Abstract
A new program (Multiple Motif Scanning) was developed to scan the Saccharomyces cerevisiae proteome for Class I S-adenosylmethionine-dependent methyltransferases. Conserved Motifs I, Post I, II, and III were identified and expanded in known methyltransferases by primary sequence and secondary structural analysis through hidden Markov model profiling of both a yeast reference database and a reference database of methyltransferases with solved three-dimensional structures. The roles of the conserved amino acids in the four motifs of the methyltransferase structure and function were then analyzed to expand the previously defined motifs. Fisher-based negative log statistical matrix sets were developed from the prevalence of amino acids in the motifs. Multiple Motif Scanning is able to scan the proteome and score different combinations of the top fitting sequences for each motif. In addition, the program takes into account the conserved number of amino acids between the motifs. The output of the program is a ranked list of proteins that can be used to identify new methyltransferases and to reevaluate the assignment of previously identified putative methyltransferases. The Multiple Motif Scanning program can be used to develop a putative list of enzymes for any type of protein that has one or more motifs conserved at variable spacings and is freely available (www.chem.ucla.edu/files/MotifSetup.Zip). Finally hidden Markov model profile clustering analysis was used to subgroup Class I methyltransferases into groups that reflect their methyl-accepting substrate specificity.
Enzymes that catalyze the transfer of a methyl group from S-adenosylmethionine to protein, nucleic acid, lipid, and small molecule substrates are widely distributed in nature and function in a variety of biological pathways including metabolic regulation, gene expression, the repair of aging biomolecules, and biosynthesis (1). There are several classes of AdoMet1 dependent methyltransferases. Class I enzymes are the most abundant and share a common three-dimensional structural core that includes a seven-strand twisted β sheet (2–5). It has been estimated that about 0.6–1.6% of genes in organisms ranging from Escherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana, and humans encode Class I methyltransferases (6). These results suggest that there are some 50 species in yeast and some 300 species in humans. However, most of these assignments have not been confirmed, and only a relatively small fraction of them have been functionally identified. Previous bioinformatics studies on Class I methyltransferases have taken advantage of the fact that the structure of the AdoMet binding site is conserved in the primary sequences of four short signature motifs designated Motifs I, Post I, II, and III (6–8). These motifs are present with variable, but often conserved, spacing in the primary sequences. Initial identifications of Class I methyltransferases in the yeast proteome were based on searches with a “Motifs in Protein Data Bases” program with individual motifs to generate a list of putative methyltransferases (8). More recently, Katz et al. (6) performed a search using the Motif Alignment and Search Tool (MAST) with Multiple Em for Motif Elicitation (MEME)-generated matrices combining Motifs I/Post I as well as PSI-BLAST searches to identify methyltransferases in both yeast and a variety of other organisms. However, the success of these methods was limited by difficulties in identifying these motifs in the known methyltransferases. In fact, it was not possible to take advantage of the information content of Motifs II and III in the latter study (6).
In recent years, two developments have provided new windows to improve the screening of proteomes to identify the complete cast of Class I methyltransferases in various organisms. In the first place, three-dimensional structures are now known for a large number of Class I methyltransferases. Because the motifs are closely linked to structural features (2–4), their identification is unambiguous. Secondly secondary structure prediction algorithms can now be used as independent confirmations of the structure-linked sequence motifs in methyltransferases whose structures have not been determined. We wanted to take advantage of the improved motif identification by developing advanced software for searching proteomes for multiple motifs at varying, but partially conserved, spacing. We have now used secondary structure prediction to obtain the motifs of a reference group of 32 known yeast methyltransferases and a search of the Research Collaboratory for Structural Bioinformatics Protein Data Bank to identify the motifs of a second reference group of 33 distinct types of Class I methyltransferases with three-dimensional structures. We describe a method for generating search matrices for each of these reference groups and a program “Multiple Motif Scanning” to score these matrices in the yeast proteome. This approach not only identified new methyltransferases but allowed us to reject some of the previously described candidate methyltransferases. Additionally we used HMM profile clustering analysis to extract more information about the possible substrates of these putative methyltransferases (9).
EXPERIMENTAL PROCEDURES
Motif Identification Using Two Reference Groups of Known Methyltransferases
To search for putative methyltransferases, two reference groups of known methyltransferases were compiled to refine the consensus sequence at each motif. The first reference group included the 32 known Class I methyltransferases from S.cerevisiaesupplemental Table 1). S. cerevisiae is a good model organism for this analysis because of the low redundancy of proteins and the high degree of their characterization. Sequences were aligned using HHpred with HHsearch 1.5, which utilizes advanced HMM profile algorithms to align proteins based on both primary and predicted secondary structures (9). The HMM profile comparisons and the secondary sequences were used to objectively identify Motifs I, Post I, II, and III (Fig. 1 and supplemental Table 1) and generate the “yeast matrix” for scoring in the Multiple Motif Scanning program.
Fig. 1.
The four signature motifs (underlined) of the Class I methyltransferases shown in their primary, predicted secondary, and actual secondary structure in three known methyltransferase proteins in S.cerevisiae. The motifs were identified using HHpred with HHsearch 1.5, which utilizes HMM profile versus profile searches to align the proteins. HHpred also generates secondary structural predictions (seen here) that also aid in the alignment and identification of the motifs, denoted C for random coil, E for β sheet, and H for helical structures. Because the crystal structures have been solved for Dot1, Hmt1, and Ppm1 proteins, the secondary structures of the crystals have been used for comparison with those predicted by HHpred. Although a crystal structure is known for the Mtf1/YMR228w, the reaction that it catalyzes is unknown. As seen here, predicted secondary structures are very similar to the actual secondary structures, especially in the motif regions.
The second reference group included known non-yeast Class methyltransferases for which the three-dimensional crystal structures is known (supplemental Table 2). The Research Collaboratory for Structural Bioinformatics Protein Data Bank was searched by the 2.1.1.x EC number corresponding to methyltransferases. Proteins of Class I AdoMet-dependent methyltransferases were selected with special consideration to eliminate duplicate proteins of different EC numbers due to nomenclature confusion. Motifs were identified structurally: Motif I consists of the first conserved β strand (β1) and the following loop, Motif Post I corresponds to the second β strand (β2), Motif II describes the fourth β strand (β4), and Motif III includes the fifth β strand (β5) and the α helix preceding it (2–8) (see Fig. 2). Preference for using a particular structure from the list of homologous proteins includes, in descending order of importance, crystallization with S-adenosylhomocysteine or AdoMet, a minimal number of mutations, and derivation from a higher organism. This reference group was used to generate the “crystal matrix” for scoring in the Multiple Motif Scanning program.
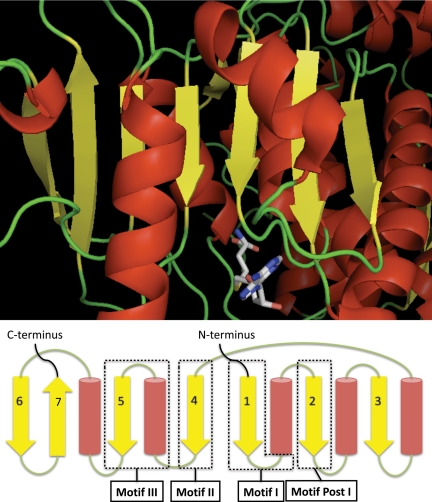
Fig. 2.
Crystal structure of Dot1 (top) and schematic (bottom) that depicts the secondary structures present in Class I methyltransferases. Class I methyltransferases are distinguished by a common three-dimensional structural core, which includes a seven-strand twisted β sheet that provides the major binding interactions for AdoMet. β strands are in yellow, helices are in red, and non-strand/non-helices are shown in green. The S-adenosylhomocysteine molecule in the crystal structure is depicted in the stick model (Protein Data Bank code 1U2Z, Ref. 29).
Scoring Matrix for Methyltransferase Motif Sequences
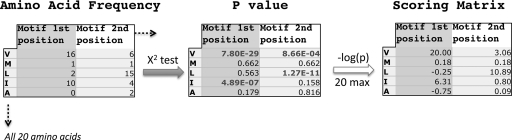
The scoring matrix for motif sequences was compiled using statistical analysis from the two known reference databases of methyltransferases described above. The number of each amino acid residue at each position of each motif was compiled and compared with the expected value that was calculated from the amino acid translation frequencies found in the Pseudogenes.org database. p values from χ2 tests were obtained by comparing the actual count of each amino acid with the expected count. p values were then converted to the absolute value of the scores by taking the negative log; scores were designated with a negative value if the actual amino acid count was less than the expected (see Fig. 3 and supplemental Table 3). This Fisher-based scoring method works well for smaller data sets because of the fact that it eliminates the needs for pseudocounts that would heavily misrepresent the composition of amino acids. We used a maximum value of 20 for the negative log to limit the contribution of individual residues present at high frequencies.
Fig. 3.
Development of a Fisher-based sequence scoring matrix used for Multiple Motif Scanning analysis. The scoring matrix for each amino acid was compiled using statistical analysis from the two known databases of methyltransferases. The number of each amino acid residue at each position of each motif was tallied. p values from χ2 tests were obtained by comparing the actual count of each amino acid with the “expected” count, which was calculated from frequencies found in the Pseudogene.org database. Values were then converted to the absolute value of the scores by taking the negative log, and scores were designated with a negative value if the actual amino acid count was less than the expected.
Scoring Matrix for Conserved Spacing between Motifs
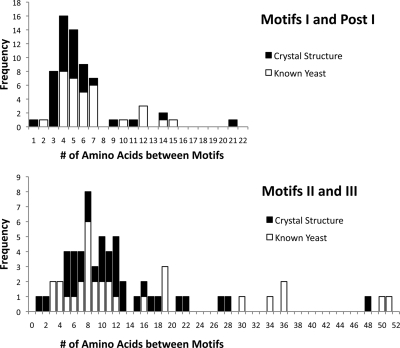
A score for the spacing between motifs was determined as the number of proteins in the combined yeast and crystal structure reference set with that spacing (Fig. 4). For spacing values that were greater than 22 residues for Motif I-Post I or 53 residues for Motif II-Motif III, a negative value of −1 point was used per additional amino acid residue present.
Fig. 4.
Conservation of spacing between motifs. The number of amino acids between the extended Motifs I and Post I and Motifs II and III for known methyltransferases are depicted. Scores for spacing were based on the frequency of yeast and structural database combined with penalties for excessive gap distance as described under “Experimental Procedures.”
Multiple Motif Scanning Program to Identify New Methyltransferases
The Multiple Motif Scanning program was developed to scan the proteome for novel methyltransferases taking advantage of all four motifs and the spacing between Motifs I and Post I and between Motifs II and III. The yeast and crystal structure sequence matrices were independently used to scan the S.cerevisiae proteome with the combined spacing matrix. The program is designed to first recognize the top five matches for the first sequence motif (here Motif I) in the amino acid sequences of the entire yeast or human proteomes (10). For each of these Motif I matches, the program then searches each protein for the best five matches, considering both sequence and spacing, for the second sequence motif (here Motif Post I). For each of these 25 combinations of the first two motifs, the program then searches for the best five third sequence motif (here Motif II). Here only sequence information was utilized, although it is possible to also use spacing as a criterion. For the 125 combinations of the three motifs in each protein, the program searched for the last motif in the sequence (here Motif III) by both sequence and spacing. The final 625 combinations were then scored.
Secondary Screen of Ranked Putative Methyltransferases by HMM Analysis
The top proteins outputted from the Multiple Motif Scanning program as well as other suspected putative methyltransferases underwent an additional HMM profile analysis. HMM primary sequence and predicted secondary structural profiles were created for these proteins with HHpred (9) to evaluate whether an unknown sequence matches the profiles of known methyltransferases in the S.cerevisiae proteome and the HMM database “SUPERFAMILY.” The putative methyltransferase designation was confirmed if the best known match was to a verified methyltransferase. On the other hand, if there was no match to a verified methyltransferase in the output, the protein was designated a false positive. If there was a verified methyltransferase in the output but a known non-methyltransferase had a higher score, we designated the protein as an unconfirmed putative methyltransferase.
Cluster Analysis
HMM profile-profile searches for all-versus-all yeast protein comparisons of yeast methyltransferases were performed (9). Profiles were built to detect relationships of proteins at the family level, resulting in “thinner” and more informative scores compared with analysis at the superfamily level or PSI-BLAST searching. Negative log p values with a cutoff of 20 were used to compile cluster groups using the program Biolayout (11).
RESULTS AND DISCUSSION
The presence of conserved short sequence motifs at similar spacings in Class I methyltransferases can allow for the identification of new enzymes from the proteomes of various organisms. We wanted to take advantage of recent developments that promised to enhance the identification of individual motifs and to take into account the spacing of the motifs in new algorithms for reiterative searches of the proteome.
Assignment of Methyltransferase Motifs from the Analysis of Known Methyltransferases
To more clearly define Class I methyltransferase motifs, the 32 known yeast methyltransferases were aligned by HHpred, which runs HMM profile-profile searches using both primary sequences and secondary structural predictions. Motifs I, Post I, II, and III of each known methyltransferase were compiled from these alignments. All motifs were identified by this method with the exception of Motif III in Ppm2/YOL141W (supplemental Table 1). The secondary structural predictions in and near the four motifs are highly accurate as evidenced by comparison with the known secondary structure for the four yeast proteins whose structures have been determined by crystallography (Fig. 1). A second reference set of methyltransferases was compiled from non-redundant, non-yeast methyltransferases with a solved crystal structure (supplemental Table 2). Here all four motifs could be identified directly from the structure (Fig. 2).
Developing Scoring Matrices for Motif Sequences and Spacing
Scoring matrices for each of the four motifs was developed by statistical analysis of the frequency of each amino acid compared with the expected value from the overall amino acid abundance as described under “Experimental Procedures” and in Fig. 3. Similar, although not identical, results were found in the yeast reference set and the crystal reference set (supplemental Tables 3 and 4). The statistical analysis revealed additional significance of certain residues adjacent to the known motifs. We utilized the expanded number of methyltransferases with known three-dimensional structures to determine whether additional residues could be added to the motifs.
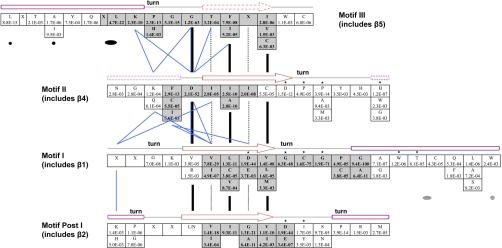
Motif I has been described as the first β strand (β1) followed by the signature GXGXG sequence that makes up an extended turn. Our secondary structure prediction indicated that the amino acid immediately before Motif I is also part of the β sheet in all of the members of the yeast methyltransferase reference set. This residue is generally aliphatic or basic and interacts with residues directly preceding β4 of Motif II, aiding in the alignment of the methyltransferase domain (Figs. 2 and 5). This conserved amino acid should therefore be considered part of Motif I. Additionally we found statistical significance in the residues following the GXGXG sequence of Motif I. These residues are in the helix that binds to the methionine carbonyl of AdoMet (Fig. 5). The helix is amphipathic, containing a hydrophobic side that comprises part of the hydrophobic pocket formed by the side chains of the four β strands of the motifs (Figs. 2 and 5).
Fig. 5.
Conserved amino acid residues in and adjacent to the four methyltransferase motifs. The motifs are arranged as they occur in the β sheet. Conserved amino acids along with the χ2 p values from the yeast data set are shown in boxes. The residues that composed the originally described motifs are highlighted in bold print with a gray background. The secondary structure as determined from both the yeast and crystal data sets is denoted by an arrow (β strand), purple box (helix), or line (non-helix or strand). Dotted representations indicate structures that are not present in all methyltransferases. Residues involved in turns are also shown. The chemical interactions of key amino acids residues within the protein (blue lines) and/or with cofactor AdoMet (*) are shown. The thick black lines between each of the β strands indicate amino acids contributing to two hydrophobic pockets created by side chains of residues coming into (solid) and out of (dashed) the plane of the β sheet. Side chains from helical regions contributing to these hydrophobic pockets are indicated similarly by solid and dashed circles and ovals.
Motif Post I contains the amino acid sequence that makes up the second β strand. Although not conserved in primary structure, the amino acid immediately preceding Motif Post I is predicted to be the first amino acid of β2 in 30 of the 32 yeast methyltransferases. However, the third residue preceding the strand has a statistically significant abundance of proline or glycine residues that may be important for the turn at that position. The amino acid position preceding this position is enhanced with basic amino acids that are involved with charge interactions with residues on the N-terminal side of β1, β4, and β5 (Fig. 5). Finally we found a statistical significance for the presence of an isoleucine or tyrosine residue immediately following Motif Post I. This residue is positioned to interact with the adenine ring of AdoMet (Fig. 5).
Motif II consists of amino acids preceding and including β4. The first two amino acids are responsible for interacting with the N terminus of β1 and β5 (Fig. 4). The three aliphatic amino acids at the end of Motif II start β4. After Motif II, a short coil sequence interacts with not only the methyl group in AdoMet that is to be transferred but also the substrate. Therefore, the specific amino acids in these sequences reflect the substrate of methylation (3). A conserved (D/N)PPY sequence is present in nucleotide and glutamine N-methyltransferases (12–15). The tyrosine residue is replaced by a cysteine residue in cytosine 5-methyltransferases (16). Other N-methyltransferases, such as the arginine methyltransferases, have a glutamic acid residue in this region (17). The CheR glutamic O-methyltransferase has a conserved arginine residue here (2). These Post Motif II residues can help situate the substrate and stabilize charge to promote the methyltransferase reaction. Following the coil, a helix exists whose N-terminal residue can form a π interaction with the adenine ring of AdoMet (Fig. 5).
The coil before Motif III is also involved in the formation of the hydrophobic pocket close to the methyl group of AdoMet. Motif III consists of a coil that is integral in forming the U-turn that aligns the rest of the residues in the motif into β5, adjacent to β4. Once again, the C-terminal residues forming the β strand are hydrophobic.
These results justify an expansion of the originally described four motifs (Fig. 6). Our analysis of the significance values of the amino acids in addition to their position and chemistry observed in the collection of crystal structures allows for the redefinition of the motif consensus sequences as well as the expansion of the motifs (Fig. 6). This expansion permits additional searching power when the motifs are used to scan the proteome and should result in a more accurate listing of putative methyltransferases.
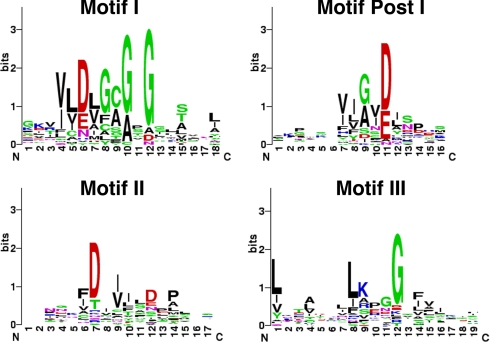
Fig. 6.
Extended Motifs I, Post I, II, and III used for Multiple Motif Scanning analysis. The amino acid frequency among known yeast methyltransferases is depicted using WebLogo (30) for the expanded Motifs I, Post I, II, and III determined in this work. A larger number of bits for each letter designates the importance of each amino acid in the motif.
Because the number of amino acids between Motifs I and Post I and between Motifs II and III are conserved (Fig. 4), separate matrices were developed to describe additional scoring parameters based on how closely the spacings between these motifs in candidate methyltransferases fit those of known methyltransferases. Here we combined data from the yeast and crystal reference sets because we observed no large differences between them (Fig. 4). The “gap” region between Motif I and Post I consists of a conserved secondary structure of a small helix followed by a random coil (supplemental Table 1 and Fig. 5). Although there is slight variance between the number of amino acids that comprise these structures, there was no correlation found between the number of helix amino acids and the number of coil amino acids (data not shown).
Multiple Motif Scanning Output of Putative Methyltransferases
Matrices generated from the newly defined methyltransferase motifs and the spacing matrices were used to scan the proteome to develop a putative list of methyltransferases. The computer program Multiple Motif Scanning was developed to perform this search based on the similarity of the primary amino acid sequence. Multiple Motif Scanning allows the user to choose the number of motifs to search, the number of residues in each motif, the score for each amino acid at each position of the motif, and the expected spacing between the motifs (Fig. 7). This program returns individual scores for each of the motifs and for each of the two spacing criteria; these scores are then combined to give a total score. We performed this search independently using matrices developed for both the yeast and the crystal structure methyltransferase data sets using the yeast proteome (10) (supplemental Tables 3 and 4). The output of this program is given in supplemental Tables 5 and 6.
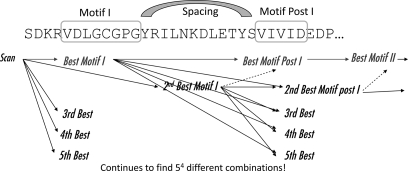
Fig. 7.
Multiple Motif Scanning software. The Multiple Motif Scanning program was developed to scan the proteome for novel methyltransferases to resolve the problems encountered by Katz et al. (6). Yeast matrix and crystal structure matrix were independently used to scan the S. cerevisiae proteome. Multiple Motif Scanning recognizes the top five best matches for the first motif entered and subsequently identifies the top five plausible second motifs for each of the matches of the first motifs. The program continues to find the top five sequences that fit the motif for every previous combination to produce 5n matches for n number of motifs. All combinations are scored, and the top 10 combinations are saved. Proteins are ranked among each other based on the top score. The extended Motifs I, Post I, II, and III were used for analysis with gap considerations between motifs.
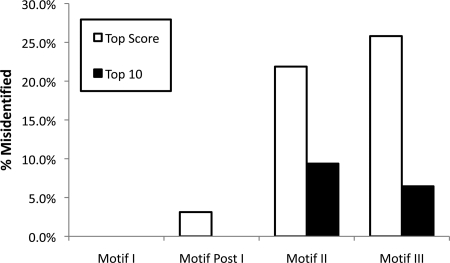
We validated the usefulness of this approach by asking how well the output described the motifs and spacings of known methyltransferases. Multiple Motif Scanning scoring with the yeast matrices resulted in the correct identification of all 32 HMM-predicted Motif I sequences as the top score (Fig. 8). In addition, all of the Motif Post I sequences were found correctly at the top score in each protein with the exception of Rrp8/YDR083C, which was found within the top 10 scores. However, for the top sequences matches, Motif II was misidentified in seven proteins, and Motif III was misidentified in eight proteins. However, when the top 10 scores are considered, only three Motif II and two Motif III sequences were not predicted. When the matrices derived from the crystal reference methyltransferases were used, the predicted motifs were less successfully identified. Here the motifs of Trm1, Hsl7, and Mtq1 were completely misidentified, and a total of 10 Motif II and 11 Motif III sequences were missed in the top ranked score. The differences seen here highlight the importance of formulating the best matrices for the Multiple Motif Scanning program. Because Motifs II and III have not been well characterized previously, the ability to identify Motifs II and III with such accuracy is beneficial for providing supplemental information in the identification of novel methyltransferases.
Fig. 8.
Accuracy of Multiple Motif Scanning.Motifs found by the Multiple Motif Scanning program using the yeast matrix were compared with the HMM profile alignments to calculate the percentage of inaccuracy in identifying the motifs. The program outputs the overall top score calculated from the combination of sequence-fitting motifs and spacing between them combined along with the next nine top scoring combinations.
In these analyses, we weighted the spacing scores arbitrarily. In initial studies, we found that a lower weighting of the spacing scores reduced the ability to correctly identify Post Motif I and Motif II; using a higher weighting resulted in known methyltransferases such as Trm11 having low scores because of large loops between the motifs.
The Multiple Motif Scanning program can be used to search for other classes of proteins containing one or more conserved motif(s). Similar to the Motif Alignment and Search Tool (MAST) program, the user is able to enter any sequence matrix that can be designed for specificity to search for the protein family of choice (18, 19). However, the Multiple Motif Scanning program has two major advantages. In the first place, it can evaluate different combinations of multiple motifs. Secondly it can determine conserved spacing elements between the motifs. These features are beneficial for correctly identifying smaller motifs that follow a motif by a conserved number of amino acids, such as Motif Post I. This program reduces the identification of false motifs that may be identified downstream due to the similarity of primary sequence despite their incorrect position.
Identification and Validations of Putative Yeast Methyltransferases
The yeast proteome was scanned separately by the yeast matrix and crystal matrix to generate a new list of enzymes ranked by their similarity to the model methyltransferase motifs (supplemental Tables 5 and 6). 94% of the known methyltransferase (30 of 32) were found within the top 2.0% of the proteome scored using the yeast matrices (Table I). Katz et al. (6) predicted that 0.8% of the yeast genome consists of methyltransferases. Therefore, proteins that score highly that are not already designated as known methyltransferases can be marked as putative methyltransferases. Several of the putative methyltransferases identified previously (6), for example YHR209W, YNL092W, and YIL064W among others, ranked highly by both the analyses with the yeast reference and crystal reference matrices (Table I and supplemental Tables 5 and 6). Interestingly some proteins scored significantly higher by the crystal matrix than by the yeast matrix. YBR141C, which is already designated as a putative methyltransferase (6), scored in the bottom half of the proteome by the yeast matrix but as number 29 according to the crystal matrix. Further analysis reveals that most of the possible motifs found between the yeast and crystal matrix sets were different. This result points out the limits of these methods.
Table I. Multiple Motif Scanning ranking of the yeast proteome.
Proteins are listed as known methyltransferases, those with unknown function, and those with a known non-methyltransferase function. MT, methyltransferase. Other confirmed putative methyltransferases not on this list include YBR141C, YGL050W, YIL110W, YLR063W, YLR137W, YMR209C, YNL022C, and YNL024C (see text).
| Rank | Score | Proteina |
Description of non-MT function | ||
|---|---|---|---|---|---|
| Known MT function | Unknown function | Known non-MT function | |||
| 1 | 390.63 | Erg6 | |||
| 2 | 386.09 | Hmt1 | |||
| 3 | 369.61 | Coq3 | |||
| 4 | 355.56 | Abd1 | |||
| 5 | 354.60 | Tgs1 | |||
| 6 | 340.97 | Bud23 | |||
| 7 | 338.95 | Trm11 | |||
| 8 | 333.69 | Mrm2 | |||
| 9 | 332.26 | Spb1 | |||
| 10 | 331.61 | Ncl1 | |||
| 11 | 327.60 | YHR209W | |||
| 12 | 326.08 | Dim1 | |||
| 13 | 324.92 | Trm7 | |||
| 14 | 320.21 | Tmt1 | |||
| 15 | 318.55 | Mtq2 | |||
| 16 | 317.13 | Trm9 | |||
| 17 | 304.22 | Trm5 | |||
| 18 | 300.76 | YNL092W | |||
| 19 | 297.53 | Coq5 | |||
| 20 | 292.94 | YIL064W | |||
| 21 | 292.64 | Nop2 | |||
| 22 | 288.28 | Trm8 | |||
| 23 | 287.33 | YDR316W | |||
| 24 | 287.12 | Nnt1 | |||
| 25 | 286.08 | Nop1 | |||
| 26 | 284.36 | YOR239W | |||
| 27 | 278.58 | Trm2 | |||
| 28 | 277.39 | YKL155C | |||
| 29 | 275.48 | YBR271W | |||
| 30 | 273.65 | (Pho8) | Repressible alkaline phosphatase | ||
| 31 | 272.61 | Trm44 | |||
| 32 | 266.75 | Spe4 | Spermine synthase (uses decarboxylated AdoMet) | ||
| 33 | 262.85 | Ppm2 | |||
| 34 | 261.45 | (Arn1) | Transporter of ferrirubin, ferrirhodin, and other ferrichromes | ||
| 35 | 261.01 | (Aif1) | Apoptosis-inducing factor | ||
| 36 | 260.58 | YPL017C | |||
| 37 | 260.18 | Fas1 | Fatty-acyl-CoA synthase, β chainb | ||
| 38 | 259.03 | YMR228W | RNA polymerase-specific factor, mitochondrial(methyltransferase-like crystal structure) | ||
| 39 | 258.47 | (Pcm1) | Phosphoacetylglucosamine mutase | ||
| 40 | 258.20 | (Fox2) | Hydratase-dehydrogenase-epimerase | ||
| 41 | 258.01 | Rrp8 | |||
| 42 | 256.52 | Spe3 | Spermidine synthase (uses decarboxylated AdoMet) | ||
| 43 | 256.40 | Adh7 | NADP(H)-dependent alcohol dehydrogenase | ||
| 44 | 254.27 | Sor1 | Sorbitol dehydrogenase | ||
| 45 | 254.27 | Sor2 | Sorbitol dehydrogenase homolog | ||
| 46 | 253.09 | Gdh3 | NADP-glutamate dehydrogenase | ||
| 47 | 252.75 | Prc1 | Carboxypeptidase Y, serine-type protease | ||
| 48 | 252.43 | Adh6 | NADPH-dependent alcohol dehydrogenase | ||
| 49 | 252.20 | (Alg13) | Catalytic component of UDP-GlcNAc-transferase | ||
| 50 | 250.42 | Mae1 | Malic enzyme | ||
| 51 | 250.23 | YJR129C | |||
| 52 | 250.19 | Trm1 | |||
| 53 | 250.01 | Gdh1 | Glutamate dehydrogenase (NADP+) | ||
| 54 | 249.90 | Rmt2 | |||
| 55 | 249.86 | YBR261C | |||
| 56 | 249.81 | (Pol2) | DNA-directed DNA polymerase ϵ, catalytic subunit A | ||
| 57 | 249.07 | Trm12 | Required for the synthesis of the hypermodified nucleoside wybutosine in tRNA (uses AdoMet?) | ||
| 58 | 248.72 | Ade5,7 | Phosphoribosylamine-glycine and phosphoribosylformylglycinamidine cyclo-ligase | ||
| 59 | 248.59 | (Sen1) | Positive effector of tRNA-splicing endonuclease | ||
| 60 | 248.31 | Mis1 | C1-tetrahydrofolate synthase precursor | ||
| 61 | 246.84 | (Sec27) | Coatomer complex β′ chain (β′-cop) of secretory pathway vesicles | ||
| 62 | 246.80 | Mtq1 | |||
| 63 | 246.53 | (Ura8) | CTP synthase 2 | ||
| 64 | 246.27 | (Gap1) | General amino acid permease | ||
| 65 | 246.01 | Dot1 | |||
| 129 | 236.85 | (Pan2) | Component of Pab1-stimulated poly(A) ribonuclease | ||
| 130 | 236.79 | YBR235W | |||
| 131 | 236.71 | Hsl7 | |||
| 132 | 236.51 | (Nus1) | Putative prenyltransferase | ||
| 133 | 236.50 | (Tom1) | E3 ubiquitin ligase required for G2/M transition | ||
| 195 | 232.20 | (Rsc2) | Member of RSC complex | ||
| 196 | 232.14 | (Ndi1) | NADH-ubiquinone-6-oxidoreductase | ||
| 197 | 232.13 | Ppm1 | |||
| 198 | 232.02 | (Lpd1) | Dihydrolipoamide dehydrogenase precursor | ||
| 199 | 232.00 | (Rgp1) | Subunit of a Golgi membrane exchange factor | ||
| 682 | 218.14 | (Aip1) | Actin cytoskeleton component | ||
| 683 | 218.14 | (Rpm2) | Ribonuclease P precursor, mitochondrial | ||
| 684 | 218.12 | Gcd14 | |||
| 685 | 218.12 | (Sec1) | Protein transport protein | ||
| 686 | 218.12 | (Isw1) | ATPase component of a four-subunit chromatin-remodeling complex | ||
a Proteins in parentheses were not confirmed by the secondary HMM profile sequence check as described under “Experimental Procedures.” Proteins that are italicized exhibit loose primary and secondary sequence homology to known methyltransferases and are considered unconfirmed putative methyltransferases. The remaining proteins are HMM-validated methyltransferases as described under “Experimental Procedures.”
b Although the mammalian fatty-acid synthase has a methyltransferase domain (28), this domain is not conserved in the yeast enzyme, and the motifs identified here do not correspond with those of the mammalian enzyme.
Previously Katz et al. (6) identified 80 yeast proteins that were not known methyltransferases as scoring highly for a search based on Motif I and Motif Post I. Because 42 of these proteins, including seven that ranked above the “75% correct” criterion (6), rank below the top 4.8% of yeast proteins in both reference sets (supplemental Tables 5 and 6), we can now focus on the remaining proteins that were also identified in our searches. Three yeast proteins, previously not identified with possible methyltransferase or AdoMet binding, ranked highly on both the yeast reference and crystal structure reference Multiple Motif Scanning searches (supplemental Tables 5 and 6). These include YMR209C, YOL060C/Mam3, and YLR063W. YOL060C, designated Mam3, has an apparent cystathionine β-synthase (CBS) domain that may bind AdoMet (20) but was not verified as a methyltransferase by our HMM analysis (see below).
Some of the proteins ranked in the top 2.0% of the proteome are known not to be methyltransferases. Many of these known “false positives” are enzymes that utilize adenosyl nucleosides and nucleotides such as dehydrogenases due to the similarity of their consensus sequence to the methyltransferase motifs (4). However, the possibility exists that some enzymes have more than one activity: CysG found in some bacteria catalyzes two methyltransferase reactions, a dehydrogenase reaction, and a ferrochelation reaction (21). Therefore, these “false positive” methyltransferases could be multifunctional enzymes. It is also important to note that some of the putative methyltransferases described here may instead transfer aminopropyl or aminobutyryl groups from AdoMet; for example Spe3, Spe4, and Trm12 all score highly by Multiple Motif Scanning (Table I). However, Spe3 and Spe4 bind decarboxylated AdoMet for aminopropyl transfer (22, 23), whereas Trm12 appears to bind AdoMet for aminobutyryltransfer (24).
To better distinguish the false positive proteins from undiscovered methyltransferases, we utilized HMM methods to provide a secondary check on each of the top scoring proteins in the ranked lists. This methodology uses predicted secondary structure as well as the primary sequence to ask whether a putative methyltransferase matches the profiles of known methyltransferases. Of the top 65 ranked proteins in Table I, 32 are known AdoMet-utilizing methyltransferases or aminopropyl/aminobutyltransferases, and 11 are HMM-validated putative methyltransferases for a total of 43 validated species. The HMM methodology used suggests that 11 of the remaining 22 species clearly are not seven-β strand methyltransferases (indicated in parentheses in Table I) and that the other 11 species are less related to the known methyltransferases than are the putative species (indicated by italics in Table I). Thus, the addition of the HMM test allows one to confirm the putative assignments and to eliminate the apparent false positives.
When the HMM analysis was extended through the 199th ranked species, a group that includes 31 of the 32 known methyltransferases, three additional species (YMR209C, YGL050W, and YNL024C) were found that passed the HMM muster. We also verified two species that scored high by the crystal matrix (YLR063W and YBR141C) as methyltransferases but did not confirm YOL060C as a methyltransferase. Importantly this analysis also did not verify HMM methyltransferase profiles for any of the other species in the 66th through 199th positions. This result suggests that our methodology can now identify almost all of the known methyltransferases while limiting the number of putative enzymes.
In reviewing the importance of the expanded Motif I, 84.4% of all known methyltransferase would be scored in the top 2.0% of the proteome using only the subscore from the expanded Motif I alone. Scoring with only the subscore of the expanded Motif Post I alone generated 50.0% of the known methyltransferases in the top 2.0% of the proteome. Surprisingly scoring using only the refined Motifs II and III together resulted in 68.8% of known methyltransferases ranked among the top 2.0% of the proteome. These statistics show that each motif does hold a portion of the methyltransferase character; however, a combination of all these motifs together best characterizes a methyltransferase.
HHM Profile Clustering Analysis Reveals Putative Substrates
Additional sequences commonalities are present in methyltransferases with similar substrates (3, 5). To discover information about the substrates of the putative methyltransferases, known and putative methyltransferases were clustered together by HMM profile-profile searches based on p values. A recent study by Ansari et al. (25) grouped putative methyltransferases proteins into nitrogen, carbon, and oxygen methyltransferases. However, we found that our clustering analysis grouped the proteins into substrate specificity rather than the nucleophile of the methyltransferases reaction.
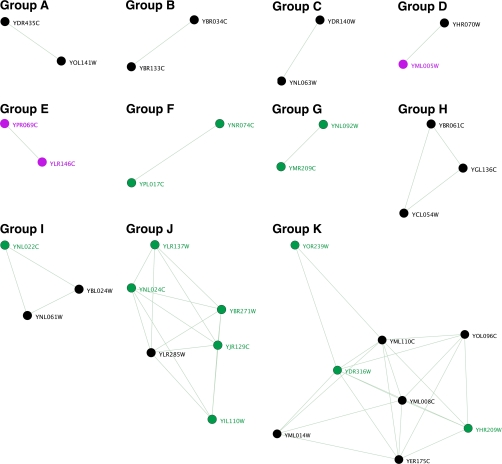
At a p value of 10−20 or below, groups of known and putative methyltransferases form seven sets of pairs, two sets of triads, one family of six, and one family of eight (Fig.9). Although 17 methyltransferases did not cluster under these conditions, the clusters that did form appear to be biologically relevant. Four pairs include known enzymes with similar functions: protein-arginine methyltransferases (Group B), protein-glutamine methyltransferases (Group C), wybutosine-forming transferases (Group D), and the spermidine/spermine synthases (Group E). Further analysis of YPL017C and YNR074C (Group F) by HMM profiling indicated that these proteins were likely to be dehydrogenases rather than methyltransferases. One set of triads consists of three known 2′-O-ribose methyltransferases (Group H). Another triad set contains two cytosine 5-methyltransferases and one unknown member, suggesting that YNL022C may be a new cytosine 5-methyltransferase (Group I). The family with six members (Group J) has one known protein, nicotinamide N-methyltransferase. This family includes two members (YIL110W and YLR137W) that were confirmed by HMM analysis but did not score highly on the yeast or crystal matrix sets. The five unknowns could possibility be small molecule methyltransferases or N-methyltransferases. The family with eight members (Group K) has five known methyltransferases and three unknowns. The known enzymes also include small molecule (Tmt1) as well as lipid methyltransferases (Coq3, Coq5, and Erg6). These may cluster together because of similar N-terminal additions as well as several residues inserted between β6 and β7 (13, 26).
Fig. 9.
HMM profile clustering to determine putative substrates of potential methyltransferases. HMM profile versus profile clustering was utilized to find the calculated homology between proteins. Proteins clustered in several groups (A\NK) that display similarity in type of substrate as well as the atomic nucleophile of the substrate in the methyltransferase reaction (carbon, nitrogen, oxygen, etc.). Known methyltransferases are depicted in black, known non-methyltransferases are depicted in magenta, and unknown ORFs are depicted in green
Although the clustering analysis did not exclusively divide the proteins based on the type of nucleophile attacking the methyl group of AdoMet, additional information about the nucleophile can be extracted by our analysis. For example, Oms1/YDR316W is most likely a lipid C-methyltransferase because it is closely surrounded by C-methyltransferases Coq5/YML110C, Erg6/YML008C, and Trm9/YML014W in Group K. In fact, tRNA methyltransferase Trm9 also clusters into the lipid/small molecule Group K perhaps because it catalyzes a carboxyl methylation reaction similar to Tmt1 (27). Alternative clustering procedures using PSI-BLAST E-values created uninformative relationships, highlighting the computational power of HMM profiling.
Acknowledgments
We are very grateful to Trevor Rangitsing for helping to develop the Multiple Motif Scanning program and to Professors Mensur Dlakić and Grace Xiao for expert help and advice.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant GM026020 and Training Grant GM008496 (to T. C. P. through the UCLA Chemistry-Biology Interface).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- AdoMet
- S-adenosylmethionine
- HMM
- hidden Markov model
- PSI-BLAST
- position-specific iterative basic local alignment search tool.
REFERENCES
- 1.Cheng X., Blumenthal R. M. ( 1999) S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions, World Scientific, Singapore [Google Scholar]
- 2.Djordjevic S., Stock A. M. ( 1997) Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure 5, 545– 558 [DOI] [PubMed] [Google Scholar]
- 3.Martin J. L., McMillan F. M. ( 2002) SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12, 783– 793 [DOI] [PubMed] [Google Scholar]
- 4.Schluckebier G., O'Gara M., Saenger W, Cheng X. ( 1995) Universal catalytic domain structure of AdoMet-dependent methyltransferases. J. Mol. Biol. 247, 16– 20 [DOI] [PubMed] [Google Scholar]
- 5.Schubert H. L., Blumenthal R. M., Cheng X. ( 2003) Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329– 335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katz J. E., Dlakić M., Clarke S. ( 2003) Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteomics 2, 525– 540 [DOI] [PubMed] [Google Scholar]
- 7.Kagan R. M., Clarke S. ( 1994) Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 310, 417– 427 [DOI] [PubMed] [Google Scholar]
- 8.Niewmierzycka A., Clarke S. ( 1999) S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 274, 814– 824 [DOI] [PubMed] [Google Scholar]
- 9.Söding J., Biegert A., Lupas A. N. ( 2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244– 248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. ( 2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985– 1988 [DOI] [PubMed] [Google Scholar]
- 11.Freeman T. C., Goldovsky L., Brosch M., van Dongen S., Mazière P., Grocock R. J., Freilich S., Thornton J., Enright A. J. ( 2007) Construction, visualisation, and clustering of transcription networks from microarray expression data. PLoS Comput. Biol. 3, 2032– 2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong W., O'Gara M., Blumenthal R. M., Cheng X. ( 1997) Structure of Pvu II DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 25, 2702– 2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert H. L., Phillips J. D., Hill C. P. ( 2003) Structures along the catalytic pathway of PrmC/HemK, an N5-glutamine AdoMet-dependent methyltransferase. Biochemistry 42, 5592– 5599 [DOI] [PubMed] [Google Scholar]
- 14.Sunita S., Purta E., Durawa M., Tkaczuk K. L., Swaathi J., Bujnicki J. M., Sivaraman J. ( 2007) Functional specialization of domains tandemly duplicated within 16S rRNA methyltransferase RsmC. Nucleic Acids Res. 35, 4264– 4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z., Shipman L., Zhang M., Anton B. P., Roberts R. J., Cheng X. ( 2004) Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase. J. Mol. Biol. 340, 695– 706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klimasauskas S., Kumar S., Roberts R. J., Cheng X. ( 1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell 76, 357– 369 [DOI] [PubMed] [Google Scholar]
- 17.Zhang X., Zhou L., Cheng X. ( 2000) Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 19, 3509– 3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey T. L., Williams N., Misleh C., Li W. W. ( 2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369– 373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey T. L., Gribskov M. ( 1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14, 48– 54 [DOI] [PubMed] [Google Scholar]
- 20.Scott J. W., Hawley S. A., Green K. A., Anis M., Stewart G., Scullion G. A., Norman D. G., Hardie D. G. ( 2004) CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Investig. 113, 274– 284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spencer J. B., Stolowich N. J., Roessner C. A., Scott A. I. ( 1993) The Escherichia coli cysG gene encodes the multifunctional protein, siroheme synthase. FEBS Lett. 335, 57– 60 [DOI] [PubMed] [Google Scholar]
- 22.Hamasaki-Katagiri N., Tabor C. W., Tabor H. ( 1997) Spermidine biosynthesis in Saccharomyces cerevisiae: polyamine requirement of a null mutant of the SPE3 gene (spermidine synthase). Gene 187, 35– 43 [DOI] [PubMed] [Google Scholar]
- 23.Hamasaki-Katagiri N., Katagiri Y., Tabor C. W., Tabor H. ( 1998) Spermine is not essential for growth of Saccharomyces cerevisiae: identification of the SPE4 gene (spermine synthase) and characterization of a spe4 deletion mutant. Gene 210, 195– 201 [DOI] [PubMed] [Google Scholar]
- 24.Noma A., Kirino Y., Ikeuchi Y., Suzuki T. ( 2006) Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 25, 2142– 2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ansari M. Z., Sharma J., Gokhale R. S., Mohanty D. ( 2008) In silico analysis of methyltransferases domains involved in biosynthesis of secondary metabolites. BMC Bioinformatics 9, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zehmer J. K., Bartz R., Liu P., Anderson R. G. ( 2008) Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J. Cell Sci. 121, 1852– 1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalhor H. R., Clarke S. ( 2003) Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 23, 9283– 9292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maier T., Leibundgut M., Ban N. ( 2008) The crystal structure of mammalian fatty acid synthase. Science 321, 1315– 1322 [DOI] [PubMed] [Google Scholar]
- 29.Sawada K., Yang Z., Horton J. R., Collins R. E., Zhang X., Cheng X. ( 2004) Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J. Biol. Chem. 279, 43296– 43306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crooks G.E., Hon G., Chandonia J. M., Brenner S. E. ( 2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188– 1190 [DOI] [PMC free article] [PubMed] [Google Scholar]