Abstract
Preimplantation genetic diagnosis (PGD), used in clinical practice, is offered to couples that may suffer from a monogenetic disorder, chromosome aneuploidy, or X-linked disease. However, blastomere biopsy, as an indispensable manipulation during the PGD procedure has not been assessed for its long term health implications. Using a mouse model, we investigated the effect of blastomere biopsy of in vitro cultured four-cell embryos on preimplantation development efficiency, postnatal growth, and physiological and behavioral activity compared with control, non-biopsied embryos. The mice generated after blastomere biopsy showed weight increase and some memory decline compared with the control group. Further protein expression profiles in adult brains were analyzed by a proteomics approach. A total of 36 proteins were identified with significant differences between the biopsied and control groups, and the alterations in expression of most of these proteins have been associated with neurodegenerative diseases. Furthermore hypomyelination of the nerve fibers was observed in the brains of mice in the biopsied group. This study suggested that the nervous system may be sensitive to blastomere biopsy procedures and indicated an increased relative risk of neurodegenerative disorders in the offspring generated following blastomere biopsy. Thus, more studies should be performed to address the possible adverse effects of blastomere biopsy on the development of offspring, and the overall safety of PGD technology should be more rigorously assessed.
Preimplantation genetic diagnosis (PGD)1 has been one of the main clinical components of assisted reproductive technologies (ARTs) since 1990 (1). At present, infertile couples experiencing recurrent miscarriage or X chromosome-linked diseases are most likely to benefit from PGD.
The treatment of human infertility by ARTs has gained widespread application, but it is disconcerting to many researchers that the clinical procedures used in ARTs are rapidly outpacing the underlying science. ART procedures are generally considered to be safe, but recent studies suggest a small increase in birth defects and low birth weights in ART children (2, 3). In addition, several clinical studies have reported an increased frequency of Beckwith-Wiedemann syndrome or Angelman syndrome caused by an imprinting defect among children conceived with ARTs (4, 5). These potential risks cause serious unease and justify more serious assessments of ARTs. However, moral, ethical, and legal issues complicate assessments of the genetic quality of ART-derived human conceptions, and significant genetic and demographic differences exist among couples participating in the ARTs, so a definitive assessment of the risks associated with this technology has been difficult to achieve. Therefore, appropriate animal models provide an important tool for studying potential effects of ARTs on the health and development of mammalian embryos (6).
In many ART procedures, embryos are kept for a short time in a synthetic culture medium before transfer into their recipient mothers. Animal data have demonstrated that in vitro embryo culture and related procedures may be associated with epigenetic changes, perturbed genomic imprinting, and alterations in fetal growth (7). Some evidence also suggested that the culture environment may produce specific abnormalities during fetal and postnatal development (8–10). In the studies using mouse models, more marked changes in adult physiology, including onset of hypertension, were observed (11).
As with other ARTs, the protocol required for PGD necessitates embryo manipulation and culture in vitro. However, in contrast to other ARTs, PGD involves embryo biopsy of one or two blastomeres at the eight-cell stage. Some studies have shown that blastomere biopsy does not have negative effects on embryo viability (12, 13), and offspring have been produced using embryo splitting technology, which is similar to the biopsy process, in at least six different domesticated species, including mouse (14), rabbit (15), sheep (16), bovine (17), goat (18), horse (19), pig (20), and in a preliminary non-human primate study one rhesus monkey (21). However, there is still a shortage of proof to convince the public that there is no potential risk to such animals conceived by this technology.
To address this issue, we developed a mouse model to study the effects of blastomere biopsy on early embryo development and on postnatal physiological phenotype and behavior. A global proteomics method was also performed to study correlative protein expression profiles in adult brains and to indicate the possibility of neural degenerative disorders in adult conceived following biopsy technology.
EXPERIMENTAL PROCEDURES
Animal Specifications
B6D2F1 mice were utilized throughout as embryo donors. Mice were either derived from biopsied embryos (biopsied group) or from in vitro cultured embryos without biopsy (control group). Ten-week-old mice were regarded as adult.
Cleavage-stage Biopsy and Embryo Transfer
Groups of zygotes were transferred into a droplet of Hepes-buffered CZB (Chatot, Ziomek, and Bavister) medium containing 5 mg/ml cytochalasin B. One blastomere in a four-cell embryo was removed randomly with an enucleation pipette as described previously for human blastomere biopsy (22). After manipulating them, the embryos were transferred back into CZB culture medium containing glucose and held there for up to 2 h at 37.5 °C. Pseudopregnant CD-1 females were used as embryo recipients after mating with vasectomized sterile CD-1 males. Biopsied “three-cell” embryos and four-cell control embryos were transferred into the oviduct of day 0.5 pseudopregnant CD-1 females.
Housing and Behavior
All mice were maintained individually under controlled temperature and lighting conditions and given food and water ad libitum. Postnatal development in biopsied mice was evaluated according to a panel of physiological indices, including body weight, reproduction ability, and forelimb force, etc. Mice in both groups were weighed weekly up to 10 weeks and once every 2 weeks thereafter. At 8–12 weeks old, 10 mice (five females and five males) in each group were cross-mated between the two groups to examine their reproductive ability, and the next generation was delivered naturally by the pregnant mice on day 19.5. To test the forelimb physiology, a puller designed by our “Pin” method was used to test the muscle strength (force index) when the mice were 6 weeks old. We connected the pin to each other as a chain, which the mice could clutch. The mice were suspended inversely and caught hold of the chain. Pins were added onto the chain until the mice discarded them by themselves. The weight of each chain was regarded as the forelimb strength of the mouse.
Intelligence and Memory Test
The water maze consisted of a circular pool (1-m diameter) with a video camera. Before mouse testing, the pool was filled to a depth of 50 cm with water at 35 ± 1 °C, and milk was added to prevent the mice from seeing the hidden platform, which consisted of a round platform (10-cm diameter) located 2 cm below the surface to serve as a refuge. The movement of each mouse was monitored by a video camera fixed to the ceiling above the center of the pool. This video camera sent a signal to 1) a video cassette recorder located in an adjacent room that produced a videotaped recording of the mouse's movements and 2) a tracking system (PolyTrak, San Diego Instruments, San Diego, CA) that produced a digitized recording of each individual swim trial for use in objective analysis of their performance of the task (23).
Spatial training occurred at 6–8 weeks of age in both the biopsied and control groups. Spatial training consisted of six trials in the water maze in a single day beginning at 13:00 h with the hidden platform in the southeast (fourth) quadrant, and a total of 36 trials were performed over the next 6 days. Mice were placed on the platform located in the fourth quadrant for 20 s and then were released facing the wall from quasirandom locations along the edge of the pool. The mice were allowed 60 s to locate the hidden platform. It was considered a successful attempt if after climbing onto the hidden platform mice remained on it for 15 s. A score of 60 s was recorded for unsuccessful attempts at locating the hidden platform. There was a 3-min intertrial interval. During these intervals, mice were placed under a heat lamp to maintain core body temperature. After the sixth trial, mice were allowed to dry off under the heat lamp and were then returned to their cages. This test was divided into three parts. 1) During the first 3 days, we checked the learning and memory ability of the mice. 2) A probe test was done on the 4th day with the platform removed; this served to ascertain whether the mice were using a spatial learning strategy that involved multiple, specific, or distal cues or some other strategy. To determine whether the mice were selectively swimming in the quadrant in which the platform had previously been located (fourth quadrant), the times that the mouse spent in each of the four quadrants was measured. 3) During the last 2 days, 12 reversal trials were administered in which the platform was placed in the opposite quadrant at the same distance from the pool wall as in the first 3 days.
The data were analyzed by tracking system software to evaluate the learning and memory ability.
Mouse Tissue Collection
Tissues were collected from three mice in each group at 10 weeks. Mice were euthanized, and the brains were exposed and removed from the body. Then each brain was cut on ice into halves; one half was snap frozen in liquid nitrogen for protein isolation, and the other was fixed in 4% polyformaldehyde or glutaraldehyde solution for histological examination.
Identification of Altered Proteins in Brain
Proteins from the brain of each mouse were extracted and separated by two-dimensional electrophoresis (2DE) as reported previously (24). Gels were silver-stained, scanned, and analyzed using ImageMaster™ 2D Platinum software (Version 5.0, GE Healthcare). The expression level was determined by the relative volume of each spot in the gel and expressed as %Vol (%Vol = (spot volume/Σvolumes of all spots resolved in the gel)). We averaged the values from the three independent experiments of biopsied and control groups, respectively; calculated the means and standard deviations; and assessed statistical significance with Student's t tests using ImageMaster 2D Platinum software. p values less than 0.05 were considered statistically significant.
Protein spots with significant differences between the two groups were excised. Gel pieces were denatured, alkylated, trypsin-digested, and analyzed by an Ultraflex II MALDI-TOF-TOF mass spectrometer (Bruker Daltonics GmbH, Bremen, Germany) under the control of FlexControl™ 2.4 software (Bruker Daltonics GmbH). MALDI-TOF spectra were recorded in the positive ion reflector mode in a mass range from 700 to 4000 Da, and the ion acceleration voltage was 25 kV. Acquired mass spectra were processed using the software FlexAnalysis™ 2.4 (Bruker Daltonics GmbH): peak detection algorithm, SNAP (Sort Neaten Assign and Place); signal-to-noise threshold, 3; quality factor threshold, 50. The tryptic autodigestion ion peaks (trypsin-(108–115), MH+ 842.509; trypsin-(58–77), MH+ 2211.104) were used as internal standards. Matrix and/or autoproteolytic trypsin fragments or known contaminant ions (keratins) were excluded. The resulting peptide mass lists were used to search International Protein Index mouse database 3.29 (53,981 sequences, 25,507,684 residues) with Mascot (v2.1.03) in automated mode using the following search parameters criteria: significant protein MOWSE (molecular weight search) score at p < 0.05; minimum mass accuracy, 100 ppm; trypsin as enzyme; one missed cleavage site allowed; alkylation of cysteine by carbamidomethylation as fixed modification; and oxidation of methionine as variable modification. Additionally the Mascot score and expectation of the first non-homologous protein to the highest ranked hit were checked. Protein identification was confirmed by sequence information obtained from MS/MS analysis in “LIFT” (laser-induced forward transfer) mode. Acquired MS/MS spectra were also processed using the software FlexAnalysis 2.4 using a SNAP (Sort Neaten Assign and Place) method set at a signal-to-noise ratio threshold of 3.0. For MS/MS spectra searching, the spectra were used to search International Protein Index mouse database 3.29 (53,981 sequences, 25,507,684 residues) automated using Mascot (v2.1.03). Search parameters for MS/MS data were as follow: 100 ppm for the precursor ion and 0.3 Da for the fragment ions. Cleavage specificity and covalent modifications were considered as described above. The score was higher than the minimal significant (p < 0.05) individual ion score. All significant MS/MS identifications by Mascot were manually verified for spectral quality and matching y and b ion series.
Bioinformatics Analysis
An analysis of diseases associated with proteins that were differentially expressed in biopsied mouse brain compared with the control mouse brain was performed using PathwayStudio (v5.0) software (Ariadne Genomics, Inc. Rockville, MD). The text-mining software uses a database assembled from scientific abstracts and a manually curated dictionary of synonyms to recognize biological terms (25). The differentially expressed proteins were converted to their corresponding gene ID and imported into PathwayStudio software; each identified relationship was confirmed manually using the relevant PubMed/Medline hyperlinked texts.
Western Blotting
Samples containing 50 μg of protein from mouse brains in biopsied and control groups were electrophoresed on 12% polyacrylamide gels and transferred to PVDF membranes (GE Healthcare). These blots were incubated for 1.5 h at room temperature in TBS containing 5% nonfat milk powder. Primary antibodies used were anti-ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1) rabbit polyclonal antibody (diluted 1:500; ab27053, Abcam), anti-glia maturation factor β (GMFB) goat polyclonal antibody (diluted 1:200; sc46999, Santa Cruz Biotechnology), anti-α/β soluble N-ethylmaleimide-sensitive factor attachment protein (α,β-SNAP) rabbit polyclonal antibody (diluted 1:1000; ab50483, Abcam), anti-17β-hydroxysteroid dehydrogenase type 10 (Hsd17b10) rabbit polyclonal antibody (diluted 1:500; ab17297, Abcam), anti-γ-enolase (ENO2) mouse monoclonal (85F11) antibody (diluted 1:2000; ab16808, Abcam), anti-peroxiredoxin 5 (PRDX5) rabbit polyclonal antibody (diluted 1:1500; ab16823, Abcam), anti-myelin basic protein (MBP) goat polyclonal antibody (diluted 1:200; sc13914, Santa Cruz Biotechnology), and anti-actin mouse monoclonal (clone C4) antibody (diluted 1:2500; MP Biomedicals). Blots were incubated with primary antibodies overnight at 4 °C. After washing three times in TBS, blots were incubated with horseradish peroxidase-conjugated secondary antibody (1:1000; Beijing ZhongShan Biotechnology Co., Beijing, China) for 1 h. Specific proteins were detected using an ECL reagent (GE Healthcare) and an AlphaImager (FluorChem5500, Alpha Innotech, San Leandro, CA).
Immunohistochemistry
Fixed brain tissues were cut into transverse sections 5 μm thick. The sections were collected on poly-l-lysine-coated slides and air-dried. After blocking, sections were incubated with polyclonal ENO2 antibody (diluted 1:75; ab53025, Abcam), polyclonal GMFB antibody (diluted 1:40; HPA002954, Sigma), polyclonal Hsd17b10 antibody (diluted 1:100; ab17297, Abcam), polyclonal PRDX5 antibody (diluted 1:100; ab16823, Abcam), polyclonal α,β-SNAP antibody (diluted 1:500; ab50483, Abcam), and polyclonal antibody against MBP (diluted 1:100; sc13914, Santa Cruz Biotechnology) in PBS for 2 h at 37 °C in a humidified chamber. After washing, slides were incubated with the secondary antibody, a rabbit anti-goat IgG conjugated with horseradish peroxidase (Beijing Zhongshan Biotechnology Co.) for 1 h at 37 °C in a humidity box. After washing, signals were viewed using an Axioskop2 microscope (Carl Zeiss, Thornwood, New York, NY). The negative controls were incubated with a solution devoid of any primary antibody.
Morphometric Analysis of MBP Immunostaining
MBP is a protein believed to be important in the process of myelination of nerves in the central nervous system (CNS). Corpora striata, as a nerve fiber-rich region, were selected for morphometric analysis of MBP immunostaining signal. Stained sections were examined under a 20× objective, and five consecutive fields in each section were captured using a charge-coupled device digital camera. Images were imported into morphometric software (CellProfiler, an open source software package). Areas of each field were counted to obtain numbers of nerve tracts, cross-sectional area of the nerve tracts, mean intensity, and integrated intensity of each nerve tract for analysis. Data were imported into a Microsoft Excel file for subsequent processing and compared between the control and biopsied groups.
Transmission Electron Microscopy
For ultrastructural examination, tissue blocks of corpora striata were postfixed with 2% OsO4 and embedded in Araldite. Ultrathin sections were stained with uranyl acetate and lead citrate and inspected using an electron microscope (JEM.1010, JEOL).
Statistics Analysis
Data were analyzed by t test for significant differences between the two groups. The nonparametric Mann-Whitney test was used for some infrequently occurring or nonparametrically distributed measures.
RESULTS
Effects of Blastomere Biopsy on Mouse Embryo Development
To evaluate the effect of manipulation in vitro on embryo developmental potential, three-cell biopsied (n = 86) and control (n = 79) four-cell embryos were placed in culture (Fig. 1). There was no difference in the ability of embryos in the two groups to develop to the blastocyst stage (p > 0.05; Table I). As expected, there was a difference (p < 0.05) in total cell numbers and in inner cell mass (ICM) numbers between the two groups; however, the ratio of ICM to total embryo number was unchanged (p > 0.05) (Table I).
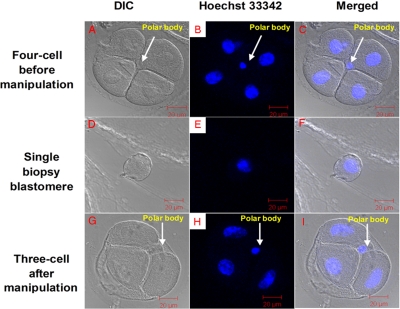
Fig. 1.
Embryo biopsy at the four-cell stage. A, under DIC; B, confocal microscopy (stained by Hoechst 33342); C, merged. A single blastomere removed from a four-cell embryo is shown under DIC (D), by confocal microscopy (stained by Hoechst 33342) (E), and merged (F). The biopsied three-cell stage embryo is shown under DIC (G), by confocal microscopy (stained by Hoechst 33342) (H), and merged (I).
Table I. Blastocyst development and cell number (mean ± S.E.) for biopsied and in vitro cultured control mouse embryos.
Every experiment was repeated at least three times.
| Group | No. of four-cell treated | No. of blastocysts | No. cells/blastocyst |
||||
|---|---|---|---|---|---|---|---|
| Embryos examined | ICM | Total | Total/blastomere | ICM/total | |||
| % | |||||||
| Biopsied | 86 | 78 (91)a | 15 | 9.8 ± 0.7a | 36.8 ± 2.8a | 12.3 ± 0.9a | 0.27 ± 1.9a |
| Control | 79 | 75 (95)a | 15 | 14.2 ± 1.1b | 51.4 ± 2.6b | 12.9 ± 0.7a | 0.28 ± 0.9a |
a Values with different superscript letters in the same column are significantly different (p < 0.05).
b Values with different superscript letters in the same column are significantly different (p < 0.05).
To assess postimplantation efficiency, biopsied (n = 225) and control (n = 98) embryos (average, 14 embryos/transfer) were transferred into recipient mothers. The birthrate and neonatal numbers following transfer of biopsied embryos were significantly lower than for the control embryos (Table II).
Table II. Morbidity and mortality of pups derived from biopsied and in vitro cultured embryos.
| Group | Embryos transferred (recipients) | Embryos/recipient (mean ± S.E.) | No. of pups | Micea examined | No. of deaths |
Survival | Miceb examined | No. of deaths |
Survival | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Maternal factors | Unexpected | Ill | Unexpected | ||||||||
| % | % | % | |||||||||
| Biopsied | 225 (17) | 13.2 ± 0.9c | 39 (17.3)c | 39 | 9 | 2 | 71.8c | 19 | 2 | 0 | 89c |
| Control | 98 (7) | 14.0 ± 0.8c | 45 (45.9)d | 44 | 20 | 0 | 54.6d | 22 | 3 | 0 | 86c |
a Nursing pups between 1 and 3 weeks of age.
b Mice bred separately between 4 and 30 weeks of age.
c Values with different superscript letters in the same column are significantly different (p < 0.05).
d Values with different superscript letters in the same column are significantly different (p < 0.05).
Adult Phenotypes Observed
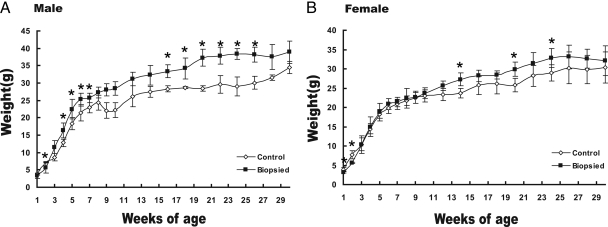
To test whether embryo biopsy produced effects on neonatal development, behavior, and physiology, neonates were separated based on treatment into male and female subgroups for weight monitoring. Male offspring generated from biopsied embryos showed higher body weights than the control group (the difference is significant at weeks 4–7 and 16–26). For females, the weight difference was minimal. However, in specific weeks, female biopsied offspring were also significantly heavier compared with control females (weeks 14, 20, and 24) (Fig. 2).
Fig. 2.
Blastomere biopsy effects on postnatal growth. Mean (±S.E.) body mass for male (A) and female (B) offspring from 1 to 30 weeks is shown. Error bars represent the standard error of the mean for each data point.*, p < 0.05 difference between the biopsied group and control group.
During the perinatal period, 39 mice in the biopsied group and 44 mice in the control group were observed for viability. About 72% of mice in the biopsied group and 55% in the control group survived to weaning, and the main reason for death was maternal factors, including being eaten and discarded by their mother; however, from weaning to 30 weeks of age when the mice were breeding in separate cages, there were no differences between the two groups in mortality (Table II).
At 6 weeks of age, forelimb strength of male and female mice was not significantly different between the two groups. Five mice of each sex were selected randomly from each group and tested for reproductive capacity. When crossed at 8–12 weeks of age, all animals were fertile, and there were no significant differences between the two groups in the number of pups produced (Table III).
Table III. Forelimb strength and fertility in mice produced from manipulated and control embryos.
| Group and gender | Test of forelimb forcea |
Reproductive abilityb |
||
|---|---|---|---|---|
| Mice examined (weeks old) | Force index (mean ± S.E.) | Mice examined (weeks old) | Average no. generated/female | |
| g | ||||
| Biopsied | ||||
| Male | 3 (6) | 60.4 ± 3.4c | 5 (8–12) | 12.3 ± 1.5c |
| Female | 3 (6) | 53.5 ± 4.3c | 5 (8–12) | |
| Control | ||||
| Male | 3 (6) | 56.1 ± 2.9c | 5 (8–12) | 8.7 ± 0.9c |
| Female | 3 (6) | 53.2 ± 3.6c | 5 (8–12) | |
a Three individual samples of both male and female mice in every group were selected to test forelimb force index, and every individual was tested three times in 1 week.
b Five individual samples of both male and female mice in every group were selected to test fertility.
c Within each column, values with same superscript letters are not statistically different at 0.05 level of significance.
Intelligence and Memory Test
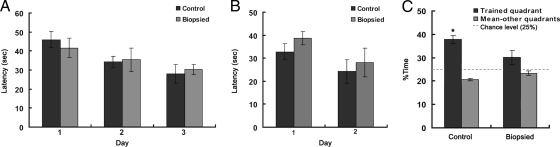
To study the intelligence and memory ability of mice produced after blastomere biopsy, a total of 36 trials were performed during six consecutive days by using a Morris water maze apparatus on five mice in the biopsied group and eight mice in the control group. The results are shown in Fig. 3. In acquisition trials performed during the first 3 days, the mice were expected to find the hidden platform located in the fourth quadrant as soon as possible within 60 s. The results showed that both control and biopsied groups displayed evident improvement over the 3 days of testing, and the mean acquisition times in the two groups did not differ significantly from each other (Fig. 3A). During the reversal trials, both groups improved their performance over the 2 days of trials with no significant difference observed between the two groups (Fig. 3B). In the probe test, both control and biopsied mice selectively stayed in the trained quadrant. Duration of stay in the trained quadrant was significant only for control mice (p < 0.001) and failed to reach an acceptable level of statistical significance for biopsied mice (p = 0.11); differences between the two groups were significant (p < 0.05) (Fig. 3C). This result implied decreased memory in biopsied mice.
Fig. 3.
Results from the Morris water task. A, average latency to find hidden platform: acquisition trials. B, average latency to find hidden platform: reversal trials. C, average time spent searching quadrant in which animal trained versus other quadrants. Error bars represent the standard error of the mean. (*, trained versus other quadrant, p < 0.001).
Analysis, Identification, and Functional Annotation of Differentially Expressed Proteins
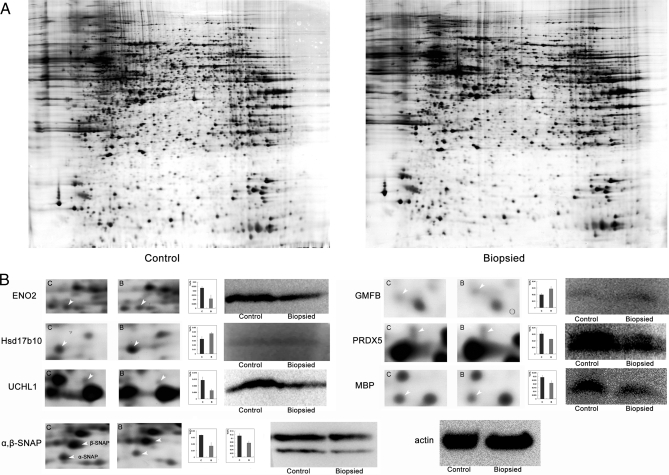
In view of the finding that the mice in the biopsied group showed slightly impaired memory, to obtain more definitive proof at the molecular level, brain tissues were studied and analyzed. We constructed triplicate 2D maps of brains from three mice from each group and compared the proteome maps between biopsied and control groups (Fig. 4). Of the protein spots with significant differences in expression level, 39 spots were identified successfully, corresponding to 36 proteins. Detailed information is listed in supplemental Table 1.
Fig. 4.
Two-dimensional electrophoresis results and validation with Western blotting. A, 2DE maps of proteins extracted from control and biopsied brains. B, magnified comparison maps of spots for ENO2, Hsd17b10, UCHL1, α,β-SNAP, GMFB, PRDX5, and MBP in the 2DE patterns of control and biopsied brains. Bars represent the relative volume (%Vol) of each of the spots on the gels. Error bars represent the standard error of the mean. Western blot analyses of each protein are shown simultaneously. Results are consistent with the results of the 2DE. Actin was used as a loading control.
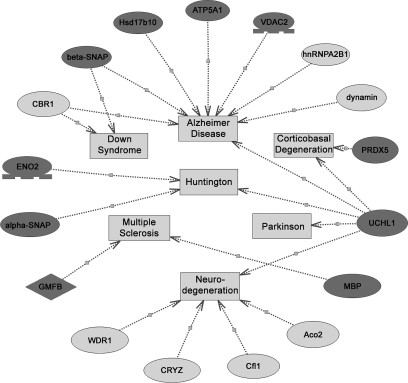
To gain a better understanding of the 36 differentially expressed proteins identified in this study, a detailed analysis of the diseases associated with these proteins was performed using PathwayStudio software. The results showed that 17 of the altered proteins are related to neurodegenerative diseases, such as Alzheimer disease, Parkinson disease, multiple sclerosis, etc. (Fig. 5; all relationships were checked manually, and the relevant literature is listed in supplemental Table 2). The nature of the differential expression of the majority of the proteins was consistent with the characteristics of neurodegenerative diseases. Considering that the linear range of silver staining is small, the spot volume obtained from 2D gels may not be well correlated to protein abundance. To avoid quantitative deviations, for these key proteins we used Western blotting to verify their expressional alterations between the biopsied and control groups (shown in Fig. 4B). Further immunohistochemical analysis also showed similar expressional tendency (Fig. 6). The altered expression profile of the other proteins might be due to a compensatory response to neuronal dysfunction.
Fig. 5.
Relationships between neurodegenerative disorders with the differentiated proteins in the biopsied brains compared with the control brains as predicted by PathwayStudio software. Proteins are shown as ovals (a dark gray oval means its altered tendency was consistent with the characteristic of neurodegenerative diseases; a light gray oval means its alteration may be compensation for neurodegenerative disorders), and neurodegenerative disorders are represented by squares. Relationships are displayed with arrows and documented by literature citations (details listed in supplemental Table 2). CRYZ, ζ-crystallin.
Fig. 6.
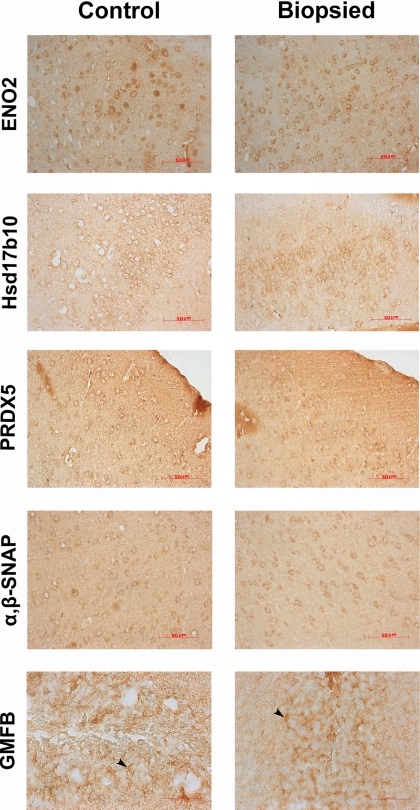
Immunohistochemistry analysis of ENO2, Hsd17b10, PRDX5, α,β-SNAP, and GMFB in control and biopsied brains. ENO2, Hsd17b10, PRDX5, and α,β-SNAP localized in the cytoplasm of neuron; GMFB localized in an astrocyte (black arrowheads indicate astrocyte in dentate gyrus). The trend in the differential expression of the proteins was in accordance with the variations obtained from 2DE and Western blotting.
Demyelination of Nerve Fibers in the Brain of Biopsied Group Mice
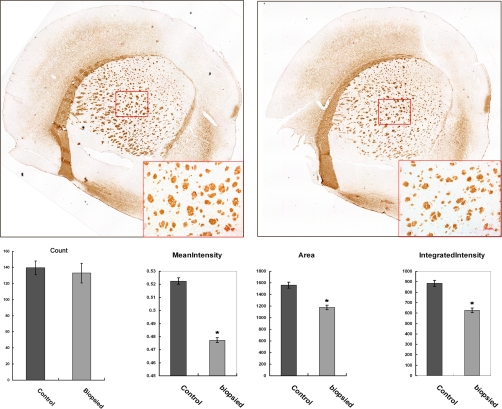
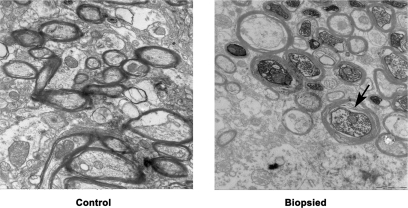
Myelinated nerve fibers were stained with antibody against MBP. The numbers of nerve tracts were comparable between the biopsied and control groups, whereas the mean intensity of MBP in each nerve tract was significantly lower in the biopsied group, further verifying the lower expression of MBP in the biopsied group and supporting the results obtained from proteomics analysis and Western blotting. In addition, the cross-sectional area and integrated intensity per nerve tract were significantly reduced, suggesting the presence of hypomyelination in the brain of mice from the biopsied group (Fig. 7). This result was also verified by ultrastructure examination, which revealed myelin sheath disruption and axonal swelling in the biopsied group (Fig. 8).
Fig. 7.
A, myelinated nerve fibers in the control and biopsied mouse striatum were stained with anti-MBP antibody. The framed areas are shown enlarged in the lower right corner. B, the myelinated nerve track numbers, cross-section area, mean intensity, and integrated intensity of each nerve track were analyzed by CellProfiler software. Error bars represent the standard error of the mean. (*, p < 1 × 10−8).
Fig. 8.
Ultrastructure examination of stratum of control and biopsied groups (magnification, 20,000×). In the biopsied mouse stratum, myelin sheath disruption and axonal swelling were observed frequently (arrow indicates the typical changes).
DISCUSSION
PGD technology plays an important role in the treatment of human infertility; however, underlying research related to its safety seems to be neglected although the technology is being widely applied. Here we used a mouse model to evaluate the long term risks of the blastomere biopsy technology used in the PGD procedure to fetal, neonatal, and adult growth and development. Our proteomics results in the brain tissue showed that some neurodegenerative disorder-related proteins were abnormally expressed in the biopsied group. We studied embryos that were biopsied at the four-cell rather than the eight-cell stage, as is normally done clinically, in an effort to compensate for differences in the timing of embryonic gene activation in the two species. This activation occurs at the two- to four-cell stage in the mouse and at the 8–16-cell stage in humans (26). Our results showed that there was no early preimplantation developmental differences between the biopsied and control groups, consistent with reported results in humans (27). However, the birthrate in the biopsied mouse group was significantly lower than that in the control group, indicating that postimplantation development might be affected by biopsy manipulation.
The physiological phenotype of an animal is regarded as one of the most important evaluation indices for assessing reproductive technologies. We tested some key phenotypes, and the results showed that there was no difference in reproductive ability or forelimb strength. However, we found body weight differences in the pups derived from the two groups of embryos. The weights of male pups significantly increased in the biopsied group compared with controls, and the body weights of female offspring in the biopsied group were also heavier than those in the control group at specific time points. Some reports have indicated that fetal overgrowth can occur when embryos are cultured in vitro (28) and also in human ART (5, 29). Studies on nuclear transfer have also shown that micromanipulations may induce fetal overgrowth (31). However, there are no comparative data on adult weights for assessing the risk of overgrowth due to embryo manipulation. Our study suggested that blastomere biopsy might affect the adult weight. This observation also supported the hypotheses that very early events in the preimplantation embryo could have a significant impact on later development (32, 33).
We also performed behavioral evaluation. In preliminary behavior experiments, neonates were tested for several parameters, namely eye opening time and cage activity, and no differences were apparent between the biopsied and control groups at several different observation times (data not shown). In an experiment testing their learning and memory ability, Ecker et al. (34) reported that culture in vitro could affect the behavior in a long term life cycle by using the zero maze and Morris water maze. In our Morris water maze study, mice were driven to find the hidden platform as soon as possible. Although there was no difference observed when we put the platform into the water maze, a significant difference was observed when the platform was hidden: mice in the biopsied group would stay in the destination quadrant for a shorter time than those in the control group, suggesting poorer memory function of the biopsied group mice.
To address the influence of blastomere biopsy on the central nervous system at a molecular level, we performed proteomics analysis of brain tissues from biopsied and control groups. Differential expression of 39 protein spots was detected between the two groups, representing 36 unique proteins. The detected proteins were annotated bioinformatically and were found to be associated with neurodegenerative state in brain development, and it is worth noting that the diversification patterns of most of these proteins are absolutely consistent with alterations reported in neurodegenerative disorders, such as UCHL1, ENO2, Hsd17b10, GMFB, MBP, etc. Considering that the linear range of silver staining is small, we used Western blotting and immunohistochemistry to verify the expressional differences of most of these key proteins. UCHL1, also known as PGP9.5, is one of the most abundant proteins in the brain (1–2% of total soluble protein), and immunohistochemical experiments demonstrate that it is exclusively localized in neurons (35). As part of the ubiquitin-proteasome system necessary for protein degradation, UCHL1 has been implicated in the death of neurons in neurodegenerative diseases (36–39). Mice with mutated UCHL1 show axonal loss and cell death (40). In human disease states, UCHL1 levels are low (37), or its function is compromised (38, 41). Alzheimer disease brains show prominent UCHL1 immunostaining associated with neurofibrillary tangles, and levels of soluble UCHL1 are inversely proportional to the number of tangles (37). UCHL1 dysfunction has also been associated with Parkinson disease (36, 38) and Huntington disease (39). In our study, a deficiency in UCHL1 expression was observed in the brains of mice derived from biopsied embryos. The low level of expression reflected depressed neuronal life expectancy. Hsd17b10, also named Alzheimer-associated β-amyloid-binding protein (ERAB), is another protein related to neurodegenerative diseases. Its expression was higher in the biopsied group brains than in the control group. ERAB, which is thought to be a hydroxysteroid dehydrogenase enzyme, is overexpressed in neurons affected in Alzheimer disease (42). Its catalytic function could weaken the protective effects of estrogen and generate aldehydes in neurons (43). Consequently the high concentration of ERAB in the brain extracts of mice from biopsied embryos would likely cause an estrogen-deficient state in neurons and may be a potential risk factor for neuronal dysfunction.
Furthermore among the altered proteins in the biopsied group brains, one protein was of particular interest, MBP. MBP is one of the major proteins of CNS myelin and constitutes as much as 30% of protein (44). The main biological function of the classic MBP is in myelin compaction in the CNS (45). Mice with mutated MBP (shiverer mice), caused by a deletion within the MBP gene, show a severe phenotype that is marked by decreased amounts of CNS myelination and a progressive disorder characterized by tremors, seizures, and early death (45, 46). In this study, the expressional level of MBP was significantly lower in the biopsied group compared with the control group. Immunostaining of MBP on the striatum also verified its lower expression in the biopsied group brains, and the cross-sectional areas of nerve tracts were clearly reduced. Furthermore the ultrastructure examination showed lamellar separation and disruption in the myelin sheaths, and axonal swelling was frequently observed in the biopsied group. These results suggested that hypomyelination was induced by blastomere biopsy. Poor and protracted myelination is also considered as a contributing factor to neurodegenerative disorders (47).
In summary, from the initial clinical applications of PGD, some individuals have warned about opening a Pandora's box of ethical problems. Many infertile couples benefit from PGD technology, and experimental studies have focused on evaluating procedure-related effects on the development of biopsied preimplantation embryos (30). However, it is clear that we must also recognize the potential risks to fetuses, neonates, and adults derived from this technology. Here we used a mouse model in efforts to evaluate the safety of blastomere biopsy as used in PGD procedures and showed a risk of defective memory function in the offspring generated from biopsied embryos. The results using proteome-based approaches further identified a high risk of neurodegeneration arising from biopsy. Our study suggested that additional efforts should be made in basic research and risk evaluation for PGD and other ART-related technologies. Also we hope that the molecular abnormalities found could be helpful in finding diagnostic protocols and in adopting multiform measures to minimize the incidence of such disorders.
Acknowledgments
We thank Dr. Barry D. Bavister for valuable editing of the manuscript.
Footnotes
* This work was supported by 973 Program Grants (2006CB701503 and 2007CB948101) and PCSIRT (No. IRT0631).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- PGD
- preimplantation genetic diagnosis
- ART
- assisted reproductive technology
- 2DE
- two-dimensional electrophoresis
- UCHL1
- ubiquitin carboxyl-terminal hydrolase isozyme L1
- GMFB
- glia maturation factor β
- α,β-SNAP
- α/β soluble N-ethylmaleimide-sensitive factor attachment protein
- Hsd17b10
- 17β-hydroxysteroid dehydrogenase type 10
- ENO2
- γ-enolase
- PRDX5
- peroxiredoxin 5
- MBP
- myelin basic protein
- CNS
- central nervous system
- ICM
- inner cell mass
- 2D
- two-dimensional
- ERAB
- Alzheimer-associated β-amyloid-binding protein
- DIC
- differential interference contrast
REFERENCES
- 1.Handyside A. H., Kontogianni E. H., Hardy K., Winston R. M. ( 1990) Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature 344, 768– 770 [DOI] [PubMed] [Google Scholar]
- 2.Kohei S., Shigehito Y. ( 2005) Assisted reproductive technologies and birth defects. Congenital Anomalies 45, 39– 43 [DOI] [PubMed] [Google Scholar]
- 3.Hansen M., Kurinczuk J. J., Bower C., Webb S. ( 2002) The risk of major birth defects after intracytoplasmic sperm injection and in vitro fertilization. N. Engl. J. Med. 346, 725– 730 [DOI] [PubMed] [Google Scholar]
- 4.Lawrence L. T., Moley K. H. ( 2008) Epigenetics and assisted reproductive technologies: human imprinting syndromes. Semin. Reprod. Med. 26, 143– 152 [DOI] [PubMed] [Google Scholar]
- 5.Bowdin S., Allen C., Kirby G., Brueton L., Afnan M., Barratt C., Kirkman-Brown J., Harrison R., Maher E. R., Reardon W. ( 2007) A survey of assisted reproductive technology births and imprinting disorders. Hum. Reprod. 22, 3237– 3240 [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Gonzalez R., Moreira P., Bilbao A., Jimenez A., Perez-Crespo M., Ramirez M. A., Rodriguez De Fonseca F., Pintado B., Gutierrez-Adan A.Proc. Natl. Acad. Sci. U. S. A. ( 2004) 101, 5880– 5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maher E. R. ( 2005) Imprinting and assisted reproductive technology. Hum. Mol. Genet. 1, R133– 138 [DOI] [PubMed] [Google Scholar]
- 8.Bavister B. D. ( 1995) Culture of preimplantation embryos: facts and artifacts. Hum. Reprod. Update 1, 91– 148 [DOI] [PubMed] [Google Scholar]
- 9.McEvoy T. G. ( 2003) Manipulation of domestic animal embryos and implications for development. Reprod. Domest. Anim. 38, 268– 275 [DOI] [PubMed] [Google Scholar]
- 10.Walker S. K., Hartwick K. M., Seamark R. F. ( 1996) Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Theriogenology 45, 111– 120 [Google Scholar]
- 11.Watkins A. J., Platt D., Papenbrock T., Wilkins A., Eckert J. J., Kwong W. Y., Osmond C., Hanson M., Fleming T. P.Proc. Natl. Acad. Sci. U. S. A. ( 2007) 104, 5449– 5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winston R. M., Hardy K. ( 2002) Are we ignoring potential dangers of in vitro fertilization and related treatments? Nat. Cell Biol. 4, ( suppl.) s14– s18 [DOI] [PubMed] [Google Scholar]
- 13.Hardy K., Martin K. L., Leese H. J., Winston R. M., Handyside A. H. ( 1990) Human preimplantation development in vitro is not adversely affected by biopsy at the 8-cell stage. Hum. Reprod. 5, 708– 714 [DOI] [PubMed] [Google Scholar]
- 14.Tarkowski A. K. ( 1959) Experiments on the development of isolated blastomeres of mouse eggs. Nature 184, 1286– 1287 [DOI] [PubMed] [Google Scholar]
- 15.Moore N. W., Adams C. E., Rowson L. E. ( 1968) Developmental potential of single blastomeres of the rabbit egg. J. Reprod. Fertil. 17, 527– 531 [DOI] [PubMed] [Google Scholar]
- 16.Willadsen S. M. ( 1979) A method for culture of micromanipulated sheep embryos and its use to produce monozygotic twins. Nature 277, 298– 300 [DOI] [PubMed] [Google Scholar]
- 17.Ozil J. P., Heyman Y., Renard J. P. ( 1982) Production of monozygotic twins by micromanipulation and cervical transfer in the cow. Vet. Rec. 110, 126– 127 [DOI] [PubMed] [Google Scholar]
- 18.Tsunoda Y., Uasui T., Sugie T. ( 1984) Production of monozygotic twins following transfer of separated half embryos in the goat. Jpn. J. Zootech. Sci. 55, 643– 647 [Google Scholar]
- 19.Allen W. R., Pashen R. L. ( 1984) Production of monozygotic (identical) horse twins by embryo micromanipulation. J. Reprod. Fertil. 71, 607– 613 [DOI] [PubMed] [Google Scholar]
- 20.Saito S., Niemann H. ( 1991) Effects of extracellular matrices and growth factors on the development of isolated porcine blastomeres. Biol. Reprod. 44, 927– 936 [DOI] [PubMed] [Google Scholar]
- 21.Chan A. W., Dominko T., Luetjens C. M., Neuber E., Martinovich C., Hewitson L., Simerly C. R., Schatten G. P. ( 2000) Clonal propagation of primate offspring by embryo splitting. Science. 287, 317– 319 [DOI] [PubMed] [Google Scholar]
- 22.Sermon K., Van Steirteghem A., Liebaers I. ( 2004) Preimplantation genetic diagnosis. Lancet 363, 1633– 1641 [DOI] [PubMed] [Google Scholar]
- 23.Vorhees C. V., Williams M. T. ( 2006) Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat. Protoc. 1, 848– 858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y. F., Cui Y. G., Guo X. J., Wang L., Bi Y., Hu Y. Q., Zhao X., Liu Q., Huo R., Lin M., Zhou Z. M., Sha J. H. ( 2006) Proteomic analysis of effect of hyperthermia on spermatogenesis in adult male mice. J. Proteome Res. 5, 2217– 2225 [DOI] [PubMed] [Google Scholar]
- 25.Nikitin A., Egorov S., Daraselia N., Mazo I. ( 2003) Pathway studio—the analysis and navigation of molecular networks. Bioinformatics 19, 2155– 2157 [DOI] [PubMed] [Google Scholar]
- 26.Majumder S., Zhao Z., Kaneko K., DePamphilis M. L. ( 1997) Developmental acquisition of enhancer function requires a unique coactivator activity. EMBO J. 16, 1721– 1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magli M. C., Gianaroli L., Ferraretti A. P., Toschi M., Esposito F., Fasolino M. C. ( 2004) The combination of polar body and embryo biopsy does not affect embryo viability. Hum. Reprod. 19, 1163– 1169 [DOI] [PubMed] [Google Scholar]
- 28.Young L. E., Sinclair K. D., Wilmu I. ( 1998) Large offspring syndrome in cattle and sheep. Rev Reprod. 3, 155– 163 [DOI] [PubMed] [Google Scholar]
- 29.Khosla S., Dean W., Brown D., Reik W., Feil R. ( 2001) Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol. Reprod. 64, 918– 926 [DOI] [PubMed] [Google Scholar]
- 30.Illmensee K., Kaskar K., Zavos P. M. ( 2005) Efficient blastomere biopsy for mouse embryo splitting for future applications in human assisted reproduction. Reprod. Biomed. Online 11, 716– 725 [DOI] [PubMed] [Google Scholar]
- 31.Tamashiro K. L., Wakayama T., Yamazaki Y., Akutsu H., Woods S. C., Kondo S., Yanagimachi R., Sakai R. R. ( 2003) Phenotype of cloned mice: development, behavior, and physiology. Exp. Biol. Med. (Maywood) 228, 1193– 1200 [DOI] [PubMed] [Google Scholar]
- 32.Barker D. J., Clark P. M. ( 1997) Fetal undernutrition and disease in later life. Rev. Reprod. 2, 105– 112 [DOI] [PubMed] [Google Scholar]
- 33.Young L. E., Fairburn H. R. ( 2000) Improving the safety of embryo technologies: possible role of genomic imprinting. Theriogenology 53, 627– 648 [DOI] [PubMed] [Google Scholar]
- 34.Ecker D. J., Stein P., Xu Z., Williams C. J., Kopf G. S., Bilker W. B., Abel T., Schultz R. M.Proc. Natl. Acad. Sci. U. S. A. ( 2004) 101, 1595– 1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson P. O., Barber P. C., Hamid Q. A., Power B. F., Dhillon A. P., Rode J., Day I. N., Thompson R. J., Polak J. M. ( 1988) The immunolocalization of protein gene product 9.5 using rabbit polyclonal and mouse monoclonal antibodies. Br. J. Exp. Pathol. 69, 91– 104 [PMC free article] [PubMed] [Google Scholar]
- 36.Dawson T. M., Dawson V. L. ( 2003) Molecular pathways of neurodegeneration in Parkinson's disease. Science 302, 819– 822 [DOI] [PubMed] [Google Scholar]
- 37.Choi J., Levey A. I., Weintraub S. T., Rees H. D., Gearing M., Chin L. S., Li L. ( 2004) Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J. Biol. Chem. 279, 13256– 13264 [DOI] [PubMed] [Google Scholar]
- 38.Giasson B. I., Lee V. M. ( 2003) Are ubiquitination pathways central to Parkinson's disease? Cell 114, 1– 8 [DOI] [PubMed] [Google Scholar]
- 39.Naze P., Vuillaume I., Destee A., Pasquier F., Sablonniere B. ( 2002) Mutation analysis and association studies of the ubiquitin carboxy-terminal hydrolase L1 gene in Huntington's disease. Neurosci. Lett. 328, 1– 4 [DOI] [PubMed] [Google Scholar]
- 40.Mukoyama M., Yamazaki K., Kikuchi T., Tomita T. ( 1989) Neuropathology of gracile axonal dystrophy (GAD) mouse. An animal model of central distal axonopathy in primary sensory neurons. Acta Neuropathol. 79, 294– 299 [DOI] [PubMed] [Google Scholar]
- 41.McNaught K. S., Olanow C. W., Halliwell B., Isacson O., Jenner P. ( 2001) Failure of the ubiquitin-proteasome system in Parkinson's disease. Nat. Rev. Neurosci. 2, 589– 594 [DOI] [PubMed] [Google Scholar]
- 42.Yan S. D., Fu J., Soto C., Chen X., Zhu H., Al-Mohanna F., Collison K., Zhu A., Stern E., Saido T., Tohyama M., Ogawa S., Roher A., Stern D. ( 1997) An intracellular protein that binds amyloid-β peptide and mediates neurotoxicity in Alzheimer's disease. Nature 389, 689– 695 [DOI] [PubMed] [Google Scholar]
- 43.He X. Y., Merz G., Mehta P., Schulz H., Yang S. Y. ( 1999) Human brain short chain l-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. Characterization of a novel 17β-hydroxysteroid dehydrogenase. J. Biol. Chem. 274, 15014– 15019 [DOI] [PubMed] [Google Scholar]
- 44.Lees M. B., Brostoff S. W. ( 1984) Proteins of myelin, in Myelin ( Morell P., ed) pp. 197– 224, Plenum Press, New York [Google Scholar]
- 45.Roach A., Takahashi N., Pravtcheva D., Ruddle F., Hood L. ( 1985) Chromosomal mapping of mouse myelin basic protein gene and structure and transcription of the partially deleted gene in shiverer mutant mice. Cell 42, 149– 155 [DOI] [PubMed] [Google Scholar]
- 46.Readhead C., Popko B., Takahashi N., Shine H. D., Saavedra R. A., Sidman R. L., Hood L. ( 1987) Expression of a myelin basic protein gene in transgenic shiverer mice: correction of the dysmyelinating phenotype. Cell 48, 703– 712 [DOI] [PubMed] [Google Scholar]
- 47.Braak H., Del T. K. ( 2004) Poor and protracted myelination as a contributory factor to neurodegenerative disorders. Neurobiol. Aging 25, 19– 23 [DOI] [PubMed] [Google Scholar]