS/MS
Fractions were injected manually (Rheodyne; manual injector with 0.5-μl internal loop) onto a 75-μm inner diameter × 160 mm column with 10-μm tip (New Objective) packed in-house with Vydac protein/peptide C18 packing material (5-μm particle size, 300 Å pore size) as described previously (28). Peptides were loaded at 2% buffer B (buffer A: 1.2% acetic acid in 98.8% water; buffer B: 1.2% acetic acid in 90% acetonitrile; flow rate: 250 nl/min) for 12.5 min before elution on a 147.5-min gradient from 2% to 40% buffer B followed by a wash out for 20 min from 40% to 60% B and returning to 2% B over 15 min and equilibrating at 2% B for 60 min. The liquid chromatography was connected to a QStar, quadrupole time-of-flight mass spectrometer (Applied Biosystems Inc.) equipped with a nanospray source (29). Data were acquired through information-dependent acquisition using Analyst 1.1 selecting for precursor ions between m/z 400 and m/z 1600 with a charge state between two and four and which exceeded 15 counts. The cycle time was 10 s, and each cycle consisted of an MS scan (m/z 400–1600) followed by three data-dependent MS/MS scans (m/z 100–1600) with ions excluded for 100 s after obtaining a single spectra. Data were collected over the entire 240-min chromatographic run with peptides typically eluting between 60 and 180 min.
Data Analysis
Data were analyzed using both Analyst QS with ProQuant 1.0 and ProGroup Report (Applied Biosystems Inc.), searching against all entries in the NCBInr (Genbank) bovine database assembled in December 2005, and the Genbank database release 153 with MS error set to ± 0.25 Da; the MS/MS error was set at ± 0.20 Da. An iTRAQ database was assembled from all peptides in above databases with presumed iTRAQ-labeled N-terminal amines, iTRAQ-labeled lysines, and carbamidomethylated cysteine modifications to each of the peptides. Peptide identification confidence was set to 90 or greater. Allowed modifications included iTRAQ-labeled tyrosine, oxidized methionine, and one missed trypsin cleavage. When multiple isoforms of a protein were identified, the highest scoring isoform is typically reported. Quantitation was performed using ProQuant by calculating area under the curve of signature ion ratios released with fragmentation of the precursor ions. Corrections were made for impurities in the iTRAQ labels based on the data provided by the manufacturer. The spectra level output was assembled by peptide ProGroup report, and the data were imported into Matlab (Mathworks), where scripts were used to combine redundant spectra, calculating means and standard deviations for each peptide, and to exclude all peptides with a ratio below 7 and an error of greater than 33%, indicative of high error in quantitation. The peptide list was then combined into a protein list, calculating the means and standard deviations of all values. Protein identifications were verified by Spectrum Mill (Agilent) and by manual validation (of both identification and quantitation). Peptide quantitation was deemed sufficient if one or more signature ion peaks had an intensity of 50 or greater. Protein identification is defined as identification of least two peptides from the protein with a confidence of greater than 90% by ProQuant. These data were normalized to the mean of the chondroitinase ABC ratios added in equal amounts to serve as the internal standard. All mathematical manipulations were done in log space to maintain symmetry about zero. Histograms and Lillifors test, a modification of Komolgorov-Smirnov test, were used to test normality. Student's t test or Wilcoxan sign rank test, both with Bonferroni correction for multiple comparisons, was performed to determine whether the protein amount was different with treatment compared with without treatment (untreated control).
Clustering Analysis
The log-transformed mean signature ion ratios from the final protein list, composed of the first iTRAQ experiment (sample set 1) with supplemented proteins identified exclusively in the second experiment, were clustered using a k-means clustering algorithm (Matlab statistics toolbox). The k-means clustering algorithm partitions the proteins into k clusters (defined by user) by iteratively minimizing the summed squared Euclidean distances of each protein within the cluster to the cluster centroid over the entire k clusters. The program was allowed to iterate as many as 1,000,000 times to achieve a minimum, given a set of starting values, and we allowed 500,000 replicates beginning with k randomly chosen proteins to achieve the actual minimum for the dataset. Between 4 and 25 clusters were examined to determine both the best fit (Silhouette plots) and especially the biologically useful number of clusters with which to interpret the system. A Student's t test was performed to determine whether the clusters were significantly different from one another and thus likely to describe unique biological phenomena.
RESULTS
Data Analysis
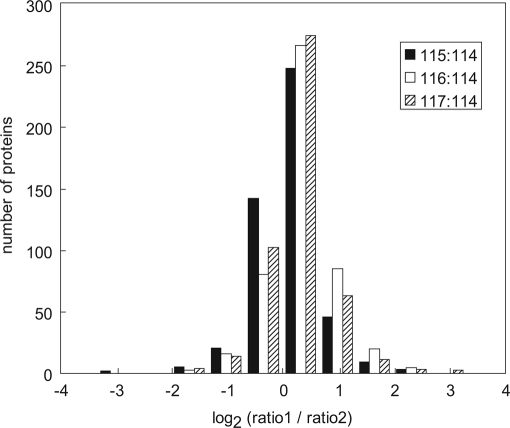
The LC/MS/MS analyses were performed on duplicate samples to provide information about the analytical variance and reproducibility of the method using an internal standard protein for correction (Fig. 2 and Supplemental Data). Two metrics were calculated to determine the data quality; error between analytical duplicates and consistency. Approximately 500 proteins released to the medium were identified and quantified by two or more peptides. The protein data from the analytical duplicates were compared by calculating the mean protein ratios (ratio1/ratio2), where “ratio1” is the geometric mean of each signature ion ratio from each protein in the first duplicate, and “ratio2” is the corresponding geometric mean from the second analytical duplicate. Overlaid histograms comparing the protein signature ion ratios prior to manual validation of both identity and quantitation are shown as log2(ratio1/ratio2) in Fig. 2. These log2(ratio1/ratio2) values were shifted slightly from zero; the overall ratio means represent a 4–19% error in quantitative agreement between the two experiments (Fig. 2). The variance around zero, represented as the standard deviation of the log transformed data, was larger than reported by Keshamouni et al. (30), and analysis indicated that both the large ratio values and poor identification/outliers contributed to the error. Therefore, to decrease error in this study, the data were subjected to manual validation and quantitation (see Supplemental Data). Finally we note a consistency (greater than 0, less than 0, or equal to 0) of greater than 90% between datasets, with most error occurring in data close to one. This metric suggests that the overall quality of the data is good, and is further improved by manual validation of protein identification and quantitation.
Fig. 2.
Histogram of errors between analytical duplicates. Overlaid histograms comparing the mean protein ratios between the first and the second analytical duplicates after internal standard correction. Data are plotted as log2 (mean protein ratio experiment 1/mean protein ratio experiment 2). The mean log2(protein ratio1/protein ratio2) mean ± S.D. were: 0.05 ± 0.46 for 115:114; 0.25 ± 0.51 for 116:114; and 0.14 ± 0.47 for 117:114.
Matrix and Synthesis in Response to Injurious Compression and Treatment with IL-1β, TNF-α
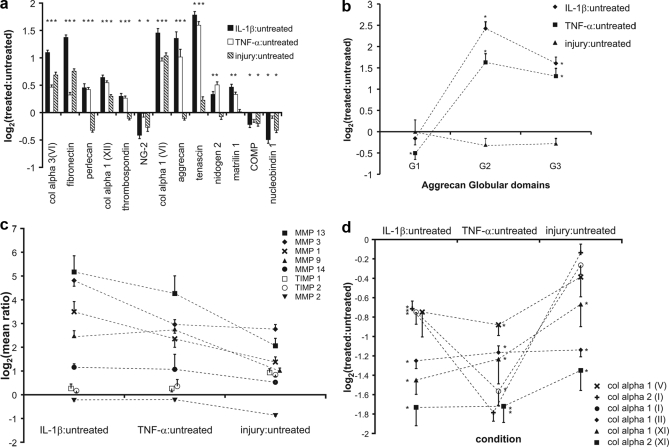
Prior experiments have suggested that loss of matrix proteins to the medium may represent new matrix synthesis, matrix degradation, or passive loss of components from the tissue. The 12 highest scoring matrix proteins released into the medium are shown in Fig. 3a, plotted as the log transformed mean ratio (± S. E.) of over 40 peptides for each protein. Proteins such as collagen VI and fibronectin showed increased release to the medium following all three treatments, whereas proteins like COMP, nucleobindin 1, and NG-2 showed a slight decrease compared with the control.
Fig. 3.
Matrix proteins and MMPs released to the medium. a, behavior of the top 12 scoring released matrix proteins. b, quantitation of released aggrecan based on peptides from each globular domain. c, MMPs and TIMPs identified in the medium with each treatment. d, released C-terminal telopeptides of fibrillar collagens, representing new fibrillar collagen synthesis. Data represented as mean ± S. E. (*, p < 0.05; Student's t test or Wilcoxan rank sum with Bonferroni correction for multiple comparisons). col alpha3(VI), collagen VI alpha 3 subunit; col alpha 1 (XII), collagen XII, alpha 1 subunit; col alpha 1 (VI), collagen VI, alpha 1 subunit; NG-2, chondroitin sulfate proteoglycan NG-2; COMP, cartilage oligomeric matrix protein.
To better characterize the anabolic-catabolic state of the tissue, we focused on two major ECM constituents, aggrecan and collagen II telopeptides, as well as metalloproteases. Aggrecan is ∼3 MDa proteoglycan consisting of a ∼300 kDa core protein containing three globular domains (N-terminal G1, C-terminal G3, and the G2 domain near G1, which defines the so-called interglobular domain spanning the G1-G2 region). The region between G2 and G3 is substituted with ∼100 chondroitin sulfate GAG chains, which are responsible for the compressive stiffness of cartilage. Cytokine treatment of cartilage is known to upregulate the production of aggrecanases ADAMTS-4 and -5, which cleave at distinct sites along the core protein including the site at aa373–374 midway within the G1-G2 domain (31). Cleavage at this site can result in the release of the entire G2-G3 region along with the substituent GAG chains. In general, aggrecanase cleavage of aggrecan in vivo and in vitro is known to cause release of various fragments that include one or more of the globular domains (32). In addition, a small percentage of newly synthesized full-length aggrecan (containing all three globular domains) can diffuse out of the tissue in vivo and in vitro. We found that the G2 and G3 domains were abundantly released to the medium in response to cytokine treatment (Fig. 3b; p < 0.001 for G2 and G3 upon both cytokine treatments); however, aggrecan G1 release was not increased and, in the case of TNF-α treatment, was actually decreased compared with the untreated sample.
Matrix remodeling and degradation in response to cytokine treatment and injurious compression has been associated with increased matrix metalloproteinase (MMP) expression and production (14, 33). We observed significant increases in release of MMP-1, MMP-3, MMP-9, and MMP-13 with all treatments (p < 0.10), whereas release of MMP-14, as well as the natural MMP inhibitors TIMP-1 and TIMP-2, were significantly increased only following mechanical injury (Fig. 3c). Only MMP-2 release was decreased significantly by mechanical injury (p < 0.05). Despite careful evaluation for peptides from aggrecanase enzymes, ADAMTS-4 and -5, only a single peptide from ADAMTS4 was identified which suggested up-regulation of the enzymes by IL-1β and TNF-α, consistent with aggrecan loss and modest elevation by injury (data not shown).
Collagen type II is a trimer of collagen alpha I (II), which contains both N-terminal and C-terminal telopeptides that are important in triple helix formation with synthesis. The N- and C-terminal telopeptides are normally cleaved post-translationally, which enables stabilization of the new formed collagen II unit. The release of these telopeptides to the medium in our experiments may represent new collagen II synthesis either as monomers or as cleavage products (34). Similarly, the other fibrillar collagens have similar telopeptides, which are released with collagen helix formation. Collagen C-terminal telopeptides released to the medium relative to untreated controls likely represented new collagen synthesis and fibril formation and were quantified for each of the fibrillar collagens detected (Fig. 3d). A decrease in all fibrillar collagen synthesis was seen in response to cytokine treatment (both IL-1β and TNF-α) and, to a lesser extent, mechanical injury (p < 0.01 for all collagen telopeptides with cytokine treatment; p < 0.05 collagen type II and collagen type XI in response to injury). Such decreases in collagen synthesis are consistent with previous reports using other methods to determine collagen production (35).
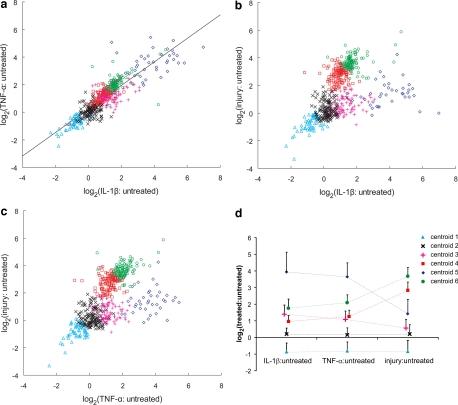
Projection Plots, Principal Component Analysis, and k-means Clustering
To gain a global understanding of the responses to the three treatment conditions, we used both projection plots of protein ratios with each treatment condition and principal component analysis (Fig. 4, a–c). Principal component analysis of the data showed that 75 and 97% of the covariance was described by one and two principal components, respectively, suggesting similarities of protein behavior among all treatments. A linear relationship is evident between the global protein response to IL-1β and TNF-α (Fig. 4a), with a Pearson product-moment correlation coefficient, R, of 0.87 (R2 = 0.7575; p < 0.0001). Plots of cytokine:untreated versus injury:untreated are also well ordered, appearing “Y”-shaped (Fig. 4, b and c). k-means clustering was used to divide the proteins into six groups based on their general proximity in space, by minimizing the sum of the protein-to-centroid distances within clusters, over all the clusters. Fig. 4d is a graph of the cluster centroid profiles represented as mean ± S. D. (p < 0.001); these profiles are color-coded to match the clusters shown in Fig. 4, a–c. Six significantly different clusters were obtained, which roughly divided each branch of the Y-shaped projections of Fig. 4, b and c into two clusters. As described below, we found that this choice helped in understanding the biological differences between those proteins that responded very strongly and those that responded more moderately to specific treatment.
Fig. 4.
Projection plots and k-means clustering profile. Two-dimensional projections of the proteins released in response to each treatment: a, log2(TNF-α: untreated) versus log2(IL-1β: untreated); b, log2(injury: untreated) versus log2(IL-1β: untreated); c, log2(injury: untreated) versus log2(TNF-α: untreated). A line was fit to the data of A with resulting slope = 0.85, an intercept of 0.22 and Pearson product moment correlation coefficient (R), 0.87 (p < 0.0001). d, the cluster profiles for each of the k = 6 clusters. A complete list of proteins divided into clusters shown is found in the Supplemental Data.
Immune Response and Cell Death
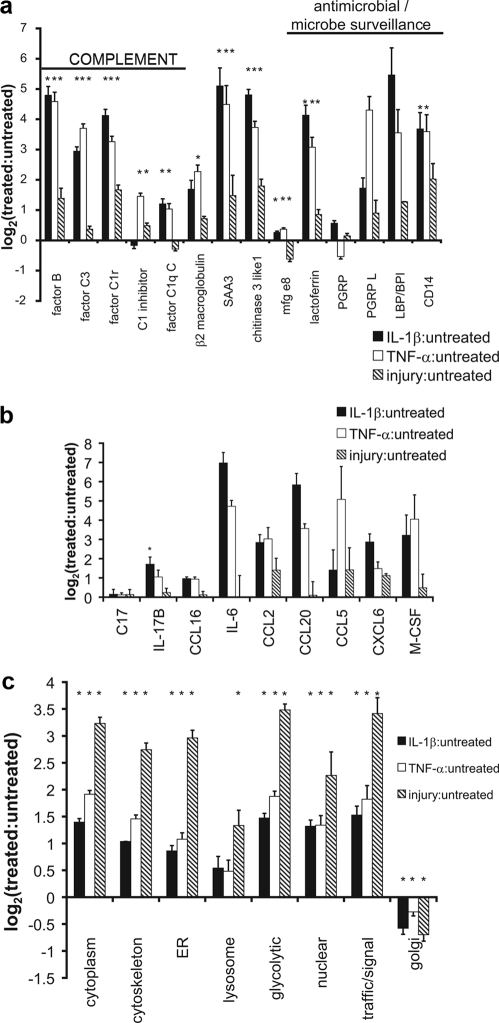
Among the most striking classes of proteins elevated in response to IL-1β, TNF-α and to a lesser extent mechanical injury, were proteins involved in innate immunity (Fig. 5, a and b), including complement, acute phase, stress response, and lipopolysaccharide (LPS)-binding proteins. Cytokines and chemokines (chemotactic cytokines) involved in modulating inflammation and recruiting immune cells were also predominantly elevated by IL-1β and TNF-α treatment (Fig. 5b). Interleukin 6 (IL-6), chemokines CCL2, CCL20, CXCL6, and macrophage colony stimulating factor (M-CSF) were all increased over 5-fold with both IL-1β and TNF-α treatments. IL-17B was significantly elevated with IL-1β treatment (p < 0.05). Although most of the changes in release of cytokines and chemokines were not statistically significant because of the low number of peptides identified, the extent of their elevation compared with the error between analytical duplicates suggests that there are real increases in protein release. In contrast to the elevation of proteins involved in immunity caused by IL-1β and TNF-α treatment, mechanical injury caused a larger increase in the release of intracellular proteins. Mechanical injury caused a ∼10-fold increase in all intracellular proteins whereas cytokine treatment caused a 2–4-fold increase (Fig. 5c). This release may be a consequence of acute lysis, necrosis, or cellular leakage following apoptosis (16).
Fig. 5.
Immunity and Cell Death. a, proteins involved in innate immunity including complements and antimicrobials, released upon treatment. b, cytokines and chemokines and their release with each treatment. c, intracellular proteins released in response to treatment separated by intracellular function or location. Data represented as mean ± S.E. (*, p < 0.05; Student's t test or Wilcoxan rank sum with Bonferroni correction for multiple comparisons). SAA3, serum amyloid A3; mfg e8, milk fat globule-EGF factor 8; PGRP, peptidoglycan recognition protein; PGRP-L, peptidoglycan recognition protein long; LBP, lipopolysaccharide-binding protein; C17, cytokine-like protein C17; IL-17B, interleukin 17B; CCL16, CC chemokine 16; IL-6, interleukin 6; CCL2, CC motif chemokine 2; CCL20, CC motif chemokine 20; CCL5, CC motif chemokine 5; CXCL6, C-X-C motif chemokine 6 (GCP-2); M-CSF, macrophage colony stimulating factor.
Proteins Involved in Growth and Differentiation Pathways
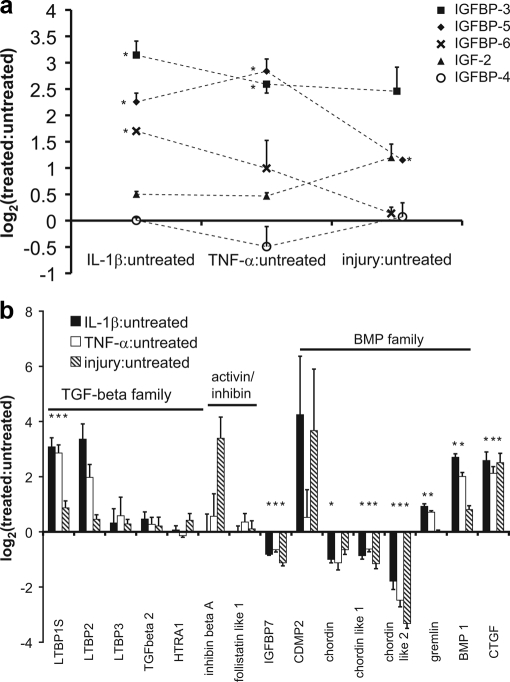
Because of the importance of both the IGF and TGF-β superfamily pathways in regulating anabolic and differentiation behavior in cartilage, we focused on the effects of cytokine and mechanical injury treatments on these pathways and their known inhibitors and modulators (Fig. 6). The TGF-β superfamily is involved in modulating chondrocyte phenotype and enhancing cartilage matrix production. Inhibin β-A was strongly elevated by mechanical injury (p ∼ 0.1), and cartilage-derived morphogenetic protein 2 (CDMP-2) was elevated by IL-1β and injury (Fig. 6a). The bone morphogenetic protein (BMP) inhibitors, chordin-like 1 and chordin-like 2, were both decreased by all treatments (p < 0.05) whereas gremlin was elevated by IL-1β and TNF-α treatment. BMP-1, a C-terminal collagen endopeptidase known to degrade chordin (36), was elevated by all treatments. Latent TGF-β-binding proteins (LTBPs), LTBP-1S (p < 0.05) and LTBP-2, were elevated only with IL-1β and TNF-α treatment. Finally, connective tissue growth factor (CTGF) was elevated with all treatments (p < 0.10) whereas serine protease (HTRA1), IGF-BP7, and inhibin β inhibitor, follistatin-like protein 1 were all unchanged with all treatments. IGF-II was elevated ∼2-fold by injury; however this was not significant because of the limited number of peptides identified. Of the four IGF-binding proteins (IGF-BPs) found in this tissue, IGF-BP3 and IGF-BP5 were both elevated by all treatments (p < 0.05 for both treatments and all proteins except IGF-BP3 with injury where p < 0.1). IGF-BP6 was also elevated with IL-1β treatment (p < 0.05), and IGF-BP4 was largely unchanged. The increased release of binding proteins may alter the tissue's ability to respond to IGF-I/II.
Fig. 6.
TGF-β superfamily and IGF family with inhibitors/modulators. a, TGF-β superfamily members and their inhibitors, released upon treatment. b, IGF family and binding proteins. Data represented as mean ± S. E. (*, p < 0.05; Student's t test or Wilcoxan rank sum with Bonferroni correction for multiple comparisons). TGF-β, transforming growth factor-β; LTBP1S, latent TGF-β-binding protein (1 short isoform); HTRA1, serine protease 11; IGFBP7, IGF-binding protein 7; CDMP, cartilage-derived morphogenetic protein 2; BMP1, bone morphogenetic protein 1; CTGF, connective tissue growth factor; IGF, insulin-like growth factor.
DISCUSSION
This study compares proteins released from free-swelling cartilage to proteins released from cartilage in response to injurious compression and to inflammatory cytokines, TNF-α and IL-1β, motivated in part by the need for better synovial fluid biomarkers and potential therapeutic targets. Proteins released from cartilage into the medium result from pre-existing proteins being actively or passively released from the extracellular matrix and from release of newly synthesized proteins, as could occur following joint injury in vivo. This study taken in conjunction with recent genomics studies (37–39) on normal and osteoarthritic tissue may help to better elucidate common proteins involved in injury as well as the progression to osteoarthritis.
Matrix Degradation and Synthesis in Response to Injurious Compression and Treatment with IL-1β and TNF-α
Release of certain ECM proteins and proteinases after mechanical injury or cytokine treatment has been well documented in previous studies (8, 9, 12, 14) and may result from increased degradation or enhanced synthesis. Interestingly, many of the released matrix proteins identified in the present study (Fig. 3a) are located primarily in the pericellular matrix (PCM), and their release may be the result of higher turnover in the PCM or increased damage to the PCM with injury. An important example of this concept is aggrecan: Quantitative autoradiography of radiolabeled explants demonstrated that aggrecan has a higher density and shorter half-life in the PCM than ECM in adult and newborn bovine cartilage explants, and that injurious compression enhanced release of fragments of newly synthesized aggrecan (i.e. radiolabeled) more rapidly from the PCM than ECM (40, 41). Previous studies have also shown rapid degradation of pre-existing cartilage tissue aggrecan via the action of aggrecanases in response to IL-1β and TNF-α treatment (25, 42), but comparably little loss of sGAG within 24 h of mechanical injury (24).
In the present study, we identified release of the specific globular domains of aggrecan, where the G2-G3 domains demark the chondroitin sulfate-GAG-bearing region, and G1 is bound to ECM-hyaluronan stabilized by link protein (43). We observed no significant loss of G1, G2, or G3 domains following injury, consistent with DiMicco et al. (24). In contrast, we found a 3–5-fold increase in the release of the G2 and G3 domains following IL-1β and TNF-α treatments, respectively (Fig. 3b; p < 0.05), characteristic of aggrecan proteolysis caused by IL-1β and TNF-α, whereas G1 was released at roughly the rate of the untreated explants (Fig. 3b). These results suggest that aggrecan G1 was retained in the tissue or released more slowly after cytokine treatment, consistent with previous reports showing accumulation of aggrecan G1 in cartilage with aging (44). Confocal microscopy studies suggested that aggrecan G1-NITEGE fragments may be internalized, a process that may be a hyaluronan-dependent event mediated by CD44 (45). Thus, aggrecan G1 possesses delayed transport compared with G2 and G3 and may be accumulated or turned over at the cell surface in response to aggrecan proteolysis.
In response to IL-1β, TNF-α and mechanical injury, chondrocytes upregulate production of MMPs, which may enhance matrix remodeling or degradation. We found a statistically significant increase in release of MMP-1, 3, 9, and 13 with cytokine and mechanical injury compared with untreated (Fig. 3c). The latter finding is consistent with the recent report of increased gene expression of MMP-1, 3, 9, and 13 by 24 h after a similar mechanical injury to bovine calf cartilage (14). Other explant and chondrocyte model systems have shown increased expression of MMP-9 and 13 expression with TNF-α treatment (46, 47), and of MMP-1, 3, 8, 9, and 13 following IL-1β treatment (33, 48–50). It is noteworthy that only mechanical injury and not cytokines, in our study, increased the release of MMP-14 (MT1-MMP) and TIMP-2, which are known to interact together to activate pro-MMP-2 (51). Although MMP-2 appeared to decrease overall, this may reflect a shift from pro to active MMP-2 or may be a sign of MMP-2 dysregulation with injury. The co-localization of the trimolecular complex of MMP-14, TIMP-2, and MMP-2 has been found in the synovium of patients with inflammatory arthritis (52). Recent work by Dean and Overall (53) to characterize new substrates for MMP-2 in fibroblasts using MMP-2 knock-outs suggest that many of the proteins identified in this study as increased in the medium are in fact substrates of MMP-2 including osteopontin, galectin 1, HSP-90α, and CTGF, all of which are shown to be elevated with injury or with all the treatments. These findings suggest that the decrease in MMP-2 with injury results in higher levels of these proteins identified in this study. Consistently, proteins that Dean and Overall (53) found decreased in the medium of fibroblasts in the absence of MMP-2 were also decreased including amyloid β A4, quiescin Q6, and NG-2. This may suggest a possible role for MMP-2 in overall regulation of cell surface-associated molecules in cartilage. Of the aggrecanases, only a single ADAMTS4 peptide was identified and the quantification of which correlated well with aggrecan loss. The difficulty identifying ADAMTS-4 and -5 is likely because of the enzymes being present at a low concentration (54), or the possibility that their association to the cells may lead to chondrocyte-controlled turnover of the enzymes (32). Taken together, the results of Fig. 3c represent a shift toward catabolic and/or remodeling behavior of the chondrocytes.
Finally, IL-1 and TNF-α are also known to decrease collagen type II and type XI synthesis (35, 46, 55), and mechanical injury has been reported to decrease collagen synthesis (11, 22). The observed decrease in release of C-terminal telopeptides of several collagen types (Fig. 3d) following both cytokine and injury treatments is also indicative of decreased collagen synthesis.
Clustering Analyses
The ∼500 released proteins were plotted in three-dimensional space based on their quantified response to each treatment, subjected to principal component analysis, and then clustered (Fig. 4d) to better understand the global similarities and differences between treatments as well as the biological interpretation of the data. To our knowledge this is the first study to compare these treatments at a global proteomics level. The linear response observed in the log2(IL-1β:untreated) versus log2(TNF-α: untreated) plot (Fig. 4a) indicates very similar protein release patterns between the two cytokines, which may reflect similarities in the activity of both cytokines. Alternatively, TNF-α and IL-1β are known to up-regulate expression of each other and of other pro-inflammatory cytokines (2), which may suggest that both conditions reflect treatments with both or the dominant of the two cytokines (likely IL-1β). The Y-shaped distribution found between either cytokine or mechanical injury (Fig. 4, b and c) indicated both similarities and differences in the response to injury versus cytokine treatment. This Y-shaped distribution indicated a population of proteins that decreased in response to all treatments; these were predominantly matrix proteins and proteins that may serve a role in normal cartilage homeostasis. The populations that increased depended on the type of treatment, cytokine, or injury.
Immune Response and Cell Death
Cartilage responds to cytokine treatment in part through the release of proteins known to play a role in the innate immune response. Complement factors B, C3, and C1r were significantly increased with all treatments, suggesting a role for alternative or lectin complement pathway (Fig. 5a). Chondrocytes have been shown to produce factor B, and synovial cells produce factors B, C2, and C3 under inflammatory conditions (20, 56). Mice deficient in factor B and complement C3 (B−/−, C3−/−) both showed an increased resistance to collagen-induced arthritis (57). CD14 and lipopolysaccharide-binding protein are both involved in the transfer of LPS to the MD2·TLR4 complex required for cell signaling (58); however, elevation of these LPS handling proteins may also inhibit LPS response (59). Peptidoglycan recognition proteins are a class of peptidoglycan interacting proteins that may serve either as antibacterial agents or control of inflammatory signals (60). Taken together, elevation of complement and LPS-binding proteins, particularly with cytokine treatment, suggests that cartilage serves as an active participant in the innate immune response against pathogens.
In addition to elevation of innate immune proteins, elevation of cytokines and chemokines (e.g. IL-6, CCL2, CCL20, CCL5, M-CSF, and CXCL6) was also noted particularly in response to IL-1β and TNF-α (Fig. 5b). IL-17B was also elevated slightly with IL-1β treatment and has been previously identified in cartilage (61). Cytokine-like protein C17 and CCL16 recently have been identified in cartilage (20, 62); however little is known of their regulation. The elevation of chemokines was also recently noted by Saas et al. (63) as well as Sandell et al. (39) in a genomics study looking at the effects of IL-1β on adult human articular cartilage including CCL2, IL-6, and CCL20. Thus, in response to IL-1β and TNF-α treatment, cartilage releases chemokines and cytokines, likely through an increase in expression at the mRNA level, which may modulate inflammatory processes and immune recruitment and may initiate tissue repair signals.
Mechanical injury caused substantial release of a large number of intracellular proteins, many by 10-fold over untreated controls, compared with 2–4-fold increases by cytokine treatment (Fig. 5c). Such release of intracellular proteins may result from mechanical disruption of the cells (lysis, necrosis) and/or loss of membrane integrity in cells undergoing apoptosis (16). Although cytokines are unlikely to cause necrosis, TNF-α in particular may induce apoptosis. Previous unpublished results indicate that treatment with 100 ng/ml TNF-α promotes apoptosis in up to 10% of the chondrocytes; elevation of apoptosis with IL-1β has not been shown. Mechanical injury in this same tissue can cause apoptosis in 20–40% of cells (10). Of note, intracellular proteins localized to the Golgi apparatus decreased in response to mechanical injury, suggesting that loss of these proteins to the medium is likely secondary to loss associated with protein trafficking, which may be increased with increased protein synthesis. Loss of intracellular proteins likely reflects cell death from a combination of apoptosis and cell lysis with passive release of intracellular proteins.
Growth and Differentiation Pathways
Although in vivo and in vitro analyses of cartilage exposed to injurious mechanical loading often show a long-term degradative phenotype, non-injurious loading may enhance matrix production (64). Little is known about the differences in signaling between injurious and non-injurious loading, which may alter long-term tissue phenotype. With respect to cytokine treatment, Arner et al. (65) showed that cartilage exposed to short term IL-1β treatment is able to recover its proteoglycan content suggesting a return of synthetic function following the treatment. Growth factors are known to be released in response to loading that may alter tissue behavior and serve as an important signal for repair — the TGF-β superfamily and IGF family of growth factors have proven important in cartilage anabolic response. The TGF-β gene superfamily and IGF pathways play a role in cartilage phenotype determination, in anabolic responses, and in modulation of inflammatory response. The TGF-β gene superfamily is composed of TGF-β 1, 2, and 3 as well as the activins/inhibins and the BMP family (66). TGF-β1–3 and inhibin/activin signal through TGF-β receptors. TGF-β1–3 is complexed with LTBPs for secretion and may be released via proteolytic cleavage by MMP-2 or MMP-9 or through interactions with thrombospondin-1 or αvβ6 integrin (67). Although LTBP-1 and -2 were substantially elevated with cytokine treatment (Fig. 6a), TGF-β2 remained nearly unchanged. The increase in LTBPs may be the result of increased latent TGF-β secretion or an increase in TGF-β turnover. Because of the importance of TGF-β in mesenchymal stem cell differentiation, wound healing, and fibrosis (68, 69), alterations in its production may represent a signal for early attempts at repair, phenotypic maintenance, or modulation of cytokine-induced degradative activity. Inhibin β-A was elevated in response to injury with a trend toward significance (p ∼ 0.1), whereas follistatin-like protein 1, an inhibitor, was unchanged (Fig. 6a). Inhibin β-Α homodimer or activin A is shown to be expressed in OA cartilage by Hermansson et al. (20). Activin A (inhibin β-A homodimer) may play a role in tissue repair as a member of the TGF-β superfamily (70) or, alternatively, a role in early inflammation and/or acute tissue injury response (71).
BMPs also play a role in chondrocyte phenotypic maintenance. Release of BMP member CDMP-2 into the medium was elevated in response to IL-1β and injury (Fig. 6a) with a concomitant decrease in BMP antagonists, gremlin, chordin, chordin-like1, and chordin-like 2. Previous studies of protein levels in human normal and OA cartilage showed that gremlin is decreased by both IL-1β and TNF-α treatment, follistatin is increased with TNF-α, and chordin is unaffected by IL-1β and TNF-α (72, 73). The differences in these reports may reflect the measuring of levels in medium rather than within tissue; alternatively, there may be differences in the ability of young versus old cartilage to respond to cytokines, which may be important in early repair signaling following injury. Overall, the increase in CDMP-2 and BMP-1, with concomitant decrease in BMP inhibitors, suggests a global increase in BMP family signaling, which can enhance growth and differentiation.
Cartilage matrix contains IGF-BPs, which regulate IGF-1 stores and activity. IGF-II was elevated slightly by mechanical injury, secondary to increased production or release of internal stores. IGF-BP3 and IGF-BP5 were elevated by both cytokines and mechanical injury compared with untreated controls (Fig. 6b), whereas IGF-BP6 was only elevated by IL-1β treatment (p < 0.05). Studies have shown that IGF-BP3, 4, and 5 are increased in human OA cartilage, and IGF-BP3 synthesis is increased in OA, suggesting a role for IGF pathway in OA (74, 75). IGF-BP3 may also enhance apoptosis in mesenchymal chondroprogenitor cells independent of IGF binding, possibly through TGF-β receptor V signaling (76, 77). IGF-β receptor V (LRP-1) was also present in the medium (Fig. 7) and appeared to be constitutively released from the cell surface regardless of treatment in this model. IGF-BP5 has also been implicated in IGF-independent functions.
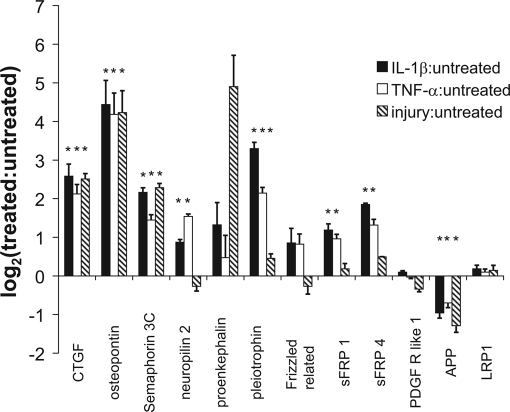
Fig. 7.
Released proteins involved in other biological pathways that may be relevant to the response of cartilage to both mechanical injury and treatment with inflammatory cytokines. Data represented as mean ± S. E. (*, p < 0.05; Student's t test or Wilcoxan rank sum with Bonferroni correction for multiple comparisons). sFRP, secreted frizzled-related proteins; PDGF R like 1, platelet-derived growth factor receptor like 1; APP, amyloid precursor protein; LRP1, low density lipoprotein receptor-related protein 1.
Release of certain proteins, including CTGF, osteopontin, semaphorin 3C, as well as proteins important in other biologically relevant pathways, were elevated by all treatments (Fig. 7). CTGF is elevated in cartilage by other factors such as BMP-2, TGF-β, and M-CSF (78, 79). Injection of CTGF into an implantable hydrogel, in a rat OA model, was found to enhance new tissue growth following experimental defect formation (80). Transfection of mouse synovial membrane cells with CTGF resulted in fibrosis (determined by collagen type I expression and matrix deposition) and an increase in TGF-β, matrix degrading enzymes, and cartilage damage (81). Similar to CTGF, semaphorin 3C was elevated 3–5-fold by all treatments (Fig. 7); its co-receptor neuropilin-2 was also identified. The expression of semaphorin 3C (sema E) in rheumatoid synovial cells has suggested that it may serve an immunosuppressive role (82). Alternatively semaphorin 3C has been shown to inhibit axon growth of sympathetic nerves, decreasing norepinephrine and thereby playing a pro-inflammatory role by enhancing TNF-α production (83). This represents the first study to identify semaphorin 3C as a product of cartilage, and we hypothesize that semaphorin 3C may play a role in either immunosuppression or early repair processes in the tissue.
Other biological pathways may also be important in understanding the response of cartilage to various types of injury. Proenkephalin, the precursor for enkephalins, was elevated by mechanical injury (Fig. 7) and has been shown to be abundantly expressed in bone and cartilage during organogenesis (84). Chondrocyte proliferation correlates well with proenkephalin gene expression (85) and modulation of cartilage growth whereas potentially antagonizing pain pathways may be an important target for ongoing therapeutics. Similarly, the wnt signaling pathway may play a role in modulating cellular activity to cytokines and mechanical compression injury given the release of different frizzled related proteins and the production of pleiotrophin particularly with cytokine treatment. Taken together, a number of important signaling proteins were noted to be elevated in response to either mechanical injury or treatment with inflammatory cytokines, and these proteins likely play a role through activation of inflammatory and/or tissue repair pathways, thus modulating the anabolic-catabolic axis of the tissue.
Limitations of This Study
Cartilage was harvested from 2–3-week-old calves, and immature cartilage was more biologically active and was still forming matrix. Thus, although this model system may not completely capture the behavior of normal adult cartilage, it is useful to note that chondrocytes in adult osteoarthritic cartilage also have higher metabolic and biosynthetic activity than normal adult cartilage. In addition, chondral lesions have been reported to be the most common injury to immature human knees (more prevalent than ACL or meniscal injuries) (86).
Because of the time- and technology-intensive nature of this study, the samples represent biologic averages from multiple animals; thus, information about the variability between animals is lacking. Therefore, it is possible that although averages may show significant differences, it is not possible to conclude whether this difference is consistent between animals and therefore represents a biologically important difference. However, given no evidence of outliers from the data by sGAG loss or by biosynthesis, it is likely that average of the data represents the behavior of the entire group.
Because of the nature of the experiment and the known up-regulation of proteases, protein proteolytic fragments were released from the tissue, and different proteolytic fragments from a single protein may possess different transport properties that govern the kinetics of release. This was shown for aggrecan and collagen by separating into globular domains (aggrecan) or telopeptides versus fibril forming regions (collagens). For collagens, peptides within the fibril forming region may be redundant compared between collagen types; therefore, we were careful about the extent of interpretation of these domains. Although there are a number of limitations to this study, the results offer the first intensive comparison between cytokine-induced versus mechanically induced cartilage damage, and the findings offer new insights into the similarities and differences in the biology that may be helpful in understanding how these two factors - inflammatory cytokine exposure and mechanical compression injury — may contribute to long term cartilage degeneration following joint injury.
In conclusion, iTRAQ labeling and 2D-LC/MS/MS analysis were performed on proteins released from cartilage that was subjected to no treatment, injurious compression, IL-1β or TNF-α, and proteins involved with injury, immunity, and possibly early repair signals were observed. Proteins released following mechanical injury included intracellular proteins associated with mechanical cell disruption or apoptosis. Proteins associated with innate immunity and immune cell recruitment were most pronounced following cytokine treatment and included complement proteins, antimicrobials, stress response proteins, LPS-binding proteins, cytokines, and chemokines. Evidence of altered anabolic pathways also suggests signs of modulation of the inflammatory signals with possible attempts at repair. TGF-β superfamily members, TGF-β2, inhibin β-Α, and CDMP-2 were identified. Inhibin β-Α was elevated by mechanical injury, whereas cytokine treatment increased LTBPs and decreased BMP antagonist, chordin-like 1 and chordin-like 2. A shift in both growth factor and growth factor inhibitor release represents a global shift of the entire TGF-β superfamily pathway toward increasing TGF-β superfamily signaling, which is typically considered a pro-anabolic signal in cartilage. A release of different IGF-binding proteins suggested a shift in the IGF pathway with the potential to increase extracellular stores of the IGFs, which may be important for repair. Alternatively, the increase in IGF-BP3 and IGF-BP5 may serve as an inhibitory mechanism for both the IGF and TGF-β pathway. Finally, we observed an increase in CTGF, semaphorin 3C, and proenkephalin, which may also play a role in modulating inflammatory signals and promoting possible repair. Despite the release of pro-growth signal in response to cytokines and to injury, the ability to respond to these signals depends on the chondrocytes health and its cell receptor repertoire. In addition, it is important to consider that these signals likely modulate the behavior of the entire joint and not simply the behavior of the cartilage itself.
The observations of protein response in this study reflect tissue damage, immunity, and pro-growth and differentiation signals that may be important for tissue repair. Cartilage possesses some ability to repair damage as long as the injury is not too severe and inflammation not too prolonged. Understanding of these pathways and their effects on cartilage may aid in identifying new therapeutic strategies to treat disease processes. Data mining to discover additional extracellular signaling pathways and proteolytic fragments may be helpful in further characterizing the effects of cytokines and injury on cartilage using this in vitro model.
Acknowledgments
We thank Dr. Jimmy Flarakos for his technical assistance with the offline 2D-LC/MS/MS experimental approach and Dr. Diana Chai for her helpful suggestions with the manuscript and data analysis.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants AR33236, AR45779, and CA26731 and by NIEHS Grant P30 ES002109 (Massachusetts Institute of Technology Centre for Environmental Health Sciences). This work was also supported by a National Defense Science and Engineering Graduate Fellowship (to A. L. S.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- OA
- osteoarthritis
- ACL
- anterior cruciate ligament
- ADAMTS4/5
- a disintegrin and metalloproteinase with thrombospondin motifs 4 and 5
- aggrecan G1
- aggrecan first globular domain
- BMP
- bone morphogenetic protein
- CCL2
- C-C chemokine 2
- M-CSF
- macrophage colony stimulating factor
- CD14
- cluster of differentiation 14
- CDMP-2
- cartilage-derived morphogenetic protein 2
- COMP
- cartilage oligomeric matrix protein
- CTGF
- connective tissue growth factor
- ECM
- extracellular matrix
- FGF-2
- fibroblast growth factor 2
- IGF
- insulin-like growth factor
- IGF-BP
- insulin-like growth factor-binding protein
- IL-1β
- interleukin-1β
- iTRAQ
- isobaric tag for relative and absolute quantitation
- ITS
- insulin transferrin and selenium supplement
- LPS
- lipopolysaccharide
- LRP-1
- low density lipoprotein-related protein 1
- LTBP-1S
- latent transforming growth factor-β-binding protein 1 short isoform
- LTBP
- latent transforming growth factor-β-binding protein
- MMP
- matrix metalloproteinase
- nano-2D-LC/MS/MS
- nano-two-dimensional liquid chromatography and tandem mass spectrometry
- PCM
- pericellular matrix
- GAG
- sulfated glycosaminoglycan
- SDS
- sodium dodecyl sulfate
- TCEP
- tris(2-carboxyethyl)phosphine hydrochloride
- TEAB
- triethylammonium bicarbonate
- TGF-β
- transforming growth factor-β
- TIMP
- tissue inhibitor of metalloproteinases
- TLR
- toll-like receptor
- TNF-α
- tumor necrosis factor-α
REFERENCES
- 1.Aigner T., McKenna L. ( 2002) Molecular pathology and pathobiology of osteoarthritic cartilage. Cell. Mol. Life Sci. 59, 5– 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldring M. B. ( 2000) The role of the chondrocyte in osteoarthritis. Arthritis Rheum. 43, 1916– 1926 [DOI] [PubMed] [Google Scholar]
- 3.Westacott C. I., Whicher J. T., Barnes I. C., Thompson D., Swan A. J., Dieppe P. A. ( 1990) Synovial fluid concentration of five different cytokines in rheumatic diseases. Ann. Rheum. Dis. 49, 676– 681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furst D. E., Breedveld F. C., Kalden J. R., Smolen J. S., Burmester G. R., Bijlsma J. W., Dougados M., Emery P., Keystone E. C., Klareskog L., Mease P. J. ( 2005) Updated consensus statement on biological agents, specifically tumor necrosis factor alpha (TNFα) blocking agents and interleukin-1 receptor antagonist (IL-1ra), for the treatment of rheumatic diseases. Ann. Rheum. Dis. 64, Suppl. 4, 2– 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roos E. M. ( 2005) Joint injury causes knee osteoarthritis in young adults. Curr. Opin. Rheumatol. 17, 195– 200 [DOI] [PubMed] [Google Scholar]
- 6.Tchetverikov I., Lohmander L. S., Verzijl N., Huizinga T. W., TeKoppele J. M., Hanemaaijer R., DeGroot J. ( 2005) MMP protein and activity levels in synovial fluid from patients with joint injury, inflammatory arthritis, and osteoarthritis. Ann. Rheum. Dis. 64, 694– 698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irie K., Uchiyama E., Iwaso H. ( 2003) Intraarticular inflammatory cytokines in acute anterior cruciate ligament injured knee. Knee 10, 93– 96 [DOI] [PubMed] [Google Scholar]
- 8.Lohmander L. S., Saxne T., Heinegård D. K. ( 1994) Release of cartilage oligomeric matrix protein (COMP) into joint fluid after knee injury and in osteoarthritis. Ann. Rheum. Dis. 53, 8– 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohmander L. S., Atley L. M., Pietka T. A., Eyre D. R. ( 2003) The release of crosslinked peptides from type II collagen into human synovial fluid is increased soon after joint injury and in osteoarthritis. Arthritis Rheum. 48, 3130– 3139 [DOI] [PubMed] [Google Scholar]
- 10.Loening A. M., James I. E., Levenston M. E., Badger A. M., Frank E. H., Kurz B., Nuttall M. E., Hung H. H., Blake S. M., Grodzinsky A. J., Lark M. W. ( 2000) Injurious mechanical compression of bovine articular cartilage induces chondrocyte apoptosis. Arch. Biochem. Biophys. 381, 205– 212 [DOI] [PubMed] [Google Scholar]
- 11.Kurz B., Jin M., Patwari P., Cheng D. M., Lark M. W., Grodzinsky A. J. ( 2001) Biosynthetic response and mechanical properties of articular cartilage after injurious compression. J. Orthop. Res. 19, 1140– 1146 [DOI] [PubMed] [Google Scholar]
- 12.Chen C. T., Burton-Wurster N., Lust G., Bank R. A., Tekoppele J. M. ( 1999) Compositional and metabolic changes in damaged cartilage are peak-stress, stress-rate, and loading-duration dependent. J. Orthop. Res. 17, 870– 879 [DOI] [PubMed] [Google Scholar]
- 13.Lin P. M., Chen C. T., Torzilli P. A. ( 2004) Increased stromelysin-1 (MMP-3), proteoglycan degradation (3B3- and 7D4) and collagen damage in cyclically load-injured articular cartilage. Osteoarthr. Cartil. 12, 485– 496 [DOI] [PubMed] [Google Scholar]
- 14.Lee J. H., Fitzgerald J. B., Dimicco M. A., Grodzinsky A. J. ( 2005) Mechanical injury of cartilage explants causes specific time-dependent changes in chondrocyte gene expression. Arthritis Rheum. 52, 2386– 2395 [DOI] [PubMed] [Google Scholar]
- 15.Vincent T. L., Hermansson M. A., Hansen U. N., Amis A. A., Saklatvala J. ( 2004) Basic fibroblast growth factor mediates transduction of mechanical signals when articular cartilage is loaded. Arthritis Rheum. 50, 526– 533 [DOI] [PubMed] [Google Scholar]
- 16.Stevens A. L., Wishnok J. S., Chai D. H., Grodzinsky A. J., Tannenbaum S. R. ( 2008) A sodium dodecyl sulfate-polyacrylamide gel electrophoresis-liquid chromatography tandem mass spectrometry analysis of bovine cartilage tissue response to mechanical compression injury and the inflammatory cytokines tumor necrosis factor alpha and interleukin-1beta. Arthritis Rheum. 58, 489– 500 [DOI] [PubMed] [Google Scholar]
- 17.Hunziker E. B. ( 1999) Articular cartilage repair: are the intrinsic biological constraints undermining this process insuperable? Osteoarthr. Cartil. 7, 15– 28 [DOI] [PubMed] [Google Scholar]
- 18.Vincent T., Hermansson M., Bolton M., Wait R., Saklatvala J.Proc. Natl. Acad. Sci. U. S. A. ( 2002) 99, 8259– 8264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruber J., Vincent T. L., Hermansson M., Bolton M., Wait R., Saklatvala J. ( 2004) Induction of interleukin-1 in articular cartilage by explantation and cutting. Arthritis Rheum. 50, 2539– 2546 [DOI] [PubMed] [Google Scholar]
- 20.Hermansson M., Sawaji Y., Bolton M., Alexander S., Wallace A., Begum S., Wait R., Saklatvala J. ( 2004) Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin β-A (activin A), a regulatory molecule for chondrocytes. J. Biol. Chem. 279, 43514– 43521 [DOI] [PubMed] [Google Scholar]
- 21.Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. ( 2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154– 1169 [DOI] [PubMed] [Google Scholar]
- 22.Sah R. L., Kim Y. J., Doong J. Y., Grodzinsky A. J., Plaas A. H., Sandy J. D. ( 1989) Biosynthetic response of cartilage explants to dynamic compression. J. Orthop. Res. 7, 619– 636 [DOI] [PubMed] [Google Scholar]
- 23.Frank E. H., Jin M., Loening A. M., Levenston M. E., Grodzinsky A. J. ( 2000) A versatile shear and compression apparatus for mechanical stimulation of tissue culture explants. J. Biomech. 33, 1523– 1527 [DOI] [PubMed] [Google Scholar]
- 24.DiMicco M. A., Patwari P., Siparsky P. N., Kumar S., Pratta M. A., Lark M. W., Kim Y. J., Grodzinsky A. J. ( 2004) Mechanisms and kinetics of glycosaminoglycan release following in vitro cartilage injury. Arthritis Rheum. 50, 840– 848 [DOI] [PubMed] [Google Scholar]
- 25.Stevens A. L., Wheeler C. A., Tannenbaum S. R., Grodzinsky A. J. ( 2008) Nitric oxide enhances aggrecan degradation by aggrecanase in response to TNF-alpha but not IL-1beta treatment at a post-transcriptional level in bovine cartilage explants. Osteoarthr. Cartil. 16, 489– 497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benton H. P., Tyler J. A. ( 1988) Inhibition of cartilage proteoglycan synthesis by interleukin I. Biochem. Biophys. Res. Commun. 154, 421– 428 [DOI] [PubMed] [Google Scholar]
- 27.Patwari P., Cook M. N., DiMicco M. A., Blake S. M., James I. E., Kumar S., Cole A. A., Lark M. W., Grodzinsky A. J. ( 2003) Proteoglycan degradation after injurious compression of bovine and human articular cartilage in vitro: interaction with exogenous cytokines. Arthritis Rheum. 48, 1292– 1301 [DOI] [PubMed] [Google Scholar]
- 28.Hsieh Y. L., Wang H., Elicone C., Mark J., Martin S. A., Regnier F. ( 1996) Automated analytical system for the examination of protein primary structure. Anal. Chem. 68, 455– 462 [DOI] [PubMed] [Google Scholar]
- 29.Bhat V. B., Choi M. H., Wishnok J. S., Tannenbaum S. R. ( 2005) Comparative plasma proteome analysis of lymphoma-bearing SJL mice. J. Proteome Res. 4, 1814– 1825 [DOI] [PubMed] [Google Scholar]
- 30.Keshamouni V. G., Michailidis G., Grasso C. S., Anthwal S., Strahler J. R., Walker A., Arenberg D. A., Reddy R. C., Akulapalli S., Thannickal V. J., Standiford T. J., Andrews P. C., Omenn G. S. ( 2006) Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J. Proteome Res. 5, 1143– 1154 [DOI] [PubMed] [Google Scholar]
- 31.Caterson B., Flannery C. R., Hughes C. E., Little C. B. ( 2000) Mechanisms involved in cartilage proteoglycan catabolism. Matrix Biol. 19, 333– 344 [DOI] [PubMed] [Google Scholar]
- 32.Patwari P., Gao G., Lee J. H., Grodzinsky A. J., Sandy J. D. ( 2005) Analysis of ADAMTS4 and MT4-MMP indicates that both are involved in aggrecanolysis in interleukin-1-treated bovine cartilage. Osteoarthr. Cartil. 13, 269– 277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koshy P. J., Lundy C. J., Rowan A. D., Porter S., Edwards D. R., Hogan A., Clark I. M., Cawston T. E. ( 2002) The modulation of matrix metalloproteinase and ADAM gene expression in human chondrocytes by interleukin-1 and oncostatin M: a time-course study using real-time quantitative reverse transcription-polymerase chain reaction. Arthritis Rheum. 46, 961– 967 [DOI] [PubMed] [Google Scholar]
- 34.Rebuck N., Croucher L. J., Hollander A. P. ( 1999) Distribution of two alternatively spliced variants of the type II collagen N-propeptide compared with the C-propeptide in bovine chondrocyte pellet cultures. J. Cell. Biochem. 75, 13– 21 [DOI] [PubMed] [Google Scholar]
- 35.Goldring M. B., Birkhead J., Sandell L. J., Kimura T., Krane S. M. ( 1988) Interleukin 1 suppresses expression of cartilage-specific types II and IX collagens and increases types I and III collagens in human chondrocytes. J. Clin. Investig. 82, 2026– 2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petropoulou V., Garrigue-Antar L., Kadler K. E. ( 2005) Identification of the minimal domain structure of bone morphogenetic protein-1 (BMP-1) for chordinase activity: chordinase activity is not enhanced by procollagen C-proteinase enhancer-1 (PCPE-1). J. Biol. Chem. 280, 22616– 22623 [DOI] [PubMed] [Google Scholar]
- 37.Aigner T., Fundel K., Saas J., Gebhard P. M., Haag J., Weiss T., Zien A., Obermayr F., Zimmer R., Bartnik E. ( 2006) Large-scale gene expression profiling reveals major pathogenetic pathways of cartilage degeneration in osteoarthritis. Arthritis Rheum. 54, 3533– 3544 [DOI] [PubMed] [Google Scholar]
- 38.Ijiri K., Zerbini L. F., Peng H., Otu H. H., Tsuchimochi K., Otero M., Dragomir C., Walsh N., Bierbaum B. E., Mattingly D., van Flandern G., Komiya S., Aigner T., Libermann T. A., Goldring M. B. ( 2008) Differential expression of GADD45beta in normal and osteoarthritic cartilage: potential role in homeostasis of articular chondrocytes. Arthritis Rheum. 58, 2075– 2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandell L. J., Xing X., Franz C., Davies S., Chang L. W., Patra D. ( 2008) Exuberant expression of chemokine genes by adult human articular chondrocytes in response to IL-1beta. Osteoarthr. Cartil. 16, 1560– 1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinn T. M., Grodzinsky A. J., Hunziker E. B., Sandy J. D. ( 1998) Effects of injurious compression on matrix turnover around individual cells in calf articular cartilage explants. J. Orthop. Res. 16, 490– 499 [DOI] [PubMed] [Google Scholar]
- 41.Quinn T. M., Maung A. A., Grodzinsky A. J., Hunziker E. B., Sandy J. D. ( 1999) Physical and biological regulation of proteoglycan turnover around chondrocytes in cartilage explants. Implications for tissue degradation and repair. Ann. N. Y. Acad. Sci. 878, 420– 441 [DOI] [PubMed] [Google Scholar]
- 42.Bonassar L. J., Sandy J. D., Lark M. W., Plaas A. H., Frank E. H., Grodzinsky A. J. ( 1997) Inhibition of cartilage degradation and changes in physical properties induced by IL-1beta and retinoic acid using matrix metalloproteinase inhibitors. Arch. Biochem. Biophys. 344, 404– 412 [DOI] [PubMed] [Google Scholar]
- 43.Dudhia J. ( 2005) Aggrecan, aging and assembly in articular cartilage. Cell. Mol. Life Sci. 62, 2241– 2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roughley P. J., White R. J., Poole A. R. ( 1985) Identification of a hyaluronic acid-binding protein that interferes with the preparation of high-buoyant-density proteoglycan aggregates from adult human articular cartilage. Biochem. J. 231, 129– 138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Embry J. J., Knudson W. ( 2003) G1 domain of aggrecan cointernalizes with hyaluronan via a CD44-mediated mechanism in bovine articular chondrocytes. Arthritis Rheum. 48, 3431– 3441 [DOI] [PubMed] [Google Scholar]
- 46.Lefebvre V., Peeters-Joris C., Vaes G. ( 1990) Modulation by interleukin 1 and tumor necrosis factor alpha of production of collagenase, tissue inhibitor of metalloproteinases and collagen types in differentiated and dedifferentiated articular chondrocytes. Biochim. Biophys. Acta 1052, 366– 378 [DOI] [PubMed] [Google Scholar]
- 47.Liacini A., Sylvester J., Li W. Q., Huang W., Dehnade F., Ahmad M., Zafarullah M. ( 2003) Induction of matrix metalloproteinase-13 gene expression by TNF-alpha is mediated by MAP kinases, AP-1, and NF-kappaB transcription factors in articular chondrocytes. Exp. Cell Res. 288, 208– 217 [DOI] [PubMed] [Google Scholar]
- 48.Hembry R. M., Bagga M. R., Dingle J. T., Thomas P. P., Reynolds J. J. ( 1994) Metalloproteinase production by rabbit articular cartilage: comparison of the effects of interleukin-1 alpha in vitro and in vivo. Virchows Arch. 425, 413– 424 [DOI] [PubMed] [Google Scholar]
- 49.Dozin B., Malpeli M., Camardella L., Cancedda R., Pietrangelo A. ( 2002) Response of young, aged and osteoarthritic human articular chondrocytes to inflammatory cytokines: molecular and cellular aspects. Matrix Biol. 21, 449– 459 [DOI] [PubMed] [Google Scholar]
- 50.Chubinskaya S., Huch K., Mikecz K., Cs-Szabo G., Hasty K. A., Kuettner K. E., Cole A. A. ( 1996) Chondrocyte matrix metalloproteinase-8: up-regulation of neutrophil collagenase by interleukin-1 beta in human cartilage from knee and ankle joints. Lab. Invest. 74, 232– 240 [PubMed] [Google Scholar]
- 51.Wang Z. M., Li X., Cocklin R. R., Wang M., Wang M., Fukase K., Inamura S., Kusumoto S., Gupta D., Dziarski R. ( 2003) Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J. Biol. Chem. 278, 49044– 49052 [DOI] [PubMed] [Google Scholar]
- 52.Goldbach-Mansky R., Lee J. M., Hoxworth J. M., Smith D., 2nd, Duray P., Schumacher R. H., Jr, Yarboro C. H., Klippel J., Kleiner D., El-Gabalawy H. S. ( 2000) Active synovial matrix metalloproteinase-2 is associated with radiographic erosions in patients with early synovitis. Arthritis Res. 2, 145– 153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dean R. A., Overall C. M. ( 2007) Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol. Cell. Proteomics 6, 611– 623 [DOI] [PubMed] [Google Scholar]
- 54.Glasson S. S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H. L., Flannery C. R., Peluso D., Kanki K., Yang Z., Majumdar M. K., Morris E. A. ( 2005) Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 434, 644– 648 [DOI] [PubMed] [Google Scholar]
- 55.Reginato A. M., Sanz-Rodriguez C., Diaz A., Dharmavaram R. M., Jimenez S. A. ( 1993) Transcriptional modulation of cartilage-specific collagen gene expression by interferon gamma and tumor necrosis factor alpha in cultured human chondrocytes. Biochem. J. 294, 761– 769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Ceulaer C., Papazoglou S., Whaley K. ( 1980) Increased biosynthesis of complement components by cultured monocytes, synovial fluid macrophages and skynovial membrane cells from patients with rheumatoid arthritis. Immunology 41, 37– 43 [PMC free article] [PubMed] [Google Scholar]
- 57.Hietala M. A., Jonsson I. M., Tarkowski A., Kleinau S., Pekna M. ( 2002) Complement deficiency ameliorates collagen-induced arthritis in mice. J. Immunol. 169, 454– 459 [DOI] [PubMed] [Google Scholar]
- 58.Triantafilou M., Triantafilou K. ( 2002) Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. 23, 301– 304 [DOI] [PubMed] [Google Scholar]
- 59.Kitchens R. L., Thompson P. A. ( 2005) Modulatory effects of sCD14 and LBP on LPS-host cell interactions. J. Endotoxin Res. 11, 225– 229 [DOI] [PubMed] [Google Scholar]
- 60.Steiner H. ( 2004) Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol. Rev. 198, 83– 96 [DOI] [PubMed] [Google Scholar]
- 61.Moseley T. A., Haudenschild D. R., Rose L., Reddi A. H. ( 2003) Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev. 14, 155– 174 [DOI] [PubMed] [Google Scholar]
- 62.Haringman J. J., Smeets T. J., Reinders-Blankert P., Tak P. P. ( 2006) Chemokine and chemokine receptor expression in paired peripheral blood mononuclear cells and synovial tissue of patients with rheumatoid arthritis, osteoarthritis, and reactive arthritis. Ann. Rheum. Dis. 65, 294– 300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saas J., Haag J., Rueger D., Chubinskaya S., Sohler F., Zimmer R., Bartnik E., Aigner T. ( 2006) IL-1beta, but not BMP-7 leads to a dramatic change in the gene expression pattern of human adult articular chondrocytes–portraying the gene expression pattern in two donors. Cytokine 36, 90– 99 [DOI] [PubMed] [Google Scholar]
- 64.Kerin A., Patwari P., Kuettner K., Cole A., Grodzinsky A. ( 2002) Molecular basis of osteoarthritis: biomechanical aspects. Cell. Mol. Life Sci. 59, 27– 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arner E. C., Di Meo T. M., Ruhl D. M., Pratta M. A.In vivo studies on the effects of human recombinant interleukin-1 beta on articular cartilage. Agents Actions ( 1989) 27, 254– 257 [DOI] [PubMed] [Google Scholar]
- 66.de Caestecker M. ( 2004) The transforming growth factor-beta superfamily of receptors. Cytokine Growth Factor Rev. 15, 1– 11 [DOI] [PubMed] [Google Scholar]
- 67.Annes J. P., Munger J. S., Rifkin D. B. ( 2003) Making sense of latent TGFβ activation. J. Cell Sci. 116, 217– 224 [DOI] [PubMed] [Google Scholar]
- 68.Leask A., Abraham D. J. ( 2004) TGF-beta signaling and the fibrotic response. FASEB J. 18, 816– 827 [DOI] [PubMed] [Google Scholar]
- 69.Roelen B. A., Dijke P. ( 2003) Controlling mesenchymal stem cell differentiation by TGFBeta family members. J. Orthop. Sci. 8, 740– 748 [DOI] [PubMed] [Google Scholar]
- 70.Luyten F. P., Chen P., Paralkar V., Reddi A. H. ( 1994) Recombinant bone morphogenetic protein-4, transforming growth factor-beta 1, and activin A enhance the cartilage phenotype of articular chondrocytes in vitro. Exp. Cell Res. 210, 224– 229 [DOI] [PubMed] [Google Scholar]
- 71.Jones K. L., de Kretser D. M., Patella S., Phillips D. J. ( 2004) Activin A and follistatin in systemic inflammation. Mol. Cell. Endocrinol. 225, 119– 125 [DOI] [PubMed] [Google Scholar]
- 72.Tardif G., Pelletier J. P., Hum D., Boileau C., Duval N., Martel-Pelletier J. ( 2006) Differential regulation of the bone morphogenic protein antagonist chordin in human normal and osteoarthritic chondrocytes. Ann. Rheum. Dis. 65, 261– 264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tardif G., Hum D., Pelletier J. P., Boileau C., Ranger P., Martel-Pelletier J. ( 2004) Differential gene expression and regulation of the bone morphogenetic protein antagonists follistatin and gremlin in normal and osteoarthritic human chondrocytes and synovial fibroblasts. Arthritis Rheum. 50, 2521– 2530 [DOI] [PubMed] [Google Scholar]
- 74.Iwanaga H., Matsumoto T., Enomoto H., Okano K., Hishikawa Y., Shindo H., Koji T. ( 2005) Enhanced expression of insulin-like growth factor-binding proteins in human osteoarthritic cartilage detected by immunohistochemistry and in situ hybridization. Osteoarthr. Cartil. 13, 439– 448 [DOI] [PubMed] [Google Scholar]
- 75.Eviatar T., Kauffman H., Maroudas A. ( 2003) Synthesis of insulin-like growth factor binding protein 3 in vitro in human articular cartilage cultures. Arthritis Rheum. 48, 410– 417 [DOI] [PubMed] [Google Scholar]
- 76.Longobardi L., Torello M., Buckway C., O'Rear L., Horton W. A., Hwa V., Roberts C. T., Jr, Chiarelli F., Rosenfeld R. G., Spagnoli A. ( 2003) A novel insulin-like growth factor (IGF)-independent role for IGF binding protein-3 in mesenchymal chondroprogenitor cell apoptosis. Endocrinology 144, 1695– 1702 [DOI] [PubMed] [Google Scholar]
- 77.Leal S. M., Liu Q., Huang S. S., Huang J. S. ( 1997) The type V transforming growth factor beta receptor is the putative insulin-like growth factor-binding protein 3 receptor. J. Biol. Chem. 272, 20572– 20576 [DOI] [PubMed] [Google Scholar]
- 78.Nakanishi T., Kimura Y., Tamura T., Ichikawa H., Yamaai Y., Sugimoto T., Takigawa M. ( 1997) Cloning of a mRNA preferentially expressed in chondrocytes by differential display-PCR from a human chondrocytic cell line that is identical with connective tissue growth factor (CTGF) mRNA. Biochem. Biophys. Res. Commun. 234, 206– 210 [DOI] [PubMed] [Google Scholar]
- 79.Nakao K., Kubota S., Doi H., Eguchi T., Oka M., Fujisawa T., Nishida T., Takigawa M. ( 2005) Collaborative action of M-CSF and CTGF/CCN2 in articular chondrocytes: possible regenerative roles in articular cartilage metabolism. Bone 36, 884– 892 [DOI] [PubMed] [Google Scholar]
- 80.Nishida T., Kubota S., Kojima S., Kuboki T., Nakao K., Kushibiki T., Tabata Y., Takigawa M. ( 2004) Regeneration of defects in articular cartilage in rat knee joints by CCN2 (connective tissue growth factor). J. Bone Miner. Res. 19, 1308– 1319 [DOI] [PubMed] [Google Scholar]
- 81.Blaney Davidson E. N., Vitters E. L., Mooren F. M., Oliver N., Berg W. B., van der Kraan P. M. ( 2006) Connective tissue growth factor/CCN2 overexpression in mouse synovial lining results in transient fibrosis and cartilage damage. Arthritis Rheum. 54, 1653– 1661 [DOI] [PubMed] [Google Scholar]
- 82.Mangasser-Stephan K., Dooley S., Welter C., Mutschler W., Hanselmann R. G. ( 1997) Identification of human semaphorin E gene expression in rheumatoid synovial cells by mRNA differential display. Biochem. Biophys. Res. Commun. 234, 153– 156 [DOI] [PubMed] [Google Scholar]
- 83.Miller L. E., Weidler C., Falk W., Angele P., Schaumburger J., Schölmerich J., Straub R. H. ( 2004) Increased prevalence of semaphorin 3C, a repellent of sympathetic nerve fibers, in the synovial tissue of patients with rheumatoid arthritis. Arthritis Rheum. 50, 1156– 1163 [DOI] [PubMed] [Google Scholar]
- 84.Rosen H., Polakiewicz R. D., Benzakine S., Bar-Shavit Z.Proenkephalin A in bone-derived cells. Proc. Natl. Acad. Sci. U. S. A. ( 1991) 88, 3705– 3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Villiger P. M., Lotz M. ( 1992) Expression of prepro-enkephalin in human articular chondrocytes is linked to cell proliferation. EMBO J. 11, 135– 143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oeppen R. S., Connolly S. A., Bencardino J. T., Jaramillo D. ( 2004) Acute injury of the articular cartilage and subchondral bone: a common but unrecognized lesion in the immature knee. AJR Am. J. Roentgenol. 182, 111– 117 [DOI] [PubMed] [Google Scholar]